Abstract
Candida albicans causes both mucosal and disseminated infections, and its capacity to grow as both yeast and hyphae is a key virulence factor. Hyphal formation is a type of polarized growth, and members of the SR (serine-arginine) family of RNA-binding proteins influence polarized growth of both Saccharomyces cerevisiae and Aspergillus nidulans. Therefore, we investigated whether SR-like proteins affect filamentous growth and virulence of C. albicans. BLAST searches with S. cerevisiae SR-like protein Npl3 (ScNpl3) identified two C. albicans proteins: CaNpl3, an apparent ScNpl3 ortholog, and Slr1, another SR-like RNA-binding protein with no close S. cerevisiae ortholog. Whereas ScNpl3 was critical for growth, deletion of NPL3 in C. albicans resulted in few phenotypic changes. In contrast, the slr1Δ/Δ mutant had a reduced growth rate in vitro, decreased filamentation, and impaired capacity to damage epithelial and endothelial cells in vitro. Mice infected intravenously with the slr1Δ/Δ mutant strain had significantly prolonged survival compared to that of mice infected with the wild-type or slr1Δ/Δ mutant complemented with SLR1 (slr1Δ/Δ+SLR1) strain, without a concomitant decrease in kidney fungal burden. Histopathology, however, revealed differential localization of slr1Δ/Δ hyphal and yeast morphologies within the kidney. Mice infected with slr1Δ/Δ cells also had an increased brain fungal burden, which correlated with increased invasion of brain, but not umbilical vein, endothelial cells in vitro. The enhanced brain endothelial cell invasion was likely due to the increased surface exposure of the Als3 adhesin on slr1Δ/Δ cells. Our results indicate that Slr1 is an SR-like protein that influences C. albicans growth, filamentation, host cell interactions, and virulence.
INTRODUCTION
The opportunistic pathogen Candida albicans is a multimorphic fungus that is a leading cause of nosocomial bloodstream infections in the United States (1). C. albicans can adopt at least three morphologies, a budding yeast form and two filamentous forms, hyphae and pseudohyphae, which result from polarized cell growth. The ability of C. albicans to switch between yeast and hyphal morphologies has been implicated in virulence in various model systems (2, 3). Compared to C. albicans yeast cells, hyphae have a greater capacity to adhere to and invade host cells (4). Epithelial cell invasion leads to the establishment of mucosal infections, as seen in oropharyngeal and vulvovaginal candidiasis, whereas internal organ infection during disseminated disease requires C. albicans cells to penetrate through the endothelial lining of the vasculature. Given the link between the yeast-hyphal transition and virulence, many studies have explored changes in gene expression during the switch to polarized hyphal growth and the signaling pathways and transcription factors required for this transition (5–9). To date, however, less attention has been focused on posttranscriptional regulation of gene expression in C. albicans morphological switching (10–13).
In numerous eukaryotic systems, posttranscriptional processes are critical for establishing polarity and are frequently controlled by mRNA sequences outside the open reading frame (14–16). Transcriptome analysis of C. albicans recently revealed numerous long 5′ and 3′ untranslated regions (UTRs) and confirmed the presence of more than 400 introns (7, 8, 17), which indicate opportunities for posttranscriptional regulation of gene expression. Three proteins involved in mRNA decay have been linked to C. albicans morphogenesis to date. Strains lacking the 5′-to-3′ exonuclease Kem1/Xrn1 display altered hyphal growth (10) and defects in intermediate stages of biofilm formation (11). Deletion of the mRNA deadenylase components Ccr4 and Pop2 also results in filamentation defects (13). In addition, the She3 mRNA-binding protein binds to and directs hyphal tip localization of specific mRNAs, and absence of this protein affects filamentous growth on solid media (12).
The SR (serine-arginine) family of RNA-binding proteins has been implicated in many steps of mRNA metabolism across eukaryotes. First identified as constitutive and regulated splicing factors, SR proteins are now known to have multiple roles, including linking transcription to splicing, nuclear mRNA export, and translational regulation (18–20). Fungal genomes encode proteins that are structurally similar to SR proteins, and the major SR-like protein in Saccharomyces cerevisiae, Npl3, plays numerous roles in mRNA metabolism (21–26). Interestingly, deletion of Npl3 causes polarity defects—in diploid bud-site selection and in the switch from budding to pseudohyphal growth in haploid cells (27, 28). We therefore sought to identify SR-like proteins in C. albicans that might be involved in posttranscriptional processes that affect morphogenesis. Our search for SR-like proteins similar to Npl3 in C. albicans revealed that the protein most similar to S. cerevisiae Npl3 contained only a single SR dipeptide (29). A second protein, which we have named SR-like RNA-binding protein 1 (Slr1), shows sequence similarity not only to Npl3 but also to the Schizosaccharomyces pombe SR protein Srp1 and to the Aspergillus nidulans protein SwoK, the latter of which has been implicated in polarized growth (30).
Here we describe the characterization of Slr1 in C. albicans and its importance for C. albicans growth, filamentation, and virulence. Decreased growth and filamentation of cells lacking Slr1 in vitro correlated with reduced virulence in a mouse model of disseminated infection, yet this strain displayed a kidney fungal burden equivalent to that for wild-type C. albicans, as well as a greater fungal burden in the brain. The increased ability of cells lacking Slr1 to invade brain endothelial cells in vitro and greater surface exposure of the hypha-specific Als3 adhesin protein on slr1Δ/Δ cells likely contributed to its increased brain tropism in mice. These results indicate that Slr1 is involved in processes that are important for both filamentation and virulence of C. albicans in the mammalian host.
MATERIALS AND METHODS
Strain construction.
Genotypes of strains used in this study are found in Table 1. All C. albicans strains in this study are derived from strain BWP17 and were created using a lithium acetate-based transformation protocol (31). All strains used in growth and virulence studies were prototrophic for arginine and histidine and contained the URA3 gene at its native locus (32). Strain construction is described in the supplemental material and important features of plasmids and oligonucleotides are found in Tables S1 and S2.
Table 1.
Strains used in this study
| Strain | Genotypesa | Parental strain or reference |
|---|---|---|
| AMC8 | npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | BWP17 |
| NPL3 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC18 | npl3Δ::HIS1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC37 |
| npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC20 | npl3Δ::HIS1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC38 |
| npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC22 | npl3Δ::HIS1::NPL3 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC18 |
| npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC24 | npl3Δ::HIS1::NPL3 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC20 |
| npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC37 | npl3Δ::HIS1 ura3Δ::λimm434 arg4::hisG his1::hisG | AMC8 |
| npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC38 | npl3Δ::HIS1 ura3Δ::λimm434 arg4::hisG his1::hisG | AMC8 |
| npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC67 | slr1Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | BWP17 |
| SLR1 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC68 | slr1Δ::HIS1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC90 |
| slr1Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC70 | slr1Δ::HIS1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC89 |
| slr1Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC71 | slr1Δ::hisG npl3Δ::HIS1 ura3Δ::λimm434 arg4::hisG his1::hisG | AMC37 |
| SLR1 npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC72 | slr1Δ::hisG npl3Δ::HIS1 ura3Δ::λimm434 arg4::hisG his1::hisG | AMC71 |
| slr1Δ::hisG npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC81 | ura3Δ::λimm434::URA3-IRO1 arg4::hisG::ARG4 his1::hisG::HIS1 | BWP17 |
| ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC82 | slr1Δ::HIS1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC89 |
| slr1Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC83 | slr1Δ::hisG npl3Δ::HIS1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC72 |
| slr1Δ::hisG npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC87 | slr1Δ::HIS1 ura3Δ::λimm434 arg4::hisG his1::hisG | BWP17 |
| SLR1 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC89 | slr1Δ::HIS1 ura3Δ::λimm434 arg4::hisG his1::hisG | AMC87 |
| slr1Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC90 | slr1Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | AMC67 |
| slr1Δ::HIS1 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC103 | slr1Δ::ARG4::SLR1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC89 |
| slr1Δ::HIS1 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC104 | slr1Δ::ARG4::SLR1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC89 |
| slr1Δ::HIS1 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC105 | slr1Δ::ARG4::SLR1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC90 |
| slr1Δ::HIS1 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC110 | slr1Δ::hisG npl3Δ::HIS1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC72 |
| slr1Δ::hisG npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC111 | slr1Δ::hisG::SLR1 npl3Δ::HIS1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC72 |
| slr1Δ::hisG npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC112 | slr1Δ::hisG::SLR1 npl3Δ::HIS1 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC72 |
| slr1Δ::hisG npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC113 | slr1Δ::hisG npl3Δ::HIS1::NPL3 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC72 |
| slr1Δ::hisGnpl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| AMC114 | slr1Δ::hisG npl3Δ::HIS1::NPL3 ura3Δ::λimm434::URA3-IRO1 arg4::hisG his1::hisG | AMC72 |
| slr1Δ::hisG npl3Δ::ARG4 ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| BWP17 | ura3Δ::λimm434 arg4::hisG his1::hisG | 31 |
| ura3Δ::λimm434 arg4::hisG his1::hisG | ||
| DAY185 | ura3Δ::λimm434 ARG4::URA3::arg4::hisG HIS1::his1::hisG | 59 |
| ura3Δ::λimm434 arg4::hisG his1::hisG |
Boldface indicates differences from the parental strain, BWP17. For each strain, each gene is underlined and the two alleles of the gene are listed in the upper and lower rows.
