Abstract
Several prominent bacterial pathogens secrete nuclease (Nuc) enzymes that have an important role in combating the host immune response. Early studies of Staphylococcus aureus Nuc attributed its regulation to the agr quorum-sensing system. However, recent microarray data have indicated that nuc is under the control of the SaeRS two-component system, which is a major regulator of S. aureus virulence determinants. Here we report that the nuc gene is directly controlled by the SaeRS two-component system through reporter fusion, immunoblotting, Nuc activity measurements, promoter mapping, and binding studies, and additionally, we were unable identify a notable regulatory link to the agr system. The observed SaeRS-dependent regulation was conserved across a wide spectrum of representative S. aureus isolates. Moreover, with community-associated methicillin-resistant S. aureus (CA MRSA) in a mouse model of peritonitis, we observed in vivo expression of Nuc activity in an SaeRS-dependent manner and determined that Nuc is a virulence factor that is important for in vivo survival, confirming the enzyme's role as a contributor to invasive disease. Finally, natural polymorphisms were identified in the SaeRS proteins, one of which was linked to Nuc regulation in a CA MRSA USA300 endocarditis isolate. Altogether, our findings demonstrate that Nuc is an important S. aureus virulence factor and part of the SaeRS regulon.
INTRODUCTION
Staphylococcus aureus is one of the most frequent causes of nosocomial and community-associated (CA) bacterial infections. The increasing frequency of methicillin-resistant S. aureus (MRSA) infections and the emergence of CA MRSA have added to the recent concern over this deadly pathogen (1, 2). In particular, the CA MRSA strains of the USA300 pulsed-field gel electrophoresis type have expanded clonally across the United States, and these USA300 isolates can cause severe skin and soft tissue infections and necrotizing pneumonia (3–5).
To be an effective pathogen and survive in the host, S. aureus secretes an array of tissue-degrading enzymes, toxins, and superantigens (6). One of these enzymes, the secreted nuclease (Nuc), was first identified by Cunningham et al. in 1956 (7), but it was only recently demonstrated that Nuc is important for S. aureus pathogenesis. Berends et al. determined that Nuc contributes to the severity of CA MRSA lung infection and evasion of neutrophil extracellular traps (8). The production of Nuc is conserved across methicillin-susceptible S. aureus and MRSA strains (9), and thus, it has been used as a marker for the direct detection of S. aureus (10). We recently demonstrated that Nuc has an additional role in S. aureus as a modulator of biofilm formation (9).
The regulation of S. aureus nuc gene expression has received only limited attention. While Nuc enzyme levels are known to be higher in sarA and sigB mutants (9), it is often stated in the S. aureus literature that production of Nuc is under agr quorum-sensing system control because diminished Nuc activity was observed in agr-defective strains (11–13). However, multiple reports have provided preliminary observations that Nuc is actually regulated by SaeRS, a two-component system responsible for the production of a diverse array of secreted toxins, immunomodulators, and enzymes (14–17). Initial proteomic and microarray observations were reported by Rogasch et al. (17) that demonstrated the influence of saeS on Nuc expression in strains COL and Newman, and we made similar observations through microarray analysis of CA MRSA saeRS deletion mutants (14, 15). However, these previous studies did not establish whether diminished Nuc production in sae deletion strains is direct or indirect and whether it occurs in vivo in an SaeRS-dependent manner. In the present study, we add to our knowledge of Nuc regulation in S. aureus and demonstrate that the SaeRS system is the dominant regulator of nuc transcription and enzyme production in vitro and in vivo. Additionally, we present evidence that Nuc contributes to CA MRSA pathogen survival during invasive S. aureus infection.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains used in this study are described in Table 1. Strains of S. aureus were grown and maintained in tryptic soy broth (TSB) or on tryptic soy agar (TSA). The antibiotic concentrations (in μg/ml) used for selection of chromosomal markers or maintenance of S. aureus plasmids were the following: chloramphenicol, 10; erythromycin (Erm), 10; spectinomycin (Spc), 150; tetracycline (Tet), 5. Recombinant DNA and genetic techniques for plasmid and strain constructions were used as previously described (18, 23). Reagents were purchased from New England BioLabs, Fisher Scientific, and Sigma unless otherwise indicated.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| S. aureus strains | ||
| AH1263 | USA300 CA MRSA Erms (LAC*) | 18 |
| AH1292 | AH1263 Δagr::tetM | This work |
| AH1483 | AH1263 ΔsigB | 9 |
| AH1558 | AH1263 ΔsaeQRS::spc | This work |
| AH1589 | AH1263 hla::Tn551-Erm | This work |
| AH1680 | AH1263 nuc::LtrB | 9 |
| AH1621 | AH1263 agrA::Tn551-Erm | This work |
| MW2 | CA MRSA USA400 | 19 |
| AH1552 | MW2 ΔsaeQRS::spc | 15 |
| AH1942 | MW2 Δagr::tetM | This work |
| AH759 | UAMS-1 | 20 |
| AH2333 | UAMS-1 ΔsaeQRS::spc | This work |
| AH1081 | UAMS-1 Δagr::tetM | 20 |
| AH847 | COL | 21 |
| AH2330 | COL ΔsaeQRS::spc | This work |
| AH2331 | COL Δagr::tetM | This work |
| AH2334 | Newman ΔsaeQRS::spc | This work |
| AH2380 | Newman Δagr::tetM | This work |
| BK2039 | RN4282 | 22 |
| BK9868 | Newman | |
| BK11490 | LAC | |
| BK18805 | USA300-0114 | 3 |
| BK18806 | USA300-0180 | 3 |
| BK18807 | USA300-0114 | 3 |
| BK18808 | USA300-0047 | 3 |
| BK18809 | USA300-0114 | 3 |
| BK18810 | USA300-0114 | 3 |
| BK18811 | USA300-0247 | 3 |
| BK18813 | USA300-0045 | 3 |
| BK19069 | USA300-0114 (FPR3757) | 3 |
| BK19321 | USA300-0120 | 3 |
| BK21141 | Mu50 | |
| BK21157 | USA600 (NRS022) | |
| BK21189 | RN1 | |
| BK21195 | Cowan | |
| BK22030 | 1962 pandemic CC30 | |
| BK22251 | 1965 pandemic CC30 | |
| Plasmids | ||
| pALC2073-sigB | sigB-complementing clone | 23 |
| pAgrAC-1 | Complementing clone for agrA | 24 |
| pCM20 | pCE with Pnuc-sGFP | 9 |
| pCM28-hla | hla-complementing clone | 25 |
| pCM28-nuc | nuc-complementing clone | 9 |
| pKOR1-sigB | sigB complementing clone | 23 |
| pRB473-saeRS | saeRS-complementing clone | 15 |
| pSKerm-saeLac | saePQRS-complementing clone | This work |
| pSKerm-saeMW2 | saePQRS-complementing clone | This work |
Strain and plasmid construction.
