Abstract
Salmonella enterica serotype Gallinarum is the causative agent of fowl typhoid, a disease characterized by high morbidity and mortality that causes major economic losses in poultry production. We have reported that S. Gallinarum harbors a type VI secretion system (T6SS) encoded in Salmonella pathogenicity island 19 (SPI-19) that is required for efficient colonization of chicks. In the present study, we aimed to characterize the SPI-19 T6SS functionality and to investigate the mechanisms behind the phenotypes previously observed in vivo. Expression analyses revealed that SPI-19 T6SS core components are expressed and produced under in vitro bacterial growth conditions. However, secretion of the structural/secreted components Hcp1, Hcp2, and VgrG to the culture medium could not be determined, suggesting that additional signals are required for T6SS-dependent secretion of these proteins. In vitro bacterial competition assays failed to demonstrate a role for SPI-19 T6SS in interbacterial killing. In contrast, cell culture experiments with murine and avian macrophages (RAW264.7 and HD11, respectively) revealed production of a green fluorescent protein-tagged version of VgrG soon after Salmonella uptake. Furthermore, infection of RAW264.7 and HD11 macrophages with deletion mutants of SPI-19 or strains with genes encoding specific T6SS core components (clpV and vgrG) revealed that SPI-19 T6SS contributes to S. Gallinarum survival within macrophages at 20 h postuptake. SPI-19 T6SS function was not linked to Salmonella-induced cytotoxicity or cell death of infected macrophages, as has been described for other T6SS. Our data indicate that SPI-19 T6SS corresponds to a novel tool used by Salmonella to survive within host cells.
INTRODUCTION
The Salmonella genus includes over 2,500 known serotypes distributed between two species, S. enterica and S. bongori (1). Salmonella infections can range from subclinical colonization to serious systemic disease, depending both on the serotype and the infected host. While some serotypes can infect and cause systemic disease in a wide range of animal hosts, others are restricted to a specific host. S. enterica serotype Gallinarum has a host range restricted to birds and causes a severe systemic disease called fowl typhoid. This disease is characterized by high morbidity and mortality, causing major economic losses in poultry production in several parts of the world (2, 3). S. Gallinarum has been eradicated from most developed countries, such as the United States, Australia, and many Western European countries, as a result of improved surveillance and slaughter practices; however, it is still a major concern in developing countries (2).
The ability of Salmonella to invade and survive within host cells depends on the delivery of specialized effector proteins into the host cell cytosol by two different type III secretion systems (T3SS), each encoded in distinct genomic islands known as Salmonella pathogenicity island 1 (SPI-1) and SPI-2. The SPI-1 T3SS confers the ability to invade nonphagocytic cells and to colonize and breach the gastrointestinal tract of their hosts. On the other hand, SPI-2 T3SS allows Salmonella to survive within eukaryotic cells and spread to internal organs, such as the liver and spleen (4). Both T3SS are present in every S. enterica serotype and correspond to the most critical virulence determinants in Salmonella. Nevertheless, S. enterica serotypes differ greatly in terms of pathogenicity and host specificity, suggesting that other virulence factors are involved in the adaptation of different serotypes to specific hosts and/or environments.
The type VI secretion system (T6SS) corresponds to a newly described mechanism for protein translocation that is widespread among Gram-negative bacteria (5) and has been linked to a wide variety of cellular processes, including virulence in several bacterial pathogens (6). We have reported that the Salmonella genus comprises five phylogenetically distinct T6SS, each encoded in differentially distributed SPIs, including SPI-6, SPI-19, SPI-20, SPI-21, and SPI-22 (7, 8). S. Gallinarum harbors one T6SS encoded in SPI-19 (SPI-19 T6SS), which is required to efficiently colonize the avian host (9, 10). The requirement of a fully functional SPI-19 T6SS for systemic infection suggested a role for this system in the interaction of Salmonella with cells of the reticuloendothelial system (9).
Interaction of S. Gallinarum with the avian immune system has been reported to be critical for the ability of this serotype to colonize and cause disease in chicks (11). This interaction occurs in each of the different stages of the progression of avian systemic salmonellosis, from intestinal invasion and systemic infection to the immune clearance/persistence of the pathogen (reviewed in reference 12). Nevertheless, despite the critical aspects of this Salmonella-immune cell relationship, there is little information regarding the bacterial virulence determinants important for the interaction of S. Gallinarum with macrophages, especially those of avian origin. In this context, even though several SPIs have been shown to contribute to infection in vivo (3, 9, 13), only SPI-1 and SPI-2 T3SS have been evaluated in vitro (14, 15).
In the present work, we aimed to further characterize SPI-19 T6SS and the molecular mechanisms behind the phenotypes previously observed in vivo. To accomplish this, we analyzed the expression, production, and secretion profiles of selected T6SS components under different in vitro growth conditions and analyzed the role played by SPI-19 T6SS in the interaction of S. Gallinarum with murine and avian macrophages. We determined that SPI-19 T6SS components are expressed and produced, but not secreted, under different growth conditions. On the other hand, we determined intracellular production of the cell-puncturing protein VgrG once the bacteria are internalized by macrophages and that SPI-19 T6SS contributes to S. Gallinarum survival within murine and avian macrophages.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in the present study are listed in Table 1. Bacteria were routinely grown in Luria-Bertani (LB) medium (10 g/liter tryptone, 5 g/liter yeast extract, 5 g/liter NaCl) at 37°C with aeration. When required, LB medium was supplemented with ampicillin (Amp; 100 mg/liter), chloramphenicol (Cam; 20 mg/liter), and kanamycin (Kan; 50 mg/liter). Media were solidified by the addition of agar (15 g/liter). For SPI-1-inducing conditions, bacteria were grown in LB medium supplemented to reach final concentrations of 0.171 M NaCl, 0.3 M NaCl, or 0.2 M sucrose, as described previously (16). LB medium (0.085 M NaCl) was used as a control condition. For SPI-2-inducing conditions, bacteria were grown in a modified N-minimal medium [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 0.1 mM Tris-HCl, pH 5.5, 38 mM glycerol, 0.1% Casamino Acids] adjusted to pH 5.5 and supplemented to reach a final concentration of 10 μM MgCl2 (17). N-minimal medium, adjusted to pH 7.4 and supplemented to achieve/reach a final concentration of 10 mM MgCl2, was used as a control.
Table 1.
Strains used in this study
| Strain | Feature(s) | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F− Φ80lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk− mk+) phoA supE44 thi-1 gyrA96 relA1 λ− | Laboratory collection |
| S. Typhimurium | ||
| 14028s | Wild-type, sequenced isolate | Laboratory collection |
| ΔSPI-1::Kan | ΔSTM2865-STM2899 mutant strain, Kanr | This work |
| pSipA/FT pmCherry | Wild-type strain harboring plasmids pSipA/FT and pmCherry | This work |
| ΔSPI-1 pSipA/FT pmCherry | ΔSPI-1::Kan mutant strain harboring plasmids pSipA/FT and pmCherry | This work |
| S. Gallinarum | ||
| 287/91 | Wild-type, sequenced isolate | Laboratory collection |
| WT/pWRKC | attG1::pWRKC | This work |
| ΔssaG | ΔSG1710 mutant strain | This work |
| ΔaroA | ΔSG0290 mutant strain | This work |
| ΔSPI-19::Kan | ΔSG1024-SG1054::Kan mutant strain, Kanr | This work |
| ΔSPI-19 | ΔSG1024-SG1054 mutant strain | 9 |
| ΔclpV | ΔSG1034 mutant strain | 9 |
| Δhcp1 | ΔSG1029 mutant strain | This work |
| Δhcp2 | ΔSG1043 mutant strain | This work |
| ΔvgrG | ΔSG1044 mutant strain | This work |
| ΔicmF | ΔSG1031 mutant strain | This work |
| ΔclpV/clpV | ΔSG1034 mutant strain harboring plasmid pGEM-T::SG1034 | This work |
| hcp1-3×FLAG | SG1029-3XFLAG gene fusion | This work |
| hcp2-3×FLAG | SG1043-3XFLAG gene fusion | This work |
| vgrG-3×FLAG | SG1044-3XFLAG gene fusion | This work |
| vgrG-GFP/pmCherry | SG1044-GFP gene fusion harboring plasmid pmCherry | This work |
| hcp1-lacZ | SG1029-lacZ operon fusion | This work |
| hcp2-lacZ | SG1043-lacZ operon fusion | This work |
| vgrG-lacZ | SG1044-lacZ operon fusion | This work |
| icmF-lacZ | SG1031-lacZ operon fusion | This work |
| clpV-lacZ | SG1034-lacZ operon fusion | This work |
| pSipA/FT pmCherry | Wild-type strain harboring plasmids pSipA/FT and pmCherry | This work |
| pVgrG/FT pmCherry | Wild-type strain harboring plasmids pVgrG/FT and pmCherry | This work |
Standard DNA techniques.
Total genomic DNA was obtained from overnight bacterial cultures using the GenElute bacterial genomic DNA kit (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. Plasmid DNA was obtained from overnight cultures using either the QIAprep Spin Miniprep kit or the QIAprep Spin plasmid Maxikit (Qiagen, Germantown, MD) according to the manufacturer's instructions. PCR products were purified using the QIAquick PCR purification kit (Qiagen, Germantown, MD) as recommended. The multicopy plasmid pGEM-T Easy (Promega, Madison, WI) was used for complementation studies. DNA samples were routinely analyzed by electrophoresis in 1% agarose gels (prepared in 1× Tris-acetate-EDTA buffer) and visualized under UV light after ethidium bromide staining.
Mutant constructions and analyses.
Mutant strains with deletions of SPI-19 (SG1024 to SG1054), clpV (SG1034), icmF (SG1031), hcp1 (SG1029), hcp2 (SG1043), and vgrG (SG1044) were constructed using the Red-swap method (18). PCR primers 60 bases long were synthesized with 40 nucleotides (nt) on the ends corresponding to the extreme of the desired deletion (Table 2) and 20 nt that anneal to the 5′ or 3′ end of a Cam resistance cassette flanked by the FRT sites (FLP recombinase target sequence) present in plasmid pCLF2 (GenBank accession number HM047089), which was used as the template DNA. S. Gallinarum strain 287/91 (10), carrying the temperature-sensitive plasmid pKD46, which expresses the Lambda-Red recombinase system, was grown to an optical density at 600 nm (OD600) of 0.5 at 30°C in LB medium containing Amp and l-arabinose (10 mM). Bacteria were made electrocompetent by sequential washes with ice-cold, sterile 10% glycerol and transformed with approximately 500 ng of the corresponding purified PCR product. Transformants were selected on LB Cam agar at 37°C. The presence of each mutation was confirmed by PCR amplification using primers flanking the sites of substitution. Also, each mutant was assayed for Amp sensitivity to confirm the loss of pKD46. Finally, mutant alleles were transferred to the wild-type background by generalized transduction using P22 phage.