In vitro growth assays.
To determine the maximal growth rate in liquid culture, indicated strains (5 to 10 replicates, including independently isolated mutant and reconstituted strains) were inoculated into 300 μl yeast extract-peptone-dextrose (YPD) in a 96-deep-well plate and grown overnight (23 to 27 h). Cells were diluted 1,000-fold into 150 μl YPD in flat-bottom 96-well dishes, and growth was monitored in a Tecan Sunrise (San Jose, CA) microplate reader. Cultures were shaken at 30°C or 37°C for 24 h, and optical density at 600 nm was monitored every 15 min. Optical density data were exported to the Excel software program using Magellan data analysis software and subsequently analyzed using growth curve analysis software, kindly provided by Darren Abbey and Sven Bergmann. This Matlab script fits the logistic growth curve to the data for a single well and then calculates the generation time for each sample at its maximal growth rate based on this fit (33, 34). Mean generation times were calculated for each strain.
To test for sensitivity to cell wall and cell membrane stressors, strains were grown overnight in YPD broth (1% yeast extract, 2% Bacto peptone, and 2% glucose), diluted into fresh YPD broth the following morning, and plated after 6 h of growth. Cells were washed and resuspended in phosphate-buffered saline (PBS), and 5 × 104 and 5 × 103 cells were spotted onto YPD agar with or without 0.2% sodium dodecyl sulfate (SDS) or 20 μM calcofluor white and grown for 1 day at 30°C.
Filamentation on solid medium was tested by plating 107 stationary-phase cells on plates containing 2% agar and 5% fetal bovine serum or 1.5% agar and 1× RPMI 1640 medium (Invitrogen 31800 with l-glutamine; pH adjusted to 7.5 with HEPES) or on solid Spider medium (data not shown) (35). Colonies were photographed after 6 days of incubation at 37°C. To test filamentation in liquid culture, cells from overnight cultures grown in YPD broth at 30°C were washed, diluted to 105 cells/ml in prewarmed RPMI 1640 medium without l-glutamine (Irving Scientific), and added to sterile, 12-mm, gelatin-coated glass coverslips in a 24-well tissue culture plate. The cells were incubated in 5% CO2 at 37°C for 1.5 or 3 h, fixed with 3% paraformaldehyde, and imaged by differential-interference contrast microscopy.
Host cells.
The FaDu oral epithelial cell line was obtained from the American Type Culture Collection and grown as recommended by the supplier. Human umbilical vein endothelial cells (HUVECs) were harvested by the method of Jaffe et al. (36) and grown as described previously (37). Human brain microvascular endothelial cells (HBMECs) were grown as described previously (38, 39).
Host cell damage assay.
The capacities of the different C. albicans strains to damage the FaDu oral epithelial cell line, HUVECs, and HBMECs were determined by a 51Cr release assay as previously outlined (37). Briefly, the host cells were grown to 90% confluence in 96-well tissue culture plates, loaded with 51Cr overnight, and then inoculated with yeast-phase C. albicans cells. For experiments with FaDu cells, the inoculum was 1 × 105 C. albicans cells per well suspended in RPMI 1640 medium without l-glutamine (Irvine Scientific), and the incubation period was 3 h. For HUVECs, the inoculum was 4 × 104 C. albicans cells per well in RPMI 1640 medium, and the incubation period was 3 h (40). Because HBMECs were resistant to C. albicans-induced damage, the inoculum was 1 × 105 C. albicans cells per well in Ham's F-12K medium, and the incubation period was increased to 16 h. At the end of the incubation period, the medium above the cells was collected, and the 51Cr content was determined by γ counting. Damage assays for each host cell type were performed in triplicate on three separate occasions with the wild-type (AMC81), slr1Δ/Δ mutant (AMC82), and slr1Δ/Δ mutant complemented with SLR1 (slr1Δ/Δ+SLR1) (AMC104) strains.
Endocytosis assay.
Endocytosis by HUVECs and HBMECs of the different C. albicans strains was assessed using a differential fluorescence assay as described previously (39). Each type of endothelial cell was grown on fibronectin-coated glass coverslips in 24-well tissue culture plates and then infected with 105 C. albicans yeast cells in RPMI 1640 medium. After 3 h of incubation, the cells were fixed with 3% paraformaldehyde, after which the noninternalized cells were stained with anti-C. albicans rabbit serum (Biodesign International) conjugated with Alexa 568 (Invitrogen). Next, the endothelial cells were permeabilized in 0.1% (vol/vol) Triton X-100 in PBS, and then both the internalized and noninternalized organisms were stained with anti-C. albicans rabbit serum conjugated with Alexa Fluor 488 dye (Invitrogen). The coverslips were viewed using an epifluorescence microscope (Axiovert 10; Carl Ziess, Inc.), and the number of organisms endocytosed by the endothelial cells was determined by subtracting the number of noninternalized organisms (which fluoresced red) from the total number of organisms (which fluoresced green). More than 100 organisms were counted on each coverslip, and the experiments were performed in triplicate on three separate occasions.
Virulence studies.
The relative virulences of the various C. albicans strains were determined using mouse models of hematogenously disseminated candidiasis (HDC) (41) and oropharyngeal candidiasis (OPC) (42, 43) using male ICR mice (Taconics Farms). In the HDC model, eight mice per strain were injected via the tail vein with 7.5 × 105 yeast and monitored for survival over a 3-week period. Seven or eight additional mice were similarly infected and sacrificed after 4 days, after which the fungal burdens of the kidney, liver, and brain were analyzed by quantitative culture as previously described (39). The histopathology of a kidney from each mouse was examined by periodic acid-Schiff staining (44). A second survival study was also performed with an slr1Δ/Δ mutant generated from an independent slr1Δ/SLR1 heterozygous strain and a corresponding reconstituted strain, as well as the wild-type strain, using 10 mice per strain and the same inoculum.
In the OPC model, 6 to 7 mice per C. albicans strain were immunosuppressed with cortisone acetate (200 mg/kg of body weight; Sigma-Aldrich) administered subcutaneously on days −1, 1, and 3 relative to infection. On the day of infection, the mice were sedated with ketamine and xylaxine, after which a calcium alginate swab saturated with 106 C. albicans yeast cells was placed sublingually for 75 min. After 5 days of infection, the mice were sacrificed, and then their tongues were harvested, weighed, and homogenized. C. albicans cells in homogenized tissue were quantified by culture on solid on Sabouraud dextrose agar containing 10 μg/ml chloramphenicol (34).
Flow cytometry.
To assess the level of Als3 expression on the surface of the various strains, the organisms were germinated for 90 min in liquid RPMI 1640 medium containing 10% fetal bovine serum at 37°C. The resulting germ tubes were fixed in 3% paraformaldehyde and blocked with normal goat serum. Surface-exposed Als3 was detected by staining with a polyclonal rabbit anti-Als3 primary antibody (37) followed by an Alexa Fluor 488-labeled goat anti-mouse secondary antibody. The amount of Als3 staining was quantified by flow cytometry.
Statistics.