PCR amplification of the saePQRS operon was performed on genomic DNA isolated from MW2 (AH843), Newman (AH1178), LAC (AH1263), and 18805 (AH1685) with primers CEF16 and CEF17. PCR products were gel purified with a gel extraction kit from IBI (Peosta, IA), digested with HindIII and XmaI, and ligated into pSKerm+MCS digested with the same enzymes. Plasmid pSKerm+MCS was generated by digesting the agrCA genes from pAgrC1agrA (24) with HindIII and XmaI. A multiple cloning site was synthesized with primers MEO63 and MEO64 and subsequently ligated into the digested vector at the HindII and XmaI sites. Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA).
Reporter fusion experiments.
For GFP reporter construct analysis, deletions in the agr (Δagr::tetM) and sae (ΔsaeQRS::spc) systems were constructed in an erythromycin-sensitive strain of USA300 (strain AH1263 [18]). A deletion in sigB was constructed in the same genetic background with pKOR1-sigB as described previously (23). Reporter plasmid pCM20 was transformed into AH1263 and the mutant strains, and cultures were grown overnight in TSB containing Erm. To measure promoter activity, the overnight cultures were diluted 1:100 in 5 ml of the same medium and grown at 37°C with shaking (200 rpm). At designated times, 200 μl of culture was removed from each tube and transferred to a 96-well microtiter plate (Corning 3096). Fluorescence readings were recorded with a Tecan Infinite M200 plate reader with excitation and emission wavelengths of 480 and 515 nm, respectively. Cell growth was also measured on the basis of the optical density at 600 nm (OD600) to calculate relative fluorescence values.
TaqMan real-time reverse transcription-PCR analysis.
Relative quantification of S. aureus genes was done by measuring the difference between the expression of target transcripts and that of the housekeeping gene gyrB as previously described (14, 15, 26). S. aureus total RNA was harvested during growth in TSB in the mid-exponential (OD600 of 1.5) and early stationary (OD600 of 2.0) phases of growth by lysis in RLT buffer (Qiagen), followed by RNeasy (Qiagen) column purification as described elsewhere (15, 26, 27). For analysis during growth in TSB, the n-fold change was determined by comparing the expression of wild-type (WT) LAC and the sae-complemented strain to that of the sae mutant.
Nuc activity measurements and Western immunoblot assays.
For protein immunoblotting and Nuc activity measurements, strains were grown in TSB for 15 h at 37°C. Cells were removed by filtration through 0.22-μm filters, and spent medium was used for immunoblot assays and enzyme assays. Immunoblot assays for Nuc were performed with sheep anti-Nuc IgG conjugated to horseradish peroxidase (anti-Nuc–HRP conjugate; Toxin Technology, Sarasota, FL) as previously described (9). Immunoblot assays for alpha-toxin (Hla) in spent medium were performed with sheep anti-Hla–HRP conjugate (Toxin Technology) as previously described (28). Nuc enzyme activity was measured with a florescence resonance energy transfer (FRET) assay as previously described (9).
5′ RACE.
Rapid amplification of 5′ cDNA ends (5′ RACE) analysis was performed as previously described (14, 29), with the use of SuperScript III reverse transcriptase (Invitrogen Life Technologies), terminal transferase (New England BioLabs), and gene-specific primers (Table 1). 5′ RACE products were visualized by 1.5% DNA agarose gel electrophoresis, excised, purified with a QIAquick PCR purification kit (Qiagen), and cloned into pCR2.1Topo (Invitrogen). Subsequent plasmids were purified with the QIAprep Spin Miniprep kit (Qiagen) and sequenced with a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems). Sequence alignment of 5′ RACE products against the USA300 genome (GenBank accession no. NC_007793) was performed with the nucleotide basic local alignment search tool.
Purification of SaeR and EMSAs.
Purification of recombinant SaeR (SaeRHis) was performed as previously described (14). Briefly, expression of SaeRHis was achieved with saeR-specific primers and a Champion pET Directional TOPO Expression kit (Invitrogen). Escherichia coli BL21 cells transformed with the pET100D vector containing saeR were grown at 37°C with agitation in 4 liters of LB to an OD600 of 0.8, induced with 0.5 mmol/liter isopropyl-β-d-thiogalactopyranoside (IPTG), and grown for 6 more h. Cells were harvested by centrifugation (4,000 × g at 4°C for 10 min), resuspended in 20 mmol/liter Tris at pH 8 (Tris-HCl), and then lysed via sonication on ice for 15 min. After lysed cells underwent centrifugation (15,000 × g at 4°C for 10 min), samples were resuspended in 60 ml of 8 mol/liter urea–0.5 mol/liter sodium chloride–Tris-HCl, sonicated on ice for 15 min, and purified by nickel-nitrilotriacetic acid affinity (Qiagen) and DEAE cellulose anion-exchange chromatography (GE Healthcare) with a BioLogic LP chromatography system (Bio-Rad). Purified SaeRHis underwent dialysis with 3 liters of Tris-HCl and was concentrated with a Centricon Plus 20 centrifugal filter (Millipore). The concentration of SaeRHis was determined with a Micro BCA protein assay kit (Pierce). Electrophoretic mobility shift assays (EMSAs) were performed as described previously (14, 30), with primers and probes in Table 2. The indicated concentrations of SaeRHis were combined with 0.5 pmol of 6-carboxyfluorescein (FAM)/nuc promoter, 500 ng of salmon sperm DNA, and (when indicated) either 2.5 or 25 pmol of the unlabeled probe sequence in 20 μl of binding buffer (100 mmol/liter potassium chloride, 1 mmol/liter EDTA, 0.1 mmol/liter dithiothreitol, 5% [vol/vol] glycerol, 10 ng/ml bovine serum albumin) and incubated at room temperature for 15 min. Samples were run on a 10% native gel in 1× Tris-acetate-EDTA with 50 mM KCl (pH 7.8) and visualized with a Kodak Image Station 2000MM with excitation at 465 nm and a 535-nm filter.
