Abstract
The Ros-type regulator MucR is one of the few transcriptional regulators that have been linked to virulence in Brucella. Here, we show that a Brucella abortus in-frame mucR deletion strain exhibits a pronounced growth defect during in vitro cultivation and, more importantly, that the mucR mutant is attenuated in cultured macrophages and in mice. The genetic basis for the attenuation of Brucella mucR mutants has not been defined previously, but in the present study the genes regulated by MucR in B. abortus have been elucidated using microarray analysis and real-time reverse transcription-PCR (RT-PCR). In B. abortus 2308, MucR regulates a wide variety of genes whose products may function in establishing and maintaining cell envelope integrity, polysaccharide biosynthesis, iron homeostasis, genome plasticity, and transcriptional regulation. Particularly notable among the MucR-regulated genes identified is arsR6 (nolR), which encodes a transcriptional regulator previously linked to virulence in Brucella melitensis 16 M. Importantly, electrophoretic mobility shift assays (EMSAs) determined that a recombinant MucR protein binds directly to the promoter regions of several genes repressed by MucR (including arsR6 [nolR]), and in Brucella, as in other alphaproteobacteria, MucR binds to its own promoter to repress expression of the gene that encodes it. Overall, these studies have uncovered the diverse genetic regulon of MucR in Brucella, and in doing so this work has begun to define the MucR-controlled genetic circuitry whose misregulation contributes to the virulence defect of Brucella mucR mutants.
INTRODUCTION
Human infections by Brucella spp. represent the most common zoonosis worldwide (1). These Gram-negative bacteria naturally infect a variety of wild and domestic animals, and humans become infected through exposure to infected animals and animal products (2). The Brucella spp. are members of the α2 subclass of proteobacteria, which includes bacteria that are symbionts and pathogens of plants and mammalian pathogens (3). Very often, the bacteria in this group reside within or in close association with the cells of their host, and these interactions with the eukaryotic host cell are essential for the life of the bacteria. Due to the close phylogenetic relatedness of the α2-proteobacteria, these organisms use common genes and strategies for facilitating their interactions with their specific host (4), and the gene encoding the transcriptional regulator Ros/MucR is one of the genes conserved in the α2-proteobacteria that is important for host-bacterium interactions.
In Agrobacterium tumefaciens, ros (for rough outer surface) was identified as a gene whose inactivation results in small, nonmucoid colonies (compared to the normally larger, mucoid colonies of the wild-type strain) (5), and while virulence-associated genes (e.g., virC and virD) are disregulated in this mutant, a ros mutant maintains wild-type virulence (6, 7). In Sinorhizobium meliloti, mutation of the mucR gene leads to a slight growth defect compared to the parental strain, and overexpression of mucR in the parental strain results in a significant increase in colony mucoidy; however, the S. meliloti mucR mutant strain is not defective in nodule occupancy or its ability to fix nitrogen (8). Conversely, a Rhizobium etli rosR mutant exhibits altered colony morphology compared to the parental strain, and this mutant is defective in nodulation competitiveness and competitive growth in the rhizosphere (9, 10).
While early studies genetically linked ros and mucR mutations to growth defects and differences in colony mucoidy, the mechanism of action of the Ros/MucR proteins was not known at the time; however, it has since been determined that Ros/MucR proteins are transcriptional repressors that regulate numerous genes, including those involved in polysaccharide synthesis, motility, and quorum sensing (11, 12, 13, 14, 15, 16, 17, 18). Ros/MucR-type regulators are unusual in that they contain a Zn finger motif that is uncommon in prokaryotes, whereas transcriptional regulators with this type of motif are commonly found in eukaryotes (19). In fact, the origin of the Zn finger motif-containing proteins has been the source of some debate in recent years. Due to the close association of the alphaproteobacteria with eukaryotic host cells, it has been suggested that an ancestral alphaproteobacterium acquired a gene encoding the Zn finger protein from a eukaryotic host (19, 20); however, others have proposed that Zn finger proteins are of bacterial origin (21, 22). Regardless of their origin, the Zn finger motif-containing proteins, such as Ros and MucR, are essential for the biology of many members of the alphaproteobacteria.
A MucR ortholog was recently identified in the Brucella spp., and this regulator is essential for the virulence of Brucella melitensis 16 M (23). Additionally, a B. melitensis mucR mutant shows promise as a potential candidate vaccine against Brucella infections (24). While it is clear that mucR is important for the pathogenesis of Brucella, the genes regulated by MucR in Brucella remain undefined. In the present study, an isogenic mucR deletion strain was derived from Brucella abortus 2308 in an attempt to define the Brucella MucR regulon, as well as to assess the phenotype of a B. abortus mucR mutant.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Brucella abortus 2308 and derivative strains were routinely grown on Schaedler blood agar (SBA), which is Schaedler agar (Becton, Dickinson and Co., Franklin Lakes, NJ) containing 5% defibrinated bovine blood (Quad Five, Ryegate, MT), or in brucella broth (Becton, Dickinson and Co., Franklin Lakes, NJ). For cloning and recombinant protein production, Escherichia coli strains (DH5α and BL21) were grown routinely on tryptic soy agar or in Luria-Bertani broth. When appropriate, growth media were supplemented with ampicillin (100 μg/ml) or kanamycin (45 μl/ml).
Construction and genetic complementation of a mucR mutant.
The mucR locus (bab1_0594) in Brucella abortus 2308 was mutated using a nonpolar, unmarked gene excision strategy described previously (25). An approximately 1-kb fragment representing the region upstream of the gene extending to the second codon of the coding region was amplified by PCR using primers ΔmucR-Up-For and ΔmucR-Up-Rev (Table 1), genomic DNA from Brucella abortus 2308 as a template, and Pfx polymerase (Invitrogen). Similarly, a fragment containing the last two codons of the coding region extending to approximately 1 kb downstream of mucR was amplified with primers ΔmucR-Down-For and ΔmucR-Down-Rev (Table 1). The upstream fragment was digested with BamHI, while the downstream fragment was digested with PstI, and both fragments were treated with polynucleotide kinase in the presence of ATP. Both of the DNA fragments were included in a single ligation mix with BamHI/PstI-digested pNPTS138 (Table 2) (26), which contains a kanamycin resistance marker and sacB gene for counterselection with sucrose. The resulting plasmid (pC3029) (Table 2) was introduced into B. abortus 2308, and merodiploid transformants were obtained by selection on SBA plus kanamycin. A single kanamycin-resistant clone was grown for ∼6 h in brucella broth and then plated onto SBA containing 10% sucrose. Genomic DNA from sucrose-resistant, kanamycin-sensitive colonies was isolated and screened by PCR for loss of the mucR gene, and an isogenic mucR mutant derived from B. abortus 2308 was named CC092. The mucR mutation in this strain was verified by DNA sequence analysis and Southern hybridization.
Table 1.
Oligonucleotide primers used in this study
| Designation | Sequence (5′→3′)a |
|---|---|
| ΔmucR-Up-For | GCGGATCCGATGAAATAGGGCTGTTCGC |
| ΔmucR-Up-Rev | GGTGCTTTCGTCGTTCGTT |
| ΔmucR-Down-For | TGATTCTTCAGCGAGTGAATCACG |
| ΔmucR-Down-Rev | GCCTGCAGCATTCCCAGCGATGTATGGG |
| mucR-comp-For | CTCAATTTTCTTGCGGTGC |
| mucR-comp-Rev | AGGCAGGACTGTCAGGAGAA |
| mucR-lacZ-For | GCAAGCTTTTCTATAAATCATATTTGTCTTGG |
| mucR-lacZ-Rev | GCGGATCCTTCCATAAGTTTTCCTTTT |
| rMucR-For | GCGGTCTCCGCGCGAAAATCTGGAAACGAACGACGAAA |
| rMucR-Rev | GCGGTCTCCTATCAGGCGTCCTTCGGCTTGCGG |
| babR-RT-For | AAGAATTATGCGCATGACCT |
| babR-RT-Rev | GTTCCCACCCATCTGGAAAT |
| BAB1_0746-RT-For | AGGGAAGCAAAAGCGACATGT |
| BAB1_0746-RT-Rev | AAAGCTAGACTGGTGCCGGTACG |
| BAB1_0747-RT-For | CATGGAAACGTTTGGCAAATG |
| BAB1_0747-RT-Rev | CTGAAGTTCACTGCAGCTGTAATGG |
| BAB1_1035-RT-For | GTTCTTTGTCGGCTCCTTAACCTT |
| BAB1_1035-RT-Rev | TCCGAAATCAAGATGCTTGAGG |
| BAB1_1605-RT-For | TGGCAAAAGCTATTGATCTCAGCC |
| BAB1_1605-RT-Rev | GGCGTGGTGTGGGACTCATATA |
| BAB1_1893-RT-For | CATATGACCTGCCAGCCGC |
| BAB1_1893-RT-Rev | CCTGGTCGTCGCATCCTATG |
| BAB1_2012-RT-For | AACGGGAGAAACTTGTAAGGCAG |
| BAB1_2012-RT-Rev | GCAGCCTATGGTGTTTGTAGCG |
| BAB1_2041-RT-For | TCTTCTTCAGCCAGACATTCGAC |
| BAB1_2041-RT-Rev | TCAGCAGGCCGGATGTAAGCT |
| BAB2_0143-RT-For | GTTTTCTCGTTCCACTTCGCC |
| BAB2_0143-RT-Rev | CGGGTACAACCACGCTTGAA |
| BAB2_0807-RT-For | TGCTGGAGCTTGGTTTCAGG |
| BAB2_0807-RT-Rev | GGTTGTCTTCCCGCAATTCC |
| BAB2_1072-RT-For | GGTCGTGATCTTGTCCACCATAA |
| BAB2_1072-RT-Rev | GACGATCGATTTGATGCTACGG |
| BAB1_0746-EMSA-For | TTGAAATTGCCAACGAGCTTG |
| BAB1_0746-EMSA-Rev | GCCTGTTATTTCTTCATGGTCGC |
| BAB1_1035-EMSA-For | CGCCATAAAACGAACCTCA |
| BAB1_1035-EMSA-Rev | CGTCGGCAGAAGTAATTTT |
| BAB1_1605-EMSA-For | ATGATTTAGTAGAAAACGCAGA |
| BAB1_1605-EMSA-Rev | TGGCCAGATGTTGTGAAAG |
| BAB1_1893-EMSA-For | GCTGTCTTGTTCATTCTGTC |
| BAB1_1893-EMSA-Rev | CGGAATCATCTGTACCACC |
| BAB2_0840-EMSA-For | GCGGATCCGACAAAAATTGTTTAGAA |
| BAB2_0840-EMSA-Rev | TAAGATCACTCTTTCAAAGGCGGCT |
| mucR-EMSA-For | CTCAATTTTCTTGCGGTGCCCTG |
| mucR-EMSA-Rev | CGCAGCGGCTGACAATGGCAA |
Underlined sections indicate restriction endonuclease recognition sequences.