Differences in the generation times between different C. albicans strains in vitro were determined by analysis of variance with a Tukey posttest. Differences in the host cell interactions among the various C. albicans strains in vitro were evaluated by analysis of variance. Differences in the survival of mice infected with the different strains were analyzed by the log rank test, and differences in organ fungal burden were assessed using the Wilcoxon rank sum test.
RESULTS
Identification of SR-like proteins in C. albicans.
A BLAST search of the C. albicans genome with the ScNpl3 sequence revealed two proteins with strong similarity to Npl3 (Fig. 1). The apparent C. albicans Npl3 ortholog (CaNpl3) shares a core domain structure with SpSrp2 and with ScNpl3: two RNA recognition motifs (RRMs), followed by an arginine-rich domain (Fig. 1). Each protein, however, contains features not seen in the other two proteins. ScNpl3 has an additional N-terminal domain that the other two proteins lack. The arginine-rich domain of SpSrp2 is less repetitive than those of the Npl3 proteins and is followed by a higher-complexity C-terminal region not found in the Npl3 proteins. Whereas the arginine-rich domains of SpSrp2 and ScNpl3 contain multiple serine-arginine (SR) or arginine-serine (RS) dipeptides, CaNpl3 has only a single SR dipeptide, which is found at the C terminus of the protein in the heptapeptide RERSPTR.
Fig 1.
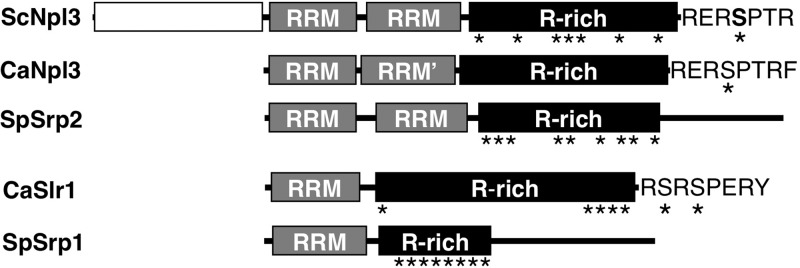
C. albicans proteins similar to S. cerevisiae and S. pombe SR-like proteins. In domain comparison of two SR-like proteins in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Candida albicans, RNA recognition motifs (RRM) and arginine-rich (R-rich) domains, as well as nonconserved N- and C-terminal domains, are shown. Asterisks indicate locations of SR/RS dipeptides. The second domain of the C. albicans Npl3-like protein (RRM′) has sequence similarity with the other two atypical RRMs in ScNpl3 and SpSrp2 but is not recognized as an RRM by protein signature recognition searches (InterProScan). GenBank accession numbers for these proteins are as follows: NP_010720.1 (ScNpl3), XP_715038.1 (CaNpl3), NP_594570.1 (SpSrp2), CAA22007.1 (CaSlr1), and NP_596398.1 (SpSrp1).
The S. cerevisiae genome contains no apparent ortholog of the S. pombe SR-like protein (SpSrp1) gene SRP1 (45), but ORF19.1750 in C. albicans encodes an SR-like protein with a domain structure similar to that of SpSrp1: a single RRM followed by a long arginine-rich region (Fig. 1). Reciprocal BLASTP searches of the C. albicans and S. pombe proteomes using the RRM sequences from each protein indicate a strong sequence similarity in these regions. Based on this similarity, we have named ORF19.1750 SR-like RNA-binding protein 1 (SLR1). Both the SpSRP1 and C. albicans SLR1 (CaSLR1) genes contain introns, and the location of the 5′ splice site for the first intron is the same in the two genes, occurring 44 nucleotides downstream of the final codon of the RNP-2 motif within the RRM, suggesting an evolutionary relationship between these genes. BLASTP searches with the RRM domains of CaSlr1 and SpSrp1 revealed similar proteins in numerous fungal lineages, but no apparent orthologs were found in Zygosaccharomyces rouxii, Vanderwaltozyma polyspora, or Saccharomyces species.
Although CaSlr1 has these structural similarities to SpSrp1, its sequence reveals distinct differences from SpSrp1. Similar to SpSrp2, SpSrp1 contains an additional domain C-terminal to the arginine-rich region (Fig. 1). The amino acid composition of the arginine-rich region of Slr1 more closely resembles those of the Npl3 proteins than that of SpSrp1. In addition, the C terminus of CaSlr1 (RSRSPER) resembles that of ScNpl3 and CaNpl3 (RERSPTR) (identical residues are underlined) (Fig. 1), a known target for ScNpl3 serine phosphorylation (46, 47). In contrast to the Npl3 proteins, however, most of the SR/RS peptides in Slr1 are grouped in a 16-residue region at the C terminus.
Slr1 is more important for C. albicans growth than Npl3.
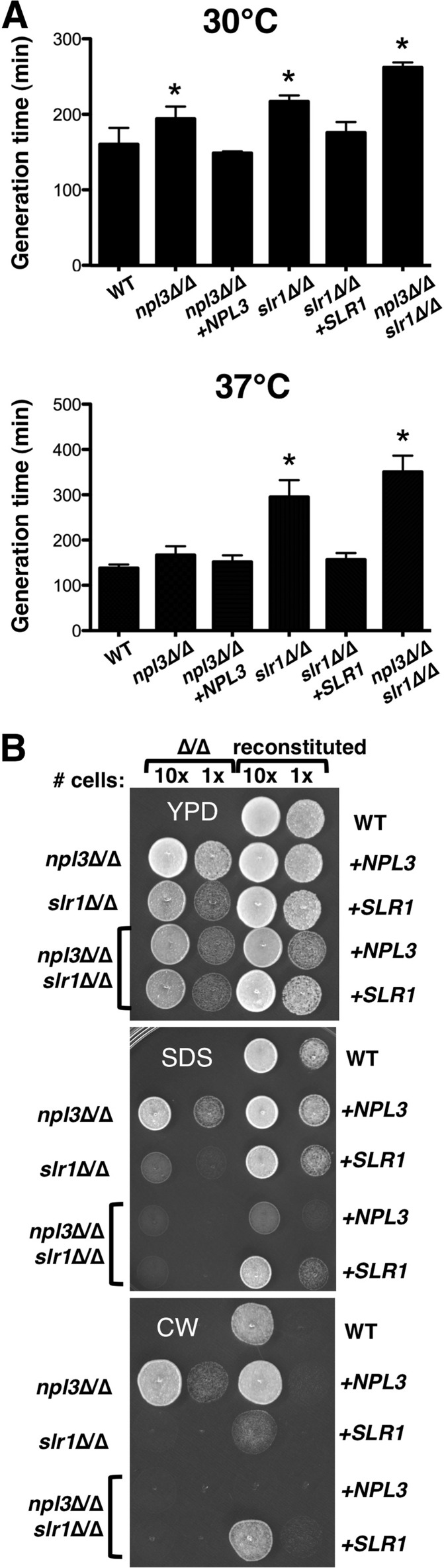
To investigate the roles of SR-like proteins in C. albicans, strains that lacked Npl3, Slr1, or both proteins were constructed. Wild-type copies of each gene were then reintegrated at their native loci in the homozygous deletion mutants to create reconstituted, heterozygous control strains. In contrast to the importance of Npl3 and Srp2 for growth of S. cerevisiae and S. pombe, deletion of CaNPL3 only slightly slowed growth of C. albicans in rich medium at 30°C, and the generation time at maximal growth rate for npl3Δ/Δ strains was not significantly different from that of a wild-type strain at 37°C (Fig. 2A; see also Fig. S1A in the supplemental material). Deletion of SLR1, however, did increase the generation time in YPD broth at both temperatures, with a greater difference from that of the wild type at 37°C (Fig. 2A). C. albicans was viable in the absence of both SR-like proteins, and the absence of Npl3 slowed growth of slr1Δ/Δ cells at both temperatures (npl3Δ/Δ slr1Δ/Δ versus slr1Δ/Δ: 30°C, P < 0.01; 37°C, P < 0.05). In addition to altering the maximal growth rate, deletion of SLR1 increased the lag phase at 37°C but not at 30°C (see Fig. S1B).
Fig 2.