Table 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea |
|---|---|
| 5′ RACE probes | |
| Poly adapter | 5′-CACCTGAGCAGAGTGACGAGGACTACGGCTTCAGCTTTTTTTTTTTTTTTTTT-3′ |
| Poly outer adapter | 5′-CACCTGAGCAGAGTGACG-3′ |
| nuc PCR/5′ RACE | 5′-ACATATGCCAGCACTTAATAAGTATTCTG-3′ |
| nuc cDNA/5′ RACE | 5′-TACTGATAGCCATCCCTATAAGTAATATTG-3′ |
| nuc promoter top | 5′-TTTATTATAATTATTAAATTTTTATTAATTAATTGTAAAAATGTAGAATT-3′ |
| nuc promoter bottom | 5′-AATTCTACATTTTTACAATTAATTAATAAAAATTTAATAATTATAATAAA-3′ |
| EMSA probes | |
| agrB promoter top | 5′-ATTTAACAGTTAAGTATTTATTTCCTACAGTTAGGCAATATAATGATAAA-3′ |
| agrB promoter bottom | 5′-TTTATCATTATATTGCCTAACTGTAGGAAATAAATACTTAACTGTTAAAT-3′ |
| sbi promoter top | 5′-CACTAGTTAATCTATTAGTTAACATTAGTTAATAATTAGTTAATTTCCAT-3′ |
| sbi promoter bottom | 5′-ATGGAAATTAACTAATTATTAACTAATGTTAACTAATAGATTAACTAGTG-3′ |
| hla probe top | 5′-AAATCAATTAATTAACTATTAAATAAAAATTAACTATATATTAACTAGTG-3′ |
| hla probe bottom | 5′-CACTAGTTAATATATAGTTAATTTTTATTTAATAGTTAATTAATTGATTT-3′ |
| FAM/nuc promoter bottom | 5′-FAM/AATTCTACATTTTTACAATTAATTAATAAAAATTTAATAATTATAATAAA-3′ |
| Cloning | |
| MEO63 | 5′-AGCTTGGATCCATCGATGTCGACGCTAGCGGTACCGATATCC-3′ |
| MEO64 | 5′-CCGGGGATATCGGTACCGCTAGCGTCGACATCGATGGATCCA-3′ |
| CEF16 | 5′-GTTGTTAAGCTTTTATTGTGGCAAAAGGTTT-3′ |
| CEF17 | 5′-GTTGTTCCCGGATTATTAGGCGGCATACAG-3′ |
| TaqMan | |
| gyrB fwd | 5′-CAAATGATCACAGCTTTGGTACAG-3′ |
| gyrB rvs | 5′-CGGCATCAG TCATAATGACGAT-3′ |
| gyrB probe | 5′-AATCGGTGGCGACTTTGATCTAGCGAAAG-3′ |
| hla fwd | 5′-CAACAACACTATTGCTAGGTTCCATATT-3′ |
| hla rvs | 5′-CCTGTTTTTACT GTAGTATTGCTTCCA-3′ |
| hla probe | 5′-ATGAATCCTGTCGCTAATGCCGCAGA-3′ |
| nuc fwd | 5′-ATATGGACGTGGCTTAGCGT-3′ |
| nuc rev | 5′-TGAATCAGCGTTG TCTTCGCTCCA-3′ |
| nuc probe | 5′-ACGAAGCTTTAGTTCGTCAAGGCTTGGC-3′ |
HindIII and BamHI restriction digest sites are underlined.
Mouse infection models.
All animal studies were performed in accordance with the National Institutes of Health guidelines and were approved by the Animal Care and Use Committee at Montana State University—Bozeman. For the peritonitis model, male and female BALB/c mice (8 to 10 weeks old) were purchased from commercial sources and the Montana State University Animal Resource Center. All mice were inoculated via the intraperitoneal route with ∼5 × 107 S. aureus bacteria harvested at the mid-exponential growth phase as described previously (31). To determine bacterial burdens, mice were sacrificed at 8 h postinfection and the peritoneal cavity was washed with 10 ml of sterile Hanks' balanced salt solution with an 18-gauge needle and a 10-ml syringe, diluted in distilled water (dH2O), and plated on TSA plates. To determine S. aureus burdens in tissues, hearts and kidneys were aseptically removed (8 h postinfection), washed in dH2O, homogenized in dH2O, and then plated on TSA. For enumeration of CFU, TSA plates were incubated overnight in 37°C and CFU were counted the following day. Separate experiments were used to assess Nuc activity following S. aureus infection by FRET analysis (described above) of peritoneal wash fluids.
Statistical analyses.
All data sets were analyzed with GraphPad Prism, version 5.0 (GraphPad Software, San Diego, CA). All mouse data sets were analyzed with paired t tests on a log scale. A one-way analysis of variance (ANOVA) with Tukey's posttest was used to analyze the expression of the nuc gene by using superfolder green fluorescent protein (sGFP) promoter expression. In bar graphs, error bars represent the standard errors of the means.
RESULTS
nuc expression is controlled by SaeRS.
