Abstract
Infection by the chronic periodontitis-associated pathogen Porphyromonas gingivalis activates a Toll-like receptor 2 (TLR2) response that triggers inflammation in the host but also promotes bacterial persistence. Our aim was to define ligands on the surfaces of intact P. gingivalis cells that determine its ability to activate TLR2. Molecules previously reported as TLR2 agonists include lipopolysaccharide (LPS), fimbriae, the lipoprotein PG1828, and phosphoceramides. We demonstrate that these molecules do not comprise the major factors responsible for stimulating TLR2 by whole bacterial cells. First, P. gingivalis mutants devoid of the reported protein agonists, PG1828 and fimbriae, activate TLR2 as strongly as the wild type. Second, two-phase extraction of whole bacteria resulted in a preponderance of TLR2 agonist activity partitioning to the hydrophilic phase, demonstrating that phosphoceramides are not a major TLR2 ligand. Third, analysis of LPS revealed that TLR2 activation is independent of lipid A structural variants. Instead, activation of TLR2 and TLR2/TLR1 by LPS is in large part due to copurifying molecules that are sensitive to the action of the enzyme lipoprotein lipase. Strikingly, intact P. gingivalis bacterial cells treated with lipoprotein lipase were attenuated in their ability to activate TLR2. We propose that a novel class of molecules comprised by lipoproteins constitutes the major determinants that confer to P. gingivalis the ability to stimulate TLR2 signaling.
INTRODUCTION
Porphyromonas gingivalis is a Gram-negative anaerobic bacterium associated with chronic periodontitis, an inflammatory disease that results in tooth loss (1). P. gingivalis is typically found in diseased subgingival sites (2). Mouse models of P. gingivalis infection have demonstrated its capacity to elicit periodontitis, as measured by bone loss (3–5). A recent study revealed a key role played by P. gingivalis infection in altering the composition, and increasing the abundance, of oral commensal bacteria in conventionally grown mice (4). The interaction between an increasing bacterial load and the host innate immune system triggers a proinflammatory response, which is considered a major factor in causing periodontitis.
The Toll-like receptor (TLR) family of innate immune receptors plays a pivotal role in host surveillance and in initiating an immediate immune response that is designed to neutralize microbial threats to the host (6). P. gingivalis infection triggers activation of TLR2, which leads to production of proinflammatory cytokines. One impact of these cytokines is a damaging effect on alveolar bone, resulting in bone loss, as demonstrated by studies using TLR2−/− mice (3, 7). Paradoxically, the robust TLR2 proinflammatory response does not clear P. gingivalis infection in wild-type mice. Instead, the infection is cleared more efficiently in TLR2−/− mice and by macrophages from TLR2−/− mice (3, 5, 7), indicating that TLR2 stimulation confers enhanced persistence to this chronic pathogen. A lack of bacterial clearance illustrates subversion of the host TLR2 response by P. gingivalis. Hence, activation of TLR2 is modulated by P. gingivalis to its benefit. Recently identified mechanisms by which TLR2 stimulation results in suppression of a bactericidal response, but not in suppression of inflammation, include cross talk between TLR2 and other receptors, including CXCR4 (8), CR3 (9) and C5aR (7). TLR2 has also been shown, by structural and functional studies, to heterodimerize with either TLR1 or TLR6 (10, 11). These heterodimers engage distinct ligands, as observed with TLR2/TLR1 responding to triacylated lipoproteins, a structure that typifies Gram-negative bacterial lipoproteins, and TLR2/TLR6 engaging diacylated lipoproteins such as those found in Gram-positive bacteria (12–14).
In contrast to the TLR2 response to P. gingivalis, it is well established that the TLR4 response mounted by the host to counter P. gingivalis infection is strikingly low (15–18). On a molecular level, this is due to the structure of the P. gingivalis TLR4 agonist, lipid A, a moiety of the lipopolysaccharide (LPS) macromolecule. P. gingivalis synthesizes a heterogeneous population of structurally distinct lipid A molecules, which range in function from inert agonists to mild agonists to antagonists of TLR4 stimulation (19–21). The chronic disease-associated bacterium P. gingivalis, therefore, potentially uses its unusual lipid A repertoire to modulate activation of TLR4, hence promoting survival of both P. gingivalis and other bacteria in the milieu.
P. gingivalis molecules reported to date that stimulate TLR2 are LPS, fimbriae, the lipoprotein PG1828, and phosphoceramides. LPS preparations from P. gingivalis stimulate TLR2 potently, as shown by us previously (22) and by other laboratories (23–26). In contrast to Escherichia coli LPS, whose capacity to stimulate TLR2 was eliminated by repurification of LPS using phenol extraction (27), a similarly repurified preparation of P. gingivalis LPS retained its capacity for TLR2 stimulation (22, 24). This led us to speculate that TLR2 activation by LPS could be triggered by specific P. gingivalis lipid A structures (22). However, an evaluation of a range of synthetic molecules modeled upon distinct P. gingivalis lipid A structures has shown that they act through TLR4 and not TLR2 (28–31), raising the possibility that TLR2 stimulation by LPS is due to tightly bound copurifying molecules. Adding weight to this hypothesis, and similar to observations made with E. coli LPS preparations (32, 33), a lipoprotein with TLR2 agonist activity was identified from P. gingivalis LPS preparations (34). A mutant lacking this lipoprotein, PG1828, rendered its LPS attenuated for TLR2 activation (35).
In this study, we addressed the contributions made by reported P. gingivalis agonists in triggering TLR2 and TLR2/TLR1 activation by intact bacterial cells. We show that in contrast to LPS derived from Δ1828 mutants, whole bacterial P. gingivalis Δ1828 mutants are not attenuated for TLR2 or TLR2/TLR1 activation. Additionally, mutants with deletions in both PG1828 and fimA, the gene encoding the major subunit in fimbriae, activate TLR2 and TLR2/TLR1 as potently as wild-type P. gingivalis. A closer examination of LPS preparations revealed a class of TLR2/TLR1 activating hydrophilic molecules that exhibit sensitivity to the enzyme lipoprotein lipase. In short, it is demonstrated that the four reported P. gingivalis TLR2 agonists do not account for the majority of TLR2 stimulation triggered by viable bacteria. Instead, a novel class of agonists, likely comprised by lipoproteins, determines TLR2 stimulation by both P. gingivalis LPS preparations and intact bacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis 33277, Bacteroides thetaiotaomicron VPI5482, and E. coli DH10b and JM83 strains were obtained from our culture collection. P. gingivalis A7436 and 381 were obtained from Caroline Genco's laboratory (36), and P. gingivalis W50 was obtained from Michael Curtis' laboratory (37). Prevotella intermedia ATCC 5611 and Fusobacterium nucleatum ATCC 51656 were obtained from the American Type Culture Collection (ATCC). P. gingivalis and P. intermedia strains were grown on blood agar plates containing 5% sheep's blood and in TYHK broth (30 g/liter Trypticase soy broth, 5 g/liter yeast extract, and 1 mg/liter vitamin K3). Following sterilization by autoclaving, filter-sterilized hemin was added to TYHK broth, just prior to inoculation, to a final concentration of 1 μg/ml. TYHK agar plates were also used for growth of P. gingivalis on solid medium. Hemin (1 μg/ml) and antibiotics were added following sterilization. Antibiotics were added to the following concentrations: erythromycin, 5 μg/ml; tetracycline, 1 μg/ml. E. coli strains were grown in L broth (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl), and 100 μg/ml ampicillin was added when required for selection. B. thetaiotaomicron was grown on TYHK agar plates and TYHK broth containing 1 μg/ml hemin, and F. nucleatum was grown in the same medium but without hemin. Anaerobic strains, which included all those mentioned above except for E. coli, were grown in an anaerobic growth chamber (5% H2, 5% CO2, 90% N2) at 37°C.
Construction of P. gingivalis mutants.
The genome sequences of P. gingivalis strains W83 and 33277 were obtained from the NCBI (National Center for Biotechnology Information) and KEGG (Kyoto Encyclopedia of Genes and Genomes) databases. Genes from W83 have a PG designation, and genes from 33277 have a PGN designation. We use the PG nomenclature for describing genes but used the PGN sequence information for designing gene manipulations, conducted primarily in 33277. P. gingivalis single mutants with deletions in fimA (PG2132 in W83, PGN_0180 in 33277) and the gene encoding PG1828 lipoprotein (PG1828 in W83, PGN_1739 in 33277) were constructed by allelic exchange. Genes deleted were replaced with either the erythromycin resistance-encoding ermF-AM (38) cassette or the tetracycline resistance-encoding tetQ cassette (39). DNA fragments ∼700 to 1,000 bp up- and downstream of the targeted gene were amplified by PCR from 33277 genomic DNA. The flanking fragments were coligated, with the antibiotic resistance cassette between them, into pGem-TEz (Promega, Madison, WI). Primers used for construction of fimA and PG1828 deletion plasmids are listed in Table 1. tetQ was amplified from pYT646b (39), and ermF-AM was obtained as a BamHI fragment from prtT::erm (40). The gene disruption plasmids pfimA 5′flank:tetQ:3′flank and p1828 5′flank:ermF-AM:3′flank were electroporated into 33277 in a GenePulser Xcell (Bio-Rad, Hercules, CA). Mutant colonies arising due to homologous recombination between the flanking segments on the plasmid and chromosome were selected for by plating on TYHK agar plates containing the appropriate antibiotic. They took 4 to 5 days to appear and were confirmed for deletion of the targeted gene by PCR analysis. PG1828 and fimA mutants were similarly constructed in P. gingivalis strains A7436, W83, and 381. The PG1828 fimA double mutant was constructed by electroporating pfimA 5′flank:tetQ:3′flank into 33277ΔPG1828::erm, followed by selection for both tetracycline and erythromycin resistance. 33277ΔfimA mutants were confirmed to lack FimA protein, as assessed by immunoblotting with anti-FimA antibodies (41, 42).