Table 2.
Oligonucleotides used in this study
| Primer | Sequencea |
|---|---|
| SPI-19_(H1+P1) | TAGCTGAATTGCAATATGCGAAAAAAGCCGAGCTTGATGACAAACGTGTAGGCTGGAGCTGCTTC |
| SPI-19_(H2+P2) | AAGCATCTTCAATAATCACGGGTATAAATGCTTACACTCTTTATCCATATGAATATCCTCCTTAG |
| SEN0997_SEN1009_(H1+P1) | TAGCTGAATTGCAATATGCGAAAAAAGCCGAGCTTGATGACAAACGTGTAGGCTGGAGCTGCTTC |
| SEN0997_SEN1009_(H2+P2) | AAGCATCTTCAATAATCACGGGTATAAATGCTTACACTCTTTATCCATATGAATATCCTCCTTAG |
| SG1029_(H1+P1) | GGAGATTAACATGGCCAATTTAATTTATTTAACACTGAACGTGTAGGCTGGAGCTGCTTC |
| SG1029_(H2+P2) | AAAATTTTTACTAAAACACCCTCTCATCCCATAAACTGAACATATGAATATCCTCCTTAG |
| SG1031_(H1+P1) | GTGTTCAGATTACCGACCCCCCGATTATTCAGCACACTGAGTGTAGGCTGGAGCTGCTTC |
| SG1031_(H2+P2) | TCAGTACAGGGTATCCGGCAGCCGGAACTGACTGAACAGCCATATGAATATCCTCCTTAG |
| SG1034_(H1+P1) | ATGATCCAGATTGACTTAGCCACCCTGGTAAAACGGCTTGGTGTAGGCTGGAGCTGCTTC |
| SG1034_(H2+P2) | TCATAGAACGGCTTCGTCCTCTGCTTCCGCTACGGGCTGACATATGAATATCCTCCTTAG |
| SG1043_(H1+P1) | GAGTTTATAAATGCCAACACCGTGTTACATCTCAATTACGGTGCAGGCTGGAGCTGCTTC |
| SG1043_(H2+P2) | TTGATTCCGTTTACGCTTCCAGCGGCGCGCGCCAGTCATCCATATGAATATCCTCCTTAG |
| SG1044_(H1+P1) | GAGGTTATTCATGTCAACAGGATTACGTTTCACACTGGAGGTGCAGGCTGGAGCTGCTTC |
| SG1044_(H2+P2) | ATTATGTTATTCAGTACATGATCAGATCCGATGGATCGGGCATATGAATATCCTCCTTAG |
| Ssag_(H1+P1) | CCAGTATCCTTACGATGTATTTATTTTAAGGAAAAGCATTGCGTAGGCTGGAGCTGCTTC |
| Ssag_(H2+P2) | GATTTCCAGCAGCAACCGTCGAACATCGTCGCTAATAACTCATATGAATATCCTCCTTAG |
| Hcp1_FLAG_FWD | CGGGACCAGTGCATTCAGTTTATGGGATGAGAGGGTGTTTGACTACAAAGACCATGACGG |
| Hcp1_FLAG_REV | TTTACCATGTGCAACCCTCCATTCATTTGTAAAATTTTTACATATGAATATCCTCCTTAG |
| Hcp2_FLAG_FWD | CACCTCTGGCGCGGATGACTGGCGCGCGCCGCTGGAAGCGGACTACAAAGACCATGACGG |
| Hcp2_FLAG_REV | TTTACCTCCGTCTCCAGGCAAAAACAGCACCCTGCCCGTCCATATGAATATCCTCCTTAG |
| VgrG_FLAG_FWD | GACTACAAAGACCATGACGG |
| VgrG_FLAG_REV | CATATGAATATCCTCCTTAG |
| SG1044_GFP_fwd | CAGCAGGATGACCCCGATCCATCGGATCTGATCATGTACAGTAAAGGAGAAGAACTTTTC |
| SG1044_blatem_rev | CCCGGTGGCTCCTGCTGTAGTCCGGTATTCATTATGTTATCGTGTAGGCTGGAGCTGCTTC |
| SG1034_OUT5 | ATCCGGCATGTTCTTGCG |
| SG1043_OUT5 | CATGGTGAATGAGGTTTACG |
| SG1034_out3_BamHI | CGGGATCCCAGGAGCGTCTGCATTCTGC |
| SG1034_out5_BamHI | CGGGATCCGGCTGCTTGCCAGGTAAGGA |
| ssag_OUT5 | ATAGCATTAACAGTGCTAAG |
| SP6 | ATTTAGGTGACACTATAGAAT |
| T7 | TAATACGACTCACTATAGGG |
| Lac36 | GACCATTTTCAATCCGCA |
| SPI19_OUT_UP | CGCCTTACATAGCTACGATCTCAGG |
| SPI19_OUT_DOWN | CAGCTCAGGCAAAGAACCTATGC |
| FlagTEM_MCS_OUT5 | TGTTGCCCGTCTCACTGGTG |
| FlagTEM_MCS_OUT3 | CCACTCGTGCACCCAACTGA |
| SG1044_NdeI | CATATGAGGTTATTCATGTCAAC |
| SG1044_XhoI_OUT3 | CTCGAGGTA CATGATCAGA TCCGATG |
| LepA_F | CGTAATCATGGTCATAGCTGTTTCC |
| LepA_R | ACTTCAAGATCCATTTCCATCG |
Italics indicate the region that anneals to the 5′ or 3′ end of the antibiotic resistance cassette used for the mutagenesis. Underlining indicates XbaI, BamHI, XhoI, or NdeI restriction sites.
To obtain nonpolar deletions, the Cam resistance gene was removed by transforming each mutant with pCP20, which encodes the FLP recombinase. Transformants were plated on LB Amp agar at 30°C. Individual colonies were replica plated on LB agar, LB Amp agar, and LB Cam agar at 37°C. Transformants that had lost the resistance cassette and pCP20 were selected as those colonies that were unable to grow in the presence of Amp and Cam. The absence of the antibiotic resistance gene cassette was confirmed for each mutant by PCR analysis.
To obtain the S. Typhimurium 14028s ΔSPI-1 mutant strain, a P22 phage lysate from ΔSPI-1::Kan S. Typhimurium SL1344 (NCTC 13347) (19) was used to transduce this mutation to the 14028s background.
Construction of lacZ operon fusions.
To construct lacZ operon fusions, the method developed by Ellermeier and collaborators was used (20). This method consists of the specific integration of a suicide plasmid (pCE36) that has an FRT site directly upstream of promoterless lacZY into a chromosomal FRT site from a previous Red-swap mutation. First, S. Gallinarum mutant strains Δhcp1::FRT, Δhcp2::FRT, ΔicmF::FRT, ΔclpV::FRT, and ΔvgrG::FRT were transformed with the pCP20 plasmid and selected for Amp resistance at 30°C. These strains were then grown at 30°C and transformed by electroporation with plasmid pCE36. Transformants were recovered in LB, and dilutions were plated on LB Kan 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) agar at 37°C. Growth at 37°C allows for the optimal production of FLP recombinase and blocks the replication of the temperature-sensitive plasmid pCP20, resulting in the stable integration of pCE36 and the loss of pCP20. Fusions were checked by PCR with the appropriate set of primers (Table 2) to ensure integration of pCE36 in the correct chromosomal location and to rule out the presence of multiple plasmid integrants.
Construction of 3×FLAG gene fusions.
Epitope tagging of T6SS components was performed by constructing gene fusions between the hcp1, hcp2, and vgrG open reading frames (ORFs) and the 3×FLAG epitope by a modification of the Red-swap method described by Uzzau and collaborators (21) using primers listed in Table 2. The PCR products were amplified from template plasmid pSUB11 (Table 3), and the Red-swap protocol was similar to that described above. Each fusion was checked by PCR and transduced into a clean wild-type background, and then the resistance marker was excised from the chromosome as described above.
Table 3.
Plasmids used in this study
| Plasmid | Feature(s) | Source or reference |
|---|---|---|
| pKD46 | bla PBAD gam bet exo pSC101 ori(Ts), Ampr | 18 |
| pGEM-T Easy | Cloning vector, Ampr | Promega |
| pGEM-T::clpV | pGEM-T Easy harboring clpV, Ampr | This work |
| p3174 | Plasmid encoding the gfpmut3 gene | 22 |
| pSUB11 | 3×FLAG gene fusions plasmid, oriR6K, Kanr | 21 |
| pCP20 | bla cat cI857 λPR flp pSC101 ori(Ts), Camr, Ampr | 69 |
| pCE36 | aph FRT lacZY+ this oriR6K, Kanr | 20 |
| pCLF2 | Red-swap-redesigned vector, Camr | 70 |
| pCLF4 | Red-swap-redesigned vector, Kanr | 71 |
| pWRKC | Derivative of pir-dependent plasmid pNFB9, integrates at Gifsy-1 attachment site attG1, Camr | This work |
| pmCherry | Constitutive production of the mCherry fluorescent protein, Ampr | C. Marolda |
| pFlagTEM-1 | Plasmid used to construct translational fusions to the mature TEM-1 β-lactamase reporter | 26 |
| pSipA/FT | Plasmid which encodes the translational fusion between the mature TEM-1 β-lactamase reporter and SipA (STM2882) | 26 |
| pVgrG/FT | Plasmid which encodes the translational fusion between the mature TEM-1 β-lactamase reporter and VgrG (SG1044) | This work |
Construction of a VgrG-GFP gene fusion.
A gene fusion between the green fluorescent protein (GFP) ORF and the ORF of the VgrG T6SS component (termed VgrG-GFP) was constructed by a modification of the Red-swap method described by Gerlach and collaborators (22). The PCR product was amplified using primers listed in Table 2 and plasmid p3174 as the template (Table 3) and included the ORF encoding the GFP reporter followed by a Kan resistance cassette flanked by FRT sites. Proper integration of the reporter cassette was confirmed by PCR. Positive clones were moved into a wild-type background by generalized transduction using P22, and the resistance marker was excised from the chromosome as described above.