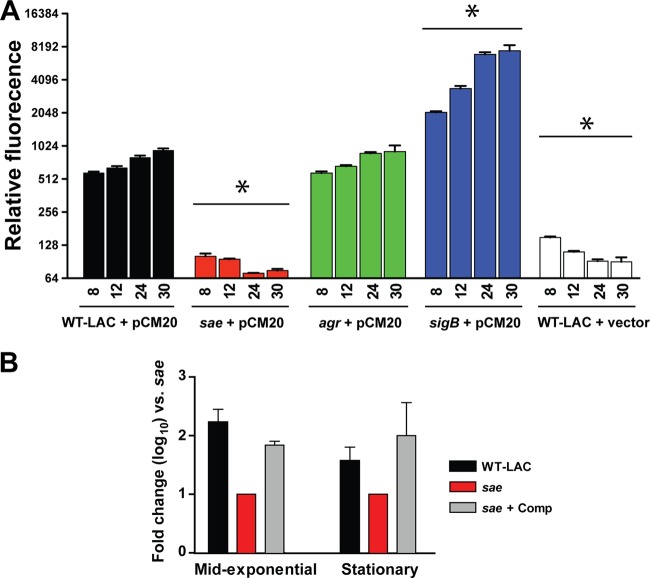
We recently presented preliminary observations that S. aureus nuc gene expression was not under agr quorum-sensing system control at the transcriptional and posttranscriptional levels (9). Microarray studies have suggested that the SaeRS two-component regulatory system controls the transcription of nuc (14, 15, 17). To gain insight into the role of SaeRS in nuc gene regulation, a time course study was performed in the WT LAC, ΔsaeQRS::Spc (sae mutant), and Δagr::Tet (agr mutant) backgrounds containing a nuc promoter-GFP fusion plasmid (pCM20). At each time point, nuc gene expression was significantly reduced in the sae mutant, but not in the agr mutant, compared to that in the WT (Fig. 1A), providing a strong indication that regulation of nuc occurs through SaeRS. A sigB mutant was included as a control since nuc transcription and Nuc enzyme activity are increased in this mutant (9, 32), and indeed, GFP expression was increased in this mutant. To further confirm the influence of SaeRS on nuc gene expression, a TaqMan real-time PCR assay was used to compare nuc transcription during the in vitro growth of the WT LAC, sae mutant, and sae-complemented strains (Fig. 1B). Similar to the reporter fusion experiments (Fig. 1A), nuc expression was differentially regulated by saeRS in vitro during both exponential-phase growth and early stationary-phase growth.
Fig 1.
SaeRS is required for expression of the nuc gene. (A) A plasmid containing the nuc promoter coupled to sGFP (pCM20) was transformed into the USA300 WT LAC and sae, agr, and sigB mutant strains. The WT strain with the empty vector was included as a negative control. GFP fluorescence was measured at 8, 12, 24, and 30 h and normalized to cell growth. Each value is the mean of three independent replicates, and error bars indicate the standard error of the mean. For statistical analysis, a one-way ANOVA was performed; statistical significance (P < 0.01) is indicated by an asterisk. (B) Relative nuc gene expression in USA300 strains grown in vitro. Data are normalized to gyrB transcript abundance, and n-fold change is relative to sae. WT LAC (black bars), the sae mutant (red bars), and the complemented (Comp) mutant strain (gray bars) were grown in TSB, and nuc expression was analyzed during mid-exponential-phase and early stationary-phase growth. Data are from two biological repetitions assayed in triplicate by TaqMan reverse transcription-PCR.
Nuc enzyme levels are dependent on SaeRS and not agr.
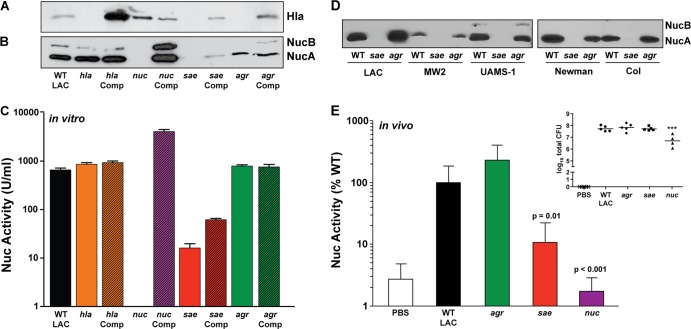
To extend the nuc regulation studies to functional protein assays, immunoblot assays and enzyme activity measurements were performed in parallel with the WT LAC, mutant, and complemented strains. Alpha-toxin (Hla), a well-described secreted virulence determinant that is posttranscriptionally regulated in S. aureus (33), was used as a control in the immunoblot assay experiments (Fig. 2A). In accordance with previously published data (25, 28), both the sae and agrA mutants did not produce alpha-toxin unless complemented. Similar to the observations in Fig. 1, Nuc protein was absent in the sae mutant immunoblot assay (Fig. 2B), and enzyme activity was down 80-fold compared to that of the WT (Fig. 2C). With an saeRS-complementing clone, Nuc protein levels and activity were partially restored (Fig. 2B and C). In contrast, Nuc activity levels were unaffected by an agrA mutation or complementation of the mutation (Fig. 2C).
Fig 2.
SaeRS is required for Nuc enzyme production. Protein immunoblot assays for Hla (A) and Nuc (B) were performed with spent medium from WT LAC and strains containing hla, nuc, sae, and agr mutations grown to stationary phase. The long and processed forms of Nuc are designated NucB and NucA, respectively. A complemented (Comp) version of each mutant was included as a control. (C) The Nuc enzyme activity of these samples was determined with a FRET assay. Statistical analyses were performed with a two-tailed Student t test. (D) Nuc immunoblot assays of sae and agr mutants of strains LAC, MW2, UAMS-1, Newman, and COL were performed, and the NucA and NucB bands are indicated. Variation in Nuc protein levels across S. aureus isolates is SaeRS dependent. (E) Nuc activity in vivo following intraperitoneal infection with S. aureus. Mice were infected with 2 × 107 CFU of WT USA300 or isogenic agr, sae, and nuc mutants. Nuc activity in peritoneal fluids was assayed at 8 h postinfection with a FRET assay (n = 5 mice per strain for the WT and the agr and sae mutants and 4 mice for the nuc mutant). P values were determined by paired t test. PBS, phosphate-buffered saline. The inset shows the number of CFU recovered from mice at the time of peritoneal fluid harvest. ***, P = 0.001 (determined by paired t test).
To further demonstrate that SaeRS influences Nuc protein production in diverse strains, immunoblot assays were performed with a series of representative isolates containing sae and agr mutations. The strain backgrounds included were WT LAC, USA400 (MW2), USA200 (UAMS-1), Newman, and COL (Fig. 2D). In every strain background tested, the production of Nuc was dependent on SaeRS and independent of the agr system, providing additional evidence that Nuc production is under SaeRS control. Collectively, these data, in combination with gene expression studies (Fig. 1), demonstrate that the SaeRS regulatory system is essential for the production of the functional Nuc enzyme across S. aureus genetic backgrounds.
In vivo Nuc production is dependent on SaeRS.