Table 2.
Plasmids used in this study
| Plasmid name | Description | Reference or source |
|---|---|---|
| pBBR1MCS-4 | Broad-host-range cloning vector; Ampr | 27 |
| pNPTS138 | Cloning vector; contains sacB gene; Kanr | 26 |
| pMR15 | Broad-host-range vector containing a promoterless lacZ gene; Kanr | 29 |
| pC3029 | In-frame deletion of mucR plus 1 kb of each flanking region in pNPTS138 | This study |
| pC3030 | mucR locus including the entire promoter region in pBBR1MCS-4 | This study |
| pC3031 | mucR promoter region cloned into pMR15 | This study |
Genetic complementation of the mucR mutation in CC092 was achieved by expressing the wild-type mucR allele from its native promoter in pBBR1MCS-4 (Table 2) (27). The mucR gene, along with the native mucR promoter, was amplified by PCR using primers mucR-comp-For and mucR-comp-Rev (Table 1) and Pfx polymerase (Invitrogen). The resulting DNA fragment was treated with polynucleotide kinase and then ligated into SmaI-digested pBBR1MCS-4. This construct, pC3030 (Table 2), was introduced into the B. abortus mucR mutant strain CC092 by electroporation.
All Brucella strains generated during this study were tested by the crystal violet exclusion assay in order to assess whether a given strain produced a smooth or rough form of lipopolysaccharide (LPS) (28). Briefly, Brucella strains were grown on tryptic soy agar for 72 to 96 h, and the plates were flooded with a dilute (1:1,000) solution of crystal violet for ∼25 s. The parental strains B. abortus 2308 and B. melitensis 16 M were included as smooth LPS-producing controls, while B. abortus RB51 served as a rough LPS-producing control.
Construction of a transcriptional mucR-lacZ promoter fusion and β-galactosidase assays.
The promoter region of the B. abortus 2308 mucR gene was fused to a lacZ reporter as a transcriptional fusion. Approximately 400 bp of the region upstream of mucR was amplified by PCR using primers mucR-lacZ-For and mucR-lacZ-Rev (Table 1) and Brucella abortus 2308 genomic DNA as a template. The amplified DNA fragment was sequentially digested with BamHI and HindIII and subsequently ligated into BamHI/HindIII-digested pMR15 (Table 2) (29), which contains a promoterless lacZ gene. The lacZ promoter fusion plasmid (pC3031) (Table 2) was then electroporated into Brucella abortus 2308 and its derivative strains. Brucella abortus strains harboring the lacZ fusion construct were grown in brucella broth, and β-galactosidase assays were performed as described previously (30).
Microarray analysis.
Total RNA was isolated from Brucella cultures grown to late exponential phase as described previously (25), and contaminating genomic DNA was removed by treatment with RNase-free DNase I (Ambion). RNA was prepared from at least two biological replicates for each strain. Ten micrograms of each RNA sample was reverse transcribed, fragmented, and 3′ biotinylated as described previously (31). The labeled cDNA (1.5 μg) was hybridized to custom-made Brucella abortus GeneChips (PMD2308a520698F) by following the manufacturer's recommendations for antisense prokaryotic arrays (Affymetrix, Inc., Santa Clara, CA). Signal intensities were normalized to the median signal intensity value for each GeneChip, averaged, and analyzed with GeneSpring software, version 11.3. Transcripts exhibiting ≥2-fold changes in expression between the parental strain 2308 and the mucR mutant strain (CC092), as determined by Affymetrix algorithms for statistically significant differential expression (t test; P ≤ 0.05), were reported. The microarrays used in this study were developed based on the B. abortus 2308 genome sequence (accession numbers AM040264 [chromosome 1] and AM040265 [chromosome 2]) and all B. abortus GenBank entries that were available at the time of design. In total, 3,019 predicted Brucella open reading frames and 1,892 intergenic regions greater than 50 bp in length are represented on PMD2308a520698F.
Real-time RT-PCR.
Real-time reverse transcription-PCR (RT-PCR) was performed as described previously (25). Briefly, total Brucella RNA was isolated as above and treated with RNase-free DNase I (Ambion) to remove genomic DNA. cDNA was generated from the final RNA preparation using the SuperScript III cDNA synthesis system (Invitrogen, Carlsbad, CA) by following the manufacturer's protocol, and this cDNA was used for real-time PCR employing a SYBR green PCR supermix (Roche, Mannheim, Germany). For these experiments, primers for 16S RNA were used as a control, while gene-specific primers were used for evaluating relative mRNA levels (Table 1). Parameters for PCR included a single denaturing step for 5 min at 95°C, followed by 40 cycles (denature for 15 s at 95°C, anneal for 15 s at 50°C, and extend for 15 s at 72°C) of amplification. Fluorescence from SYBR green incorporation into double-stranded DNA was measured with an iCycler machine (Bio-Rad), and the relative abundance of mRNA was determined using the Pfaffl equation (32).
Purification of recombinant MucR.
The Strep-tagII system (IBA, Göttingen, Germany) was used to produce recombinant Brucella MucR in E. coli strain BL21. The coding region of the mucR gene (bab1_0594) was amplified using the primers rMucR-For and rMucR-Rev (Table 1), B. abortus 2308 chromosomal DNA as a template, and Taq polymerase. The amplified DNA fragment was digested with BsaI and ligated into BsaI-digested pASK-IBA6, which encodes an amino-terminal Strep-tagII on the protein of interest. The resulting plasmid, prMucR, was transformed into E. coli strain BL21, and the strain harboring this plasmid was grown to an optical density at 600 nm (OD600) of approximately 0.6 before recombinant gene expression was induced by addition of anhydrotetracycline (200 μg/ml, final concentration). Following 3 h of incubation at 37°C, the cells were collected by centrifugation (4,200 × g for 10 min at 4°C) and lysed by treatment with CelLytic B cell lysis reagent (Sigma, St. Louis, MO) in the presence of the protease inhibitor phenylmethanesulfonyl fluoride. The supernatant from the suspension of lysed cells was collected by centrifugation (14,000 × g for 10 min at 4°C), and the collected supernatant was passed through an affinity column packed with Strep-Tactin Sepharose. The column was washed extensively with buffer W (100 mM Tris-HCl and 300 mM NaCl), and recombinant protein was eluted with 2.5 mM desthiobiotin in buffer W. The degree of purity of recombinant MucR was high as judged by SDS-PAGE.
EMSAs.
All recombinant MucR (rMucR) electrophoretic mobility shift assay (EMSA) experiments were carried out in a 20-μl total reaction volume containing binding buffer composed of 10 mM Tris-HCl (pH 7.4), 50 mM KCl, 1 mM dithiothreitol, 6% glycerol, 50 μg/ml bovine serum albumin, and 50 μg/ml salmon sperm DNA. DNA fragments corresponding to the promoter regions of various genes were amplified by PCR from Brucella abortus 2308 chromosomal DNA using specific primer sets (Table 1). The amplified DNA fragments were purified by agarose gel electrophoresis, and the fragments were end labeled with [γ-32P]ATP (PerkinElmer, San Jose, CA) and polynucleotide kinase (Promega, Madison, WI). Increasing amounts of recombinant MucR were mixed with the radiolabeled DNA fragments in binding buffer, and the reaction mixtures were incubated at room temperature for 20 min. As controls, 25× molar concentrations of nonradiolabeled specific DNA (specific competitor) or nonradiolabeled nonspecific DNA (nonspecific competitor; coding region of bab2_0350 [ohrR]) were added to some reaction mixtures. In other reaction mixtures, 0.5 mM chelator EDTA was included. The binding reaction mixtures were subjected to electrophoresis on 6% native polyacrylamide gels in 0.5× Tris-borate-EDTA (TBE) running buffer for approximately 1 h. Following electrophoresis, gels were dried onto 3-mm Whatman paper using a vacuum gel dryer system and visualized by autoradiography.
Analysis of Brucella LPS.
Brucella strains were grown in brucella broth to stationary phase, and LPS was isolated by hot phenol extraction as described previously (33). Briefly, 50 ml of hot phenol (45%) was added to 50 ml of Brucella culture, and the mixture was incubated at ∼65°C for 20 min. The organic phase was isolated and was incubated overnight with 3 volumes of saturated sodium acetate in methanol at −20°C. Following precipitation, the samples were subjected to centrifugation at 10,000 × g for 20 min, and the pellet was suspended in 50 mM sodium phosphate buffer containing 20 mM MgCl2 and 5 mM EDTA. The samples were then treated with DNase and RNase (2 μg/ml of each) at 37°C for 2 h. Subsequently, proteinase K (20 μg/ml) was added, and the samples were incubated at ∼65°C for 8 h. The LPS samples were then extracted with an equal volume of phenol, and the interface was removed. Both the organic and aqueous phases were incubated overnight with 3 volumes of saturated sodium acetate in methanol at −20°C. Following centrifugation at 10,000 × g for 20 min, the pellet was suspended in distilled water. Purified LPS was separated in 15% SDS-PAGE gels containing 3 M urea, and the gels were silver stained as described previously (34).
Western blot analyses of LPS were performed using monoclonal antibodies specific for the O chain (A78 12G12 F12) or the LPS core (A68 24G12 A8). Following separation by SDS-PAGE as described above, the LPS samples were transferred to a nitrocellulose membrane. Membranes were blocked at room temperature in 5% skim milk in TBST (Tris-buffered saline [50 mM Tris, 150 mM NaCl, pH 7.4], 0.05% Tween 20), and primary antibodies were incubated with the membranes at room temperature in 5% skim milk. Secondary antibodies (anti-mouse antibodies conjugated to horseradish peroxidase [HRP]) were used at a 1:20,000 dilution. All washing steps were performed with TBST. Development of the HRP signal was performed using WestPico (Pierce [Thermo Scientific], Rockford, IL).