Protein sample preparations.
For whole-cell lysates, bacteria were grown overnight in an orbital shaker at 37°C and subcultured in the appropriate growth media. When the culture reached an OD600 of ∼0.3 to 0.4, the cells were collected by centrifugation (13,000 rpm for 2 min). The supernatant was removed and the pellet was weighted in order to normalize samples. Each mg of bacterial pellet was resuspended in 10 μl of 1× protein loading buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.01% bromophenol blue, 5% β-mercaptoethanol). Finally, the samples were boiled for 10 min and stored at −20°C.
For protein secretion analyses, bacterial cells were removed by centrifugation (13,000 rpm for 2 min). The culture supernatant was filter sterilized and mixed with trichloroacetic acid (TCA) to a final concentration of 10%. The samples were chilled on ice for 30 min and centrifuged at 13,000 rpm for 15 min. The protein pellet was washed twice with ice-cold acetone, air dried, and resuspended in 1× protein loading buffer. Finally, the samples were boiled for 5 min and stored at −20°C.
SDS-PAGE analyses and immunoblotting.
Polyacrylamide gels (12%) were prepared and mounted using a Mini-Protean III (Bio-Rad, Richmond, CA) PAGE system. The samples were run at 100 V (constant) with 1× protein running buffer (1.44% glycine, 0.3% Tris, 0.1% SDS). Transfer from the polyacrylamide gel was performed in a Mini Trans-Blot system (Bio-Rad, Richmond, CA). A polyvinylidene difluoride (PVDF) membrane was activated by immersion in 100% methanol for 15 s. The transfer was performed for 65 min at 230 mA in 1× transfer buffer (1.44% glycine, 0.3% Tris, 0.01% SDS, and 20% methanol). The membrane was blocked by incubation in a solution containing 5% nonfat milk in TBS (25 mM Tris, pH 8, 125 mM NaCl) for 90 min at room temperature. Later, the membrane was incubated at 4°C for 2 h with the M2 anti-FLAG primary antibody (Sigma-Aldrich, St. Louis, MO) diluted at 1:10,000 in TBS. For sigma 70 detection, an anti-RNA polymerase sigma 70 antibody (ab12088) was used (Abcam, Cambridge, United Kingdom) at a 1:1,000 dilution in TBS. After five washes with a TBS–0.05% Tween solution, the membrane was incubated at room temperature for 2 h with a 1:20,000 dilution of the secondary antibody conjugated with horseradish peroxidase. For immunodetection, the Supersignal West-Pico solution (Pierce, Rockford, IL) was used, exposing the membrane on a CL-Xposure (Pierce, Rockford, IL) photographic film.
β-Galactosidase assays.
Bacteria were grown overnight in LB broth and then subcultured and grown in 50 ml of the appropriate medium at 37°C in an orbital shaker. Every 30 min, a 1-ml sample was withdrawn to evaluate bacterial growth (OD600 and CFU/ml), and the β-galactosidase activity was measured as described previously (20). Enzyme activities (in Miller units), normalized for cell density (OD600), were calculated using the following equation: [A420 − (1.75 × A550)] × 1,000/(reaction time × culture volume × OD600), where reaction time was measured in minutes and culture volume in ml. Each sample was analyzed in triplicate in at least three independent experiments.
Bacterial killing assays.
Bacterial strains used for the interbacterial competition studies included Enterobacter cloacae (ATCC 700323), Shigella sonnei (ATCC 25931), Escherichia coli (ATCC 35218), Escherichia coli K-12 (ATCC 25922), Serratia marcescens, and Staphylococcus saprofiticus (ATCC BAA750). Each of these strains was tested for competition against antibiotic-resistant derivatives of the wild-type and ΔSPI-19 mutant strains. A Cam-resistant derivative of S. Gallinarum was generated by transformation with plasmid pWRKC (Camr), a derivative of plasmid pNFB9 which can integrate at the Gifsy-1 attachment site (attG1) in the chromosome (23). The chromosomal insertion of pWRKC, flanking the attG1 site, was confirmed by PCR amplification using primers LepA_F and LepA_R (Table 2). For competition assays, each strain was grown overnight in LB media and then collected and washed three times in sterile phosphate-buffered saline (PBS; 13,000 rpm for 2 min). The cell suspensions were normalized to an OD600 of 0.5 and mixed at a 1:1 ratio with either the S. Gallinarum pWRKC (Camr) or the ΔSPI-19::Kan (Kanr) strain. Fifty μl of the mixture then was spotted onto a prewarmed brain heart infusion (BHI) agar plate and incubated for 1 week at 37°C. The initial inoculum was serially diluted and plated to determine the initial CFU/ml. After 1 week, cells were recovered from the plates and resuspended in 1 ml of sterile PBS, and serial dilutions were plated out on BHI agar with or without kanamycin. The amount of prey CFU was determined by subtracting the Kan-resistant CFU counts from the total CFU counts.
Cell culture conditions.
The murine macrophage cell line RAW264.7 and the avian macrophage line HD11 were routinely maintained in Dulbecco's modified Eagle medium (DMEM) high glucose (high glucose is composed of 584 mg/liter l-glutamine, 4,500 mg/liter glucose, 110 mg/liter sodium pyruvate) supplemented with 10% inactivated bovine fetal serum (FBS) and antibiotic/antimycotic solution (10,000 nU/ml penicillin, 10,000 mg/liter streptomycin, 25 mg/liter amphotericin B) at 37°C in a 5% CO2 atmosphere.
In vitro infection assays.
Gentamicin protection assays were performed according to Schwan and collaborators (24). RAW264.7 and HD11 macrophages were seeded in 48-well cell culture plates at a density of ∼2.5 × 105 cells per well. Bacterial strains were grown overnight at 37°C in an orbital shaker, and on the day of the assay they were subcultured in LB at a 1:30 dilution and grown until they reached an OD600 of ∼0.6. The bacterial cells were collected by centrifugation (13,000 rpm for 2 min), washed three times with sterile PBS, and suspended in prewarmed 10% FBS-DMEM to reach a final concentration of 2.5 × 107 CFU/ml. This suspension was serially diluted, and bacteria were quantified by plating on LB agar plates. Macrophages were washed once with sterile PBS, and then 100 μl of the bacterial suspension was added (multiplicity of infection [MOI] of 10). Plates were centrifuged at 250 × g for 5 min at room temperature to synchronize infection, and invasion was allowed to proceed for 45 min (time point 0). After this time, the wells were washed 3 times with warm, sterile PBS and the cells were further incubated for 1 h with 10% FBS-DMEM supplemented with gentamicin (200 μg/ml) to kill extracellular bacteria. The gentamicin concentration then was lowered to 10 μg/ml for the rest of the experiment. At 2 and 20 h postuptake, different wells were washed 3 times with warm, sterile PBS, and the macrophages were lysed with 0.5% sodium deoxycholate (DOC) in PBS. Serial dilutions were prepared and plated on LB agar plates to determine viable intracellular bacteria. For each assay, viable macrophages were determined by trypan blue exclusion. The intracellular survival of mutant strains was calculated as CFU per 105 viable macrophages.
Viability and cytotoxicity assays.
Salmonella-induced cytotoxicity was determined by the CytoTox 96 nonradioactive cytotoxicity assay (Promega, Madison, WI). For each assay, RAW264.7 and HD11 cells were seeded in 96-well plates at a density of ∼30,000 cells per well. The next day, the cells were infected with the bacterial strains as described for the infection assays. At 20 h postuptake, the supernatant of infected cells was collected and transferred to a new 96-well plate. The cytotoxicity reagent then was added following 30 min of incubation at room temperature protected from light. After incubation, absorbance at 562 nm was measured and the cytotoxicity levels were calculated as a percentage of lactate dehydrogenase (LDH) release normalized to the spontaneous release from noninfected cells.
For viability and sub-G1 population analyses, RAW264.7 and HD11 macrophages were seeded at a density of ∼300,000 cells per well. Sub-G1 analysis allows the detection of hypoploid cells, which are cells with lower DNA content due to apoptotic cell death. Gentamicin protection assays were performed as described above. At 20 h postuptake, cells from both the supernatant and the monolayer were recovered (to obtain a complete infected cell population), washed, and resuspended in 1 ml of warm 10% FBS-DMEM. For viability analysis, 5 μl of a 1 mg/ml stock of propidium iodide (PI) was added to 500 μl of the cell suspension, and the samples were analyzed by flow cytometry. For sub-G1 analysis, a volume of 500 μl of the cell suspension was used. Cells were collected by centrifugation and then treated with 100% methanol for 1 h at −20°C. After incubation, cells were collected and resuspended in 500 μl of PBS containing RNase at a concentration of 50 μg/ml. After incubation for 1 h, 5 μl of a 1 mg/ml stock of PI was added. The levels of PI incorporation were quantified on a FACScan flow cytometer (Becton, Dickinson). Cell size was evaluated by forward-angle light scattering (FSC). PI-negative cells of normal size were considered alive. A total of 5,000 cells per sample were analyzed.
Epifluorescence microscopy.
For epifluorescence analysis of VgrG-GFP production upon internalization of S. Gallinarum into RAW264.7 and HD11 macrophages, cells were seeded on 25-mm coverslips in 6-well plates at a density of ∼375,000 cells per well. After 20 h, the cells were infected with the bacterial strains as described for the infection assays. At either 45 min or 2 h postuptake, the cells were washed 3 times with warm, sterile PBS and fixed with a 4% formaldehyde solution for 20 min at room temperature. After incubation, the coverslips were washed with sterile PBS and mounted in a 4′,6-diamidino-2-phenylindole (DAPI)-containing solution from VectaShield (Vector Laboratories, Burlington, CA). For Cytochalasin D analysis, cells were incubated for 1 h with 10 μg/ml Cytochalasin D before infection and maintained with this concentration throughout the experiment. The samples were stored at 4°C and visualized under an epifluorescence microscope. Fluorescence microscopy analysis was performed using a Spinning Disk Olympus IX 81 fluorescence microscope, and fluorescence micrographs were captured using a Zeiss Axiocam MRC5 and CellR software.
VgrG translocation assays.