To determine if Nuc activity is controlled in vivo in an SaeRS-dependent manner, we analyzed Nuc activity in mice infected intraperitoneally with USA300 (WT LAC) and isogenic agr, sae, and nuc mutants. Nuc activity was detected by the FRET activity assay in peritoneal fluids from all mice infected with WT LAC, as well as the agr mutant, at 8 h postinfection (Fig. 2E). In contrast, the Nuc activity in the peritoneal fluid of mice infected with the sae mutant was only 10% of the WT LAC level. Decreased Nuc activity in the sae mutant was not due to a reduced bacterial burden at the time of harvest (see inset). As a control, we included the nuc mutant and, as expected, this strain had 1.7% of the activity of the WT (lower than that of the phosphate-buffered saline control), demonstrating the specific nature of the FRET assay in host samples. Taken together, these findings demonstrate that SaeRS is essential for Nuc production in vivo during infection.
SaeR binds directly to the nuc promoter.
To determine if nuc expression is directly regulated by SaeR, promoter mapping and EMSAs were performed. 5′ RACE identified the transcriptional start site of nuc 22 bp upstream of the methionine start codon (Fig. 3A). With the SaeR recognition sequence obtained from previous studies (14), the putative binding site was identified upstream of the transcription start site (yellow in Fig. 3A). EMSA analysis demonstrated that recombinant SaeR binds the nuc promoter specifically and in a concentration-dependent manner. Unlabeled probes containing the defined SaeR-binding sites for the nuc, hla, and sbi promoters (14) successfully competed for SaeR versus the nuc promoter. As expected, titration of the unlabeled agrB promoter, which does not contain the SaeR-binding site, had no effect on the SaeR shift of the nuc promoter (Fig. 3C). The observed binding of SaeR to the nuc promoter is weaker than its binding to the hla and sbi promoter regions. This reduced affinity may be due to the binding conditions tested since SaeR was not phosphorylated in our assays and phosphorylation can affect binding affinity (34). Alternatively, reduced binding could be due to lower homology to the SaeR consensus sequence in the nuc promoter region (14, 34).
Fig 3.
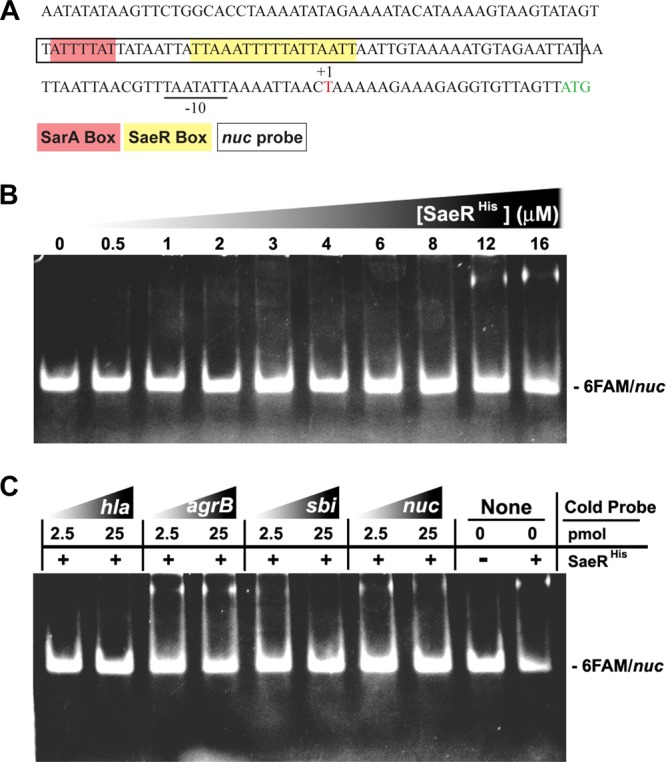
The nuc transcript start site and SaeR binding to the nuc promoter. (A) Sequence results generated by 5′ RACE with nuc-specific primers in Table 2. The nuc transcript start site (+1) and putative −10 and −35 promoter regions are underlined. The Nuc start codon is in green, and the nuc promoter probe sequence (50 bp) used for EMSA experiments is boxed. Putative SaeR (yellow) and SarA (red) boxes are shown. (B) EMSA with increasing concentrations of SaeRHis (0, 0.5, 1, 2, 3, 4, 6, 8, 12, and 16 μmol/liter), 0.5 pmol of the FAM-labeled nuc promoter probe, and 500 ng of salmon sperm DNA. (C) Binding of SaeRHis (16 μmol/liter) to the nuc promoter probe (0.5 pmol) in the presence of 2.5 or 25 pmol of the indicated unlabeled promoter and 500 ng of salmon sperm DNA.
Nuc contributes to pathogen survival during invasive disease.
To date, the role of Nuc in staphylococcal pathogenesis is incompletely defined. Secreted DNase produced by Streptococcus pyogenes has been shown to be important in pathogen dissemination (35), and we addressed the potential role of Nuc in USA300 pathogenesis by using a mouse model of peritonitis. Mice were infected intraperitoneally with WT and nuc mutant USA300. Bacterial burdens in the heart, kidneys, and peritoneum were determined after 8 h. The nuc mutation resulted in statistically significant decreases in the numbers of CFU recovered from all of the locations sampled (Fig. 4A to C), demonstrating that Nuc contributes to S. aureus pathogenesis by promoting bacterial survival. These data support the earlier observation made during analyses of Nuc expression in the peritoneum that demonstrated reduced bacterial recovery (from the peritoneum) in mice infected with the nuc mutant (Fig. 2E, inset).
Fig 4.