Sensitivity of B. abortus strains to SDS and polymyxin B in a disk assay.
Brucella strains were grown on SBA at 37°C under 5% CO2 for 48 to 72 h, and the bacterial cells were harvested into phosphate-buffered saline (PBS) and suspended at a concentration of ∼3.33 × 107 CFU/ml in brucella broth containing 0.5% agar (maintained at 55°C). Four milliliters of this suspension was overlaid onto brucella agar plates, and after solidification of the overlay, a sterile 7-mm Whatman disk was placed in the center of each plate. Seven microliters of either a 20% sodium dodecyl sulfate (SDS) or 10-mg/ml polymyxin B solution was applied to each filter disk, and the plates were incubated at 37°C with 5% CO2 for 72 h. The zones of inhibition around each disk were then measured in millimeters.
Iron utilization assay.
Brucella strains were grown in low-iron minimal medium (35) from a starting inoculum of 106 CFU/ml. Following incubation for 72 h at 37°C, bacterial suspensions were prepared in phosphate-buffered saline (pH 7.2) to a final concentration of 109 CFU/ml. Aliquots of 100 μl of the bacterial cell suspensions were mixed with 4 ml of 0.8% Noble agar, and the mixtures were overlaid onto tryptic soy agar (TSA) plates containing 300 μM ethylenediamine-N,N′-bis(2-hydroxyphenylacetic acid) (EDDHA). In addition, plates used for the ferrous iron utilization assay contained 2 mM sodium ascorbate in both the Noble agar overlay and the TSA plates to ensure that the iron was maintained in the ferrous form. Seven-millimeter sterile filter paper (Whatman no. 3) disks were placed on the plates, and 10 μl of a 50 mM solution of FeCl3 or 50 mM (NH4)2Fe(SO4)2 · 6H2O was added to the filter disks. The plates were subsequently incubated for 72 h at 37°C with 5% CO2, and following this incubation period, the diameter (in millimeters) of the zone of bacterial growth around each filter disk was measured and recorded.
Virulence studies.
Experiments to test the intracellular survival and replication of Brucella strains in primary peritoneal murine macrophages were carried out as described previously (36). Briefly, resident peritoneal macrophages were isolated from mice and seeded in 96-well plates in Dulbecco's modified Eagle's medium with 5% fetal bovine serum, and the following day, the macrophages were infected with opsonized brucellae at a multiplicity of infection (MOI) of 50:1. After 2 h of infection, extracellular bacteria were killed by treatment with gentamicin (50 μg/ml). In some wells, the macrophages were then lysed with 0.1% deoxycholate in PBS, and serial dilutions were plated on SBA. In other wells, the macrophages were washed with PBS following gentamicin treatment, and fresh cell culture medium containing gentamicin (20 μg/ml) was added to the monolayer. After 24 and 48 h of infection with Brucella strains, the macrophages were lysed, and serial dilutions were plated on SBA. Triplicate wells were used for each Brucella strain tested.
The experimental methods for assessing the chronic infection of mice by Brucella strains were described previously (36). C57BL/6 mice (5 per Brucella strain) were infected intraperitoneally with ∼5 × 104 CFU of each Brucella strain in sterile PBS. The mice were sacrificed at 1 and 4 weeks postinfection, and serial dilutions of spleen homogenates were plated on SBA.
Microarray data accession number.
The GenBank accession number for the microarray data described in this report is GSE40532.
RESULTS
The Brucella abortus mucR mutant exhibits a significant growth defect during in vitro cultivation.
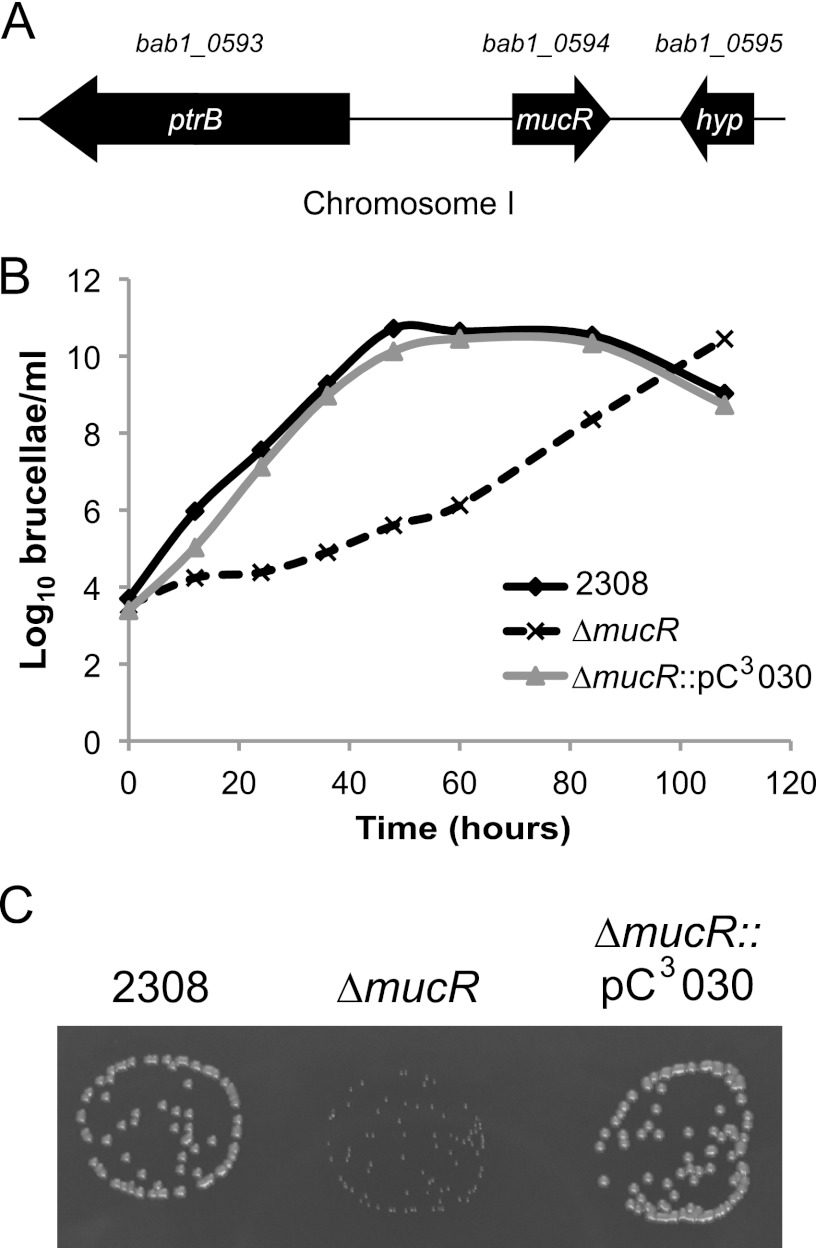
The mucR gene of Brucella abortus 2308 is located on chromosome I and designated bab1_0594 in the genome sequence, and mucR is flanked by the ptrB gene, encoding oligopeptidase B, and a hyp gene, encoding a small hypothetical protein (Fig. 1A). Importantly, this genetic organization is conserved among other Brucella spp., including B. melitensis, B. suis, and B. ovis. The most striking feature of the B. abortus mucR mutant observed during its phenotypic characterization was the slow growth of this mutant in brucella broth compared to that displayed by the parental 2308 strain (Fig. 1B) and the smaller-size colonies it produced when grown on SBA (Fig. 1C). The generation time for B. abortus 2308 grown in brucella broth was calculated to be 2.3 h, while the mucR mutant grown in the same medium exhibited a generation time of 8.0 h. Neither of the growth defects is observed in a derivative of the B. abortus mucR mutant carrying a copy of mucR on a pBBR1MCS-based plasmid (Fig. 1B and C). Despite the delayed growth exhibited by the B. abortus mucR mutant, this strain eventually attains the same cell density as the parental strain as it reaches stationary phase in brucella broth, and even with its growth defect, the B. abortus mucR mutant exhibits the same cellular morphology as parental strain 2308, as shown when these strains are examined by phase-contrast microscopy following growth on SBA (data not shown).
Fig 1.
Deletion of mucR in Brucella abortus 2308 results in a growth deficiency. (A) Genetic organization of the mucR locus. The mucR gene is found on chromosome I and is designated bab1_0594 in the B. abortus 2308 genome sequence. mucR is flanked by ptrB (bab1_0593), encoding oligopeptidase B, and by a small, hypothetical-protein-encoding gene. (B) Growth curves of Brucella abortus strains in a rich medium. B. abortus 2308, the isogenic mucR mutant (ΔmucR), and the complemented mucR mutant (ΔmucR::pC3030) strains were grown in brucella broth, and the growth of each culture was monitored by plating serial dilutions on SBA to determine the number of CFU/ml. (C) Photograph of Brucella abortus colonies on blood agar after 60 h of growth.