The vgrG ORF (SG1044), without the corresponding stop codon, was amplified by PCR using as the template a preparation of S. Gallinarum 287/91 genomic DNA and cloned into plasmid pFlagTEM-1 (25, 26). The production of the VgrG-BlaTEM recombinant protein was assessed by SDS-PAGE and Western blot analysis using anti-3×FLAG monoclonal antibodies. To demonstrate T6SS-dependent VgrG translocation, a modified gentamicin protection assay was performed as described previously (8). RAW264.7 and HD11 macrophages were seeded in 96-well optical plates (black walls and clear, flat bottoms) and infected with each of the bacterial strains expressing the corresponding BlaTEM fusions.
Bacteria were grown to an OD600 of 0.3 and induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) until they reached an OD600 of 0.6. The cells were collected and washed three times with sterile PBS (13,000 rpm for 2 min) and resuspended in the appropriate volume of prewarmed 10% FBS-DMEM. Infections were performed for 45 min at an MOI of 50. Noninternalized bacteria were removed by three washes with sterile PBS, and cells were further incubated with 200 μl 10% FBS-DMEM supplemented with 1 mM IPTG and 200 μg/ml gentamicin. After 1 h of incubation, the cell supernatant was replaced with 100 μl Hanks' buffered salt solution (HBSS; Gibco) supplemented with 20 mM HEPES and 3 mM probenecid (pH 7.4; Sigma-Aldrich, St. Louis, MO) (designated HBSS-HP), and 20 μl freshly prepared CCF4-AM β-lactamase substrate (LiveBLAzer FRET-B/G loading kit; Invitrogen) was added. After 90 min of incubation at room temperature in the dark, the cells were washed five times with HBSS-HP. Fluorescence emission at 450 and 520 nm was measured from the bottom using a plate reader fluorometer (Synergy H1; BioTek) (410-nm excitation wavelength, 10-nm band pass). The translocation level was calculated as recommended in the LiveBLAzer FRET-B/G loading kit manual. Briefly, emission values were first corrected by subtraction of the average background signals recorded for empty wells, and the mean 450-nm/520-nm emission ratio of a triplicate of wells was calculated for each sample. The translocation level is expressed as fold increase of the mean 450-nm/520-nm emission ratio of each infected sample relative to the mean emission ratio of uninfected cells. Epifluorescence microscopy analysis was performed using a Spinning Disk Olympus IX 81 fluorescence microscope. Fluorescence micrographs were captured using a Zeiss Axiocam MRC5 and CellR software. Images were imported into the ImageJ software (v.1.44).
Statistical analysis.
The statistical significance of differences in the data was determined using one-way analysis of variance (ANOVA) with Dunnett's posttest (GraphPad Prism v5.0).
RESULTS
SPI-19 T6SS components are expressed and produced under in vitro bacterial growth conditions.
A recent report from our group provided genetic evidence of SPI-19 T6SS functionality (9). Nevertheless, no study regarding expression, production, and secretion of its components has been performed to date. In order to further characterize SPI-19 T6SS in S. Gallinarum, operon fusions to the lacZ reporter were constructed for the conserved core components hcp1, hcp2, vgrG, clpV, and icmF, and gene fusions to the 3×FLAG epitope were constructed for hcp1, hcp2, and vgrG, as they encode secreted components of T6SS (Fig. 1A and Table 1). Different in vitro growth conditions were tested to study the expression and production of these components, including (i) standard bacterial culture conditions (growth in LB medium at 37°C with agitation); (ii) SPI-1-inducing growth conditions modified to mimic the conditions found by S. Gallinarum in the intestinal lumen of the chicken (42°C, high osmolarity, anaerobiosis) (16); and (iii) SPI-2-inducing growth conditions intended to mimic those found by the bacterium inside macrophages (pH 5.5 and 10 μM MgCl2) (17). Even though these media cannot fully replicate the conditions encountered by Salmonella in the host, they have proven useful for identifying underlying regulatory networks in Salmonella (16, 17, 27–29).
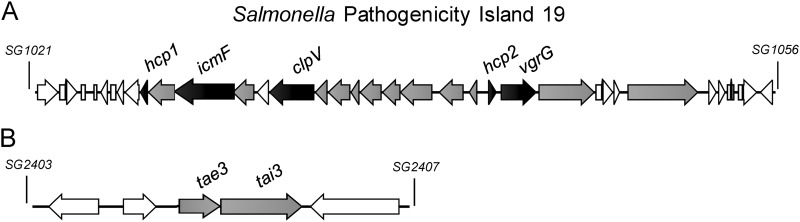
Fig 1.
Salmonella pathogenicity island 19 and the T6SS-related toxin/antitoxin gene cluster. (A) Schematic representation of the SPI-19 T6SS gene cluster in S. Gallinarum. (B) The putative toxin/antitoxin module identified by Russell and colleagues (35). T6SS core components are shown in gray. Components studied in this work are shown in black.
Expression analyses revealed a growth phase-dependent regulation of each gene analyzed under standard bacterial culture conditions, with the hcp1 gene showing the highest expression levels of all the genes tested (Fig. 2A). Gene expression analyses under SPI-1- and SPI-2-inducing conditions showed a significant increase only for hcp1 under SPI-2-inducing conditions (data not shown).
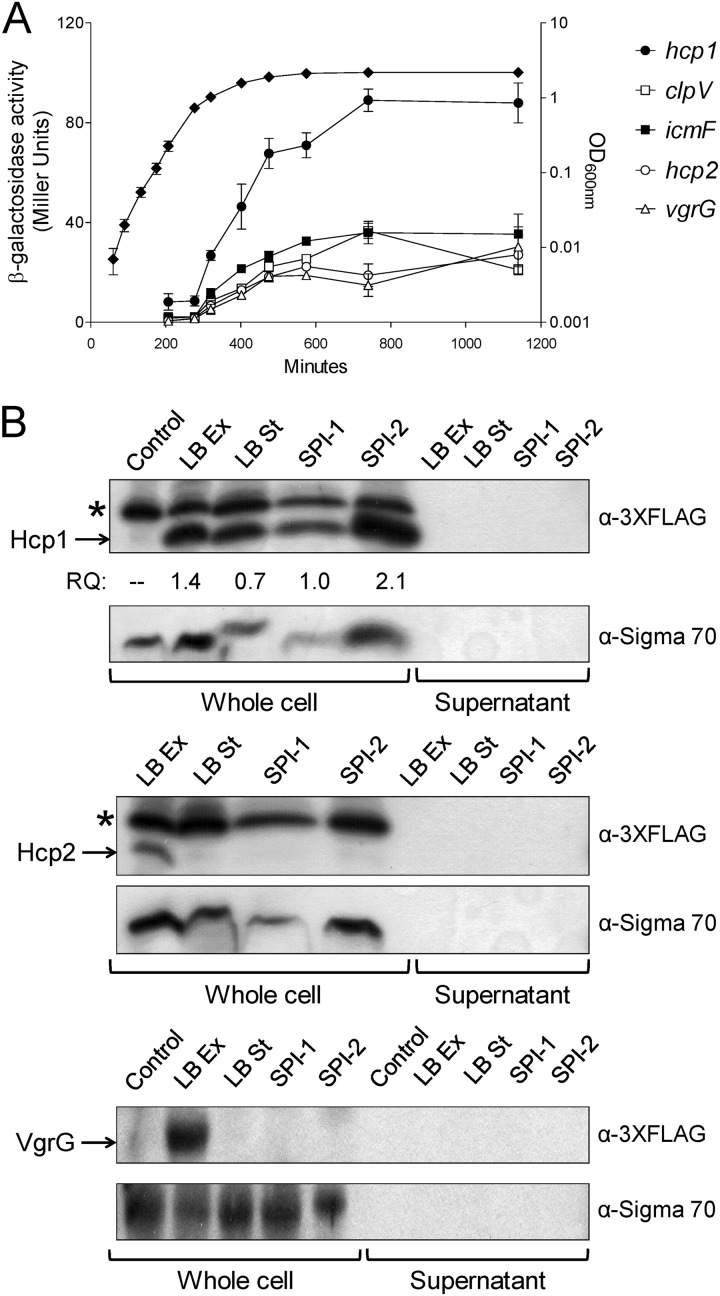
Fig 2.
Expression, production, and secretion analysis of selected SPI-19 T6SS components under different in vitro conditions. (A) Activity of lacZ operon fusions to hcp1, clpV, icmF, hcp2, and vgrG during the bacterial growth curve. Expression was measured as β-galactosidase activity (expressed as Miller units). (B) Production and secretion analyses of 3×-FLAG epitope-tagged versions of Hcp1, Hcp2, and VgrG from whole-cell lysates and supernatant fractions. The asterisk indicates an unspecific band detected with the 3×-FLAG antibody. Relative quantification (RQ) of the Hcp1 protein was determined by densitometric analysis and is expressed as the ratio against the production levels of the nonspecific band detected with the 3×-FLAG antibody. Detection of the RNA polymerase by an α-sigma 70 antibody was used as a control to analyze protein secretion in the cell culture supernatants. Under standard bacterial growth conditions (LB), samples were acquired at both exponential (Ex; OD600 of 0.3) and stationary (St; OD600 of 1.8) phases of growth.
Production of the structural/secreted proteins Hcp1, Hcp2, and VgrG revealed that these three proteins are produced in vitro (Fig. 2B). The Hcp1 protein was detected under all of the conditions tested, and relative quantification by gel densitometric analysis of its production revealed that increased levels of the protein were present under SPI-2-inducing conditions (Fig. 2B). In contrast to expression analyses, the Hcp2 and VgrG proteins were detected only at the exponential phase of bacterial growth and were barely detected at stationary phase or under SPI-1- and SPI-2-inducing conditions (Fig. 2B).
Regardless of the differences observed for the expression and production of T6SS core components, it was not possible to detect secretion of either Hcp1, Hcp2, or VgrG to the culture media under the different growth conditions tested (Fig. 2B). These findings suggest that additional signals are required for full activation of SPI-19 T6SS in S. Gallinarum.
SPI-19 T6SS is not involved in interbacterial killing under different in vitro growth conditions.
An increasing number of T6SSs have been linked to interbacterial killing against different Gram-negative bacteria through the delivery of different toxins which target the peptidoglycan of susceptible bacterial species (30–34). A heuristic informatic approach was recently developed for large-scale identification of T6SS antibacterial effectors (35). In Salmonella, this approach identified several toxin/antitoxin modules inside the SPI-6 T6SS gene cluster and one additional module in a genomic region not associated with T6SS loci (tae3 and tai3 genes in Fig. 1B) (35). Interestingly, this module is present in every Salmonella enterica genome we have analyzed (data not shown), so it is tempting to speculate that even though the SPI-19 T6SS gene cluster does not seem to encode a toxin/antitoxin module, it could be involved in interbacterial killing through translocation of the toxin encoded in this novel cluster (Fig. 1B).