Growth defects associated with SLR1 deletion in C. albicans. (A) Generation times for indicated strains (5 to 10 replicates) were calculated from 24-h growth curves as described in Materials and Methods. Complete genotypes are presented in Table 1. WT, wild type. (B) Two dilutions (10× = 5 × 104 cells; 1× = 5 × 103 cells) of strains were grown at 30°C for 1 day on YPD without or with 0.02% SDS or 20 μM calcofluor white (CW). Relevant genotypes of deletion strains tested (2 left spots) are denoted on the left; the wild-type gene reintroduced into each deletion strain to create reconstituted strains (2 right spots) is denoted on the right.
To determine whether SR-like proteins affected the stress sensitivity of C. albicans, cells were grown at low (16°C) temperature and in the presence of cell membrane and wall stressors, sodium dodecyl sulfate (SDS) and calcofluor white, respectively. Growth of both npl3Δ/Δ and slr1Δ/Δ mutant strains was reduced at low temperature (see Fig. S2 in the supplemental material), as is seen with many S. cerevisiae mutants with defects in RNA processing (48). The npl3Δ/Δ mutant did not display increased sensitivity to SDS or calcofluor white, whereas slr1Δ/Δ cells were extremely sensitive to both of these stressors (Fig. 2B). The double mutant strain was slightly more sensitive to SDS than the slr1Δ/Δ strain. Reconstitution of the double mutant strain with either SLR1 or NPL3 resulted in a phenotype equivalent to that of the single mutant strains. Thus, Slr1 is more important than Npl3 for growth of C. albicans in vitro, particularly during cell wall and membrane stress.
Slr1 is required for normal hyphal formation.
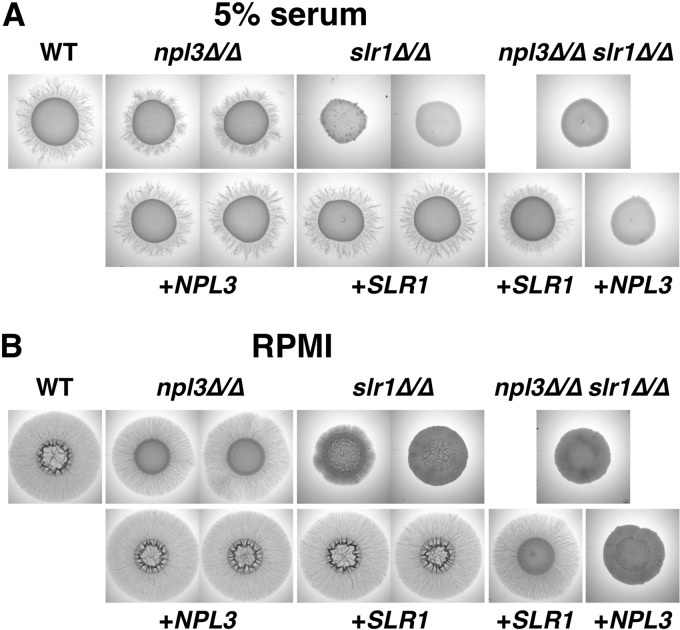
SR-like proteins have been implicated in polar growth of fungi, including the budding yeast S. cerevisiae (27, 28) and the filamentous fungus A. nidulans (30). We therefore tested the ability of C. albicans cells lacking SR-like proteins to filament under different inducing conditions. On solid medium, deletion of SLR1 decreased filamentous colony growth. Wild-type and npl3Δ/Δ cells formed colonies with peripheral filaments on serum and RPMI agar plates, whereas these radiating filaments were absent from slr1Δ/Δ colonies (Fig. 3). Cells lacking both SR-like proteins formed colonies similar to slr1Δ/Δ colonies on these media (Fig. 3), and reconstitution with wild-type copies of NPL3 or SLR1 restored the WT or single mutant phenotypes (Fig. 3).
Fig 3.
Absence of Slr1 decreases filamentation on solid medium. Stationary-phase cells (107) with the indicated genotypes were spotted onto plates containing 5% serum (A) or onto RPMI agar plates (B) and grown for 6 days at 37°C.
Filamentation in liquid culture allows the relative speed and form of filamentation to be compared among strains. Although slr1Δ/Δ cells showed slower filamentation after incubation in RPMI at 37°C for 1.5 h (Fig. 4A), by 3 h, all strains formed hyphae (Fig. 4B). Cells lacking NPL3 formed hyphae similar to those of wild-type cells under both conditions (Fig. 4). Thus, Slr1 has a stronger effect than Npl3 on C. albicans filamentation in vitro, and this effect is more pronounced on solid medium.
Fig 4.
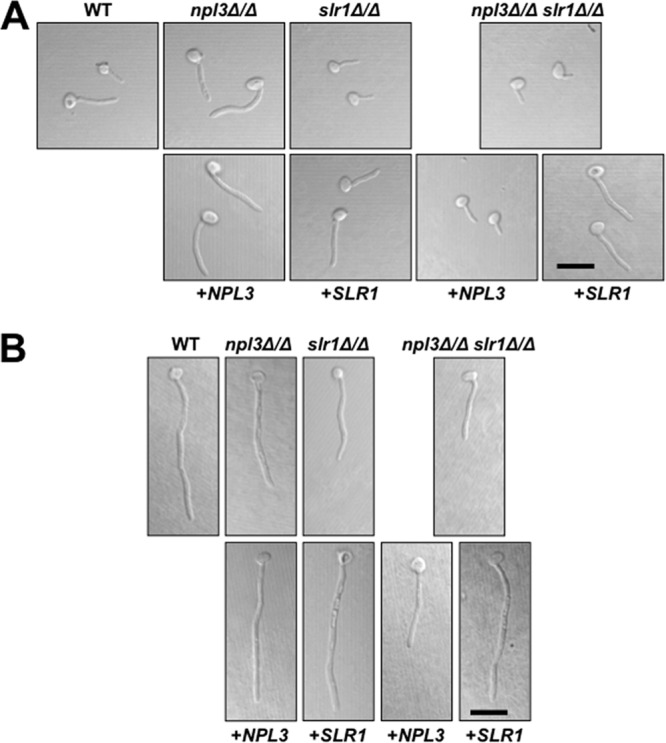
Cells lacking Slr1 can form hyphae in liquid RPMI. Overnight YPD cultures of cells with the indicated genotypes were diluted to 105 cells/ml in RPMI and incubated on gelatin-coated coverslips for 1.5 (A) or 3 (B) h at 37°C and 5% CO2. Paraformaldehyde-fixed cells were imaged by differential interference contrast (DIC) microscopy. Scale bars = 5 μm.
Deletion of SLR1 reduces C. albicans damage to host cells in vitro.