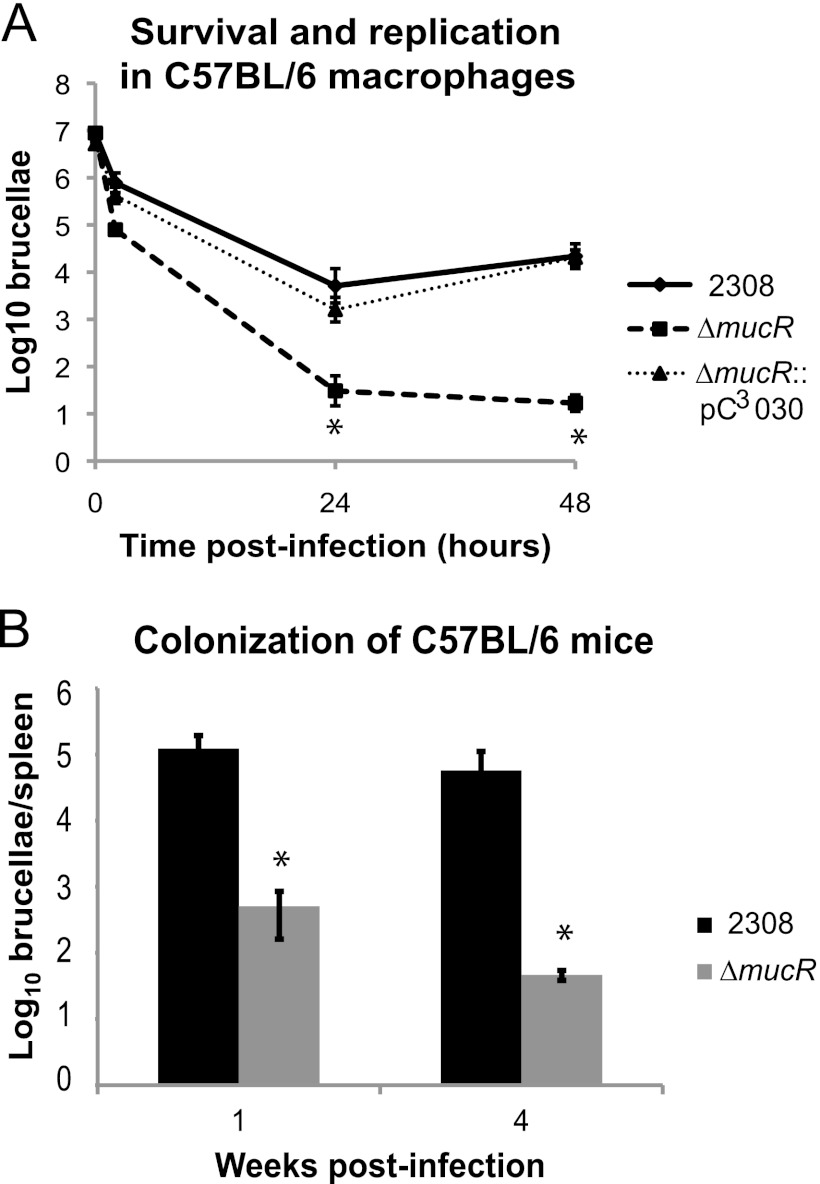
Deletion of mucR leads to severe attenuation of B. abortus 2308 in mice.
The mucR gene was previously reported to be important for the virulence of Brucella melitensis 16 M (23, 24); however, the role of mucR in B. abortus 2308 virulence has not been reported. Therefore, the B. abortus mucR mutant was tested for its ability to survive and replicate in cultured murine macrophages and for its capacity to produce chronic infections in C57BL/6 mice (Fig. 2). Compared to the parental strain, 2308, the mucR mutant strain was attenuated at both 24 and 48 h postinfection in macrophages. Complementation of mucR expression in the mutant strain restored its virulence in these cultured phagocytes to parental levels (Fig. 2A). Similarly, the B. abortus mucR mutant exhibited significantly reduced spleen colonization levels at 1 week and 4 weeks postinfection compared to 2308 when C57BL/6 mice were infected with these strains via the intraperitoneal route (Fig. 2B). These data are in line with the significant attenuation of a B. melitensis mucR mutant described previously (23) and suggest that the link between MucR and virulence is a conserved relationship in Brucella strains.
Fig 2.
A Brucella abortus mucR mutant is significantly attenuated in the mouse model. (A) Brucella abortus 2308, the mucR isogenic mutant strain (ΔmucR), and the complemented mucR mutant strain (ΔmucR::pC3030) were tested for survival and replication in primary murine macrophages. Macrophages were isolated from the peritoneal cavities of C57BL/6 mice, and the cells were infected with opsonized Brucella strains. Extracellular bacteria were killed by gentamicin treatment. At the indicated times, the macrophages were lysed, and serial dilutions were plated on blood agar to determine the number of viable intracellular brucellae. The asterisks indicate significant differences in survival and replication between parental strain 2308 and the mucR mutant strain (t test; P < 0.05). (B) Spleen colonization of mice experimentally infected with Brucella strains. C57BL/6 mice were infected intraperitoneally with approximately 5 × 104 CFU of B. abortus 2308 or the mucR mutant. At 1 and 4 weeks postinfection, the mice were sacrificed, and the serial dilutions of spleen homogenates were plated on blood agar to determine the number of brucellae colonizing the spleen. The asterisks indicate significant differences in spleen colonization between parental strain 2308 and the mucR mutant strain (t test; P < 0.05).
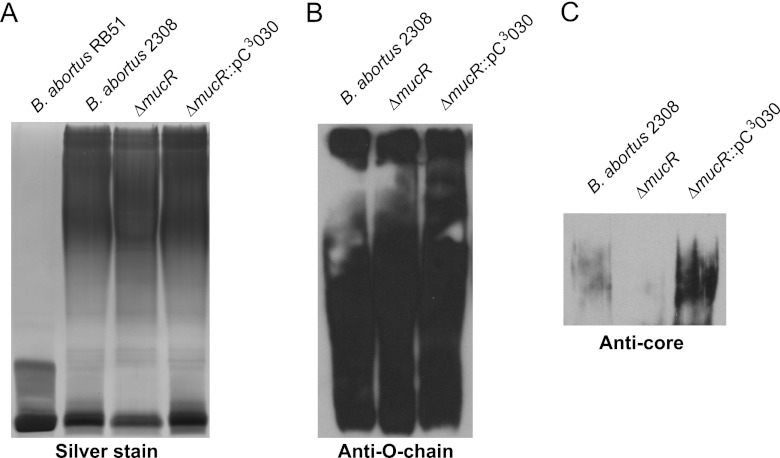
In Brucella strains, such as B. melitensis and B. abortus, that naturally produce the smooth form of lipopolysaccharide (S-LPS), the maintenance of an intact LPS O chain is critical for virulence (37). One of the prominent characteristics of the B. abortus mucR mutant that was observed during its initial phenotypic characterization was that the colonies produced by this mutant take up crystal violet, suggesting that it might have a rough (e.g., O-chain-deficient) LPS (28). Genetic complementation of the B. abortus mucR mutant, however, converted it to an S-LPS phenotype in the crystal violet assay, but as shown in Fig. 3A, the silver-stained SDS-PAGE gel profile of an LPS preparation from the B. abortus mucR mutant indicates that the O chain is intact. Additionally, immunoblot analysis with O-chain-specific antibodies confirmed that the B. abortus mucR mutant produces an intact O antigen (Fig. 3B). Conversely, immunoblot analysis with LPS core-specific antibodies revealed that the LPS core of the mucR mutant is altered compared to those of parental strain 2308 and the mucR complemented strain (Fig. 3C). It is currently unknown why the B. abortus mucR mutant displays a “false” rough LPS phenotype in the crystal violet assay, but the results of the SDS-PAGE and immunoblot analyses of the LPS from this strain suggest that complete loss of the LPS O chain is not a major contributor to the attenuation it exhibits in the mouse model. On the other hand, modification of the LPS core may contribute to the attenuation of the mucR mutant, but more work is needed to fully characterize the defect in the LPS core of the B. abortus mucR mutant strain.
Fig 3.
The lipopolysaccharide (LPS) core is modified in the B. abortus mucR mutant. Brucella strains were grown in brucella broth to stationary phase, and LPS was isolated by hot phenol extraction as described previously (33). Purified LPS from the specified Brucella strains was separated on an SDS-PAGE gel containing 3 M urea, and the LPS was then visualized by silver staining (A), Western blot analysis with an anti-O-chain monoclonal antibody (B), and Western blot analysis with an anticore monoclonal antibody (C).
The B. abortus mucR mutant exhibits a cell envelope defect.
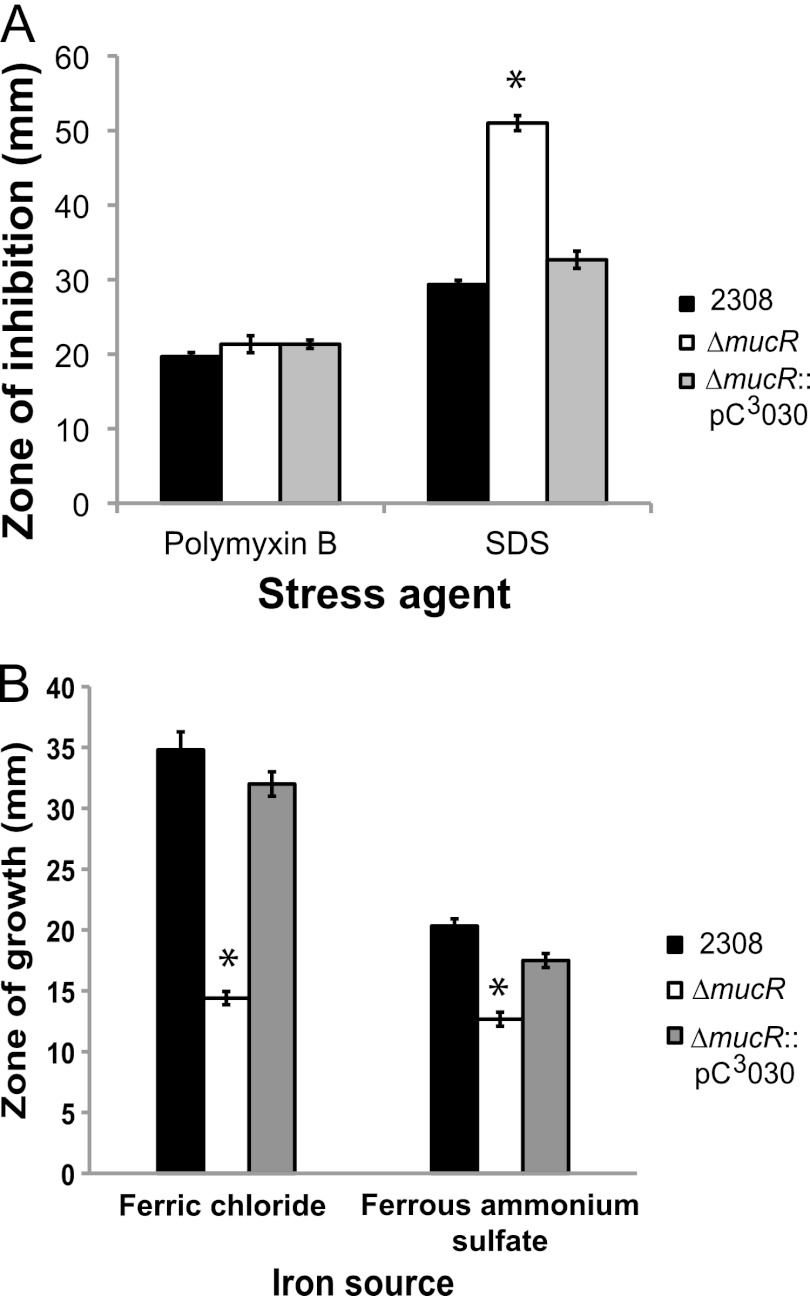
Due to the results obtained with crystal violet assays showing that the B. abortus mucR mutant aberrantly takes up crystal violet, we hypothesized that the mucR mutant has a generalized envelope defect that allows for the increased uptake of the dye. To test this hypothesis, we employed a disk diffusion assay to assess the sensitivity of the B. abortus mucR mutant to polymyxin B and sodium dodecyl sulfate (SDS) (Fig. 4A). All of the strains tested exhibited similar sensitivities to polymyxin B. However, compared to parental strain 2308, the mucR mutant showed a significantly increased sensitivity when exposed to SDS, and this defect was reversed when mucR was expressed from a plasmid in the mucR mutant strain. Altogether, these data indicate that MucR is important for the stability of the cell envelope in B. abortus 2308, but it remains unclear how deletion of mucR results in cell envelope abnormalities.