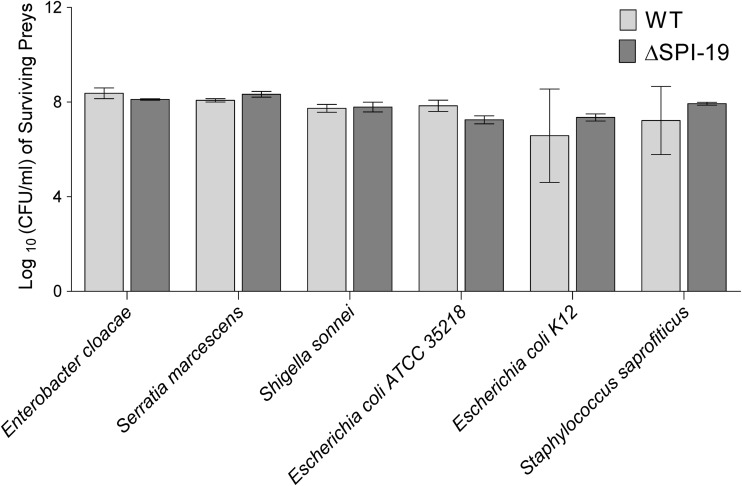
To test this, an in vitro competition assay was performed using the wild type or a ΔSPI-19 derivative of S. Gallinarum against a panel of 6 Gram-negative bacteria, including bacteria previously shown to be susceptible to T6SS killing. A Gram-positive strain was included in the assay as a control. As shown in Fig. 3, each prey strain tested grew at similar levels after coculture with either the wild-type or the SPI-19 mutant strain. When the same experiment was performed under different in vitro growth conditions (30, 37, and 42°C and SPI-1-inducing conditions), no differences were observed (data not shown). These results indicate that S. Gallinarum SPI-19 T6SS is not involved in interbacterial killing under the conditions studied.
Fig 3.
SPI-19 T6SS is not involved in interbacterial killing. Survival of prey strains listed below the x axis was determined by measuring the corresponding CFU after 1 week of exposure to either the wild type or a ΔSPI-19 mutant of S. Gallinarum. Data represent CFU from three independent experiments. Bars represent mean values, and error bars denote standard deviations. No statistically significant differences between the mutant and the wild type were determined by one-way ANOVA with Dunnett's posttest.
VgrG is produced after S. Gallinarum internalization by murine and avian macrophages.
The contribution of the SPI-19 T6SS to S. Gallinarum colonization of orally infected chickens suggested a role for this system in the interaction of Salmonella with eukaryotic cells, especially those of the reticuloendothelial system (7). Because expression of T6SS components in other bacteria is induced within eukaryotic cells (reviewed in reference 36), we analyzed the production of the VgrG protein within macrophages. To date, most studies regarding the ability of Salmonella to infect and survive within macrophages have been performed in murine macrophages. However, since S. Gallinarum corresponds to an avian-adapted serotype, we chose to use both murine (RAW264.7) and avian (HD11) macrophages and compare the results obtained for each cell line.
A chromosomal gene fusion was constructed between the vgrG ORF and the ORF encoding the GFP reporter using the method described by Gerlach and colleagues (22). The vgrG gene was chosen because (i) it is an essential T6SS component, (ii) production of VgrG was not detected under the conditions we tested, including those used prior to infection (either LB medium, 37°C, and an OD600 of 0.6 or DMEM, 5% CO2, and 37°C) (Fig. 4A), and (iii) it has been shown that VgrG C-terminal fusions can be constructed without affecting the functionality of the corresponding T6SS (37).
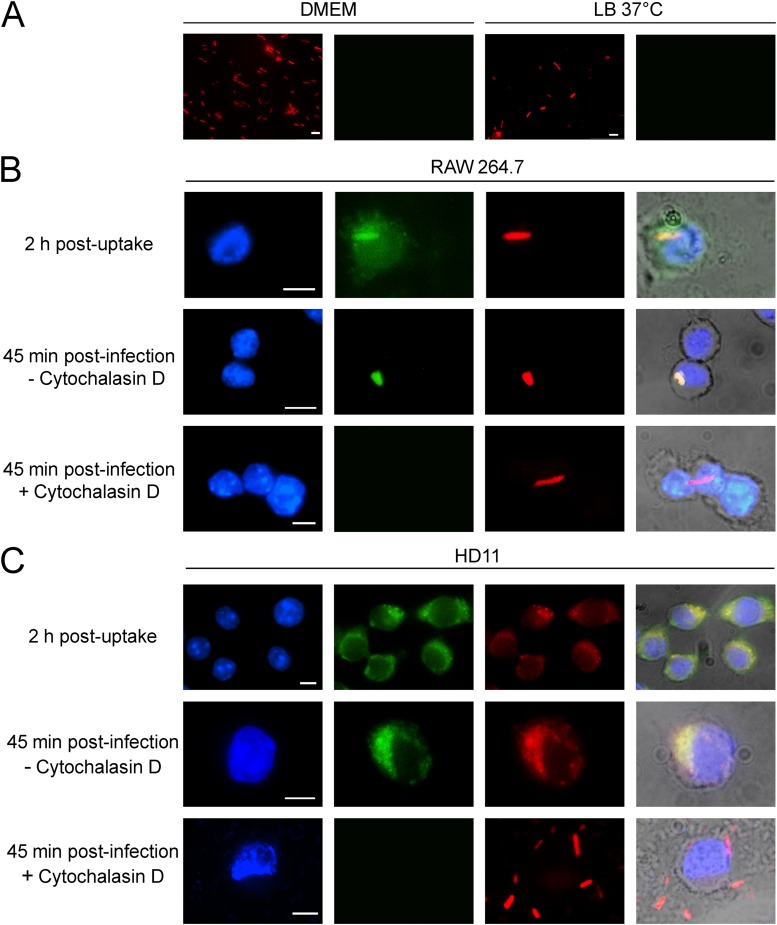
Fig 4.
Intracellular production of the VgrG-GFP fusion protein. (A) Production of VgrG-GFP under cell culture (10% FBS-DMEM, 5% CO2, 37°C) and standard bacterial growth conditions (OD600 of 0.6, LB, and 37°C). (B and C) Production of VgrG-GFP after interaction in RAW264.6 (B) and HD11 (C) macrophages. Samples were obtained after 45 min of infection and after 2 h postuptake. Samples were prepared in the presence or absence of the phagocytosis inhibitor Cytochalasin D. Bacterial cells were visualized through constitutive production of the mCherry fluorescent protein from plasmid pmCherry. Macrophages were visualized by DAPI staining and bright-field observation. The white scale bar represents 5 μm.
To analyze the production of the VgrG-GFP fusion protein after Salmonella internalization by macrophages, gentamicin protection assays were performed. RAW264.7 and HD11 cells were seeded onto sterile glass coverslips and infected with an S. Gallinarum strain harboring the VgrG-GFP fusion in its chromosome. This strain also carries the pmCherry plasmid, encoding the red fluorescent protein mCherry under the control of a constitutive promoter, to allow bacterial visualization. After 45 min of infection, cells were washed and cell culture media added with a final concentration of 100 μg/ml gentamicin to kill extracellular bacteria. Two hours postuptake, the cells were washed, fixed, and mounted for their analysis by epifluorescence microscopy. As shown in Fig. 4B and C, intracellular production of the VgrG-GFP fusion was detected in bacteria expressing cytosolic mCherry in both murine and avian macrophages at 2 h postuptake.
To determine whether production of VgrG was induced by the initial cell contact between Salmonella and the macrophages or occurred after bacterial internalization, cells were pretreated with Cytochalasin D to inhibit Salmonella uptake. As shown in Fig. 4B and C, pretreatment with Cytochalasin D prevented VgrG production in both murine and avian macrophages infected with S. Gallinarum. Interestingly, infections performed with the HD11 macrophage cell line revealed that S. Gallinarum was detected in granule/vacuolar structures after internalization. This was not seen in infected murine macrophages (Fig. 4C). Further studies will be needed to fully characterize the intracellular fate of S. Gallinarum in HD11 macrophages compared to results from previous studies performed in murine macrophages. Altogether, our results indicate that production of VgrG is induced once S. Gallinarum is internalized by macrophages of either murine or avian origin.
SPI-19 T6SS contributes to S. Gallinarum intracellular survival within murine and avian macrophages.
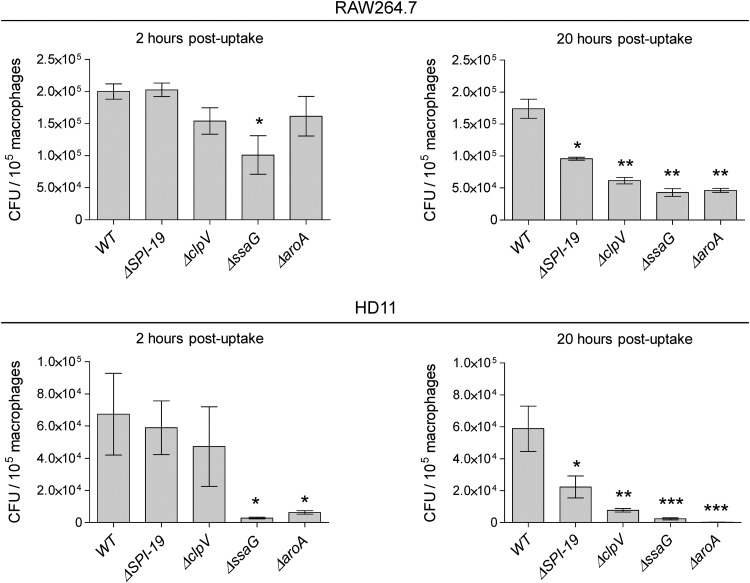
Intracellular production of VgrG in RAW264.7 and HD11 cells suggested that SPI-19 T6SS is involved in the interaction of Salmonella with macrophages. To test this, a set of defined S. Gallinarum mutants was constructed and tested in intracellular survival assays in both murine and avian macrophages. Gentamicin protection assays were performed, infecting each macrophage cell line with either the wild-type, ΔSPI-19, or ΔclpV mutant strain. As controls, ΔssaG and ΔaroA mutant strains were also included, as both genes encode proteins required for Salmonella survival within macrophages. The ssaG gene encodes an essential SPI-2 T3SS structural component (38), while aroA corresponds to an auxotrophic mutant for aromatic amino acids with impaired ability to survive and grow within macrophages (39).