Table 1.
Primers used for construction of mutants
| Gene | Orientation | Sequencea |
|---|---|---|
| fimA, up-flank | Forward | AGATCAGCATGCGATGGTAAAGCGTCGCAAC |
| fimA, up-flank | Reverse | CATCACCTCGAGTTAGAGATTGTCTTGCATATAGC |
| fimA, down-flank | Forward | AGATCTAAGCTTAGGCTGCTACTTGGTAATCGAC |
| fimA, down-flank | Reverse | ACTCACGCTAGCTGTCGGATTAGTATTCTGC |
| tetQ | Forward | AGATCACTCGAGCAACGAATTATCTCCTTAACG |
| tetQ | Reverse | ACTCACAAGCTTCCAACTGTATTGCCTTATAG |
| PG1828, up-flank | Forward | TCATTTGTCATCATGGTGCCTC |
| PG1828, up-flank | Reverse | TCGAGGATCCAGTGTAATTAATGTTCTATAACG |
| PG1828, down-flank | Forward | TCGAGGATCCAAGTCTGACTTCAAAAGAGTCG |
| PG1828, down-flank | Reverse | AGCATATAATACAGAGTCAGCAC |
Restriction sites used for constructing the deletion plasmids are underlined.
Preparation of bacterial hydrophobic and hydrophilic extracts.
We used a protocol similar to the initial steps used for extraction of phosphoceramides (43). Specifically, 150 ml P. gingivalis 33277 culture grown for 48 h (∼2 × 109 bacteria/ml) was centrifuged and lyophilized, yielding an ∼100-mg dried pellet. The pellet was dissolved in 1 ml water, followed by addition of 4 ml 2:1 methanol-chloroform and mixed by vortexing. After 6 h, 750 μl of 2 N KCl plus 0.5 M K2HPO4 solution and 750 μl chloroform were added, vortexed, and spun at low speed to facilitate clear separation of the lower hydrophobic chloroform phase from the upper hydrophilic aqueous phase. The lower phase was pipetted out and 750 μl chloroform was added to the hydrophilic phase. Following vortexing and a spin, the hydrophobic phases were pooled. The aqueous hydrophilic phase was frozen, lyophilized, weighed, and resuspended in water. The hydrophobic phase was dried in a fume hood, weighed, and resuspended in 70% ethanol. The weights of dried hydrophilic and hydrophobic fractions were ∼25 to 50 mg and ∼15 to 30 mg, respectively.
Preparation of LPS and isolation of lipid A.
P. gingivalis bacteria were grown for 48 h in TYHK broth containing 1 μg/ml hemin. LPS was isolated from 150 to 200 ml culture using the Tri-reagent protocol, as previously described (36). Following precipitation of lyophilized LPS (the last step of Tri-reagent procedure) with 0.375 M magnesium chloride, the pellet was washed twice with cold 95% ethanol and once with cold 100% ethanol. Phospholipids were removed by adding 400 μl monophasic 2:1 chloroform-methanol solution, centrifuging, and discarding the supernatant. The pellet, which contains LPS, was lyophilized, weighed, and resuspended in water.
Lipid A was isolated from LPS as described previously (20). The rationale for using the final chloroform-methanol-water (1:1:0.9, vol/vol/vol) extraction step was to separate lipid A from residual carbohydrate contaminants. The chloroform phase, containing lipid A, was dried, weighed, and resuspended in 0.1% triethylamine. The aqueous layer was frozen, lyophilized, weighed, and resuspended in water for further analysis.
HEK293 TLR activation assays.
The assays were performed as previously described (20). Briefly, HEK293 cells were plated in 96-well plates and transfected the following day with plasmids encoding NF-κB-dependent firefly luciferase reporter, β-actin promoter-dependent Renilla luciferase reporter, and human TLRs. In the case of human TLR2, TLR2/TLR1, or TLR1 alone, 0.001 μg plasmid encoding the indicated TLR was cotransfected with 0.002 μg plasmid encoding human mCD14. In the case of human TLR4, 0.002 μg plasmid encoding human TLR4 was cotransfected with 0.0025 μg plasmid encoding human MD-2. At 18 to 20 h posttransfection, test wells were stimulated in triplicate for 4 h at 37°C with various doses of sample, which were suspended in Dulbecco's modified Eagle medium (DMEM) containing 10% human serum. For stimulation with intact bacteria, 1-ml cultures of the indicated strains were first washed with TYHK, and their concentration estimated by measuring the optical density at 600 nm. Luciferase activity was assayed using a dual luciferase assay reporter system (Promega, Madison, WI). NF-κB activity was measured as the ratio of NF-κB-dependent firefly luciferase activity to β-actin promoter-dependent Renilla luciferase activity, which served as an internal standard. The data were plotted as the fold difference of NF-κB activity of the sample over unstimulated control. The carrier solvents (water, 70% ethanol, 0.1% triethylamine, and TYHK) were not included in the unstimulated control. However, each solvent individually was shown not to contribute toward stimulation of any of the TLRs tested.
Lipoprotein lipase treatment.
The synthetic lipoprotein PAM3CSK4 was purchased from InvivoGen (San Diego, CA). Lipoprotein lipase from a Burkholderia sp. was purchased from Sigma-Aldrich (St. Louis, MO). The amount of lipoprotein lipase added to LPS preparations and the subsequent aqueous and chloroform fractions, as well as to whole bacteria, is indicated in the figure legends. The enzyme and substrate were incubated at 37°C overnight in the case of LPS or fractions derived from LPS and for 2 h in the case of whole bacteria.
Statistical analysis.
The significance of all described comparisons was established using two-tailed unpaired t tests on triplicate samples with a significance level of 0.05, unless otherwise stated.
RESULTS
P. gingivalis stimulates TLR2/TLR1 more potently than TLR2 alone.
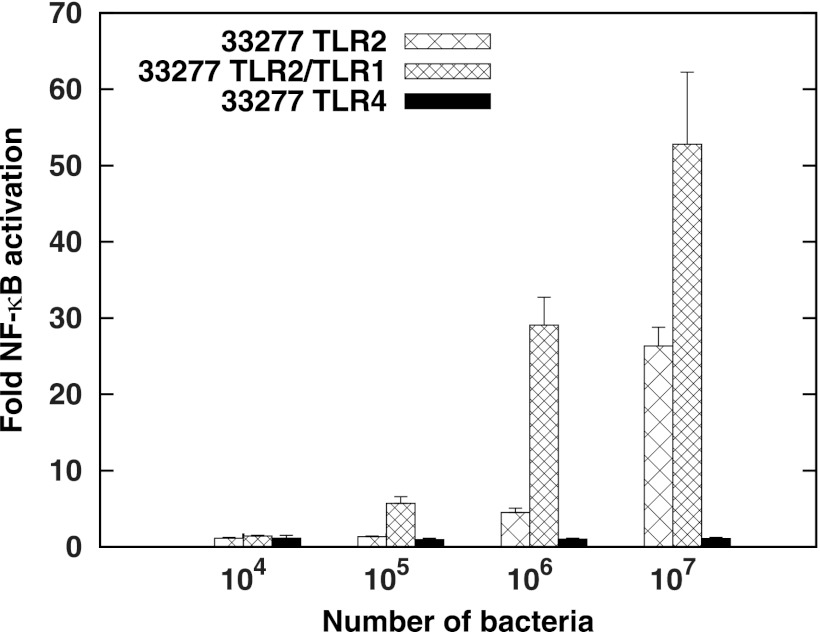
TLR2-ligand functional studies in P. gingivalis have largely involved examining purified ligands in TLR2−/− mice or macrophages/monocytes from TLR2−/− mice. These assay systems express a wide range of innate immune receptors besides TLR2. In order to assess the function of individual TLRs in mounting an innate immune response, we and other groups have utilized nonimmune human embryonic kidney (HEK) 293 cells transfected with a plasmid encoding the TLR of interest and measured its activity by cotransfecting an NF-κB-responsive reporter (44). TLR2-expressing or TLR4-expressing HEK293 cells were stimulated with whole live bacteria, instead of individual agonists, to determine the collective activation by all ligands present on the surfaces of P. gingivalis cells. Consistent with previous observations, use of this system demonstrated potent TLR2, but not TLR4, activation by P. gingivalis (Fig. 1). Since lipoproteins from Gram-negative bacteria are typically triacylated, and given the functional and structural evidence demonstrating the important role TLR1 plays as a coreceptor in initiating a response to these triacylated ligands (10, 13, 14), we tested cells transfected with both TLR2 and TLR1. Figure 1 demonstrates an increased signaling potency of TLR2/TLR1 compared to that of TLR2 alone. These data suggest that lipoproteins contribute to TLR2 activation by P. gingivalis whole cells.
Fig 1.
P. gingivalis whole bacteria stimulate TLRs as follows: TLR2/TLR1 > TLR2 > TLR4. HEK293 cells were transfected with human TLR4 and MD-2, with human TLR2 and CD14, or with human TLR2, TLR1, and CD14, followed by infection with 104, 105, 106, or 107 intact 33277 bacteria, as indicated on the x axis. The fold NF-κB stimulation of infected cells relative to unstimulated controls is plotted on the y axis. The results are means ± standard deviations (SD) for triplicate samples from one of three independent experiments.
PG1828 and FimA are not the major ligands contributing to TLR2 and TLR2/TLR1 activation.