Fig 4.
The Brucella abortus mucR mutant exhibits a cell envelope defect and an iron acquisition defect in vitro. (A) Brucella strains were tested in a disk diffusion assay for their comparative susceptibilities to polymyxin B and sodium dodecyl sulfate (SDS). The results are plotted as the average diameters (±standard deviations) of the zones of inhibition around disks containing the indicated stress agents, and the results are from single experiment that was repeated in triplicate. Asterisks denote statistically significant differences (t test; P < 0.05) between a mutant strains and parental strain 2308. (B) Brucella abortus 2308, the mucR mutant strain (ΔmucR), and the complemented mucR mutant strain (ΔmucR::pC3030) were tested for their ability to utilize ferric (Fe3+) or ferrous (Fe2+) iron in a disk diffusion assay. Iron sources (50 mM FeCl3 or 50 mM ferrous ammonium sulfate) were applied to sterile Whatman paper disks on plates containing the chelator EDDHA, and following incubation at 37°C for 72 h, the diameter (in millimeters) of the zone of bacterial growth around each filter disk was measured. The data are represented as the average and standard deviation of the zones of growth recorded for each strain in triplicate, and asterisks denote statistically significant differences (t test; P < 0.05) between the mucR mutant strain and parental strain 2308.
Definition of the MucR regulon in B. abortus 2308.
To gain insight into the spectrum of Brucella genes regulated by MucR, as well as the genetic basis for the distinctive phenotypic properties exhibited by the B. abortus mucR mutant strain, microarray analysis was performed using RNA isolated from parental strain 2308 and the mucR mutant strain grown in brucella broth to early stationary phase. In all, 91 genes that exhibited altered expression (>2-fold difference) were identified in the mucR mutant (Table 3). The majority (76/91) of the altered genes were upregulated in the mutant strain, suggesting that MucR may serve predominately as a transcriptional repressor in Brucella strains, as it does in other alphaproteobacteria (16). The link between MucR and altered expression of a selected subset of the Brucella genes identified in the microarray analysis was independently confirmed by real-time PCR (Table 3).
Table 3.
Genes regulated by MucR in Brucella abortus 2308
| Function and designation | Description | Fold change (ΔmucR strain vs 2308) in gene expression by: |
|
|---|---|---|---|
| Microarraya | RT-PCR | ||
| Membrane proteins and transport systems | |||
| BAB1_0115 | Outer membrane protein Omp25d | 3.97 | |
| BAB1_0793 | Membrane-bound proton-translocating pyrophosphatase | 2.02 | |
| BAB1_1893 | Hypb (DMEc family transporter) | 14.69 | |
| BAB1_2138 | Rare lipoprotein A | −2.07 | |
| BAB2_0055 | Amino acid transporter | 3.27 | |
| BAB2_0837 | Hyp (polyferredoxin) | −2.34 | |
| BAB2_0838 | Iron permease FTR1 | −3.58 | |
| BAB2_0839 | Hyp | −4.55 | |
| BAB2_0840 | Membrane antigen (iron transport) | −3.03 | −7.10 |
| Transcription and translation | |||
| BAB1_0190 | Transcriptional regulator, LuxR family protein BabR | 21.06 | 73.82 |
| BAB1_0460 | Ribosomal protein P2 | 4.24 | |
| BAB1_0594 | Transcriptional regulatory protein MucR | −99.95 | |
| BAB1_1605 | Transcriptional regulator, LysR family protein ArsR6/NolR | 2.10 | 2.36 |
| BAB2_0143 | AsnC family regulatory protein | 2.02 | 2.62 |
| BAB2_0806 | LuxR family regulatory protein | 2.69 | |
| BAB2_0807 | Crp family regulatory protein | 2.04 | 1.53 |
| Polysaccharide biosynthesis and modification | |||
| BAB1_0326 | Glycosyl transferase family protein | 5.07 | |
| BAB1_0560 | Phosphoglucomutase/phosphomannomutase | 2.34 | |
| BAB1_1465 | Glycoside hydrolase family protein | 2.37 | |
| BAB1_1973 | ExsB protein | −3.75 | −3.31 |
| BAB1_1974 | Putative 6-pyruvoyl tetrahydropterin synthase | −2.98 | |
| BAB1_1975 | Proline-rich extensin; radical SAMd family protein | −2.73 | |
| BAB2_0133 | Glycosyltransferase family protein | 2.96 | |
| BAB2_0134 | Glu/Leu/Phe/Val dehydrogenase | 2.99 | |
| BAB2_0135 | Dolichyl-phosphate-mannose-protein mannosyltransferase | 2.57 | |
| Genome plasticity | |||
| BAB1_0554 | IS5 family transposase OrfA | 2.81 | 104.68 |
| BAB1_0555 | IS5 family transposase OrfB | 3.68 | |
| BAB1_0746 | Hyp | 91.29 | 1841.68 |
| BAB1_0747 | Integrase catalytic subunit | 61.91 | |
| Metabolism, signaling, and enzymatic processes | |||
| BAB1_0459 | Transglycosylase-associated protein | 7.96 | |
| BAB1_0512 | EALe domain-containing protein BpdB | 4.30 | |
| BAB1_0655 | Antifreeze protein | 2.08 | |
| BAB1_0738 | l-Lactate permease | −2.07 | |
| BAB1_1035 | Proline-rich extensin | 12.81 | 23.55 |
| BAB1_1099 | Patatin | 3.20 | |
| BAB1_1206 | 7-Cyano-7-deazaguanine reductase | −2.38 | |
| BAB1_1511 | Nudix hydrolase | 2.13 | |
| BAB1_1535 | C5 cytosine-specific DNA methylase | 4.11 | |
| BAB1_2001 | Aquaporin Z | 3.28 | |
| BAB1_2010 | Glyceraldehyde-3-phosphate dehydrogenase | 14.23 | |
| BAB1_2041 | ATP/GTP-binding domain-containing protein | 21.01 | 120.32 |
| BAB2_0196 | Nickel-dependent hydrogenase b-type cytochrome subunit | 2.82 | |
| BAB2_0257 | Beta-lactamase | 12.72 | |
| BAB2_0348 | Protein kinase | −2.07 | |
| BAB2_0607 | Twin-arginine translocation pathway signal | 6.94 | |
| BAB2_0846 | Frizzled protein | 2.54 | |
| BAB2_0865 | Pyridoxal-dependent decarboxylase | 2.52 | |
| BAB2_0866 | Glutamate decarboxylase alpha | 2.21 | |
| BAB2_1072 | PemK family protein | 7.23 | |
| BAB2_1107 | Aminoacyl-tRNA synthetase class I | 9.88 | 7.21 |
| Hypothetical | |||
| BAB1_0013 | Hyp | 3.29 | |
| BAB1_0043 | Hyp | 3.68 | |
| BAB1_0069 | Hyp | 6.57 | |
| BAB1_0070 | Hyp | 3.79 | |
| BAB1_0087 | Hyp | 2.19 | |
| BAB1_0189 | Hyp | 27.68 | |
| BAB1_0198 | Hyp | 2.31 | |
| BAB1_0265 | Hyp | 2.85 | |
| BAB1_0271 | Hyp | 4.47 | |
| BAB1_0324 | Hyp | 9.18 | |
| BAB1_0745 | Hyp | 7.09 | |
| BAB1_0750 | Hyp | 10.25 | |
| BAB1_0751 | Hyp | 2.31 | |
| BAB1_0893 | Hyp | 2.06 | |
| BAB1_1125 | Hyp | 5.91 | |
| BAB1_1352 | Hyp | 2.21 | |
| BAB1_1398 | Hyp | −2.65 | |
| BAB1_1487 | Hyp | 2.62 | |
| BAB1_1489 | Hyp | 2.22 | |
| BAB1_1509 | Hyp | 3.47 | |
| BAB1_1529 | Hyp | 19.56 | |
| BAB1_1536 | Hyp | 2.48 | |
| BAB1_1604 | Hyp | 4.88 | |
| BAB1_1689 | Hyp | 2.34 | |
| BAB1_1854 | Hyp | 2.73 | 11.41 |
| BAB1_2000 | Hyp | 3.15 | |
| BAB1_2002 | Hyp | 3.22 | |
| BAB1_2011 | Hyp | 10.97 | 573.77 |
| BAB1_2012 | Hyp | 74.13 | |
| BAB1_2015 | Hyp | −3.80 | |
| BAB1_2021 | Hyp | 7.01 | |
| BAB2_0092 | Hyp | 3.85 | |
| BAB2_0132 | Hyp | 2.79 | |
| BAB2_0258 | Hyp | 7.81 | |
| BAB2_0450 | Hyp | 2.84 | |
| BAB2_0613 | Hyp | 2.55 | |
| BAB2_0861 | Hyp | 6.12 | |
| BAB2_0867 | Hyp | 2.61 | |
| BAB2_0873 | Hyp | 2.04 | |
| BAB2_0887 | Hyp | 5.23 | |
| BAB2_1151 | Hyp | −2.56 | |
Microarray analysis was performed using total cellular RNA from Brucella strains grown in brucella broth to late exponential phase, and values for genes whose expression was shown to be more than 2-fold altered in the mucR mutant compared to strain 2308 are shown.
Hyp, hypothetical.