At 2 and 20 h postuptake, intracellular bacteria were recovered and bacterial loads were determined and expressed as CFU per 105 macrophages. As shown in Fig. 5, none of the T6SS mutants showed a significant intracellular survival defect compared to the wild-type strain at 2 h postuptake. However, at 20 h postuptake, each T6SS mutant strain showed a statistically significant defect in intracellular survival compared to the wild-type strain (Fig. 5). Bacterial loads per macrophage at 20 h postuptake were similar to those observed for the ssaG and aroA mutant strains.
Fig 5.
Contribution of SPI-19 T6SS to Salmonella intracellular survival. Gentamicin protection assays were performed using RAW264.7 and HD11 macrophages infected with the corresponding wild-type and mutant strains. At 2 and 20 h postuptake, cells were lysed and intracellular bacteria were determined as described in Materials and Methods. Data correspond to the averages from at least three independent assays performed in triplicate. Bars represent mean values, and error bars denote standard deviations. Asterisks indicate statistically significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001) between each mutant and the wild type as determined by one-way ANOVA with Dunnett's posttest.
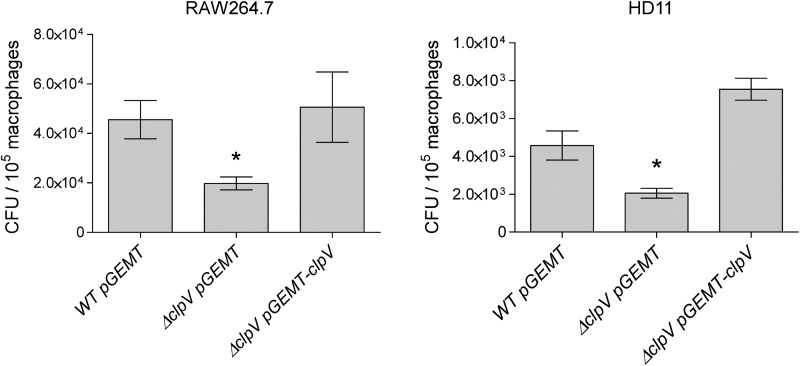
To directly link the phenotype observed for T6SS mutant strains to T6SS functionality, the ΔclpV mutant was transformed with a plasmid harboring a wild-type copy of the clpV gene and gentamicin protection assays were performed. As shown in Fig. 6, introduction of the clpV gene in trans restored the ability of the S. Gallinarum ΔclpV strain to survive within RAW264.7 and HD11 macrophages.
Fig 6.
Restoration of clpV gene in trans complements the survival defect of a ΔclpV mutant strain. Gentamicin protection assays were performed by infecting RAW264.7 and HD11 macrophages with the wild-type, ΔclpV mutant, and ΔclpV complemented strains. At 20 h postuptake, cells were lysed and intracellular bacteria determined as described in Materials and Methods. Data correspond to the averages from at least three independent assays performed in triplicate. Bars represent mean values, and error bars denote standard deviations. Asterisks indicate statistically significant differences (*, P < 0.05) between the mutant and the wild type as determined by one-way ANOVA with Dunnett's posttest.
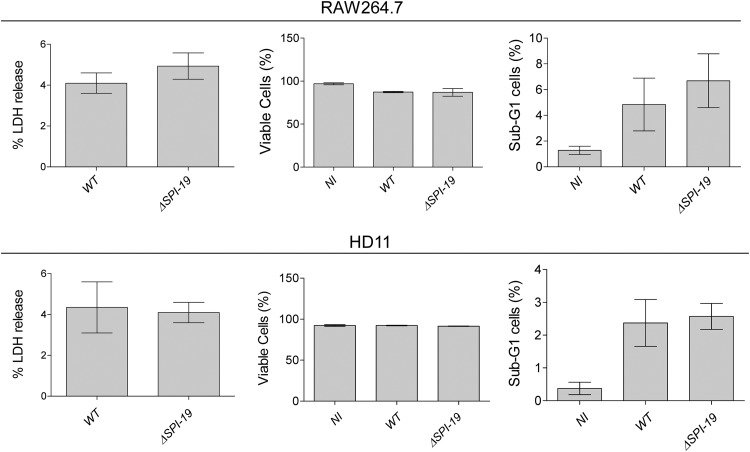
To determine if SPI-19 T6SS contributes to cytotoxicity and host cell death, as described for other T6SS (40–42), macrophages were infected with either the wild-type strain or the ΔSPI-19 mutant. Cytotoxicity was evaluated by release of lactate dehydrogenase (LDH) to the culture media, and cell viability was assayed by propidium iodide (PI) incorporation and analysis of the sub-G1 population by flow cytometry. The sub-G1 population is commonly used as an index of apoptotic cell death (43). As shown in Fig. 7, no differences in cytotoxicity, viability, and cell death were observed between the wild-type and mutant strains in either murine or avian macrophages. Altogether, these results clearly indicate that SPI-19 and the T6SS encoded therein contribute to S. Gallinarum intracellular survival in both murine and avian macrophages, and that this T6SS does not contribute to Salmonella-induced cytotoxicity and host cell death of infected macrophages.
Fig 7.
SPI-19 T6SS does not contribute to Salmonella-induced cell death of infected macrophages. RAW264.7 and HD11 macrophages were infected with either the wild-type or ΔSPI-19 mutant strain, and gentamicin protection assays were performed. After 20 h postuptake, cytotoxicity was evaluated by LDH release, and cell death by was determined by propidium iodide incorporation and cell cycle analysis of infected macrophages. NI, noninfected. Bars represent mean values, and error bars denote standard deviations. No statistical significant differences between the mutant and the wild type were determined by one-way ANOVA with Dunnett's posttest.
Finally, to assess if the VgrG protein is translocated into the cytosol of infected macrophages, a FRET-based fluorescent assay was performed using the CCF4/AM substrate. This method has been used recently to monitor VgrG translocation in Vibrio cholerae (37). A reporter gene fusion was constructed by cloning the vgrG ORF into plasmid pFlagTEM-1, which allows the production of β-lactamase fusion proteins under the control of an IPTG-inducible promoter (25). VgrG-BlaTEM translocation into the host cytosol allows processing of the FRET fluorescent substrate CCF4/AM. RAW264.7 and HD11 macrophages were infected with S. Gallinarum strains harboring plasmids encoding either VgrG-BlaTEM or SipA-BlaTEM fusion proteins. The SipA-BlaTEM fusion was used as a positive control, as it corresponds to a known SPI-1 T3SS translocated effector protein (26). As a control, infection experiments were also performed using the S. Typhimurium 14028s strain. Translocation was assessed by both fluorometric plate reader quantification and epifluorescence microscopy. As expected, translocation of SipA was observed for S. Typhimurium in both macrophages by fluorometric quantification (see File S1 in the supplemental material). However, in the case of S. Gallinarum, we could not detect translocation of SipA or VgrG fusion proteins into either macrophage cell line (see File S1). In spite of this, epifluorescence microscopy analysis showed that a very small number of cells per field presented substrate processing in macrophages infected with the S. Gallinarum strains harboring pSipA/FT or pVgrG/FT (see File S1).
DISCUSSION
The recent discovery of at least 5 different T6SSs in Salmonella has generated many questions of whether these systems contribute to Salmonella host and environmental adaptation. We have previously shown that SPI-19 T6SS contributes to efficient colonization of chicks infected with S. Gallinarum, suggesting a role for this T6SS in the interaction of Salmonella with cells of the reticuloendothelial system (7). In this study, we characterized SPI-19 T6SS by analyzing the expression, production, and translocation of selected components/effectors and by defining its role in the interaction of S. Gallinarum with murine and avian macrophages.
Expression analysis of five SPI-19 T6SS core components revealed a growth phase-dependent regulation, with an increase in their expression when the bacteria reached the onset of stationary phase (Fig. 2A). Interestingly, analysis of the production of the structural/secreted proteins Hcp1, Hcp2, and VgrG showed an inverse relationship regarding growth phase regulation, with increased protein levels detected at exponential phase (Fig. 2B). This discrepancy between gene expression and protein production suggests that these components are under posttranscriptional regulatory mechanisms, as described for other T6SS (44, 45). Growth phase-dependent expression and production of T6SS components was not unexpected. Core components of the T6SSs of Yersinia pestis and Pseudomonas aeruginosa have been shown to be regulated by the bacterial phase of growth (46, 47), and T6SSs of Aeromonas hydrophila, Vibrio cholerae, and Pseudomonas aeruginosa have been linked to quorum-sensing regulatory mechanisms (48–50).
Analysis of culture supernatants failed to demonstrate secretion of either Hcp1, Hcp2, or VgrG, even under the growth conditions that favored their production (Fig. 2B). In this context, it is possible that secretion of the SPI-19 T6SS is regulated by a posttranslational mechanism responding to an unidentified environmental/host signal. In agreement with this notion, it has been reported that T6SS activation in Pseudomonas aeruginosa requires a serine-threonine kinase and a phosphatase that regulate the phosphorylation status of a forkhead-associated (FHA) inner membrane protein (51). Even though the SPI-19 T6SS locus does not encode putative serine-threonine kinase and a phosphatase, it does encode a putative FHA domain-containing protein (SG1038) (7). Therefore, it is tempting to speculate that activation of the SPI-19 T6SS depends on a similar posttranslational regulatory mechanism.
In order to dissect the putative roles of the SPI-19 T6SS, we tested its contribution to interaction with bacterial cells and macrophages. Interbacterial killing assays failed to demonstrate antibacterial activity of SPI-19 T6SS under different growth conditions (Fig. 3). Nevertheless, it is important to mention that this assay is not comprehensive, as other unidentified Gram-negative bacteria can be the target of SPI-19 T6SS, and other inducing signals might be important for activating SPI-19 T6SS-dependent killing. Furthermore, the presence of a toxin/antitoxin module (Fig. 1B) suggests that the SPI-19 T6SS could indeed be involved in interbacterial killing.
Cell culture-based assays revealed that the VgrG protein was produced after Salmonella internalization by cultured murine and avian macrophages (Fig. 4). Preferential production of VgrG upon phagocytosis was not unexpected, because expression of T6SS components within macrophages has been described for other T6SS, including those encoded in the tss5 locus of Burkholderia pseudomallei, the FPI island of Francisella tularensis, the T6SS1 locus of Burkholderia mallei, and the vas cluster of Vibrio cholerae, all of which are upregulated upon phagocytosis (reviewed in reference 36).