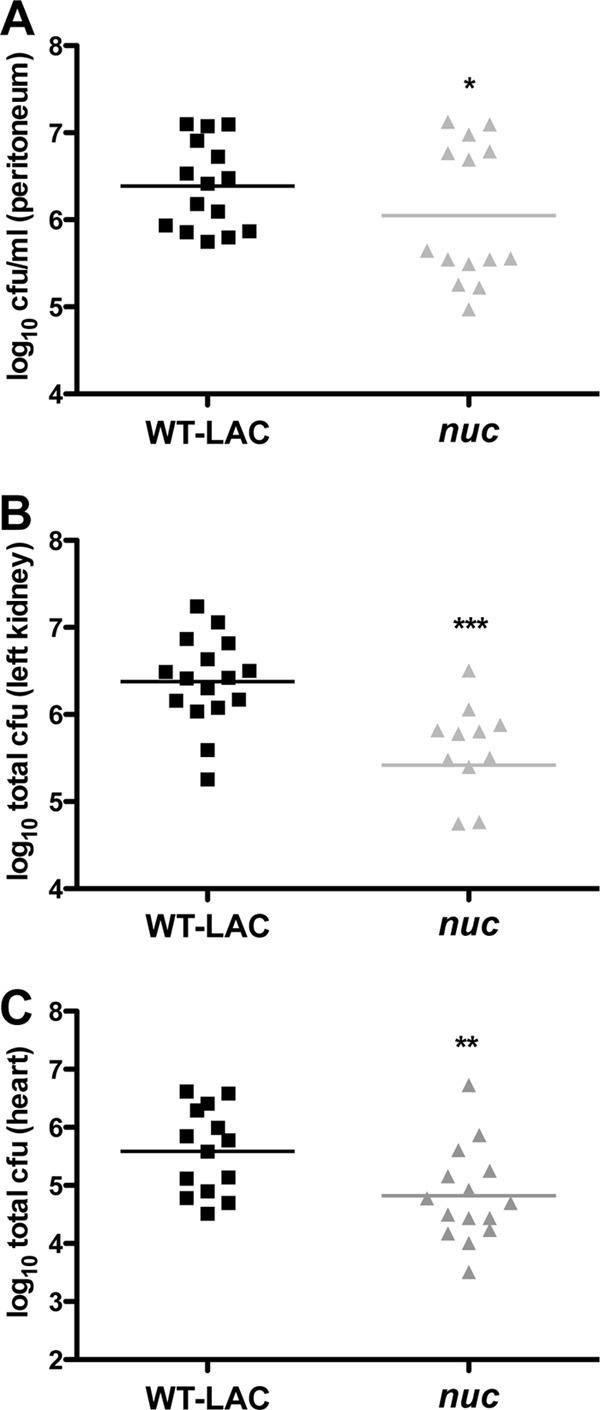
Nuc promotes bacterial survival in a mouse model of peritonitis. Mice were infected intraperitoneally with ∼5 × 107 WT LAC or nuc mutant bacteria for 8 h. Peritoneum (A), kidney (B), and heart (C) bacterial burdens were enumerated. Data are from two separate experiments with seven or eight mice per group. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (determined by paired t test).
Contribution of SaeRS to Nuc variance across S. aureus strains.
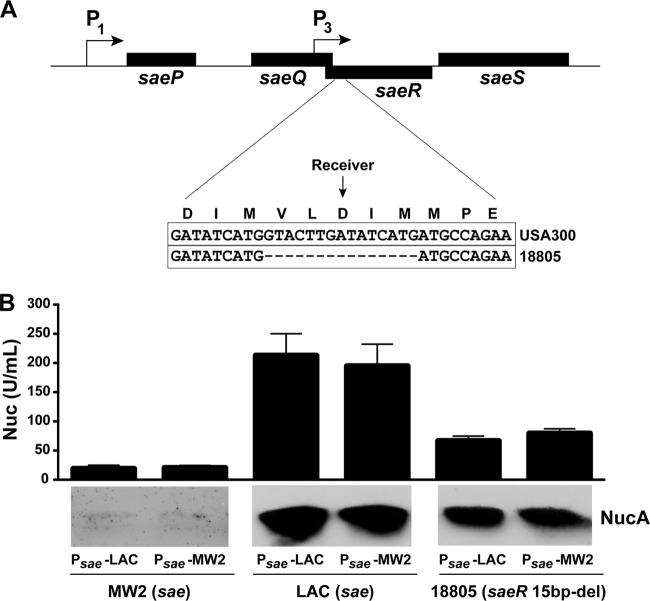
Knowing that nuc expression is dependent on the SaeRS regulatory system, we extended our analysis to investigate whether we could identify strains with altered levels of Nuc production among a collection of clinical isolates. A set of 11 USA300 strains that have been characterized by epidemiological, virulence, and molecular profiles (described in reference 3) were subjected to Nuc activity assays and protein immunoblot assays (Fig. 5), and the WT LAC and nuc mutant strains served as controls. Ten of these USA300 strains possessed high Nuc activity (750 to 1,250 U/ml) and were indistinguishable in the immunoblot assay. The lone exception was strain 18805, an endocarditis isolate with an unusual spa type of 168 (3). Nuc activity in 18805 was ∼20-fold lower than that of other USA300 strains, and the protein was not detectable by immunoblot assay (Fig. 5). Even though this strain was isolated from an endocarditis patient, 18805 has been shown to be attenuated in a model of invasive infection (16), which supports our finding that nuc contributes to pathogen survival following peritonitis (Fig. 4). To determine if reduced Nuc activity in 18805 was due to saeRS, we sequenced the saeRS genes. Surprisingly, we identified a 15-bp deletion within the 18805 saeR gene, from nucleotide 145 to nucleotide 159, deleting five codons that encode SaeR residues VLDIM (Fig. 6A). This VLDIM deletion encompasses the predicted aspartate-receiving residue (D51) on the basis of response regulator alignments (22), presumably rendering SaeR nonfunctional. When a plasmid expressing a USA300 WT sae locus (from strain LAC) was transformed into 18805, the Nuc expression phenotypes were repaired (Fig. 6B), indicating that the defect in Nuc production is due to the saeR 15-bp deletion.
Fig 5.
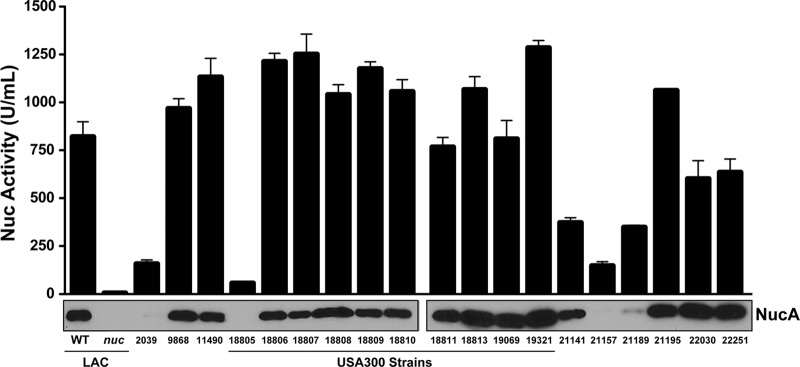
Evaluation of Nuc activity and protein levels across S. aureus strains. A collection of USA300 (11490, 18805 to 19321) and other S. aureus strains were grown in TSB for 15 h, and the cells were removed by filtration. As controls, WT LAC and the nuc mutant were included. Nuc enzyme activity and immunoblot assays of the spent medium were performed. Nuc activity was plotted with errors showing standard deviations, and immunoblot assay results for each sample are shown below the plot. See Table 1 for more information on the strains.