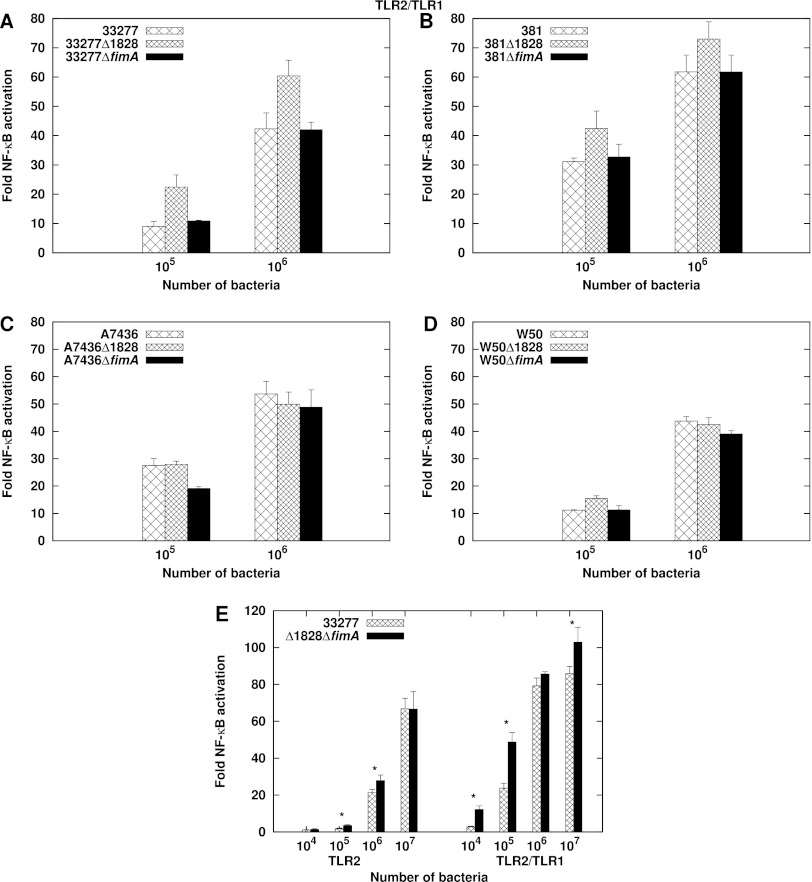
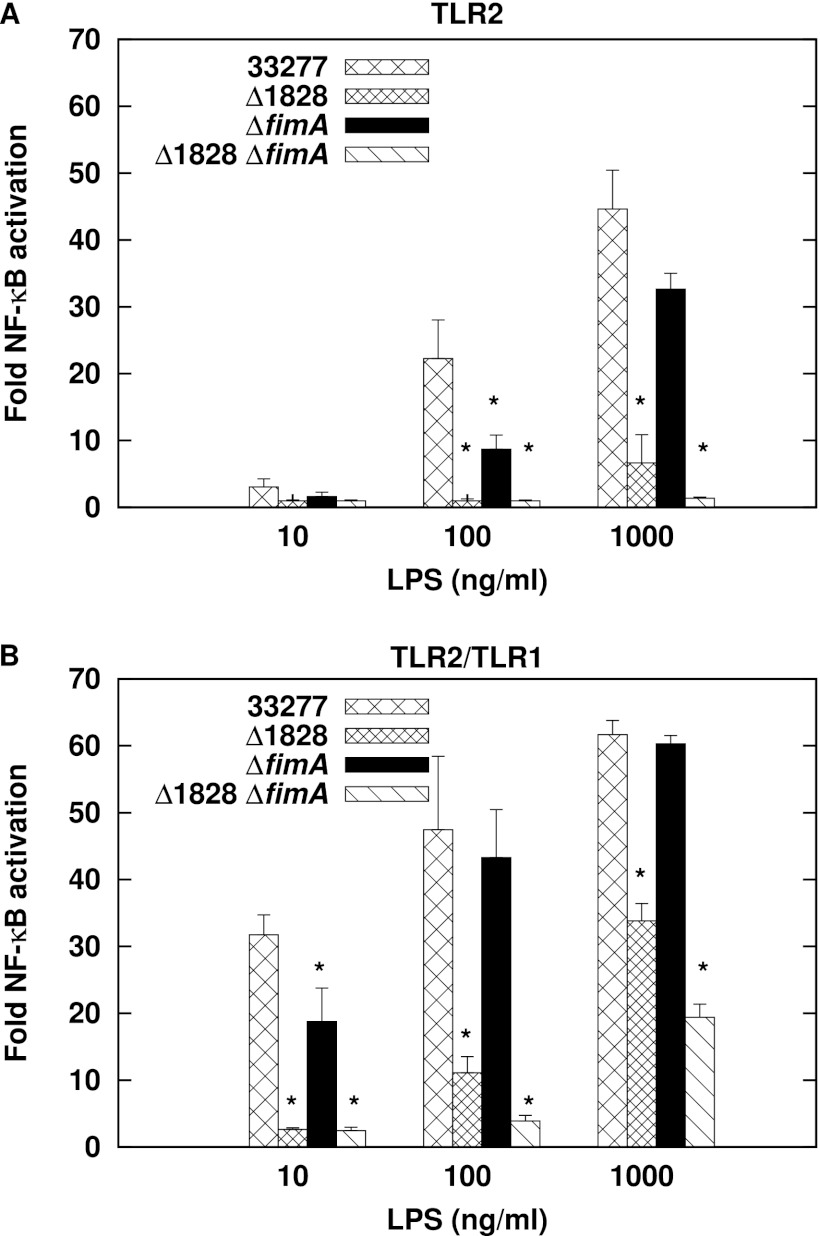
The two P. gingivalis proteins known to activate TLR2 are fimbriae (23, 26, 45, 46) and the lipoprotein PG1828 (34, 47). We determined whether these ligands contribute significantly to TLR2 and TLR2/TLR1 activation by whole bacterial cells by testing P. gingivalis mutants that are deficient for either FimA, the major fimbrial subunit, or PG1828. Previous studies have demonstrated that recombinant FimA stimulates TLR2 to an extent similar to that of fimbriae (23). Single ΔfimA::tetQ and Δ1828::ermF-AM mutants were constructed in the commonly studied P. gingivalis strains 33277, A7436, 381, and W50. Figure 2A to D demonstrate no decrease in the magnitude of TLR2/TLR1 stimulation by the single mutant strains relative to the wild type. We next tested a P. gingivalis 33277 double mutant with both PG1828 and fimA deleted. Intact double-mutant bacteria were also not attenuated for TLR2 and TLR2/TLR1 stimulation (Fig. 2E). Interestingly, the Δ1828 mutation conferred a higher stimulating capacity in strains 33277 and 381 for reasons that are not understood. We conclude that FimA and PG1828 do not significantly contribute to TLR2 and TLR2/TLR1 activation by whole cells. These data indicate the presence of additional ligands on the bacterial surface that activate TLR2.
Fig 2.
P. gingivalis ΔPG1828 and ΔfimA mutants are not attenuated for TLR2/TLR1 activation. (A to D) HEK293 cells expressing human TLR2/TLR1/CD14 were stimulated with wild-type bacterial cells or the isogenic ΔPG1828 or ΔfimA single-mutant cells of strains 33277 (A), 381 (B), A7436 (C), and W50 (D). (E) HEK293 cells expressing human TLR2/CD14 or human TLR2/TLR1/CD14 were infected with 33277 or 33277Δ1828ΔfimA double-mutant cells. Asterisks denote significant differences in stimulation potency between 33277 and Δ1828ΔfimA (P < 0.05). Fold NF-κB stimulation of infected cells relative to unstimulated controls is plotted on the y axis. The results are means ± SD for triplicate samples from one of two independent experiments.
Hydrophilic molecules mediate TLR2 and TLR2/TLR1 activation by P. gingivalis.
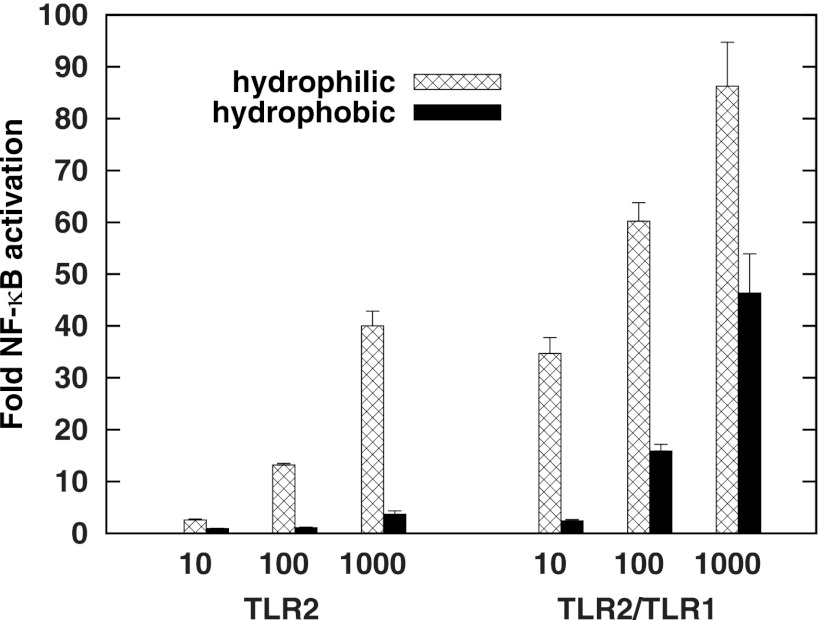
P. gingivalis LPS and phosphoceramides are two other ligands known to stimulate TLR2. These molecules can be separated by a biphasic water-methanol-chloroform extraction to obtain a hydrophobic fraction containing phospholipids such as phosphoceramides (43) and a hydrophilic fraction containing LPS and other water-soluble compounds. Intact P. gingivalis were subjected to this extraction procedure, and activation of TLR2 and TLR2/TLR1 was examined (Fig. 3). It was found that the hydrophilic fraction was a significantly more potent stimulator of TLR2 and TLR2/TLR1 signaling than the hydrophobic fraction. Hence, agonists that partition to the hydrophilic phase contribute substantially to TLR2 and TLR2/TLR1 activation.