When intracellular production of the VgrG-GFP fusion protein was determined, GFP fluorescence was observed in the cytoplasm of infected macrophages, suggesting that the VgrG protein could be translocated into the host cell cytosol. To test this, a VgrG-BlaTEM fusion was constructed, and CCF4/AM processing was evaluated in murine and avian macrophages. Even though visual inspection through epifluorescence microscopy revealed translocation in a small percentage of the total cell population, fluorometric plate reader quantification did not determine a significant increase in the fluorescent substrate processing. Taking into account that S. Gallinarum is internalized significantly less efficiently by macrophages than S. Typhimurium, and that translocation of the positive-control SipA was observed by epifluorescence microscopy but not detected by fluorescence quantification in S. Gallinarum, it is plausible to think that VgrG is indeed translocated under these conditions. Nevertheless, further experiments will be needed to clarify this issue, including the construction of different reporter fusions attaching to the VgrG protein.
Unexpectedly, fluorescence microscopy analysis showed that the VgrG-GFP protein was detected throughout the bacterial cell and not in punctate foci in the bacterial membrane, as seen for other T6SS structural components (52). One possible explanation is that the GFP reporter affects the normal function of the VgrG protein, including its subcellular localization. To test this hypothesis, we performed gentamicin protection assays in murine and avian macrophages infected with either the wild-type or VgrG-GFP strain. Results from these assays suggest that the fusion protein is partially impaired in T6SS function (see File S2 in the supplemental material).
The SPI-19 T6SS expression data are reminiscent of what has been described for the Salmonella SPI-2 T3SS. The genes encoding structural components of the SPI-2 T3SS are upregulated upon phagocytosis (53, 54), including the ssaG gene, which encodes a translocon component of the needle complex. Expression of the ssaG gene responds to the DNA supercoiling status of the bacterial cell chromosome, as it adapts to the intracellular environment. This activation depends on the direct binding of the Fis protein to the ssaG promoter region (55). Several Fis-regulated genes show a growth phase-dependent gene expression (56, 57) due to enhanced levels of the Fis protein at exponential phase. Therefore, the increased levels of Hcp2 and VgrG proteins in the exponential phase of growth could be due to increased levels of Fis. In support of this notion, we have identified the presence of several Fis binding sites in the promoter regions of hcp2 and vgrG (see File S3 in the supplemental material). Further work will be needed to clarify this hypothesis and to identify the global regulators that seem to be regulating the expression, production, and function of SPI-19 T6SS in Salmonella under in vitro growth and cell culture conditions.
Intracellular production of VgrG indicated a possible role of the SPI-19 T6SS in the interaction of S. Gallinarum with macrophages. Gentamicin protection assays revealed that SPI-9 T6SS contributes to Salmonella intracellular survival in both murine and avian macrophages. Upon phagocytosis, Salmonella is engulfed in a compartment known as the Salmonella-containing vacuole (SCV), and through the orchestrated action of SPI-1 and SPI-2 T3SS-secreted effector proteins, the bacterium ensures its survival by avoiding phagosome maturation (58). SCV biogenesis is usually divided in early (<30 min), intermediate (30 min to 5 h), and late stages (>5 h), with a defined set of T3SS effectors involved in each stage (59, 60). By altering phagosome biogenesis, Salmonella is able to reside in mature SCVs that localize in a perinuclear region associated with the Golgi network late after infection (60, 61).
In S. Gallinarum, the SPI-2 T3SS has been shown to be important for the ability of the bacteria to survive within murine and avian macrophages from 1 h postuptake (14, 15). In agreement with this, we observed that the SPI-2 T3SS ΔssaG mutant strain showed a significant defect on intracellular survival at 2 h postuptake. In contrast, the ΔSPI-19 and ΔclpV mutants showed a significant defect only at 20 h postuptake. These observations suggest that either (i) the T3SS and T6SS contribute to Salmonella survival by modulating different stages of SCV biogenesis, or (ii) the T6SS promotes replication of Salmonella once it has escaped from the phagolysosomal fate, as has been shown for other virulence-related loci, including the PhoP/Q regulon (62). Further experiments regarding the kinetics of phagolysosomal fusion and replication rates of T6SS mutant strains will be needed to clarify these issues.
Despite the role of SPI-19 in intracellular survival, there was no significant impact on host cell death and cytotoxicity of infected macrophages in vitro, as seen for other T6SSs that are involved in the interaction of the bacteria with cells of the immune system (6). Notwithstanding this, it is plausible to think that the SPI-19 T6SS could influence other processes triggered after infection of macrophages, including the production and secretion of proinflammatory cytokines and chemokines. In this context, it has been shown that immune responses generated by infection with S. Gallinarum are critical for the virulence and pathogenicity of this serotype both in vitro and in vivo (11, 12, 63, 64). Therefore, contribution of the SPI-19 T6SS to in vivo colonization of infected chicks is mediated by these processes in addition to intracellular survival within macrophages. Future work will be needed to determine these issues.
In agreement with our findings, other authors have pointed out a role for T6SSs in the interaction of Salmonella with host cells. Libby and colleagues (65) identified several genes linked to the SPI-6 T6SS of S. Typhi which are required for systemic infection in a humanized mouse model. In addition, Wang and colleagues (66) recently reported that a mutant in an essential SPI-6 T6SS core component enhanced the ability of S. Typhi to invade epithelial cells and reduced Salmonella-induced cytotoxicity toward these cells. More recently, Liu and colleagues (67) have shown that lack of SPI-6 T6SS in S. Typhimurium leads to reduced Salmonella survival and proliferation within infected murine macrophages and a defect in mice colonization. Finally, Mulder and colleagues (68) reported that several SPI-6 T6SS core components and SPI-6 T6SS-associated gene clusters contribute to S. Typhimurium intracellular survival within murine macrophages and systemic dissemination in mice.
Since the evidence described above shows that both SPI-6 and SPI-19 T6SSs are involved in the ability of Salmonella to survive within macrophages, it is tempting to speculate that despite having distinct phylogenetic origins, these two T6SSs are functionally redundant.
In the context of Salmonella evolution, the functional implications of this potential redundancy are intriguing. Comparative genomic analysis of T6SSs in Salmonella have suggested that a common ancestor of SPI-19-containing serotypes harbored both SPI-6 and SPI-19 T6SSs in their genomes. During evolution, some serotypes maintained both T6SSs (e.g., Dublin and Weltevreden), others have lost SPI-6 T6SS while maintaining SPI-19 T6SS (e.g., Gallinarum and Agona), and still others lost both T6SS loci (e.g., Enteritidis) (7). Therefore, it would be of interest to determine if these systems are indeed redundant or if there are subtle differences in their function that could explain why some Salmonella serotypes retained one of these systems over the other, and if they differentially contribute to the adaptation of Salmonella serotypes to different hosts and environments.
In conclusion, in this work we have demonstrated that SPI-19 T6SS contributes to Salmonella intracellular survival and that the VgrG protein is preferentially produced once the bacteria have been internalized by the cells. Altogether, our work expands the known set of virulence factors employed by Salmonella to subvert the immune system of its hosts.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to R. Tsolis for the generous gift of plasmids pFlagTEM-1 and pSipA/FT. We thank C. Marolda for the generous gift of plasmid pmCherry.
This work was supported by FONDECYT grant 1100092. C.J.B. is supported by FONDECYT Postdoctoral Fellowship 3120175. C.A.S. is supported by FONDECYT grant 1110172. D.P. is supported by fellowships from Fulbright, Beca Doctorado Nacional 2009 CONICYT (no. 21090041), Beca de Apoyo a la Realización de Tesis Doctoral 2012 CONICYT (no. AT-21121297), and Beca de Pasantías Doctorales en el Extranjero 2011 (no. 75110062 BCH-3).
Footnotes
Published ahead of print 28 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01165-12.