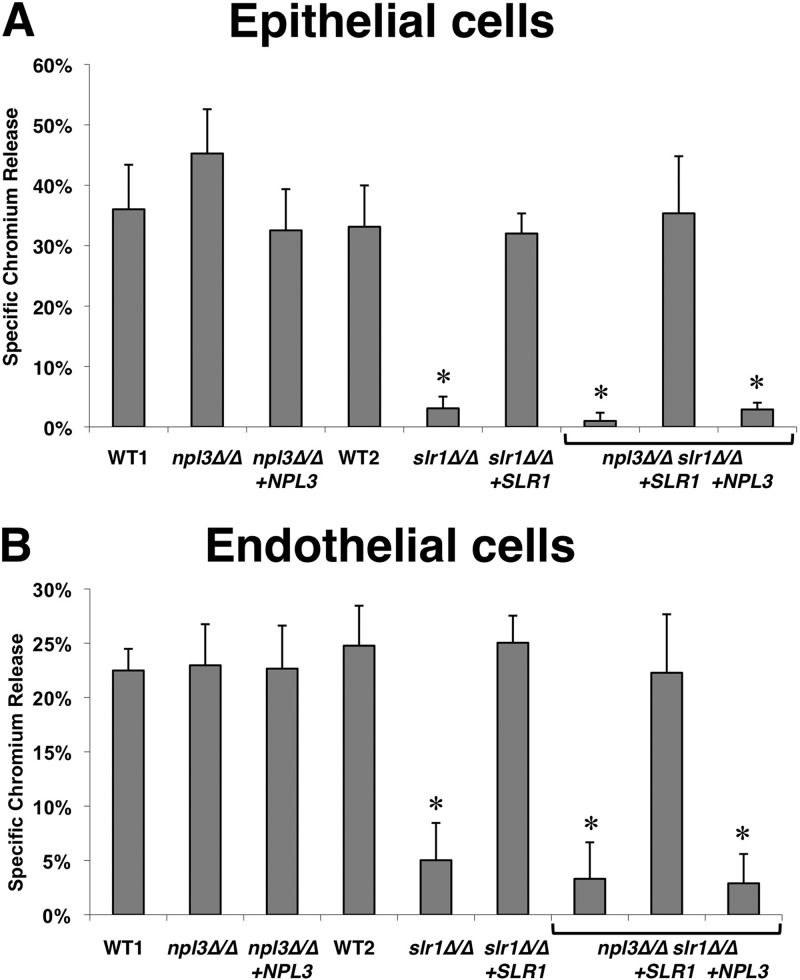
Given the effects of SLR1 and NPL3 deletion on filamentation, we tested whether these genes also affected interaction of C. albicans with the FaDu oral epithelial cell line and HUVECs (49, 50). Whereas the epithelial cell damage caused by npl3Δ/Δ cells was slightly but not significantly greater than that induced by wild-type or reconstituted npl3Δ/Δ+NPL3 cells, deletion of SLR1 significantly decreased the capacity of C. albicans to damage oral epithelial cells in vitro (Fig. 5A). The epithelial cell damage defect of with slr1Δ/Δ was so great that it was not possible to determine if deletion of both SLR1 and NPL3 caused a further reduction in damage. C. albicans-induced damage to HUVECs was also decreased by deletion of SLR1 (Fig. 5B). On both epithelial and endothelial cells, mutants in which SLR1 was deleted grew as a mixture of short true hyphae and pseudohyphae (data not shown). This defect in hyphal formation likely contributed to the reduced damage caused by the slr1Δ/Δ mutants. Collectively, these results indicate that SLR1 but not NPL3 is required for C. albicans to cause maximal damage to host cells in vitro.
Fig 5.
Deletion of SLR1 decreases cell damage by C. albicans in vitro. FaDu oral epithelial cells (A) or human umbilical vein endothelial cells (B) were preloaded with 51Cr and incubated for 3 h at 37°C with the following C. albicans strains: the wild type (WT1, DAY185; WT2, AMC81), the npl3Δ/Δ mutant (AMC18), the npl3Δ/Δ+NPL3 strain (AMC22), the slr1Δ/Δ mutant (AMC82), the slr1Δ/Δ+SLR1 strain (AMC104), the npl3Δ/Δslr1Δ/Δ mutant (AMC83), the npl3Δ/Δ slr1Δ/Δ+SLR1 strain (AMC111), and the npl3Δ/Δ+NPL3 slr1Δ/Δ strain (AMC113). Results are the means ± SD of data from 3 experiments, each performed in triplicate. *, P < 0.01 compared to results for the wild type.
SLR1 is important for maximal C. albicans virulence during OPC and HDC.
The defects of the slr1Δ/Δ mutant in damaging oral epithelial cells and HUVECs suggested that this mutant would have attenuated virulence in animal models of infection (41). To test this hypothesis, the virulence of the wild-type strain, the slr1Δ/Δ mutant, and the slr1Δ/Δ+SLR1 reconstituted strain was tested in mouse models of OPC and HDC. In the OPC model, the median oral fungal burden of mice infected with the slr1Δ/Δ mutant was more than 10-fold lower than that of mice infected with either the homozygous SLR1/SLR1 wild-type strain or the slr1Δ/Δ+SLR1 reconstituted strain (Fig. 6). However, this reduction in oral fungal burden in the mice infected with the slr1Δ/Δ mutant was statistically significant compared with that of mice infected with the wild-type strain (P = 0.02) but not the slr1Δ/Δ+SLR1 reconstituted strain (P = 0.07). The oral fungal burden in mice infected with wild-type and reconstituted strains was not significantly different (P = 0.48).
Fig 6.
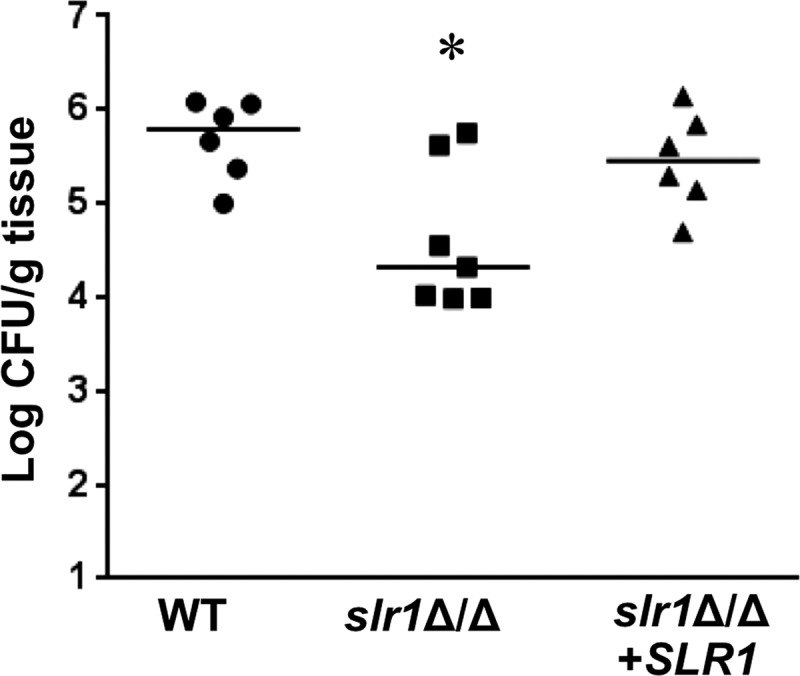
Effects of deletion of SLR1 on C. albicans virulence in the mouse model of OPC. Immunocompromised mice were infected orally with the wild type (AMC81), the slr1Δ/Δ mutant (AMC82), or the slr1Δ/Δ mutant complemented with SLR1 (AMC104). The oral fungal burden was determined after 5 days of infection for 6 or 7 mice per strain. Each symbol represents the results for an individual mouse, and the horizontal lines indicate the median values. *, P = 0.02 compared to results for the wild-type strain.
In the HDC model, the median survival of mice injected with the slr1Δ/Δ mutant was more than twice as long as that of mice infected with either the wild-type strain or the slr1Δ/Δ+SLR1 reconstituted strain (Fig. 7A). Therefore, SLR1 is necessary for maximal virulence during HDC. To verify this finding, we repeated the survival experiment with a second, independent pair of slr1Δ/Δ mutant and slr1Δ/Δ+SLR1 reconstituted strains and obtained similar results (see Fig. S3 in the supplemental material). These data indicate that the presence of Slr1 affects expression of genes that are important for virulence during HDC.
Fig 7.