Fig 3.
Hydrophilic whole bacterial fractions from P. gingivalis 33277 stimulate TLR2 and TLR2/TLR1 more potently than the hydrophobic fraction. HEK293 cells expressing either TLR2/CD14 or TLR2/TLR1/CD14 were stimulated by the indicated amounts of aqueous or organic fractions (ng/ml). Fold NF-κB stimulation of infected cells over unstimulated controls is shown on the y axis. The results are means ± SD for triplicate samples from one of three independent experiments. A significant difference in activation potency was observed within each hydrophilic-hydrophobic pair shown (P < 0.01).
TLR2 and TLR2/TLR1 stimulation by P. gingivalis LPS is independent of lipid A structure.
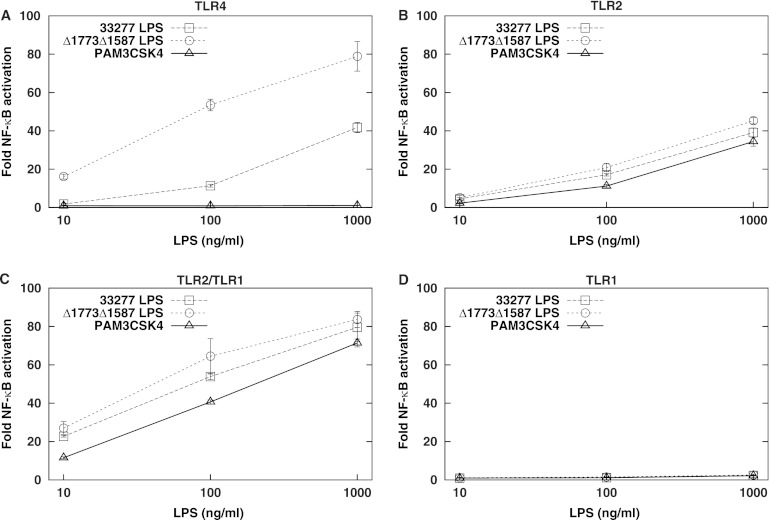
LPS macromolecules partition to the hydrophilic phase of the biphasic extraction described above, of which the lipid A moiety has been proposed to activate TLR2 (22, 24). Since P. gingivalis synthesizes multiple lipid A subtypes that differ in number of acyl chains and phosphate groups, we evaluated whether LPSs possessing distinct lipid A structures differ in their capacity to activate TLR2. We tested LPSs containing two different lipid A populations that exhibit wide differences in structure, with a concomitant difference in their abilities to activate the lipid A receptor TLR4. The first population is mostly comprised by lipid A that is tetra-acylated and nonphosphorylated, a weak TLR4 agonist. It is the major lipid A structure synthesized by wild-type P. gingivalis grown under restricted-hemin conditions (20). The second lipid A structure, penta-acylated and diphosphorylated, synthesized by a P. gingivalis mutant bearing deletions in the two lipid A phosphatases, PG1773 and PG1587 (20), is a relatively potent TLR4 activator. Figure 4 recapitulates the difference in TLR4 activation by the two lipid A species. Importantly, however, they activate TLR2 and TLR2/TLR1 to similar extents. TLR1 alone was not activated by P. gingivalis LPS preparations. The synthetic triacylated lipopeptide PAM3CSK4 served as a positive control for TLR2 and TLR2/TLR1 activation (10) and was shown to not activate either TLR4 or TLR1 alone. These data indicate that activation of TLR2 and TLR2/TLR1 by P. gingivalis LPS preparations is independent of lipid A structure. We also tested LPS from a P. gingivalis mutant that lacks O antigen due to a deletion in the O-antigen ligase-encoding gene, PG1051. (37). LPS from this mutant was not attenuated for TLR2 or TLR2/TLR1 stimulation (data not shown), indicating that O antigen also does not contribute to activation.
Fig 4.
P. gingivalis lipid A structure affects TLR4 activation but not TLR2 and TLR2/TLR1 activation. HEK293 cells expressing human TLR4/MD-2 (A), human TLR2/CD14 (B), human TLR2/TLR1/CD14 (C), or human TLR1/CD14 (D) were infected with LPS from wild-type 33277 or the ΔPG1587 ΔPG1773 isogenic mutant. The synthetic TLR2/TLR1 ligand PAM3CSK4 was used as a control, in which case HEK293 cells were stimulated with 0.1, 1, and 10 ng/ml PAM3CSK4, 100× less than that used for stimulation by LPS. Fold NF-κB stimulation of infected cells compared to unstimulated controls is plotted on the y axis. The results are means ± SD for triplicate samples from one of three independent experiments. Significant differences in stimulation potency between 33277 LPS and Δ1773Δ1587 LPS (P < 0.01) were observed for TLR4 but not for TLR2, TLR2/TLR1, and TLR1 activation at each concentration tested.
The results so far suggest that TLR2 activation by LPS is mediated by impurities present in the preparation. PG1828 is a known TLR2-activating lipoprotein contaminant found in LPS preparations and, interestingly, was demonstrated to act independently of TLR1 (47). We assessed whether TLR2 and TLR2/TLR1 stimulation by P. gingivalis LPS can be largely attributed to PG1828. Accordingly, LPS from both Δ1828 and ΔfimA single and double mutants were tested. Figure 5 demonstrates that LPS from Δ1828 mutants is severely attenuated for TLR2 stimulation, consistent with previous results (35). It was also attenuated for TLR2/TLR1 stimulation, though to a lesser extent than for TLR2 alone. Figure 5 further shows that FimA also contributes to a small extent to TLR2 activation, indicating that LPS preparations could be contaminated by fimbriae as well. The Δ1828 mutant LPS is further attenuated by a fimA mutation, at both TLR2 and TLR2/TLR1. The fact that LPS from the double ΔPG1828 ΔfimA mutant did not entirely lack the ability to stimulate TLR2/TLR1 indicates the presence of additional agonists that copurify with P. gingivalis LPS.
Fig 5.
PG1828 is the principal contributor to TLR2 activation, and a partial contributor to TLR2/TLR1 activation, by P. gingivalis LPS preparations. HEK293 cells transfected with human TLR2/CD14 (A) or human TLR2/TLR1/CD14 (B) were exposed to LPS preparations from 33277 (wild type), ΔPG1828, ΔfimA, or Δ1828 ΔfimA mutants. Inducible NF-κB stimulation of infected cells relative to unstimulated controls is plotted on the y axis. The results are means ± SD for triplicate samples from one of three independent experiments. *, P < 0.05 versus wild-type control.
Novel hydrophilic lipoprotein lipase-sensitive molecules mediate TLR2/TLR1 activation by P. gingivalis LPS preparations.
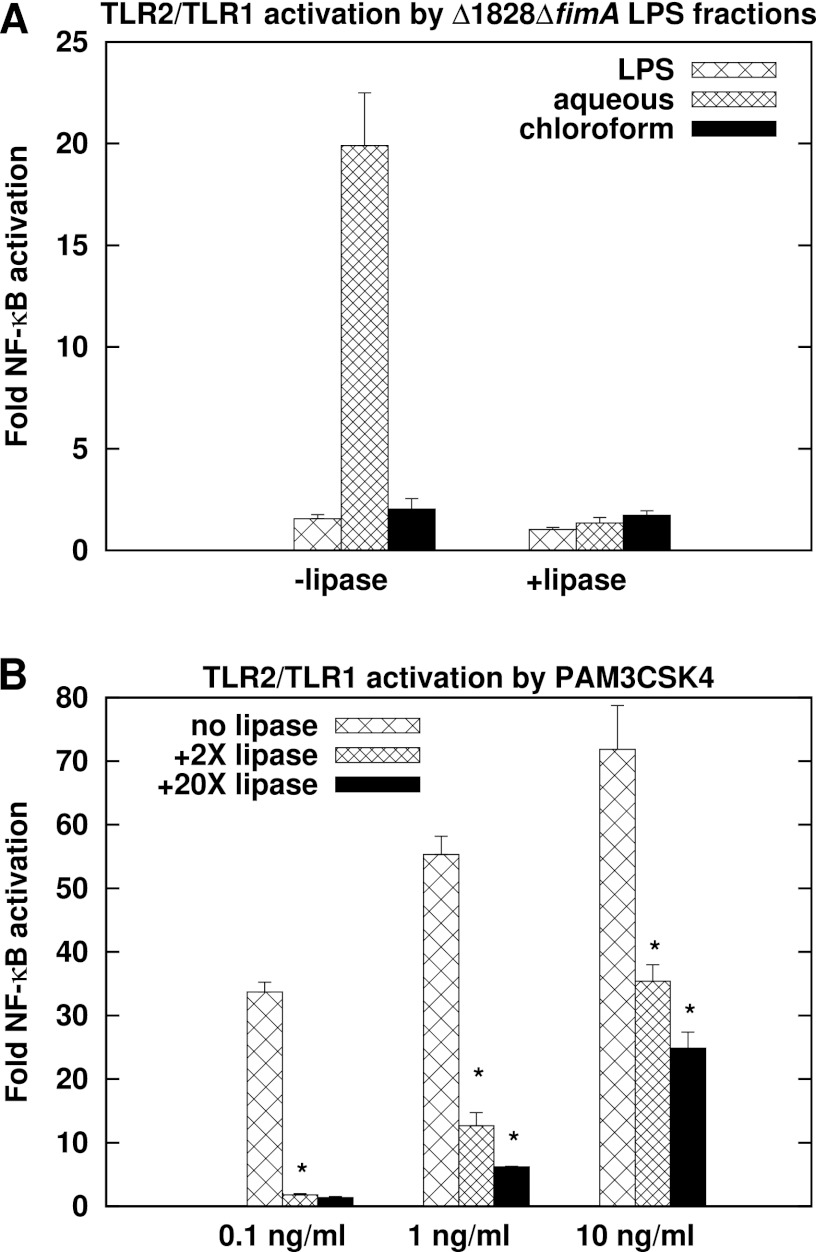
LPS from Δ1828 ΔfimA mutants was hydrolyzed to separate lipid A from the other two moieties of LPS, core oligosaccharide and O antigen, and subjected to biphasic water-methanol-chloroform fractionation (20, 48) to separate hydrophobic lipid A from potential aqueous contaminants. The chloroform phase is routinely used to assess lipid A structure by MALDI-TOF (matrix-assisted laser desorption ionization–time of flight) analysis (20) and was recently reported to contain phosphorylated dihydroceramides as well (49). Figure 6a demonstrates that the aqueous phase stimulates TLR2/TLR1 potently. The higher level of stimulation relative to the parent LPS could be due to a larger amount of ligand in similar amounts, measured as dry weight, of both preparations. The chloroform phase did not elicit a potent TLR2/TLR1 response, consistent with lipid A not being a TLR2 agonist and suggesting contaminant phosphoceramides do not contribute significantly to TLR2 activation by LPS preparations.