REFERENCES
- 1. Grimont PD, Weill FX. 2007. Antigenic formulae of the Salmonella serovars, 9th ed WHO Collaborating Centre for Reference and Research on Salmonella, Institute Pasteur, Paris, France [Google Scholar]
- 2. Shivaprasad HL. 2000. Fowl typhoid and pullorum disease. Rev. Sci. Tech. 19:405–424 [DOI] [PubMed] [Google Scholar]
- 3. Barrow PA, Freitas Neto OC. 2011. Pullorum disease and fowl typhoid–new thoughts on old diseases: a review. Avian Pathol. 40:1–13 [DOI] [PubMed] [Google Scholar]
- 4. Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66 [DOI] [PubMed] [Google Scholar]
- 5. Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learnt from available microbial genomic resources? BMC Genomics 10:104 doi:10.1186/1471-2164-10-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pukatzki S, McAuley SB, Miyata ST. 2009. The type VI secretion system: translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12:11–17 [DOI] [PubMed] [Google Scholar]
- 7. Blondel CJ, Jimenez JC, Contreras I, Santiviago CA. 2009. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10:354 doi:10.1186/1471-2164-10-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fookes M, Schroeder GN, Langridge GC, Blondel CJ, Mammina C, Connor TR, Seth-Smith H, Vernikos GS, Robinson KS, Sanders M, Petty NK, Kingsley RA, Baumler AJ, Nuccio SP, Contreras I, Santiviago CA, Maskell D, Barrow P, Humphrey T, Nastasi A, Roberts M, Frankel G, Parkhill J, Dougan G, Thomson NR. 2011. Salmonella bongori provides insights into the evolution of the salmonellae. PLoS Pathog. 7:e1002191 doi:10.1371/journal.ppat.1002191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Blondel CJ, Yang HJ, Castro B, Chiang S, Toro CS, Zaldivar M, Contreras I, Andrews-Polymenis HL, Santiviago CA. 2010. Contribution of the type VI secretion system encoded in SPI-19 to chicken colonization by Salmonella enterica serotypes Gallinarum and Enteritidis. PLoS One 5:e11724 doi:10.1371/journal.pone.0011724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, Quail MA, Stevens M, Jones MA, Watson M, Barron A, Layton A, Pickard D, Kingsley RA, Bignell A, Clark L, Harris B, Ormond D, Abdellah Z, Brooks K, Cherevach I, Chillingworth T, Woodward J, Norberczak H, Lord A, Arrowsmith C, Jagels K, Moule S, Mungall K, Sanders M, Whitehead S, Chabalgoity JA, Maskell D, Humphrey T, Roberts M, Barrow PA, Dougan G, Parkhill J. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barrow PA, Huggins MB, Lovell MA. 1994. Host specificity of Salmonella infection in chickens and mice is expressed in vivo primarily at the level of the reticuloendothelial system. Infect. Immun. 62:4602–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chappell L, Kaiser P, Barrow P, Jones MA, Johnston C, Wigley P. 2009. The immunobiology of avian systemic salmonellosis. Vet. Immunol. Immunopathol. 128:53–59 [DOI] [PubMed] [Google Scholar]
- 13. Shah DH, Lee MJ, Park JH, Lee JH, Eo SK, Kwon JT, Chae JS. 2005. Identification of Salmonella gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology 151:3957–3968 [DOI] [PubMed] [Google Scholar]
- 14. Jones MA, Wigley P, Page KL, Hulme SD, Barrow PA. 2001. Salmonella enterica serovar Gallinarum requires the Salmonella pathogenicity island 2 type III secretion system but not the Salmonella pathogenicity island 1 type III secretion system for virulence in chickens. Infect. Immun. 69:5471–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jeong JH, Song M, Park SI, Cho KO, Rhee JH, Choy HE. 2008. Salmonella enterica serovar Gallinarum requires ppGpp for internalization and survival in animal cells. J. Bacteriol. 190:6340–6350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mizusaki H, Takaya A, Yamamoto T, Aizawa S. 2008. Signal pathway in salt-activated expression of the Salmonella pathogenicity island 1 type III secretion system in Salmonella enterica serovar Typhimurium. J. Bacteriol. 190:4624–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Groisman EA, Kayser J, Soncini FC. 1997. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J. Bacteriol. 179:7040–7045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bravo D, Blondel CJ, Hoare A, Leyton L, Valvano MA, Contreras I. 2011. Type IV(B) pili are required for invasion but not for adhesion of Salmonella enterica serovar Typhi into BHK epithelial cells in a cystic fibrosis transmembrane conductance regulator-independent manner. Microb. Pathog. 51:373–377 [DOI] [PubMed] [Google Scholar]
- 20. Ellermeier CD, Janakiraman A, Slauch JM. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153–161 [DOI] [PubMed] [Google Scholar]
- 21. Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 98:15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gerlach RG, Holzer SU, Jackel D, Hensel M. 2007. Rapid engineering of bacterial reporter gene fusions by using Red recombination. Appl. Environ. Microbiol. 73:4234–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ho TD, Figueroa-Bossi N, Wang M, Uzzau S, Bossi L, Slauch JM. 2002. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:5234–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwan WR, Huang XZ, Hu L, Kopecko DJ. 2000. Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages. Infect. Immun. 68:1005–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun YH, Rolan HG, Tsolis RM. 2007. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J. Biol. Chem. 282:33897–33901 [DOI] [PubMed] [Google Scholar]
- 26. Raffatellu M, Sun YH, Wilson RP, Tran QT, Chessa D, Andrews-Polymenis HL, Lawhon SD, Figueiredo JF, Tsolis RM, Adams LG, Baumler AJ. 2005. Host restriction of Salmonella enterica serotype Typhi is not caused by functional alteration of SipA, SopB, or SopD. Infect. Immun. 73:7817–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hebrard M, Kroger C, Sivasankaran SK, Handler K, Hinton JC. 2011. The challenge of relating gene expression to the virulence of Salmonella enterica serovar Typhimurium. Curr. Opin. Biotechnol. 22:200–210 [DOI] [PubMed] [Google Scholar]
- 28. Ellermeier JR, Slauch JM. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24–29 [DOI] [PubMed] [Google Scholar]
- 29. Deiwick J, Nikolaus T, Erdogan S, Hensel M. 1999. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol. Microbiol. 31:1759–1773 [DOI] [PubMed] [Google Scholar]
- 30. Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. 2011. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J. Bacteriol. 193:6057–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U. S. A. 107:19520–19524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. English G, Trunk K, Rao VA, Srikannathasan V, Hunter WN, Coulthurst SJ. 2012. New secreted toxins and immunity proteins encoded within the type VI secretion system gene cluster of Serratia marcescens. Mol. Microbiol. [Epub ahead of print.] doi:10.1111/mmi.12028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD. 2012. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11:538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bernard CS, Brunet YR, Gueguen E, Cascales E. 2010. Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192:3850–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. 2009. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5:234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chakravortty D, Rohde M, Jager L, Deiwick J, Hensel M. 2005. Formation of a novel surface structure encoded by Salmonella pathogenicity island 2. EMBO J. 24:2043–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239 [DOI] [PubMed] [Google Scholar]
- 40. Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103:1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suarez G, Sierra JC, Sha J, Wang S, Erova TE, Fadl AA, Foltz SM, Horneman AJ, Chopra AK. 2008. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44:344–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. 2010. A type VI secretion system effector protein VgrG1 from Aeromonas hydrophila that induces host cell toxicity by ADP-ribosylation of actin. J. Bacteriol. 192:155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hacker H, Furmann C, Wagner H, Hacker G. 2002. Caspase-9/-3 activation and apoptosis are induced in mouse macrophages upon ingestion and digestion of Escherichia coli bacteria. J. Immunol. 169:3172–3179 [DOI] [PubMed] [Google Scholar]
- 44. Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72:612–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73:434–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pieper R, Huang ST, Robinson JM, Clark DJ, Alami H, Parmar PP, Perry RD, Fleischmann RD, Peterson SN. 2009. Temperature and growth phase influence the outer-membrane proteome and the expression of a type VI secretion system in Yersinia pestis. Microbiology 155:498–512 [DOI] [PubMed] [Google Scholar]
- 47. Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. 2009. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One 4:e6734 doi:10.1371/journal.pone.0006734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khajanchi BK, Sha J, Kozlova EV, Erova TE, Suarez G, Sierra JC, Popov VL, Horneman AJ, Chopra AK. 2009. N-acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology 155:3518–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lesic B, Starkey M, He J, Hazan R, Rahme LG. 2009. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 155:2845–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. 2007. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat. Cell Biol. 9:797–803 [DOI] [PubMed] [Google Scholar]
- 52. Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordonez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. 2006. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312:1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Valdivia RH, Falkow S. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007–2011 [DOI] [PubMed] [Google Scholar]
- 54. Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol. Microbiol. 30:175–188 [DOI] [PubMed] [Google Scholar]
- 55. Lim S, Kim B, Choi HS, Lee Y, Ryu S. 2006. Fis is required for proper regulation of ssaG expression in Salmonella enterica serovar Typhimurium. Microb. Pathog. 41:33–42 [DOI] [PubMed] [Google Scholar]
- 56. Bradley MD, Beach MB, de Koning AP, Pratt TS, Osuna R. 2007. Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology 153:2922–2940 [DOI] [PubMed] [Google Scholar]
- 57. Xu J, Johnson RC. 1995. Identification of genes negatively regulated by Fis: Fis and RpoS comodulate growth-phase-dependent gene expression in Escherichia coli. J. Bacteriol. 177:938–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McGhie EJ, Brawn LC, Hume PJ, Humphreys D, Koronakis V. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr. Opin. Microbiol. 12:117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Steele-Mortimer O. 2008. The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11:38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brumell JH, Grinstein S. 2004. Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 7:78–84 [DOI] [PubMed] [Google Scholar]
- 61. Bakowski MA, Braun V, Brumell JH. 2008. Salmonella-containing vacuoles: directing traffic and nesting to grow. Traffic 9:2022–2031 [DOI] [PubMed] [Google Scholar]
- 62. Thompson JA, Liu M, Helaine S, Holden DW. 2011. Contribution of the PhoP/Q regulon to survival and replication of Salmonella enterica serovar Typhimurium in macrophages. Microbiology 157:2084–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Setta AM, Barrow PA, Kaiser P, Jones MA. 2012. Early immune dynamics following infection with Salmonella enterica serovars Enteritidis, Infantis, Pullorum and Gallinarum: cytokine and chemokine gene expression profile and cellular changes of chicken cecal tonsils. Comp. Immunol. Microbiol. Infect. Dis. 35:397–410 [DOI] [PubMed] [Google Scholar]
- 64. Setta A, Barrow PA, Kaiser P, Jones MA. 2012. Immune dynamics following infection of avian macrophages and epithelial cells with typhoidal and non-typhoidal Salmonella enterica serovars; bacterial invasion and persistence, nitric oxide and oxygen production, differential host gene expression, NF-kappaB signalling and cell cytotoxicity. Vet. Immunol. Immunopathol. 146:212–224 [DOI] [PubMed] [Google Scholar]
- 65. Libby SJ, Brehm MA, Greiner DL, Shultz LD, McClelland M, Smith KD, Cookson BT, Karlinsey JE, Kinkel TL, Porwollik S, Canals R, Cummings LA, Fang FC. 2010. Humanized nonobese diabetic-scid IL2rgamma null mice are susceptible to lethal Salmonella Typhi infection. Proc. Natl. Acad. Sci. U. S. A. 107:15589–15594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang M, Luo Z, Du H, Xu S, Ni B, Zhang H, Sheng X, Xu H, Huang X. 2011. Molecular characterization of a functional type VI secretion system in Salmonella enterica serovar Typhi. Curr. Microbiol. 63:22–31 [DOI] [PubMed] [Google Scholar]
- 67. Liu J, Guo JT, Li YG, Johnston RN, Liu GR, Liu SL. 2012. The type VI secretion system gene cluster of Salmonella typhimurium: required for full virulence in mice. J. Basic Microbiol. [Epub ahead of print.] doi:10.1002/jobm.201200047 [DOI] [PubMed] [Google Scholar]
- 68. Mulder DT, Cooper CA, Coombes BK. 2012. Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar Typhimurium. Infect. Immun. 80:1996–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 70. Santiviago CA, Blondel CJ, Quezada CP, Silva CA, Tobar PM, Porwollik S, McClelland M, Andrews-Polymenis HL, Toro CS, Zaldivar M, Contreras I. 2010. Spontaneous excision of the Salmonella enterica serovar Enteritidis-specific defective prophage-like element ϕSE14. J. Bacteriol. 192:2246–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Santiviago CA, Reynolds MM, Porwollik S, Choi SH, Long F, Andrews-Polymenis HL, McClelland M. 2009. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 5:e1000477 doi:10.1371/journal.ppat.1000477 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.