DME, drug/metabolite exporter.
SAM, S-adenosylmethionine.
EAL, cyclic di-GMP phosphodiesterase.
The large majority of the genes whose expression levels were increased in the mucR mutant are predicted to encode hypothetical proteins. Other genes displaying increased expression in the mucR mutant included those encoding membrane proteins and transporters (bab1_0115, bab1_0793, and bab2_0055), putative polysaccharide biosynthesis and modification proteins (bab1_0326, bab1_0560, bab1_1465, and bab2_0133 to bab2_0135), proteins potentially linked to genomic plasticity (i.e., transposases and phage remnants; bab1_0554, bab1_0555, bab1_0746, and bab1_0747), and transcriptional regulators (babR [blxR; bab1_0190], arsR6 [nolR; bab1_1605], bab2_0143, bab2_0806, and bab2_0807). While the expression of the Crp family regulator encoded by bab2_0807 was shown to be significantly disregulated in the mucR mutant strain by microarray analysis, an independent assessment of gene expression by real-time PCR determined that expression of bab2_0807 is increased by only ∼1.5-fold in the mucR mutant compared to parental strain 2308 (Table 3). It is noteworthy that the gene bab1_0512, encoding a cyclic di-GMP phosphodiesterase, was overexpressed by more than 4-fold in the mucR mutant strain. This gene in B. melitensis 16 M (bmeI1448) was recently named bpdB, and it was reported that deletion of bpdB results in attenuation of B. melitensis in mice (38). It is equally notable that the gene bab1_0115, encoding Omp25d, displays altered expression (∼4-fold increase) in the B. abortus mucR mutant. The Omp25 family of proteins is important for the wild-type properties of the Brucella cell envelope (39), but to date, clear-cut associations between these proteins and virulence have been established only for B. ovis, which naturally lacks its LPS O chain (37).
The genes that were downregulated in the mucR mutant include genes putatively involved in polysaccharide biosynthesis and iron transport. The Brucella abortus genes bab1_1973, bab1_1974, and bab1_1975 are orthologs of the Sinorhizobium meliloti genes SMb20940, SMb20939, and SMb20938, respectively, and in S. meliloti, these genes are involved in succinoglycan synthesis (40). While the function of the bab1_1973-bab1_1975 operon in B. abortus 2308 is not currently known, it is interesting that MucR regulates the expression of these genes, because the MucR regulator in other alphaproteobacteria is known to control the production of succinoglycan (12, 41).
MucR indirectly regulates iron acquisition genes in B. abortus 2308.
One set of four genes that displayed reduced expression in the B. abortus mucR mutant is designated bab2_0837-0840 in the B. abortus 2308 genome sequence. Experimental evidence indicates that these genes encode a ferrous iron (Fe2+) transporter (A. Elhassanny, unpublished data; 42), and when the B. abortus mucR mutant was examined for its ability to use FeCl3 (a ferric iron source) and (NH4)2Fe(SO4)2 (a ferrous iron source) in an in vitro iron source utilization assay, this strain exhibited a reduced ability to use these iron sources compared to the parental 2308 strain or a complemented version of this mutant (Fig. 4B). A recombinant version of MucR did not bind to the promoter region of bab2_0840, which is the first gene of this four-gene operon, in an EMSA (Fig. 5A), indicating that the regulatory link between MucR and these iron transport genes is indirect.
Fig 5.
Brucella abortus MucR binds directly to the promoter regions of MucR-regulated genes. (A) Recombinant MucR (rMucR) protein was tested for binding to the promoter regions of MucR-regulated genes using an electrophoretic mobility shift assay (EMSA). Increasing concentrations of rMucR were incubated with radiolabeled DNA corresponding to the promoter regions of several different genes, and in some binding reactions, unlabeled specific and nonspecific competitor DNA fragments were included as controls. The binding reactions were resolved in 6% native polyacrylamide gels, and the reaction products were visualized by autoradiography. (B) Effect of EDTA on the binding of MucR to the promoter regions of MucR-regulated genes. EMSAs were performed as described for panel A, but here, 0.5 mM EDTA was included in some binding reactions. hyp, hypothetical.
MucR binds directly to the promoters of bab1_0746, bab1_1035, bab1_1605, and bab1_1893.
bab1_0746, bab1_1035, and bab1_1893 are among the most strongly MucR-regulated genes identified in B. abortus 2308 by the microarray analysis, and the regulatory link between MucR and wild-type expression of these genes has been independently verified by real-time RT-PCR analysis (Table 3). Although the arsR6 (nolR; bab1_1605) gene appears to be less strongly regulated by MucR, this gene encodes an important virulence determinant in B. melitensis 16 M (BMEI0430) (43), and the regulatory relationship between MucR and wild-type arsR6 (nolR) expression in B. abortus 2308 has been verified by real-time RT-PCR analysis, similar to its verification for bab1_0746, bab1_1035, and bab1_1893 (Table 3). Recombinant MucR exhibited specific interactions with the promoter regions of all four of these genes in EMSAs (Fig. 5A), indicating that the regulatory links between MucR and these genes are direct.
MucR requires divalent cations for its binding to the bab1_0746 and nolR promoters.
Divalent cations are required for binding of the S. meliloti MucR to DNA. Bertram-Drogatz and colleagues reported that several different divalent cations, including Mg2+, Ca2+, Mn2+, Fe2+, Co2+, Ni2+, Cu2+, and Zn2+, could be incorporated in EMSAs to facilitate binding between S. meliloti MucR and promoter DNA, but exclusion of metal ions from the binding reactions abolished these interactions (12). Accordingly, the addition of the chelator EDTA results in inhibition of S. meliloti MucR binding to DNA (16). Zn2+ was included in the binding reactions shown in Fig. 4A, and no chelator, such as EDTA, was used in these experiments. Therefore, to determine if MucR from Brucella abortus requires divalent cations for binding to DNA, EMSAs were performed in the presence of EDTA (Fig. 5B). Binding of MucR to DNA fragments corresponding to the promoter regions of bab1_0746 and nolR was abolished by the addition of 0.5 mM EDTA. From this, it is concluded that, similar to MucR-type regulators from other bacteria, the Brucella MucR also requires metal ions in order to bind to DNA and regulate gene expression.
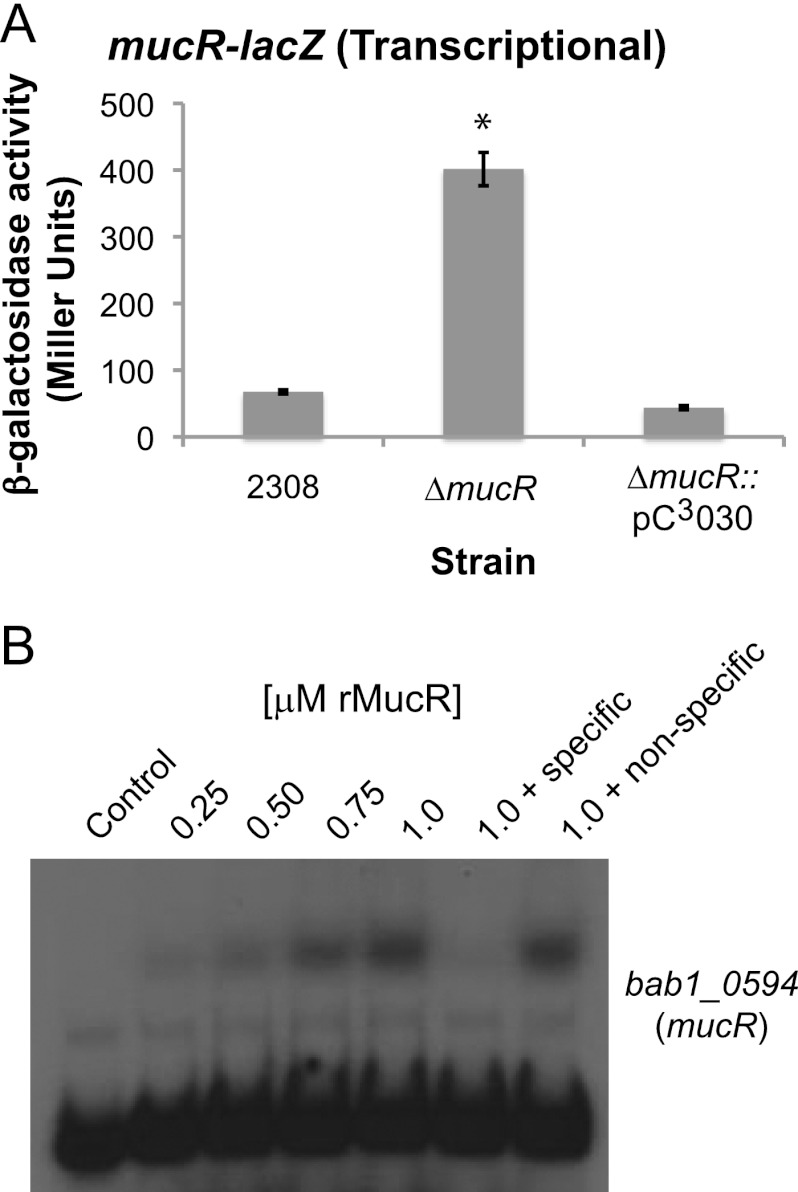
MucR regulates the expression of mucR in B. abortus 2308.
In other alphaproteobacteria, including Sinorhizobium meliloti and Agrobacterium radiobacter, MucR orthologs regulate the expression of the mucR genes, and this autoregulation is mediated by MucR binding directly to the mucR promoter (12, 16, 44). Similarly, a plasmid-borne mucR-lacZ transcriptional fusion displayed elevated mucR expression the B. abortus mucR mutant compared to the parental 2308 strain or a derivative of this mutant carrying mucR on a plasmid (Fig. 6A), and recombinant MucR bound in a specific manner to the B. abortus mucR promoter region in an EMSA (Fig. 6B). These experimental findings indicate that, like its counterparts in the other alphaproteobacteria, expression of the Brucella mucR gene is regulated by an autoregulatory mechanism.
Fig 6.