Fig 6.
TLR2/TLR1 ligands in P. gingivalis LPS preparations are hydrophilic and sensitive to lipoprotein lipase. (A) LPS, the aqueous (hydrophilic) fraction, or the chloroform (hydrophobic) fraction, each 100 ng/ml, from ΔPG1828 ΔfimA mutants was treated with 0 or 2× (wt/vol, relative to substrate, 200 ng here) lipoprotein lipase (lipase) and used to stimulate HEK293 cells expressing TLR2/TLR1/CD14. The aqueous and chloroform fractions were derived from LPS subjected to hydrolysis to yield lipid A. A significant difference was observed in the activation potencies of the aqueous phase with and without lipoprotein lipase (P < 0.05). (B) Synthetic triacylated lipoprotein PAM3CSK4 treated with 0, 2×, or 20× lipoprotein lipase was used to stimulate TLR2/TLR1/CD14-expressing HEK293 cells. Fold NF-κB stimulation relative to unstimulated controls is plotted on the y axis. The results are means ± SD for triplicate samples from one of three independent experiments. *, P < 0.05 for untreated versus 2×-lipase-treated and for 2×-lipase- versus 20×-lipase-treated PAM3CSK4.
We next investigated whether or not the hydrophilic TLR2/TLR1 ligands in Δ1828 ΔfimA mutant LPS are lipoproteins. Lipoprotein lipase, isolated from a Burkholderia sp., is an enzyme that hydrolyzes ester linkages in triglycerides resulting in sequential release of two acyl chains (50). Gram-negative bacterial lipoproteins have two acyl chains linked by thioester bonds and a third acyl chain linked by an amide bond to the N-terminal cysteine residue of the mature protein (51). Lipoprotein lipase has been shown to disrupt the TLR2-activating function of bacterial lipoproteins, including PG1828 (34). Treatment of the synthetic triacylated lipoprotein and potent TLR2/TLR1 agonist PAM3CSK4 with lipoprotein lipase resulted in abrogation of TLR2/TLR1 stimulation (Fig. 6B). Addition of lipoprotein lipase similarly curtailed TLR2/TLR1 stimulation by the ligand-containing aqueous phase of Δ1828 ΔfimA mutant LPS (Fig. 6A). These data indicate the presence of novel ligands in LPS preparations that are potentially lipoproteins.
Treatment of LPS preparations from a panel of Gram-negative bacteria with lipoprotein lipase curtails TLR2/TLR1, but not TLR4, activation.
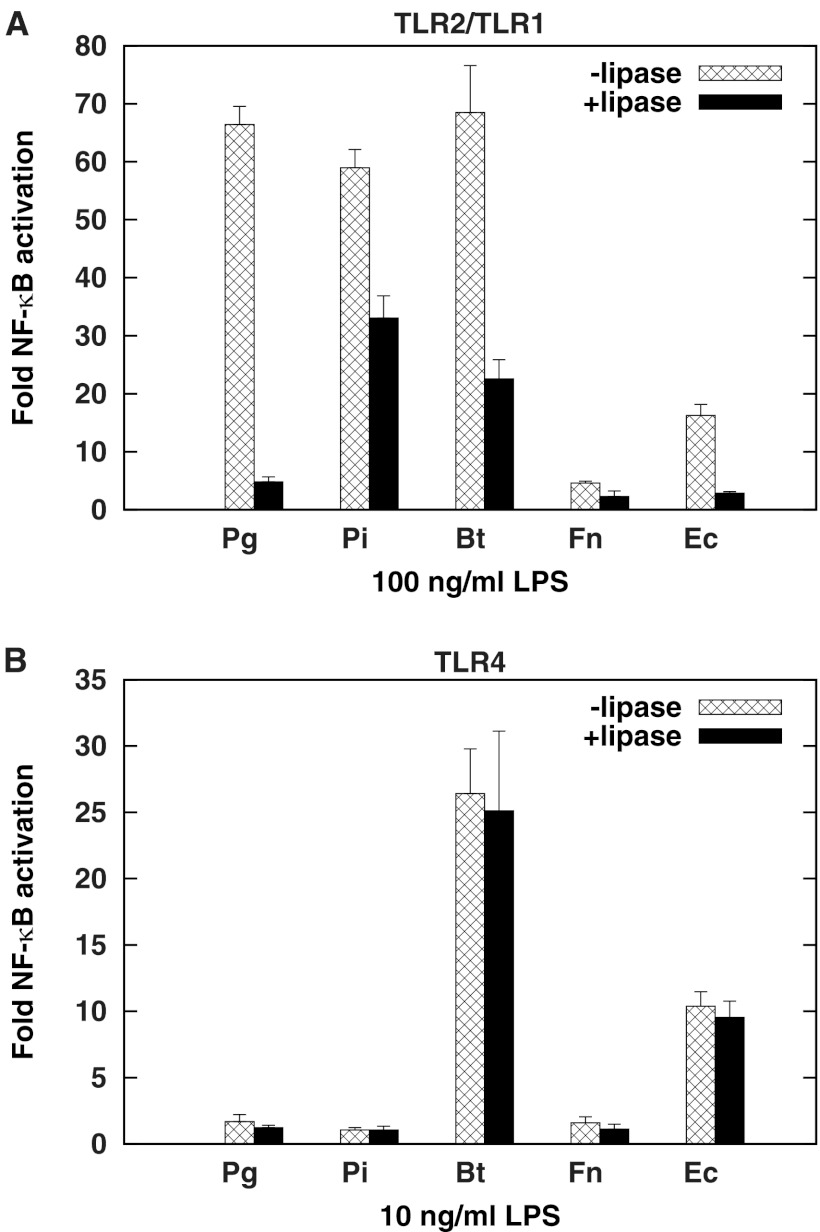
LPS from E. coli and P. gingivalis are known to contain lipoproteins that exhibit TLR2 agonist activity (33, 34). We tested LPS from a panel of Gram-negative bacteria that included P. gingivalis, the oral anaerobic periodontitis-associated species Prevotella intermedia, the oral commensal Fusobacterium nucleatum, the gut commensal Bacteroides thetaiotaomicron, and E. coli for TLR2/TLR1 activation. Figure 7A shows that LPSs from P. gingivalis, P. intermedia, and B. thetaiotaomicron stimulate TLR2/TLR1 potently. Interestingly, the addition of lipoprotein lipase significantly abrogates activation by the entire panel of LPS molecules tested. These LPS preparations, therefore, appear to contain various amounts of TLR2/TLR1 ligands that copurify with LPS. TLR4 activation by this panel of LPS molecules was also examined, both before and after lipoprotein lipase treatment. As expected, TLR4 was activated to different extents by these LPS molecules, depending on the structure of the lipid A moiety (21, 52), with B. thetaiotaomicron and E. coli JM83 LPS displaying potent activation (Fig. 7B). TLR4 activation, however, did not change significantly upon treatment with lipoprotein lipase (Fig. 7B), indicating that lipid A is not a substrate for lipoprotein lipase. These data further confirm that the structure of TLR2/TLR1 ligands in LPS preparations is distinct from that of lipid A, the TLR4 agonist.
Fig 7.
Lipoprotein lipase treatment of LPS from P. gingivalis (Pg), P. intermedia (Pi), B. thetaiotaomicron (Bt), F. nucleatum (Fn), and E. coli (Ec) attenuates TLR2/TLR1 activation (A) but not TLR4 activation (B). (A) HEK293 cells were transfected with human TLR2/TLR1/CD14 and stimulated with 100 ng/ml LPS treated with 0 or 200× (wt/vol relative to substrate, 2 μg here) lipoprotein lipase. (B) HEK293 cells were transfected with human TLR4/MD-2 and exposed to 10 ng/ml LPS treated with 0 or 200× lipoprotein lipase. Inducible NF-κB stimulation of LPS-infected cells over unstimulated controls is plotted on the y axis. The results are means ± SD for triplicate samples from one of two independent experiments. Significant differences in activation potency (P < 0.05) were observed between samples with and without lipoprotein lipase for TLR2/TLR1 but not for TLR4 stimulation.