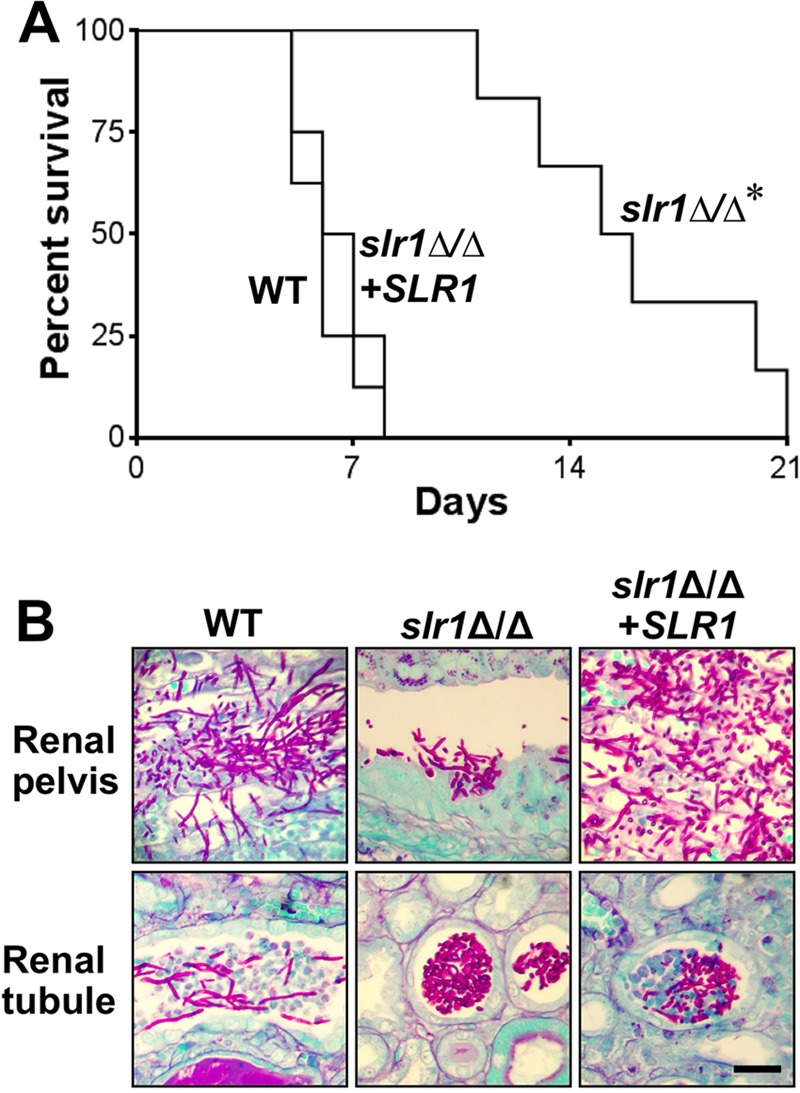
Deletion of SLR1 affects C. albicans virulence, filamentation, and intrakidney localization during disseminated candidiasis. (A) Survival of mice injected intravenously with the wild type (AMC81), the slr1Δ/Δ mutant (AMC82), or the slr1Δ/Δ mutant complemented with SLR1 (AMC104). Each strain was injected into 8 mice, and host survival was monitored over a 3-week period. *, P < 0.0001 versus results for other strains. (B) Histopathology of kidney sections after 4 days of infection with the indicated strains. Sections were stained with periodic acid-Schiff stain. Scale bar = 20 μm.
To determine whether the attenuated virulence of the slr1Δ/Δ mutant in the HDC model was solely due to slow growth in vivo, the organ fungal burden was determined 4 days postinfection. Although the slr1Δ/Δ mutant grew slower than the wild-type strain in vitro (Fig. 2A), the kidney and liver fungal burdens of mice infected with the slr1Δ/Δ strain were similar to those of mice infected with the wild-type strain (see Fig. S4 in the supplemental material). Therefore, the virulence defect of the slr1Δ/Δ mutant did not appear to be the result of impaired growth in vivo.
Next, we performed histopathologic analysis of thin sections of the infected kidneys from three mice to determine effects of SLR1 deletion on C. albicans filamentation. As expected, the wild-type cells formed extensive filaments in both the renal pelvis and renal tubules (Fig. 7B). In contrast, although the slr1Δ/Δ cells formed some filaments in the renal pelvis, they grew as large aggregates of yeast in the renal tubules. The morphology of C. albicans cells containing a single reintroduced copy of SLR1 was intermediate to that of the wild-type strain and the slr1Δ/Δ mutant. These results indicate that SLR1 influences the capacity of C. albicans to form hyphae within the renal tubules but not within the renal pelvis.
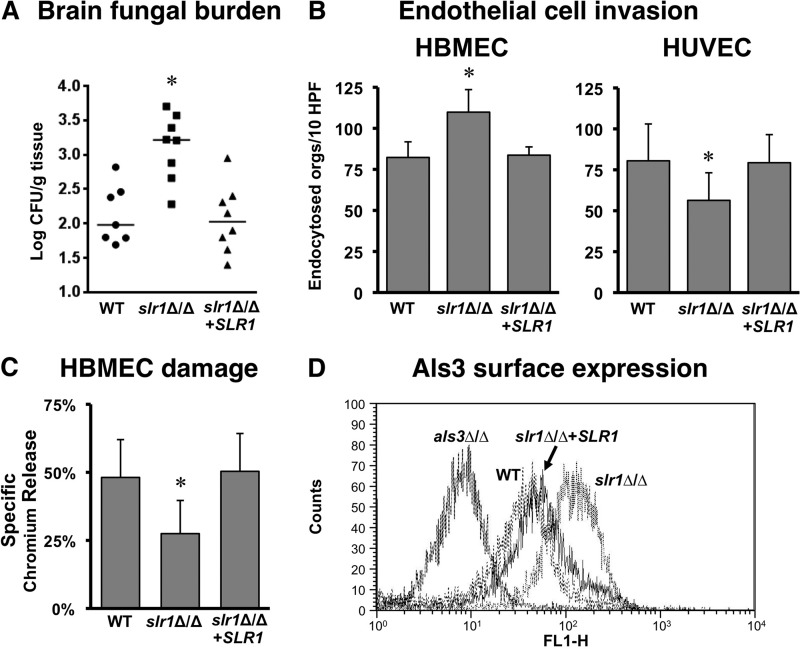
The slr1Δ/Δ mutant had increased tropism for the brain.
Interestingly, the brain fungal burden of mice infected with the slr1Δ/Δ strain was significantly higher than that of either the wild type or the reconstituted strain (Fig. 8A). This result suggested that deletion of SLR1 might increase the interaction of C. albicans with the unique endothelial cells that line the blood vessels of the brain. We therefore compared the capacity of the slr1Δ/Δ mutant to invade HBMECs, representative of brain endothelial cells, and HUVECs, representative of systemic endothelial cells. We found that the slr1Δ/Δ mutant had significantly increased invasion of HBMECs compared to that of the wild-type and reconstituted strains (Fig. 8B). In contrast, the slr1Δ/Δ mutant had significantly impaired invasion of HUVECs. Thus, it is probable that the increased capacity of the slr1Δ/Δ mutant to invade brain endothelial cells contributed to the elevated brain fungal burdens in mice infected with this strain.
Fig 8.
Deletion of SLR1 increases brain tropism of C. albicans. (A) Deletion of SLR1 increases fungal burden in the brain. Brain fungal burdens of mice 4 days after intravenous inoculation with the wild-type (AMC81), slr1Δ/Δ (AMC82), or slr1Δ/Δ+SLR1 (AMC104) strain are shown. Each symbol represents the results for an individual mouse, and the horizontal lines indicate the median values. *, P < 0.005 compared to results for the wild-type and reconstituted strains. (B and C) The strains in panel A were tested for invasion of HBMECs and HUVECs after 3 h of incubation (B) and for their capacity to cause HBMEC damage after 16 h of infection (C). Results are the means ± SD for 3 experiments, each performed in triplicate. *, P ≤ 0.02 versus results for the wild-type and reconstituted strains. (D) Level of surface expression of Als3 for the indicated strains as assessed by flow cytometry. Abbreviations: orgs, oganisms; HPF, high-powered field.
Interestingly, although the slr1Δ/Δ mutant had enhanced invasion of HBMECs, it still caused significantly less damage to these endothelial cells than the wild-type and reconstituted strains (Fig. 8C). These results suggest that brain endothelial cell invasion, rather than damage, is an important determinant of brain tropism.
We have found previously that a vps51Δ/Δ mutant also has increased capacity to invade HBMECs in vitro and traffic to the brain in the mouse model of HDC. This enhanced trafficking is due in part to increased expression of the Als3 invasin on the surface of the vps51Δ/Δ mutant (39). Interestingly, flow cytometric analysis revealed that slr1Δ/Δ germ tubes also had greater surface expression of Als3 than those of the wild-type and reconstituted strains. This enhanced expression of Als3 likely contributes to the increased trafficking of the slr1Δ/Δ mutant to the brain.