Autoregulation of mucR expression in Brucella abortus 2308. (A) β-Galactosidase activity produced by a mucR-lacZ transcriptional fusion. The activity of a mucR-lacZ transcriptional fusion was tested in B. abortus 2308, the mucR isogenic mutant strain (ΔmucR), and the mucR mutant complemented in trans (ΔmucR::pC3030). β-Galactosidase activity is shown as average Miller units ± standard deviations, and the results shown are from a single experiment that was repeated in triplicate. The asterisk indicates a significant difference in β-galactosidase activity between the parental strain 2308 and the mucR mutant strain (t test; P < 0.05). (B) Recombinant MucR (rMucR) protein was tested for binding to the mucR promoter region using an EMSA. Similar to the EMSA experiments in Fig. 3A, increasing concentrations of rMucR were incubated with radiolabeled DNA corresponding to the mucR promoter region, and in some binding reactions, unlabeled specific or nonspecific competitor DNA fragments were included as controls. The binding reactions were resolved in 6% native polyacrylamide gels and visualized by autoradiography.
DISCUSSION
One of the striking phenotypic properties exhibited by the B. abortus mucR deletion strain is its pronounced growth defect during in vitro cultivation in either broth culture or on a solid medium (Fig. 1). The slow growth of the B. abortus mucR mutant is intriguing given that a B. melitensis mucR mutant does not have the same overt growth defect (data not shown) (23). Recent experiments do, however, indicate that a B. melitensis 16 M-derived mucR mutant has a more subtle growth defect than its B. abortus counterpart. Specifically, the B. melitensis mucR mutant enters stationary phase more rapidly and at a lower cell density than the 16 M parental strain (45). The genetic basis for the growth defect exhibited by the B. abortus mucR mutant is not readily apparent from the set of MucR-regulated genes identified in this study. The bab2_1072 gene, which is >7-fold overexpressed in the B. abortus mucR mutant, is predicted to encode a PemK-like protein, and these types of toxins are involved in mRNA degradation and have been proposed to play a role in maintaining balanced growth in bacterial cell populations (46). Given this relationship between PemK family proteins and balanced cell growth, we hypothesized that overexpression of pemK in the mucR mutant could be responsible for the observed growth defect; however, deletion of pemK in the mucR mutant did not alleviate its slow-growth phenotype (data not shown). A more thorough comparative analysis of the phenotypic properties of the B. abortus and B. melitensis mucR mutants and the transcriptomes of these mutants and their parental strains will be needed to explain the basis for the differential effects that the mucR mutation has on the physiology of B. abortus 2308 and B. melitensis 16 M. Nevertheless, the experimental findings presented are consistent with the proposed role of MucR as a master transcriptional regulator of genes that perform multiple important metabolic and/or physiologic functions in Brucella.
Like the B. melitensis mucR mutant (23, 24), the B. abortus mucR mutant exhibits striking attenuation in the mouse model of chronic infection (Fig. 2). The growth defect of the B. abortus mucR mutant likely contributes to its attenuation in macrophages and mice, but there are several other genes whose altered expression in the mucR mutant may also contribute to the attenuation. The loss of either arsR6 (nolR) (43) or bpdB (38), for example, results in the attenuation of B. melitensis 16 M in mice. Like MucR, ArsR6/NolR is a transcriptional regulator that is widely conserved in the alphaproteobacteria (47), and NolR appears to be required for the proper expression of the genes encoding the type IV secretion system in B. melitensis 16 M (43). BpdB is a cyclic di-GMP phosphodiesterase that works together with other enzymes of this type and cyclic di-GMP synthases to control the levels of this important secondary signaling molecule in Brucella strains (38), and the attenuation exhibited by B. melitensis bpdA and bpdB mutants in mice indicates that cyclic di-GMP-mediated signaling plays an important role in proper virulence gene expression. Consequently, further investigation of the role that MucR plays in regulating the expression of the nolR and bpdB genes should provide us with important insight into the basis for the attenuation exhibited by the B. abortus mucR mutant.
The observation that the B. abortus mucR mutant exhibits a defect in its capacity to use nonheme iron sources in vitro is consistent with the reduced expression of a set of genes (bab2_0837-0840) predicted to encode a ferrous iron transporter (42) in this mutant. The lack of direct binding of MucR to the promoter upstream of these genes indicates that this regulatory link is indirect. Direct regulation of the bab2_0837-0840 operon may be carried out by one or more of the transcriptional regulators whose gene expression is repressed by MucR, such as BAB1_0190 (BabR/BlxR), BAB1_1605 (ArsR6/NolR), BAB2_0143, or BAB2_0806, or alternatively, the metabolic alterations in the mucR mutant may influence its iron homeostasis, leading to altered bab2_0837-0840 expression mediated by the iron-responsive regulators Irr or RirA (48). Defining the role that MucR plays in regulating bab2_0837-0840 expression in B. abortus 2308 will be an important aspect of gaining a better understanding of the basis for the attenuation of Brucella mucR mutants, because experimental evidence indicates that the ferrous iron transporter encoded by these genes plays a critical role in virulence in the mouse model (Elhassanny, unpublished).
ros and mucR mutants in other alphaproteobacteria exhibit altered mucoidy. For example, a naturally mucoid colony of Agrobacterium tumefaciens loses its mucoid appearance when ros is mutated (5), and a mucR mutation in Sinorhizobium meliloti results in a similar loss of colony mucoidy (8). Importantly, these mucoidy-related phenotypes have been linked to the disregulation of genes encoding proteins involved in exopolysaccharide synthesis (11, 12, 13, 14, 15). The B. abortus mucR mutant does not appear to be any more or less mucoid than parental strain 2308 based on macroscopic observations, but it is interesting that MucR controls the expression of genes (bab1_1973 to bab1_1975) homologous to the exsB, -C, and -D genes of S. meliloti, which are involved in succinoglycan production (40). Are these genes merely remnants from a common evolutionary ancestor, or do these genes play a yet-undefined role in exopolysaccharide synthesis in Brucella? Historically, exopolysaccharides have not been thought to play an important role in the biology of Brucella, but it has been reported that deletion of the LuxR-type regulator VjbR or overproduction of the acyl-homoserine lactone (AHL) acylase AiiD leads to production of a mannose-based exopolysaccharide in B. melitensis 16 M, which normally does not produce it (49, 50). Thus, Brucella strains apparently have the capacity to produce exopolysaccharides, but under what circumstances and for what reasons these polysaccharides are produced during the natural life of the brucellae remain unknown. Furthermore, if and how MucR is related to exopolysaccharide production in Brucella are currently not known.
Another facet of the present story that warrants further discussion is the possible relationship between MucR and proper regulation of the Brucella genes involved in lipopolysaccharide (LPS) biosynthesis. The manB gene (bab1_0560), for example, which encodes a phosphomannomutase known to be involved in the biosynthesis of the Brucella LPS O chain and linked to virulence (51), is repressed by MucR. Another MucR-regulated gene that potentially fits into this category is bab2_0132. This gene is predicted to encode a hypothetical protein with a conserved “GtrA” domain, which is notable because GtrA modifies the LPS O chain in Shigella strains (52). It is also interesting that two of the MucR-regulated genes in B. abortus 2308, bab1_0554 and bab1_0555, which are predicted to encode transposases, are situated in the wbk locus, which is a large group of genes (>20 genes) whose products function in the biosynthesis of LPS (53). Transposase-encoding genes residing in close proximity to bab1_0554 and bab1_0555 (e.g., bab1_0556 and bab1_0557) have also been implicated in the spontaneous conversion of Brucella strains from the smooth to rough LPS phenotype (54). As mentioned previously in this report, although the LPS moieties of both the B. abortus and B. melitensis mucR mutants retain their O chains, experimental evidence has been obtained with both of these mutants suggesting that these LPS molecules are not entirely “wild type” in structure, as there is a defect in the core of the LPS produced by Brucella mucR mutants (Fig. 3C) (45). Thus, another important component of understanding the role of MucR in the virulence of Brucella strains will be to determine precisely what role this transcriptional regulator plays in the proper expression of their LPS biosynthesis and modification genes.
In addition to shedding light on the genes being regulated by MucR in B. abortus 2308, these and other recent studies have also begun to provide information regarding how mucR expression and MucR activity are controlled. The studies presented here, for instance, demonstrate that mucR expression in B. abortus 2308 is controlled by repressive autoregulation, and studies by Mirabella et al. suggest that mucR expression in B. melitensis 16 M may be induced by osmotic stress (45). The results presented in this report also support the contention that the activity of MucR-type transcriptional regulators is dependent upon the availability of divalent cations (22). Given the link between MucR and iron utilization in Brucella, it is possible that iron availability plays a role in the regulation of MucR activity. However, more work will be needed to define the specific environmental stimuli that control mucR expression and MucR activity in the Brucella spp.
In conclusion, the present study provides further evidence that MucR is a major regulator of virulence genes in Brucella spp., and furthermore, it has defined the genes that comprise the MucR regulon in B. abortus 2308. Altogether, this study paves the way for future work aimed at further characterizing the role of MucR in the biology and pathogenesis of Brucella spp.
ACKNOWLEDGMENTS
We thank Tom Inzana for his helpful suggestions and advice regarding the isolation and analysis of LPS from Brucella strains. We are also very grateful to J. J. Letesson for kindly providing the anti-LPS antibodies A78 12G12 F12 and A68 24G12 A8.
This work was supported by a grant from the National Institute of Allergy and Infectious Diseases to R.M.R. (AI48499).