Lipoprotein lipase attenuates TLR2 and TLR2/TLR1 stimulation by intact P. gingivalis.
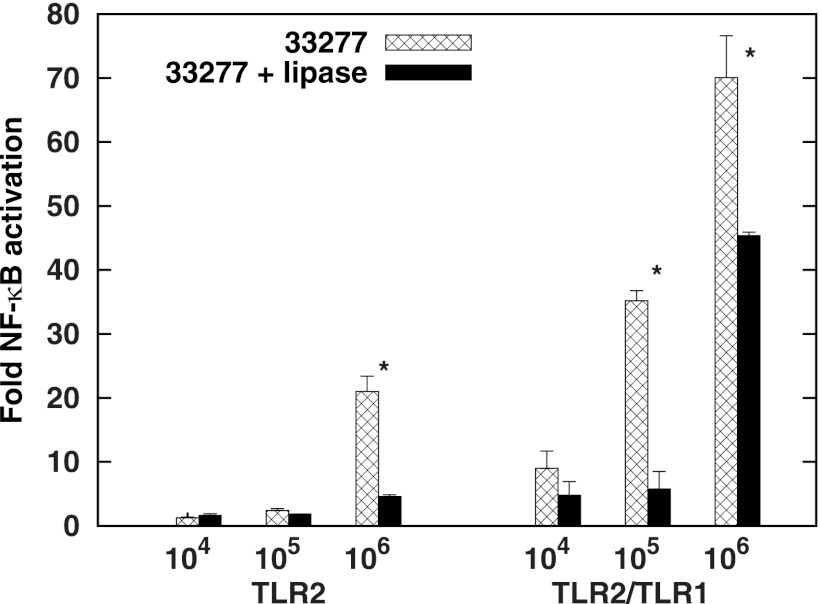
We treated whole P. gingivalis bacteria with lipoprotein lipase to assess the extent to which surface molecules sensitive to this enzyme contribute to TLR2 and TLR2/TLR1 activation. Figure 8 reveals a significant attenuation at both TLR2 and TLR2/TLR1 in response to this treatment. There was no decrease in bacterial viability as measured by colony-forming units (CFUs) following addition of lipoprotein lipase (data not shown), indicating that the membrane bilayer was not compromised. The addition of lipoprotein lipase is the first time we observed a decrease in the capacity of intact P. gingivalis to activate TLR2 and TLR2/TLR1 using a defined recombinant assay system. We conclude that this class of molecules confers on live P. gingivalis a predominant proportion of its capacity to activate TLR2 signaling.
Fig 8.
P. gingivalis whole bacteria are attenuated for TLR2 and TLR2/TLR1 activation by lipoprotein lipase. Intact cells of 33277 (105, 106, or 107) were treated with 0 or 200 μg lipoprotein lipase for 2 h at 37°C. HEK293 cells transfected with human TLR2/TLR1/CD14 were infected with 10-fold-diluted bacterial samples. Plotted on the y axis is fold NF-κB activation by bacteria relative to unstimulated controls. The results are means ± SD for triplicate samples from one of two independent experiments. *, P < 0.05 for samples with and without lipoprotein lipase.
DISCUSSION
In this work we show that the TLR2 response to P. gingivalis whole cells is not triggered solely by ligands reported to date. Mutants devoid of the two reported protein agonists, fimbriae and PG1828, are not attenuated for TLR2 activation. We are unable to use the genetic deletion approach to test the role of the reported nonprotein agonists LPS and phosphoceramides because of the nonviability or unavailability of mutants in their biosynthetic pathways. With respect to LPS, whether TLR2 activation is mediated by LPS structural moieties or contaminating agonists or both has not been amenable to straightforward resolution and is therefore a subject of controversy. Previous data demonstrated that purified preparations of P. gingivalis LPS activate TLR2 (22, 24). In our hands, repeated extractions using the method described by Manthey and Vogel (53) decreased TLR2 activation, indicating removal of contaminants, but also decreased the amount of lipid A obtained (data not shown), thereby confounding the issue. Two major P. gingivalis lipid A species, penta-acylated diphosphorylated agonist (28) and tetra-acylated monophosphorylated antagonist (31), have been chemically synthesized, and both failed to elicit TLR2 activation. It remains possible that a minor lipid A species produced by P. gingivalis contributes to TLR2 activation. However, the observation that TLR2 activation is independent of lipid A structure, as demonstrated by experiments in the present study, argues against this possibility. Taken together, the current body of evidence supports the idea that TLR2 activation by LPS preparations is predominantly attributable to copurifying ligands. The observations that LPS from Δ1828 mutants is attenuated for TLR2 and, to a lesser extent, TLR2/TLR1 activation and that lipoprotein lipase attenuates TLR2/TLR1 activation by P. gingivalis LPS preparations are consistent with the conclusions that (i) lipoprotein TLR2 agonists copurify with LPS and (ii) lipoprotein contaminants are not easily removed from P. gingivalis LPS preparations by phenol re-extraction.
Our observations with Δ1828 mutants reveal an important distinction between P. gingivalis LPS preparations and whole bacteria with respect to their TLR2 agonist composition. In contrast to the severe attenuation displayed by Δ1828 LPS, Δ1828 mutant bacteria stimulated TLR2 (with or without TLR1) as robustly as the wild type. This was confirmed in four strains (Fig. 2). Interestingly, the Δ1828 mutation in two strains, 33277 and 381, resulted in a mild increase in activation by whole bacteria. These two strains are different from the other two tested, A7436 and W50, in that they are not capsulated. Whether the absence of a capsule has any bearing on the mild hyperactivation phenotype remains to be investigated.
The distinction between the LPS preparations and whole bacteria mentioned above suggests that TLR2 agonists that coprecipitate with LPS comprise a fraction of the full repertoire of P. gingivalis TLR2 ligands. PG1828 is one such ligand, which gets separated from the other ligands in the process of LPS preparation. In this semi-isolated context, PG1828 is observed to be the main contributor to TLR2 stimulation. However, in the context of whole Δ1828 bacteria, the presence of other agonists, which do not copurify with LPS, compensates for the absence of PG1828 to stimulate TLR2.
Figure 5 suggests that FimA may also copurify with LPS. However, evaluation of P. gingivalis 33277 LPS preparations by immunoblotting with anti-FimA antibodies did not detect FimA in these preparations (data not shown). This could be because the amount of FimA falls below the level of detection. Alternatively, the fimA mutation may have an indirect effect that results in mildly decreased TLR2 activation by ΔfimA LPS preparations. For example, a perturbation of proteins associated with intact fimbriae, or the impact of ΔfimA on regulation of genes immediately downstream (54), could contribute to lowered activation. Complementation analyses of ΔfimA and Δ1828 mutants remain to be conducted. With respect to the Δ1828 mutation, annotation of the P. gingivalis genome indicates that the gene downstream is in the reverse orientation, suggesting that the mutation does not exert a polar effect.
The extents to which LPS preparations derived from a range of bacteria activate TLR2/TLR1 differ widely, suggesting various levels of contamination by TLR2 ligands in LPS preparations. The levels of attenuation achieved by the action of lipoprotein lipase also differ widely, as demonstrated by the low level of attenuation seen when the lipase was added to P. intermedia LPS, warranting further investigation into the nature of TLR2 ligands.
Lipoprotein lipase hydrolyzes ester bonds that link fatty acid chains to triglyceride, the prototype being eukaryotic very-low-density lipoproteins (50). The observation that lipoprotein lipase inactivates Gram-negative bacterial lipoproteins comes from work done with PG1828 (34), with lipoproteins in Actinomyces viscosus (55), and, in this study, with the synthetic triacylated lipoprotein PAM3CSK4. Interestingly, the TLR2-activating capacity of fimbriae was shown to be attenuated by lipoprotein lipase as well (56). Whether this is because the precursor FimA protein is lipidated (57) or is due to the presence of associated lipoprotein contaminants remains to be investigated. From a structural viewpoint, phospholipids such as phosphoceramides may also be susceptible to lipoprotein lipase. However, while isolated preparations of phosphoceramides contribute to TLR2 activation (58), our data indicate they are not major agonists, since hydrophilic fractions contribute more than hydrophobic phospholipid fractions to TLR2 activation from both whole-cell extracts and LPS preparations. The synthetic lipoprotein PAM3CSK4 is soluble in water, indicating that these lipoprotein lipase-sensitive molecules have hydrophilic properties, likely conferred by the peptide chain. Lipoproteins from the Gram-positive bacterium Listeria monocytogenes displaying TLR2 agonist activity were recently shown to be secreted, soluble, and sensitive to lipoprotein lipase as well (59). We propose that the hydrophilic lipase-sensitive TLR2 agonists in P. gingivalis whole-cell extracts and LPS preparations comprise a novel class of agonists composed by lipoproteins. Identification of members of this class is an area of active investigation.
It should be noted that the hydrophobic fraction, particularly that derived from whole P. gingivalis bacterial cells, also activates TLR2 and TLR2/TLR1, albeit to a lesser extent than the hydrophilic fraction. Potential ligands in this phase could include phosphoceramides, or even lipoproteins, which, owing to their amphipathic nature, may not be exclusively hydrophilic.
We attempted to examine the extent to which lipoproteins contribute to TLR2 activation by whole cells by constructing a P. gingivalis mutant devoid of mature lipoproteins. We targeted the gene encoding signal peptidase II, PGN_0515, for deletion analysis in 33277. Signal peptidase II cleaves the signal peptide following addition of diacylglycerol to the cysteine residue and is a prerequisite for addition of the third acyl chain (51). However, as with attempts made in other Gram-negative bacteria, we were unable to obtain mutant strains, indicating that this gene is essential for bacterial viability.