DISCUSSION
To date, studies of how gene expression influences C. albicans hyphal development and function have focused primarily on transcriptional and posttranslational signaling mechanisms (9). The prevalence of posttranscriptional processes that control polar growth in other eukaryotic systems (14–16) led us to examine the roles of two RNA-binding proteins encoded in the C. albicans genome, Npl3 and Slr1. Orthologs of NPL3 had previously been identified as essential genes in S. cerevisiae and S. pombe (51–53), and a transposon mutagenesis screen for S. cerevisiae genes required for the switch from budding to pseudohyphal growth implicated NPL3 in polar growth (28). In contrast, we found that C. albicans cells lacking NPL3 grew as robustly as wild-type cells at 37°C and displayed wild-type hyphal formation. These results were consistent with those of Bonhomme and colleagues: deletion of NPL3 did not affect hyphal formation on RPMI agar, in liquid Lee's medium, and under embedded conditions (54). The absence of SLR1, however, slowed growth at 37°C, increased sensitivity to cell wall and membrane stresses, altered filamentation on both serum and RPMI agar medium, and retarded germ tube elongation in liquid medium. Although the S. cerevisiae genome does not encode an apparent ortholog of SLR1, SLR1 gene structure and the predicted Slr1 RNA recognition motif sequence suggest an evolutionary relationship with Srp1, a nonessential SR-like protein in S. pombe, and SwoK in the filamentous fungus A. nidulans. SwoK was identified in a screen for cells with a “swollen” phenotype upon conidial germination at the restrictive temperature, indicating a switch from the polarized growth required for mycelium formation to isotropic growth (30). The filamentation phenotypes of slr1Δ/Δ C. albicans cells and the swoK1 A. nidulans mutant suggest the importance of this family of RNA-binding proteins for polar growth in multiple fungi.
Previous studies in C. albicans have linked filamentous growth to RNA-binding proteins involved in mRNA turnover and transport. Similar to Slr1, the 5′-to-3′ exonuclease Kem1, which degrades mRNAs following decapping, the deadenylase complex proteins Ccr4 and Pop2, and the mRNA-binding protein She3, which helps localize multiple mRNAs to the hyphal tip, are all required for peripheral filaments to form around colonies grown on solid Spider medium (10, 12, 13). Although hyphal formation has been linked to virulence, a recent large-scale study of strains with deletions in 674 genes has shown that almost half of all strains with attenuated infectivity in a mouse model of hematogenously disseminated candidiasis have no in vitro proliferation or filamentation defect on Spider medium (55). Conversely, two-thirds of the mutant strains with the strongest morphology defects in vitro still infected mice as well as the wild-type strain (55). Therefore, filamentation defects in vitro do not necessarily result in virulence defects in vivo. We found that the slr1Δ/Δ mutant had attenuated virulence in the mouse model of HDC. A ccr4Δ/Δ mutant also has attenuated virulence in this model system (13), whereas a she3Δ/Δ mutant has normal virulence in mice (12). Although the kem1Δ/Δ mutant has not been tested in this model, this mutant does have lower virulence in a Galleria mellonella model of infection (56).
The attenuated virulence of the slr1Δ/Δ mutant was likely multifactorial. This strain had impaired capacity to damage both endothelial and epithelial cells in vitro, a phenotype that has been associated with virulence defects in mouse models of candidiasis (40, 41). It is probable that the filamentation defect of slr1Δ/Δ cells in the kidney also contributed to its attenuated virulence. Furthermore, there were interesting differences in the localization of this mutant within different regions of the kidney. There was a greater number of slr1Δ/Δ yeast cells than wild-type cells in the renal tubules and a smaller number of slr1Δ/Δ hyphae than wild-type cells in the renal pelvis (Fig. 7B). Reconstituted cells with a single copy of SLR1 showed an intermediate phenotype. The high density of slr1Δ/Δ yeast in the tubules also raises the question of whether these yeast-form slr1Δ/Δ cells might be retained in the tubule or otherwise unable to progress to the renal pelvis. A similar phenomenon—equivalent kidney fungal burdens compared to those for wild-type cells with differences in morphology and intrakidney localization—was seen for C. albicans lacking the Rsr1 GTPase, which plays a role in hyphal tip orientation (57).
Although the slr1Δ/Δ mutant grew slowly in vitro, the kidney fungal burden of mice infected with this strain was similar to that of mice infected with the wild-type strain. Noble and colleagues similarly found that numerous mutants with growth defects in vitro achieved normal fungal density in the kidney when inoculated into mice (55). Therefore, the impaired growth of the slr1Δ/Δ mutant likely played a minor role in its reduced virulence in the mouse model of HDC. However, in the mouse model of OPC, the slr1Δ/Δ mutant achieved a lower oral fungal burden than did the wild-type strain. Based on our in vitro data, the impaired virulence of the slr1Δ/Δ mutant was likely due a combination of slower growth in the oropharynx and a reduced capacity to damage oral epithelial cells.
An unexpected finding was that the absence of SLR1 led to an increased fungal burden in the brain. This tropism suggests that Slr1 might affect the expression of genes involved in interactions with brain endothelial cells. Indeed, whereas deletion of SLR1 decreased the endocytosis of C. albicans by HUVECs, it significantly increased endocytosis by HBMECs. Recently, a similar tropism was found for C. albicans lacking the putative vacuolar sorting protein Vps51 (39). In the mouse models of HDC, the brain fungal burden of mice infected with a vps51Δ/Δ mutant was significantly greater than that of mice infected with a wild-type strain. In addition, although vps51Δ/Δ cells were poorly endocytosed by HUVECs, they were endocytosed more avidly than wild-type cells by HBMECs. Endocytosis of C. albicans by brain endothelial cells was found to be mediated in part by the binding of cell-wall-associated Als3 to heat shock protein gp96 on host cells; the brain tropism of the vps51Δ/Δ mutant resulted from greater exposure of Als3 on the C. albicans cell surface (39). We found that deletion of SLR1 also resulted in increased surface exposure of Als3, thus providing a potential mechanism for the brain tropism of slr1Δ/Δ cells.
Whereas studies of other fungal RNA-binding proteins indicate that numerous posttranscriptional steps in gene expression can affect polar growth and cell surface processes (12, 13, 16, 58), the mechanism(s) whereby Slr1 directly or indirectly affects filamentation and interactions with host cells remains to be determined. Future work to understand how Slr1 impacts C. albicans hyphal growth and virulence will focus on identifying its mRNA targets and how it influences their expression and localization during hyphal growth.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the National Institutes of Health, grants 5P20RR016463, 8P20GM103423, 2R01AI054928, and 2R01DE017088. The umbilical cords used in this study were collected by the pediatric, perinatal, and mobile unit of the UCLA Clinical and Translational Science Institute at LA BioMed/Harbor-UCLA Medical Center, funded by the National Center for Advancing Translational Sciences, grant UL1TR000124. This project was also supported by grants from the National Center for Research Resources (5P20RR016463) and the National Institute of General Medical Sciences (8 P20 GM103423) from the National Institutes of Health. C.A. was supported by undergraduate research fellowships from the American Society for Microbiology and the Howard Hughes Medical Institute.
We thank Aaron Mitchell for reagents, Anja Forche for critical reading of the manuscript and technical advice, and Darren Abbey and Sven Bergmann for providing growth curve analysis software prior to publication.
Footnotes
Published ahead of print 4 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00864-12.