Fig 6.
The 18805 strain saeR mutation and complementation studies of SaeRS polymorphisms. (A) Schematic of the natural saeR mutation in strain 18805 compared to USA300 WT LAC. (B) The WT LAC and MW2 (USA400) strains with sae mutations and natural saeR mutant 18805 were transformed with a plasmid expressing either the WT LAC or the USA400 saePQRS locus. Nuc enzyme activity was measured (top plot), and Nuc immunoblot assays were performed (bottom).
To further analyze the role of SaeRS in nuc expression, we measured Nuc activity in seven additional strains representing diverse genetic backgrounds. Within this set, 2039, 21141 (Mu50), 21157 (USA600), and 21189 (RN1) all displayed activity levels of <380 U/ml and almost no protein in the Nuc immunoblot assay (Fig. 5). After sequencing of the saeRS genes in these isolates, the Mu50 and USA600 strains were found to have N227S and E268K polymorphisms in the SaeS histidine kinase. Similarly, as shown in Fig. 2D, USA400 (MW2) has lower Nuc activity (∼200 U/ml) and its SaeS kinase possesses the same N227S and E268K polymorphisms. The USA300 strains sequenced with high Nuc production did not possess amino acid diversity in SaeRS. Therefore, we speculated that these SaeS differences could be responsible for the low Nuc activity levels in these strains. To investigate this question, low-copy-number complementation plasmids encoding the WT LAC and MW2 (USA400) versions of the saePQRS locus were constructed, and on the basis of sequencing, the resulting complementation plasmids were identical except for the polymorphisms in SaeS. The LAC sae and MW2 sae mutants were transformed with either the USA300 or the USA400 complementing plasmid and assayed for Nuc activity and protein production (Fig. 6B). However, these data did not reveal noticeable effects on Nuc levels with either plasmid construct or cross-complementation of the sae loci in each strain background. Our findings suggest that the differences in Nuc production in these strains are due to factors that are independent of the sae locus.
DISCUSSION
In this study, we investigated the regulation and in vivo function of the secreted Nuc of S. aureus. Despite numerous previous reports that Nuc is under agr quorum-sensing system control (11–13), we compiled strong evidence that the SaeRS two-component system is the dominant regulator of nuc expression both in vitro and in vivo during invasive disease and we demonstrated that SaeR binds directly to the nuc promoter. Our findings also demonstrate that Nuc is an SaeRS-dependent virulence factor important for S. aureus pathogenesis. These findings correlate well with a recent study that demonstrated a defect in the dissemination of an sae mutant in a peritoneal model similar to what we observed with nuc (31), further supporting the concept that Nuc is a key effector of invasive disease. This work also builds on an earlier report that Nuc is important for USA300 immune evasion and lung infection (8).
The discovery that secreted Nuc levels correlated with SaeRS function facilitated the testing of genetically distinct S. aureus isolates. Of 11 well-characterized USA300 isolates (3), 10 secreted high levels of Nuc enzyme (Fig. 5). Notably, one of these 10 isolates, 18811, is a known agr point mutant (3), which again supports our observation that agr is not a significant regulator of Nuc. In the USA300 strain set, the most intriguing isolate was 18805, a strain known to be defective in a murine sepsis model (3). We observed that 18805 also failed to produce Nuc (Fig. 5), and surprisingly, our studies revealed that the saeR gene contained a 15-bp deletion across the encoded aspartate receiver (D51) that likely inactivated the signal transduction system (Fig. 6A). Therefore, the saeR mutation is presumably the explanation for why strain 18805 is defective in alpha-toxin production (3) since saeRS has been shown to be essential to Hla expression (14, 36). Additionally, it is likely why 18805 is attenuated in invasive infection (3) since SaeRS is essential to S. aureus pathogenesis during invasive disease (14, 15). In the absence of a functional SaeRS system, how strain 18805 was able to cause infective endocarditis in a patient is not clear. Biofilm disease is known to increase mutation rates and result in the generation of spontaneous genetic variants (37), and it is possible that the saeR mutation arose during the course of infection. Extending our analysis to a broader strain set, strains Mu50, USA600, and USA400 (MW2) all produced noticeably less Nuc than USA300 strains did (Fig. 5 and 6). We speculated that the difference could be due to the polymorphisms identified in the SaeS sensor kinase (N227S and E268K), but follow-up complementation experiments failed to support this hypothesis. These data warrant further investigation and suggest that other factors in the genetic background, or the basal level of SaeRS function, likely contribute to Nuc expression in these strains.
Collectively, these results identify Nuc as an effector of the SaeRS virulence phenotype, furthering our understanding of the role of this two-component system during staphylococcal disease. Relating these observations to biofilm infections, we recently demonstrated that S. aureus Nuc secretion controls biofilm remodeling (9). In preliminary tests, we observed increased biofilm biomass with an sae mutant and an increase in high-molecular-weight extracellular DNA accumulation (data not shown), both paralleling the reported nuc mutant phenotypes (9) and indicating that Nuc is the biofilm effector for the SaeRS system. Multiple recent studies with Streptococcus pyogenes and Streptococcus pneumoniae demonstrated that secreted Nuc enzymes are important for immune evasion and pathogenesis (35, 38, 39). Coupled with our findings on S. aureus Nuc and those of others (8), it is evident that Nuc is a virulence factor conserved across the Gram-positive cocci. However, more studies are needed to determine its precise role in different types of staphylococcal infections.
ACKNOWLEDGMENTS
We thank C. Malone for strain constructions.
A.R.H. was supported by grant AI083211 from the National Institute of Allergy and Infectious Diseases. M.E.O. was supported by NIH training grant T32 AI07511. J.M.V., T.K.N., K.B.P., and S.G., were supported by NIH grants 5R01AI090046-02, 5R21AI088041-02, and GM103500, as well as the Montana State University Agriculture Experiment Station and an equipment grant from the M. J. Murdock Charitable Trust. A Molecular Biosciences Fellowship (P20RR16455-07) supported R.L.W. and O.W.Z.