TLR2 and TLR2/TLR1 activation, though attenuated, was observed following lipoprotein lipase treatment of P. gingivalis cells, particularly when HEK cells were exposed to high concentrations of bacteria. This could be because members of this class are not all equally accessible to efficient enzyme activity due to structural constraints. Alternatively, lipase treatment may result in limited deacylation producing diacylated lipopeptides, which may retain the capacity to engage TLR2. Finally, it remains possible that molecules not belonging to the class of fatty acids that are ester linked to a triglyceride or peptide backbone possess TLR2 agonist activity.
The level of complexity in TLR2-P. gingivalis interaction is increased by the range of coreceptors that TLR2 potentially engages. TLR1 plays an important role as the TLR2 coreceptor for binding triacylated lipoproteins, as confirmed by X-ray structural studies (10). The two ester-linked acyl chains of the lipopeptide PAM3CSK4 fit into a hydrophobic pocket in TLR2, while the third amide-linked acyl chain inserts into a similar pocket in TLR1. Hydrophobic moieties are, therefore, implicated as an important structural requirement for ligands to bind TLR2. The hydrophobic molecules lipoteichoic acid and a synthetic phosphoethanolamine derivative were also shown to bind TLR2 by X-ray crystallography. However, these ligands did not engage TLR1 or TLR6 (11). The extent of TLR2 activation by these hydrophobic ligands was low, indicating that heterodimerization of TLR2 with TLR1 or TLR6 is important to facilitate a potent response. Hence, the precise structure of hydrophobic molecules, such as the structure of the head group, may dictate whether it interacts with a coreceptor, which in turn influences the outcome of the response. P. gingivalis fimbriae were shown to activate TLR2/TLR1 heterodimers but not TLR2/TLR6 or TLR2 by itself (23). In another study, however, FimA was shown to activate both TLR2/TLR1 and TLR2/TLR6 (60). Synthetic derivatives of PG1828, on the other hand, were capable of activating TLR2 independently of TLR1 (47). Our study with Δ1828 LPS preparations shows that it is severely deficient in TLR2 activation even in the absence of TLR1 (Fig. 5A), indicating that PG1828 is, indeed, not dependent on TLR1 for TLR2 stimulation. Interestingly, however, we consistently observed a higher level of activation of TLR2/TLR1 than TLR2 alone, whether in response to LPS preparations, whole-bacterial-cell fractions, or whole bacteria. One explanation is that TLR2/TLR1 heterodimers are more sensitive to activation by the resident ligands than TLR2 alone. This, in turn, supports the concept that the ligands may be lipoproteins (10).
Another well-characterized TLR2 coreceptor is CD14, which FimA binds to activate TLR2 (23, 61, 62), and which we included in our HEK293 assays. CD11b has also been identified as a TLR2-fimbria coreceptor, and this interaction is implicated in downregulation of the antimicrobial cytokine interleukin 12 (IL-12) (9). Interaction of fimbriae with TLR2 has also been shown to induce cross talk of TLR2 with the coreceptors CR5, CXCL4, and CR3, resulting in distinct responses (5, 7, 8). The plasticity and diversity in downstream signaling when TLR2 is activated is further underscored by the observation that P. gingivalis live cells induce a different pattern of TLR2-dependent cytokines than that induced by FimA or LPS preparations alone in mouse peritoneal macrophages (26, 63). It is likely that different TLR2-activating ligands vary in their abilities to engage TLR2 and specific coreceptors. In terms of responses, TLR2 engagement has been shown to trigger production of both proinflammatory and anti-inflammatory cytokines. An emerging body of evidence suggesting that TLR2 promotes immune tolerance includes the action of B. fragilis polysaccharide A in promoting secretion of the anti-inflammatory cytokine IL-10 (64). A systematic identification of P. gingivalis TLR2 agonists, followed by structural and functional analyses, will give insights into the mechanism by which P. gingivalis fine-tunes the TLR2 response to its overall benefit.
ACKNOWLEDGMENTS
We are grateful to Pam Braham for her help in constructing Δ1828 and ΔfimA single and double mutants in P. gingivalis 33277. We thank Fuminobu Yoshimura for generously providing us with anti-FimA antibodies. We also thank Roger W. Kramer for his help with figures and statistics and critical reading of the manuscript.
This study was supported by NIH grants (T32 DE07132-26 to S.J. and DE012768 to R.P.D.) and the Douglass L. Morell Research Fund (to R.P.D.).
Footnotes
Published ahead of print 4 February 2013
REFERENCES
- 1. Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8:481–490 [DOI] [PubMed] [Google Scholar]
- 2.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134–144 [DOI] [PubMed] [Google Scholar]
- 3. Burns E, Bachrach G, Shapira L, Nussbaum G. 2006. TLR2 is required for the innate response to Porphyromonas gingivalis: activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J. Immunol. 177:8296–8300 [DOI] [PubMed] [Google Scholar]
- 4. Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. 2011. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10:497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang M, Shakhatreh MA, James D, Liang S, Nishiyama S, Yoshimura F, Demuth DR, Hajishengallis G. 2007. Fimbrial proteins of porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J. Immunol. 179:2349–2358 [DOI] [PubMed] [Google Scholar]
- 6. Medzhitov R, Janeway C., Jr 2000. The Toll receptor family and microbial recognition. Trends Microbiol. 8:452–456 [DOI] [PubMed] [Google Scholar]
- 7. Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H, Li F, Tzekou A, Lambris JD, Hajishengallis G. 2011. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J. Immunol. 186:869–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hajishengallis G, Wang M, Liang S, Triantafilou M, Triantafilou K. 2008. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc. Natl. Acad. Sci. U. S. A. 105:13532–13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hajishengallis G, Shakhatreh MA, Wang M, Liang S. 2007. Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J. Immunol. 179:2359–2367 [DOI] [PubMed] [Google Scholar]
- 10. Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. 2007. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell 130:1071–1082 [DOI] [PubMed] [Google Scholar]
- 11. Kang JY, Nan X, Jin MS, Youn Ryu S-JYH, Mah S, Han SH, Lee H, Paik S-G, Lee J-O. 2009. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity 31:873–884 [DOI] [PubMed] [Google Scholar]
- 12. Takeuchi O, Kawai T, Mühlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. 2001. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 13:933–940 [DOI] [PubMed] [Google Scholar]
- 13. Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. 2002. Role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169:10–14 [DOI] [PubMed] [Google Scholar]
- 14. Zähringer U, Lindner B, Inamura S, Heine H, Alexander C. 2008. TLR2—promiscuous or specific? A critical re-evaluation of a receptor expressing apparent broad specificity. Immunobiology 213:205–224 [DOI] [PubMed] [Google Scholar]
- 15. Darveau RP, Cunningham MD, Bailey T, Seachord C, Ratcliffe K, Bainbridge B, Dietsch M, Page RC, Aruffo A. 1995. Ability of bacteria associated with chronic inflammatory disease to stimulate E-selectin expression and promote neutrophil adhesion. Infect. Immun. 63:1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujiwara T, Ogawa T, Sobue S, Hamada S. 1990. Chemical, immunobiological and antigenic characterizations of lipopolysaccharides from Bacteroides gingivalis strains. J. Gen. Microbiol. 136:319–326 [DOI] [PubMed] [Google Scholar]
- 17. Mansheim BJ, Onderdonk AB, Kasper DL. 1978. Immunochemical and biologic studies of the lipopolysaccharide of Bacteroides melaninogenicus subspecies asaccharolyticus. J. Immunol. 120:72–78 [PubMed] [Google Scholar]
- 18. Ogawa T, Uchida H. 1996. Differential induction of IL-1 beta and IL-6 production by the nontoxic lipid A from Porphyromonas gingivalis in comparison with synthetic Escherichia coli lipid A in human peripheral blood mononuclear cells. FEMS Immunol. Med. Microbiol. 14:1–13 [DOI] [PubMed] [Google Scholar]
- 19. Chen C, Coats SR, Bumgarner RE, Darveau RP. 2007. Hierarchical gene expression profiles of HUVEC stimulated by different lipid A structures obtained from Porphyromonas gingivalis and Escherichia coli. Cell Microbiol. 9:1028–1038 [DOI] [PubMed] [Google Scholar]
- 20. Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. 2009. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell Microbiol. 11:1587–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jain S, Darveau RP. 2010. Contribution of Porphyromonas gingivalis lipopolysaccharide to periodontitis. Periodontol. 2000 54:53–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Darveau RP, Pham TT, Lemley K, Reife RA, Bainbridge BW, Coats SR, Howald WN, Way SS, Hajjar AM. 2004. Porphyromonas gingivalis lipopolysaccharide contains multiple lipid A species that functionally interact with both toll-like receptors 2 and 4. Infect. Immun. 72:5041–5051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hajishengallis G, Tapping RI, Harokopakis E, Nishiyama S, Ratti P, Schifferle RE, Lyle EA, Triantafilou M, Triantafilou K, Yoshimura F. 2006. Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell. Microbiol. 8:1557–1570 [DOI] [PubMed] [Google Scholar]