Footnotes
Published ahead of print 14 January 2013
REFERENCES
- 1. Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. 2006. The new global map of human brucellosis. Lancet Infect. Dis. 6:91–99 [DOI] [PubMed] [Google Scholar]
- 2. Pappas G, Akritidis N, Bosilkovski M, Tsianos E. 2005. Brucellosis. N. Engl. J. Med. 352:2325–2336 [DOI] [PubMed] [Google Scholar]
- 3. Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. 1990. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol. 172:3569–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Batut J, Andersson SG, O'Callaghan D. 2004. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol. 2:933–945 [DOI] [PubMed] [Google Scholar]
- 5. Close TJ, Tait RC, Kado CI. 1985. Regulation of Ti plasmid virulence genes by a chromosomal locus of Agrobacterium tumefaciens. J. Bacteriol. 164:774–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Close TJ, Rogowsky PM, Kado CI, Winans SC, Yanofsky MF, Nester EW. 1987. Dual control of Agrobacterium tumefaciens Ti plasmid virulence genes. J. Bacteriol. 169:5113–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tait RC, Kado CI. 1988. Regulation of the virC and virD promoters of pTiC58 by the ros chromosomal mutation of Agrobacterium tumefaciens. Mol. Microbiol. 2:385–392 [DOI] [PubMed] [Google Scholar]
- 8. Zhan HJ, Levery SB, Lee CC, Leigh JA. 1989. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc. Natl. Acad. Sci. U. S. A. 86:3055–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Araujo RS, Robleto EA, Handelsman J. 1994. A hydrophobic mutant of Rhizobium etli altered in nodulation competitiveness and growth in the rhizosphere. Appl. Environ. Microbiol. 60:1430–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bittinger MA, Milner JL, Saville BJ, Handelsman J. 1997. rosR, a determinant of nodulation competitiveness in Rhizobium etli. Mol. Plant Microbe Interact. 10:180–186 [DOI] [PubMed] [Google Scholar]
- 11. Becker A, Rüberg S, Küster H, Roxlau AA, Keller M, Ivashina T, Cheng HP, Walker GC, Pühler A. 1997. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J. Bacteriol. 179:1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bertram-Drogatz PA, Quester I, Becker A, Pühler A. 1998. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol. Gen. Genet. 257:433–441 [DOI] [PubMed] [Google Scholar]
- 13. Rüberg S, Pühler A, Becker A. 1999. Biosynthesis of the exopolysaccharide galactoglucan in Sinorhizobium meliloti is subject to a complex control by the phosphate-dependent regulator PhoB and the proteins ExpG and MucR. Microbiology 145:603–611 [DOI] [PubMed] [Google Scholar]
- 14. Lloret J, Martín M, Oruezabal RI, Bonilla I, Rivilla R. 2002. MucR and MucS activate exp genes transcription and galactoglucan production in Sinorhizobium meliloti EFB1. Mol. Plant Microbe Interact. 15:54–59 [DOI] [PubMed] [Google Scholar]
- 15. Janczarek M, Skorupska A. 2007. The Rhizobium leguminosarum bv. trifolii RosR: transcriptional regulator involved in exopolysaccharide production. Mol. Plant Microbe Interact. 20:867–881 [DOI] [PubMed] [Google Scholar]
- 16. Bahlawane C, McIntosh M, Krol E, Becker A. 2008. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol. Plant Microbe Interact. 21:1498–1509 [DOI] [PubMed] [Google Scholar]
- 17. McIntosh M, Krol E, Becker A. 2008. Competitive and cooperative effects in quorum-sensing-regulated galactoglucan biosynthesis in Sinorhizobium meliloti. J. Bacteriol. 190:5308–5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mueller K, González JE. 2011. Complex regulation of symbiotic functions is coordinated by MucR and quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 193:485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chou AY, Archdeacon J, Kado CI. 1998. Agrobacterium transcriptional regulator Ros is a prokaryotic zinc finger protein that regulates the plant oncogene ipt. Proc. Natl. Acad. Sci. U. S. A. 95:5293–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bouhouche N, Syvanen M, Kado CI. 2000. The origin of prokaryotic C2H2 zinc finger regulators. Trends Microbiol. 8:77–81 [DOI] [PubMed] [Google Scholar]
- 21. Moreira D, Rodríguez-Valera F. 2000. A mitochondrial origin for eukaryotic C2H2 zinc finger regulators? Trends Microbiol. 8:448–450 [DOI] [PubMed] [Google Scholar]
- 22. Baglivo I, Russo L, Esposito S, Malgieri G, Renda M, Salluzzo A, Di Blasio B, Isernia C, Fattorusso R, Pedone PV. 2009. The structural role of the zinc ion can be dispensable in prokaryotic zinc-finger domains. Proc. Natl. Acad. Sci. U. S. A. 106:6933–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu Q, Pei J, Turse C, Ficht TA. 2006. Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival. BMC Microbiol. 6:102 doi:10.1186/1471-2180-6-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arenas-Gamboa AM, Rice-Ficht AC, Kahl-McDonagh MM, Ficht TA. 2011. Protective efficacy and safety of Brucella melitensis 16MΔmucR against intraperitoneal and aerosol challenge in BALB/c mice. Infect. Immun. 79:3653–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caswell CC, Gaines JM, Roop RM., II 2012. The RNA chaperone Hfq independently coordinates expression of the VirB type IV secretion system and the LuxR-type regulator BabR in Brucella abortus 2308. J. Bacteriol. 194:3–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spratt BG, Hedge PJ, te Heesen S, Edelman A, Broome-Smith JK. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41:337–342 [DOI] [PubMed] [Google Scholar]
- 27. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 28. White PG, Wilson JB. 1951. Differentiation of smooth and nonsmooth colonies of brucellae. J. Bacteriol. 61:239–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bellaire BH, Elzer PH, Baldwin CL, Roop RM., II 2003. Production of the siderophore 2,3-dihydroxybenzoic acid is required for wild-type growth of Brucella abortus in the presence of erythritol under low-iron conditions in vitro. Infect. Immun. 71:2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miller JH. 1972. Experiments in molecular genetics, p 352–355 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 doi:10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leong D, Diaz R, Milner K, Rudbach J, Wilson JB. 1970. Some structural and biological properties of Brucella endotoxin. Infect. Immun. 1:174–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tsai CM, Frasch CE. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115–119 [DOI] [PubMed] [Google Scholar]
- 35. López-Goñi I, Moriyón I, Neilands JB. 1992. Identification of 2,3-dihydroxybenzoic acid as a Brucella abortus siderophore. Infect. Immun. 60:4496–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gee JM, Valderas MW, Kovach ME, Grippe VK, Robertson GT, Ng WL, Richardson JM, Winkler ME, Roop RM., II 2005. The Brucella abortus Cu,Zn superoxide dismutase is required for optimal resistance to oxidative killing by murine macrophages and wild-type virulence in experimentally infected mice. Infect. Immun. 73:2873–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vizcaíno N, Cloeckaert A. 2012. Biology and genetics of the Brucella outer membrane, p 133–161 In López-Goñi I, O'Callaghan D. (ed), Brucella: molecular microbiology and genomics. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 38. Petersen E, Chaudhuri P, Gourley C, Harms J, Splitter G. 2011. Brucella melitensis cyclic di-GMP phosphodiesterase BpdA controls expression of flagellar genes. J. Bacteriol. 193:5683–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guzmán-Verri C, Manterola L, Sola-Landa A, Parra A, Cloeckaert A, Garin J, Gorvel JP, Moriyón I, Moreno E, López-Goñi I. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. U. S. A. 99:12375–12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Becker A, Küster H, Niehaus K, Pühler A. 1995. Extension of the Rhizobium meliloti succinoglycan biosynthesis gene cluster: identification of the exsA gene encoding an ABC transporter protein, and the exsB gene which probably codes for a regulator of succinoglycan biosynthesis. Mol. Gen. Genet. 249:487–497 [DOI] [PubMed] [Google Scholar]
- 41. Keller M, Roxlau A, Weng WM, Schmidt M, Quandt J, Niehaus K, Jording D, Arnold W, Pühler A. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant Microbe Interact. 8:267–277 [DOI] [PubMed] [Google Scholar]
- 42. Roop RM, II, Anderson E, Ojeda J, Martinson D, Menscher E, Martin DW. 2012. Metal acquisition by Brucella strains, p 179–199 In López-Goñi I, O'Callaghan D. (ed), Brucella: molecular microbiology and genomics. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 43. Haine V, Sinon A, Van Steen F, Rousseau S, Dozot M, Lestrate P, Lambert C, Letesson JJ, De Bolle X. 2005. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect. Immun. 73:5578–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hussain H, Johnston AW. 1997. Iron-dependent transcription of the regulatory gene ros of Agrobacterium radiobacter. Mol. Plant Microbe Interact. 10:1087–1093 [DOI] [PubMed] [Google Scholar]
- 45. Mirabella A, Terwagne M, Zygmunt MS, Cloeckaert A, De Bolle X, Letesson JJ. 2013. Brucella melitensis MucR, an orthologue of Sinorhizobium meliloti MucR, is involved in resistance to oxidative, detergent, and saline stresses and cell envelope modifications. J. Bacteriol. 195:453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Condon C. 2006. Shutdown decay of mRNA. Mol. Microbiol. 61:573–583 [DOI] [PubMed] [Google Scholar]
- 47. Cren M, Kondorosi A, Kondorosi E. 1995. NolR controls expression of the Rhizobium meliloti nodulation genes involved in the core Nod factor synthesis. Mol. Microbiol. 15:733–747 [DOI] [PubMed] [Google Scholar]
- 48. Johnston AW, Todd JD, Curson AR, Lei S, Nikolaidou-Katsaridou N, Gelfand MS, Rodionov DA. 2007. Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria. Biometals 20:501–511 [DOI] [PubMed] [Google Scholar]
- 49. Uzureau S, Godefroid M, Deschamps C, Lemaire J, De Bolle X, Letesson JJ. 2007. Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J. Bacteriol. 189:6035–6047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Godefroid M, Svensson MV, Cambier P, Uzureau S, Mirabella A, De Bolle X, Van Cutsem P, Widmalm G, Letesson JJ. 2010. Brucella melitensis 16M produces a mannan and other extracellular matrix components typical of a biofilm. FEMS Immunol. Med. Microbiol. 59:364–377 [DOI] [PubMed] [Google Scholar]
- 51. Allen CA, Adams LG, Ficht TA. 1998. Transposon-derived Brucella abortus rough mutants are attenuated and exhibit reduced intracellular survival. Infect. Immun. 66:1008–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Guan S, Bastin DA, Verma NK. 1999. Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneri bacteriophage SfX. Microbiology 145:1263–1273 [DOI] [PubMed] [Google Scholar]
- 53. Godfroid F, Cloeckaert A, Taminiau B, Danese I, Tibor A, de Bolle X, Mertens P, Letesson JJ. 2000. Genetic organisation of the lipopolysaccharide O-antigen biosynthesis region of Brucella melitensis 16M (wbk). Res. Microbiol. 151:655–668 [DOI] [PubMed] [Google Scholar]
- 54. Mancilla M, López-Goñi I, Moriyón I, Zárraga AM. 2010. Genomic island 2 is an unstable genetic element contributing to Brucella lipopolysaccharide spontaneous smooth-to-rough dissociation. J. Bacteriol. 192:6346–6351 [DOI] [PMC free article] [PubMed] [Google Scholar]