Footnotes
Published ahead of print 4 February 2013
REFERENCES
- 1. Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7:629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeLeo FR, Chambers HF. 2009. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J. Clin. Invest. 119:2464–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kennedy AD, Otto M, Braughton KR, Whitney AR, Chen L, Mathema B, Mediavilla JR, Byrne KA, Parkins LD, Tenover FC, Kreiswirth BN, Musser JM, DeLeo FR. 2008. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc. Natl. Acad. Sci. U. S. A. 105:1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mediavilla JR, Chen L, Mathema B, Kreiswirth BN. 2012. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA MRSA). Curr. Opin. Microbiol. 15:588–595 [DOI] [PubMed] [Google Scholar]
- 5. Limbago B, Fosheim GE, Schoonover V, Crane CE, Nadle J, Petit S, Heltzel D, Ray SM, Harrison LH, Lynfield R, Dumyati G, Townes JM, Schaffner W, Mu Y, Fridkin SK. 2009. Characterization of methicillin-resistant Staphylococcus aureus isolates collected in 2005 and 2006 from patients with invasive disease: a population-based analysis. J. Clin. Microbiol. 47:1344–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 7. Cunningham L, Catlin BW, De Garile MP. 1956. A deoxyribonuclease of Micrococcus pyogenes. J. Am. Chem. Soc. 78:4642–4644 [Google Scholar]
- 8. Berends ET, Horswill AR, Haste NM, Monestier M, Nizet V, von Kockritz-Blickwede M. 2010. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate. Immun. 2:576–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kiedrowski MR, Kavanaugh JS, Malone CL, Mootz JM, Voyich JM, Smeltzer MS, Bayles KW, Horswill AR. 2011. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS One 6:e26714 doi:10.1371/journal.pone.0026714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lagacé-Wiens PRS, Manickam MJAK, Karlowsky JA. 2007. Thermostable DNase is superior to tube coagulase for direct detection of Staphylococcus aureus in positive blood cultures. J. Clin. Microbiol. 45:3478–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M, Payne SM. 2011. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect. Immun. 79:1927–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449 [DOI] [PubMed] [Google Scholar]
- 13. Smeltzer MS, Hart ME, Iandolo JJ. 1993. Phenotypic characterization of xpr, a global regulator of extracellular virulence factors in Staphylococcus aureus. Infect. Immun. 61:919–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. 2010. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J. Infect. Dis. 201:241–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voyich JM, Vuong C, DeWald M, Nygaard TK, Kocianova S, Griffith S, Jones J, Iverson C, Sturdevant DE, Braughton KR, Whitney AR, Otto M, DeLeo FR. 2009. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J. Infect. Dis. 199:1698–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Handke LD, Rogers KL, Olson ME, Somerville GA, Jerrells TJ, Rupp ME, Dunman PM, Fey PD. 2008. Staphylococcus epidermidis saeR is an effector of anaerobic growth and a mediator of acute inflammation. Infect. Immun. 76:141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rogasch K, Ruhmling V, Pane-Farre J, Hoper D, Weinberg C, Fuchs S, Schmudde M, Broker BM, Wolz C, Hecker M, Engelmann S. 2006. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J. Bacteriol. 188:7742–7758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146 doi:10.1371/journal.pone.0010146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819–1827 [DOI] [PubMed] [Google Scholar]
- 20. Gillaspy AF, Hickmon SG, Skinner RA, Thomas JR, Nelson CL, Smeltzer MS. 1995. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect. Immun. 63:3373–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. 2009. Interconnections between sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect. Immun. 77:1623–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jensen RO, Winzer K, Clarke SR, Chan WC, Williams P. 2008. Differential recognition of Staphylococcus aureus quorum-sensing signals depends on both extracellular loops 1 and 2 of the transmembrane sensor AgrC. J. Mol. Biol. 381:300–309 [DOI] [PubMed] [Google Scholar]
- 25. Nygaard TK, Pallister KB, Dumont AL, Dewald M, Watkins RL, Pallister EQ, Malone C, Griffith S, Horswill AR, Torres VJ, Voyich JM. 2012. Alpha-toxin induces programmed cell death of human T cells, B cells, and monocytes during USA300 infection. PLoS One 7:e36532 doi:10.1371/journal.pone.0036532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, Long RD, Dorward DW, Gardner DJ, Kreiswirth BN, Musser JM, DeLeo FR. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175:3907–3919 [DOI] [PubMed] [Google Scholar]
- 27. Voyich JM, Otto M, Mathema B, Braughton KR, Whitney AR, Welty D, Long RD, Dorward DW, Gardner DJ, Lina G, Kreiswirth BN, DeLeo FR. 2006. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194:1761–1770 [DOI] [PubMed] [Google Scholar]
- 28. Pang YY, Schwartz J, Thoendel M, Ackermann LW, Horswill AR, Nauseef WM. 2010. agr-dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J. Innate. Immun. 2:546–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scotto-Lavino E, Du G, Frohman MA. 2006. 5′ end cDNA amplification using classic RACE. Nat. Protoc. 1:2555–2562 [DOI] [PubMed] [Google Scholar]
- 30. Hellman LM, Fried MG. 2007. Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2:1849–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Watkins RL, Pallister KB, Voyich JM. 2011. The SaeR/S gene regulatory system induces a pro-inflammatory cytokine response during Staphylococcus aureus infection. PLoS One 6:e19939 doi:10.1371/journal.pone.0019939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bischoff M, Dunman P, Kormanec J, Macapagal D, Murphy E, Mounts W, Berger-Bachi B, Projan S. 2004. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J. Bacteriol. 186:4085–4099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. 2011. Peptide signaling in the staphylococci. Chem. Rev. 111:117–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sun F, Li C, Jeong D, Sohn C, He C, Bae T. 2010. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J. Bacteriol. 192:2111–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Bailey JR, Parnell MJ, Hoe NP, Adams GG, Deleo FR, Musser JM. 2005. Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc. Natl. Acad. Sci. U. S. A. 102:1679–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Montgomery CP, Boyle-Vavra S, Daum RS. 2010. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA MRSA USA300 infection. PLoS One 5:e15177 doi:10.1371/journal.pone.0015177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yarwood JM, Paquette KM, Tikh IB, Volper EM, Greenberg EP. 2007. Generation of virulence factor variants in Staphylococcus aureus biofilms. J. Bacteriol. 189:7961–7967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. 2006. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 16:401–407 [DOI] [PubMed] [Google Scholar]
- 39. Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. 2006. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16:396–400 [DOI] [PubMed] [Google Scholar]