REFERENCES
- 1. Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309–317 [DOI] [PubMed] [Google Scholar]
- 2. Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 3. Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Filler SG, Sheppard DC. 2006. Fungal invasion of normally non-phagocytic host cells. PLoS Pathog. 2:e129 doi:10.1371/journal.ppat.0020129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nantel A, Dignard D, Bachewich C, Harcus D, Marcil A, Bouin AP, Sensen CW, Hogues H, van het Hoog M, Gordon P, Rigby T, Benoit F, Tessier DC, Thomas DY, Whiteway M. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452–3465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kadosh D, Johnson AD. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell 16:2903–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sellam A, Hogues H, Askew C, Tebbji F, van Het Hoog M, Lavoie H, Kumamoto CA, Whiteway M, Nantel A. 2010. Experimental annotation of the human pathogen Candida albicans coding and noncoding transcribed regions using high-resolution tiling arrays. Genome Biol. 11:R71 doi:10.1186/gb-2010-11-7-r71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. 2010. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 20:1451–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Whiteway M, Bachewich C. 2007. Morphogenesis in Candida albicans. Annu. Rev. Microbiol. 61:529–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. An HS, Lee KH, Kim J. 2004. Identification of an exoribonuclease homolog, CaKEM1/CaXRN1, in Candida albicans and its characterization in filamentous growth. FEMS Microbiol. Lett. 235:297–303 [DOI] [PubMed] [Google Scholar]
- 11. Richard ML, Nobile CJ, Bruno VM, Mitchell AP. 2005. Candida albicans biofilm-defective mutants. Eukaryot. Cell 4:1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elson SL, Noble SM, Solis NV, Filler SG, Johnson AD. 2009. An RNA transport system in Candida albicans regulates hyphal morphology and invasive growth. PLoS Genet. 5:e1000664 doi:10.1371/journal.pgen.1000664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dagley MJ, Gentle IE, Beilharz TH, Pettolino FA, Djordjevic JT, Lo TL, Uwamahoro N, Rupasinghe T, Tull DL, McConville M, Beaurepaire C, Nantel A, Lithgow T, Mitchell AP, Traven A. 2011. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol. Microbiol. 79:968–989 [DOI] [PubMed] [Google Scholar]
- 14. Lasko P. 2011. Posttranscriptional regulation in Drosophila oocytes and early embryos. Wiley Interdiscip. Rev. RNA 2:408–416 [DOI] [PubMed] [Google Scholar]
- 15. Donnelly CJ, Fainzilber M, Twiss JL. 2010. Subcellular communication through RNA transport and localized protein synthesis. Traffic 11:1498–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vollmeister E, Feldbrugge M. 2010. Posttranscriptional control of growth and development in Ustilago maydis. Curr. Opin. Microbiol. 13:693–699 [DOI] [PubMed] [Google Scholar]
- 17. Tuch BB, Mitrovich QM, Homann OR, Hernday AD, Monighetti CK, De La Vega FM, Johnson AD. 2010. The transcriptomes of two heritable cell types illuminate the circuit governing their differentiation. PLoS Genet. 6:e1001070 doi:10.1371/journal.pgen.1001070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shepard PJ, Hertel KJ. 2009. The SR protein family. Genome Biol. 10:242 doi:10.1186/gb-2009-10-10-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Long JC, Caceres JF. 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417:15–27 [DOI] [PubMed] [Google Scholar]
- 20. Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. 2009. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell 35:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kress TL, Krogan NJ, Guthrie C. 2008. A single SR-like protein, Npl3, promotes pre-mRNA splicing in budding yeast. Mol. Cell 32:727–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dermody JL, Dreyfuss JM, Villen J, Ogundipe B, Gygi SP, Park PJ, Ponticelli AS, Moore CL, Buratowski S, Bucheli ME. 2008. Unphosphorylated SR-like protein Npl3 stimulates RNA polymerase II elongation. PLoS One 3:e3273 doi:10.1371/journal.pone.0003273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bucheli ME, Buratowski S. 2005. Npl3 is an antagonist of mRNA 3′ end formation by RNA polymerase II. EMBO J. 24:2150–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. 1994. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 126:649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee MS, Henry M, Silver PA. 1996. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 10:1233–1246 [DOI] [PubMed] [Google Scholar]
- 26. Windgassen M, Sturm D, Cajigas IJ, Gonzalez CI, Seedorf M, Bastians H, Krebber H. 2004. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Mol. Cell. Biol. 24:10479–10491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ni L, Snyder M. 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12:2147–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mosch HU, Fink GR. 1997. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics 145:671–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McBride AE, Zurita-Lopez C, Regis A, Blum E, Conboy A, Elf S, Clarke S. 2007. Protein arginine methylation in Candida albicans: role in nuclear transport. Eukaryot. Cell 6:1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shaw BD, Upadhyay S. 2005. Aspergillus nidulans swoK encodes an RNA binding protein that is important for cell polarity. Fungal Genet. Biol. 42:862–872 [DOI] [PubMed] [Google Scholar]
- 31. Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lay J, Henry LK, Clifford J, Koltin Y, Bulawa CE, Becker JM. 1998. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect. Immun. 66:5301–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ketel C, Wang HS, McClellan M, Bouchonville K, Selmecki A, Lahav T, Gerami-Nejad M, Berman J. 2009. Neocentromeres form efficiently at multiple possible loci in Candida albicans. PLoS Genet. 5:e1000400 doi:10.1371/journal.pgen.1000400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zwietering MH, Jongenburger I, Rombouts FM, Kvan 't Riet 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu H, Kohler J, Fink GR. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726 [DOI] [PubMed] [Google Scholar]
- 36. Jaffe EA, Nachman RL, Becker CG, Minick CR. 1973. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. J. Clin. Invest. 52:2745–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE, Jr, Filler SG. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5:e64 doi:10.1371/journal.pbio.0050064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stins MF, Nemani PV, Wass C, Kim KS. 1999. Escherichia coli binding to and invasion of brain microvascular endothelial cells derived from humans and rats of different ages. Infect. Immun. 67:5522–5525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Y, Mittal R, Solis NV, Prasadarao NV, Filler SG. 2011. Mechanisms of Candida albicans trafficking to the brain. PLoS Pathog. 7:e1002305 doi:10.1371/journal.ppat.1002305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Park H, Myers CL, Sheppard DC, Phan QT, Sanchez AA, Edwards JE, Filler SG. 2005. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell Microbiol. 7:499–510 [DOI] [PubMed] [Google Scholar]
- 41. Sanchez AA, Johnston DA, Myers C, Edwards JE, Jr, Mitchell AP, Filler SG. 2004. Relationship between Candida albicans virulence during experimental hematogenously disseminated infection and endothelial cell damage in vitro. Infect. Immun. 72:598–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kamai Y, Kubota M, Hosokawa T, Fukuoka T, Filler SG. 2001. New model of oropharyngeal candidiasis in mice. Antimicrob. Agents Chemother. 45:3195–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Solis NV, Filler SG. 2012. Mouse model of oropharyngeal candidiasis. Nat. Protoc. 7:637–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Phan QT, Fratti RA, Prasadarao NV, Edwards JE, Jr, Filler SG. 2005. N-cadherin mediates endocytosis of Candida albicans by endothelial cells. J. Biol. Chem. 280:10455–10461 [DOI] [PubMed] [Google Scholar]
- 45. Plass M, Agirre E, Reyes D, Camara F, Eyras E. 2008. Co-evolution of the branch site and SR proteins in eukaryotes. Trends Genet. 24:590–594 [DOI] [PubMed] [Google Scholar]
- 46. Gilbert W, Siebel CW, Guthrie C. 2001. Phosphorylation by Sky1p promotes Npl3p shuttling and mRNA dissociation. RNA 7:302–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yun CY, Fu XD. 2000. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J. Cell Biol. 150:707–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Aguilera J, Randez-Gil F, Prieto JA. 2007. Cold response in Saccharomyces cerevisiae: new functions for old mechanisms. FEMS Microbiol. Rev. 31:327–341 [DOI] [PubMed] [Google Scholar]
- 49. Zhu W, Filler SG. 2010. Interactions of Candida albicans with epithelial cells. Cell Microbiol. 12:273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Phan QT, Belanger PH, Filler SG. 2000. Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect. Immun. 68:3485–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bossie MA, DeHoratius C, Barcelo G, Silver P. 1992. A mutant nuclear protein with similarity to RNA binding proteins interferes with nuclear import in yeast. Mol. Biol. Cell 3:875–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Russell ID, Tollervey D. 1992. NOP3 is an essential yeast protein which is required for pre-rRNA processing. J. Cell Biol. 119:737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lutzelberger M, Gross T, Kaufer NF. 1999. Srp2, an SR protein family member of fission yeast: in vivo characterization of its modular domains. Nucleic Acids Res. 27:2618–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bonhomme J, Chauvel M, Goyard S, Roux P, Rossignol T, d'Enfert C. 2011. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol. Microbiol. 80:995–1013 [DOI] [PubMed] [Google Scholar]
- 55. Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fuchs BB, Eby J, Nobile CJ, El Khoury JB, Mitchell AP, Mylonakis E. 2010. Role of filamentation in Galleria mellonella killing by Candida albicans. Microbes Infect. 12:488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brand A, Vacharaksa A, Bendel C, Norton J, Haynes P, Henry-Stanley M, Wells C, Ross K, Gow NA, Gale CA. 2008. An internal polarity landmark is important for externally induced hyphal behaviors in Candida albicans. Eukaryot. Cell 7:712–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wolf JJ, Dowell RD, Mahony S, Rabani M, Gifford DK, Fink GR. 2010. Feed-forward regulation of a cell fate determinant by an RNA-binding protein generates asymmetry in yeast. Genetics 185:513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953–5959 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.