- 24. Hirschfeld M, Weis JJ, Toshchakov V, Salkowski CA, Cody MJ, Ward DC, Qureshi N, Michalek SM, Vogel SN. 2001. Signaling by Toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect. Immun. 69:1477–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Muthukuru M, Jotwani R, Cutler CW. 2005. Oral mucosal endotoxin tolerance induction in chronic periodontitis. Infect. Immun. 73:687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou Q, Desta T, Fenton M, Graves DT, Amar S. 2005. Cytokine profiling of macrophages exposed to Porphyromonas gingivalis, its lipopolysaccharide, or its FimA protein. Infect. Immun. 73:935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hirschfeld M, Ma Y, Weis JH, Vogel SN, Weis JJ. 2000. Repurification of lipopolysaccharide eliminates signaling through both human and murine toll-like receptor 2. J. Immunol. 165:618–622 [DOI] [PubMed] [Google Scholar]
- 28. Kumada H, Haishima Y, Watanabe K, Hasegawa C, Tsuchiya T, Tanamoto K, Umemoto T. 2008. Biological properties of the native and synthetic lipid A of Porphyromonas gingivalis lipopolysaccharide. Oral Microbiol. Immunol. 23:60–69 [DOI] [PubMed] [Google Scholar]
- 29. Ogawa T, Asai Y, Hashimoto M, Takeuchi O, Kurita T, Yoshikai Y, Miyake K, Akira S. 2002. Cell activation by Porphyromonas gingivalis lipid A molecule through Toll-like receptor 4- and myeloid differentiation factor 88-dependent signaling pathway. Int. Immunol. 14:1325–1332 [DOI] [PubMed] [Google Scholar]
- 30. Sawada N, Ogawa T, Asai Y, Makimura Y, Sugiyama A. 2007. Toll-like receptor 4-dependent recognition of structurally different forms of chemically synthesized lipid As of Porphyromonas gingivalis. Clin. Exp. Immunol. 148:529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y, Gaekwad J, Wolfert MA, Boons GJ. 2008. Synthetic tetra-acylated derivatives of lipid A from Porphyromonas gingivalis are antagonists of human TLR4. Org. Biomol. Chem. 6:3371–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hellman J, Tehan MM, Warren HS. 2003. Murein lipoprotein, peptidoglycan-associated lipoprotein, and outer membrane protein A are present in purified rough and smooth lipopolysaccharides. J. Infect. Dis. 188:286–289 [DOI] [PubMed] [Google Scholar]
- 33. Lee HK, Lee J, Tobias PS. 2002. Two lipoproteins extracted from Escherichia coli K-12 LCD25 lipopolysaccharide are the major components responsible for Toll-like receptor 2-mediated signaling. J. Immunol. 168:4012–4017 [DOI] [PubMed] [Google Scholar]
- 34. Hashimoto M, Asai Y, Ogawa T. 2004. Separation and structural analysis of lipoprotein in a lipopolysaccharide preparation from Porphyromonas gingivalis. Int. Immunol. 16:1431–1437 [DOI] [PubMed] [Google Scholar]
- 35. Asai Y, Hashimoto M, Fletcher HM, Miyake K, Akira S, Ogawa T. 2005. Lipopolysaccharide preparation extracted from Porphyromonas gingivalis lipoprotein-deficient mutant shows a marked decrease in toll-like receptor 2-mediated signaling. Infect. Immun. 73:2157–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Al-Qutub MN, Braham PH, Karimi-Naser LM, Liu X, Genco CA, Darveau RP. 2006. Hemin-dependent modulation of the lipid A structure of Porphyromonas gingivalis lipopolysaccharide. Infect. Immun. 74:4474–4485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paramonov NA, Aduse-Opoku J, Hashim A, Rangarajan M, Curtis MA. 2009. Structural analysis of the core region of O-lipopolysaccharide of Porphyromonas gingivalis from mutants defective in O-antigen ligase and O-antigen polymerase. J. Bacteriol. 191:5272–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fletcher HM, Schenkein HA, Morgan RM, Bailey KA, Berry CR, Macrina FL. 1995. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect. Immun. 63:1521–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen T, Dong H, Tang YP, Dallas MM, Malamy MH, Duncan MJ. 2000. Identification and cloning of genes from Porphyromonas gingivalis after mutagenesis with a modified Tn4400 transposon from Bacteroides fragilis. Infect. Immun. 68:420–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tokuda M, Chen W, Karunakaran T, Kuramitsu HK. 1998. Regulation of protease expression in Porphyromonas gingivalis. Infect. Immun. 66:5232–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fujiwara T, Morishima S, Takahashi I, Hamada S. 1993. Molecular cloning and sequencing of the fimbrilin gene of Porphyromonas gingivalis strains and characterization of recombinant proteins. Biochem. Biophys. Res. Commun. 197:241–247 [DOI] [PubMed] [Google Scholar]
- 42. Yoshimura F, Takahashi K, Nodasaka Y, Suzuki T. 1984. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J. Bacteriol. 160:949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nichols FC, Riep B, Mun J, Morton MD, Bojarski MT, Dewhirst FE, Smith MB. 2004. Structures and biological activity of phosphorylated dihydroceramides of Porphyromonas gingivalis. J. Lipid Res. 45:2317–2330 [DOI] [PubMed] [Google Scholar]
- 44. Vallabhapurapu S, Karin M. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733 [DOI] [PubMed] [Google Scholar]
- 45. Asai Y, Ohyama Y, Gen Ogawa KT. 2001. Bacterial fimbriae and their peptides activate human gingival epithelial cells through Toll-like receptor 2. Infect. Immun. 69:7387–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davey M, Liu X, Ukai T, Jain V, Gudino C, Gibson FC, Golenbock D, Visintin A, Genco CA. 2008. Bacterial fimbriae stimulate proinflammatory activation in the endothelium through distinct TLRs. J. Immunol. 180:2187–2195 [DOI] [PubMed] [Google Scholar]
- 47. Makimura Y, Asai Y, Taiji Y, Sugiyama A, Tamai R, Ogawa T. 2006. Correlation between chemical structure and biological activities of Porphyromonas gingivalis synthetic lipopeptide derivatives. Clin. Exp. Immunol. 146:159–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bligh EG, Dyer WJ. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 49. Nichols FC, Bajrami B, Clark RB, Housley W, Yao X. 2012. Free lipid A isolated from Porphyromonas gingivalis lipopolysaccharide is contaminated with phosphorylated dihydroceramide lipids: recovery in diseased dental samples. Infect. Immun. 80:860–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mead JR, Irvine SA, Ramji DP. 2002. Lipoprotein lipase: structure, function, regulation, and role in disease. J. Mol. Med. 80:753–769 [DOI] [PubMed] [Google Scholar]
- 51. Kovacs-Simon A, Titball RW, Michell SL. 2011. Lipoproteins of bacterial pathogens. Infect. Immun. 79:548–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Coats SR, Berezow AB, To TT, Jain S, Bainbridge BW, Banani KP, Darveau RP. 2011. The lipid A phosphate position determines differential host Toll-like receptor 4 responses to phylogenetically related symbiotic and pathogenic bacteria. Infect. Immun. 79:203–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Manthey CL, Vogel SN. 1994. Elimination of trace endotoxin protein from rough chemotype LPS. J. Endotoxin Res. 1:84–91 [Google Scholar]
- 54. Nishikawa K, Yoshimura F, Duncan MJ. 2004. A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Mol. Microbiol. 54:546–560 [DOI] [PubMed] [Google Scholar]
- 55. Shimada E, Kataoka H, Miyazawa Y, Yamamoto M, Igarashi T. 2012. Lipoproteins of Actinomyces viscosus induce inflammatory responses through TLR2 in human gingival epithelial cells and macrophages. Microbes Infect. 14:916–921 [DOI] [PubMed] [Google Scholar]
- 56. Aoki Y, Tabeta K, Murakami Y, Yoshimura F, Yamazaki K. 2010. Analysis of immunostimulatory activity of Porphyromonas gingivalis fimbriae conferred by Toll-like receptor 2. Biochem. Biophys. Res. Commun. 398:86–91 [DOI] [PubMed] [Google Scholar]
- 57. Shoji M, Yoshimura A, Yoshioka H, Takade A, Takuma Y, Yukitake H, Naito M, Hara Y, Yoshida Nakayama S-IK. 2010. Recombinant Porphyromonas gingivalis FimA preproprotein expressed in Escherichia coli is lipidated and the mature or processed recombinant FimA protein forms a short filament in vitro. Can. J. Microbiol. 56:959–967 [DOI] [PubMed] [Google Scholar]
- 58. Nichols FC, Housley WJ, O'Conor CA, Manning T, Wu S, Clark RB. 2009. Unique lipids from a common human bacterium represent a new class of Toll-like receptor 2 ligands capable of enhancing autoimmunity. Am. J. Pathol. 175:2430–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Machata S, Tchatalbachev S, Mohamed W, Jänsch Hain LT, Chakraborty T. 2008. Lipoproteins of Listeria monocytogenes are critical for virulence and TLR2-mediated immune activation. J. Immunol. 181:2028–2035 [DOI] [PubMed] [Google Scholar]
- 60. Bagaitkar J, Demuth DR, Daep CA, Renaud DE, Pierce DL, Scott DA. 2010. Tobacco upregulates P. gingivalis fimbrial proteins which induce TLR2 hyposensitivity. PLoS One 5:e9323 doi:10.1371/journal.pone.0009323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Eskan MA, Hajishengallis G, Kinane DF. 2007. Differential activation of human gingival epithelial cells and monocytes by Porphyromonas gingivalis fimbriae. Infect. Immun. 75:892–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hajishengallis G, Ratti P, Harokopakis E. 2005. Peptide mapping of bacterial fimbrial epitopes interacting with pattern recognition receptors. J. Biol. Chem. 280:38902–38913 [DOI] [PubMed] [Google Scholar]
- 63. Zhou Q, Amar S. 2007. Identification of signaling pathways in macrophage exposed to Porphyromonas gingivalis or to its purified cell wall components. J. Immunol. 179:7777–7790 [DOI] [PubMed] [Google Scholar]
- 64. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–977 [DOI] [PMC free article] [PubMed] [Google Scholar]