Abstract
Continuous advancements in noninvasive imaging modalities such as magnetic resonance imaging (MRI) have greatly improved our ability to study physiological or pathological processes in living organisms. MRI is also proving to be a valuable tool for capturing transplanted cells in vivo. Initial cell labeling strategies for MRI made use of contrast agents that influence the MR relaxation times (T1, T2, T2*) and lead to an enhancement (T1) or depletion (T2*) of signal where labeled cells are present. T2* enhancement agents such as ultrasmall iron oxide agents (USPIO) have been employed to study cell migration and some have also been approved by the FDA for clinical application. A drawback of T2* agents is the difficulty to distinguish the signal extinction created by the labeled cells from other artifacts such as blood clots, micro bleeds or air bubbles. In this article, we describe an emerging technique for tracking cells in vivo that is based on labeling the cells with fluorine (19F)-rich particles. These particles are prepared by emulsifying perfluorocarbon (PFC) compounds and then used to label cells, which subsequently can be imaged by 19F MRI. Important advantages of PFCs for cell tracking in vivo include (i) the absence of carbon-bound 19F in vivo, which then yields background-free images and complete cell selectivityand(ii) the possibility to quantify the cell signal by 19F MR spectroscopy.
Keywords: Molecular Biology, Issue 73, Immunology, Cellular Biology, Physiology, Anatomy, Biomedical Engineering, Hematology, nuclear magnetic resonance, NMR, Fluorine, dendritic cells, migration, lymph nodes, magnetic resonance imaging, MRI, magnetic resonance spectroscopy, MRS, spectroscopy, imaging, cell tracking, clinical techniques
Introduction
The tracking of cells in vivo is a crucial aspect in several fields of biomedicine. For this, noninvasive imaging techniques that can selectively localize cells over a period of time are extremely valuable. Prior to the development of three-dimensional magnetic resonance imaging (MRI), the tracking of immune cell migration was limited to microscopic analyses or tissue biopsies. Cell tracking with the help of MRI has gained immense attention in the past few years, not only for immunologists studying immune cell behavior in vivo, but also for clinical and stem cell researchers. During the mid-90s, the first studies on iron oxide nanoparticles 1 initiated a cascade of developments for tracking cells with MRI. Iron oxide particles shorten the MR relaxation time (T2*) of the labeled cells and thus cause signal depletion in MR images. Iron oxide particles have been employed to label macrophages 2, oligodendrocyte progenitors 3 and many other cell types. Some of these particles have also been clinically approved by the FDA for labeling cellular vaccines in melanoma patients 4. Since in vivo or ex vivo labeling of cells with iron oxide particles relies on a shortening of the T2* signal and the latter could be also brought about by in vivo susceptibility-related T2* effects such as micro bleeds, iron deposits or air bubbles, it might be difficult to identify labeled cells in vivo from other background T2* signal extinctions 5.
In this article, we describe a technique for tracking dendritic cells (DC) in vivo by employing 19F/1H magnetic resonance imaging (MRI). This cell tracking technology was only introduced in 2005 6, several years after the first recognized applications for 19F in MRI had been reported 7. One important advantage of 19F over iron oxide particle cell labeling is the low biological occurrence of 19F in tissue; this makes it possible to track cells very selectively with basically background-free images. Furthermore, it is possible to overlay the 19F MR signal from the transplanted labeled cells with anatomical images obtained from conventional 1H MRI. 19F/1H MRI is therefore considerably relevant for studies investigating cell migration in vivo. Cells studied with this method are labeled with 19F-rich particles. Synthetically-derived perfluorocarbons (PFCs) consisting primarily of carbon and fluorine atoms are commonly used to prepare the particles. These compounds are insoluble in water and need to be emulsified prior to application in vitro or in vivo. The usual size of the PFC particles that have been employed by other groups for in vivo 19F-MRI tracking experiments ranges between 100 nm and 245 nm 6,8-10. We have however shown that the efficiency in labeling dendritic cells with perfluoro-15-crown-5-ether (PFCE) particles increases with increasing particle size (>560 nm).11
Protocol
All animal procedures must be approved by the local institutional animal welfare committee prior to execution. During the MR measurements an adequate level of anesthesia and physiological monitoring (body temperature, respiratory rate) are indispensable requirements.
1. Generation of Mouse Bone Marrow-derived Dendritic Cells
Extract bone marrow cells from C57BL/6 mice as previously described 12. This protocol dates back to 1992 13 and was originally described by the group of Ralph M. Steinman (1943-2011), discoverer of the dendritic cell 14.
Briefly, extract tibia and fibula soon after sacrificing mice. Working under the laminar flow hood (to prevent microbial contamination), flush the bone marrow into a small Petri dish with wash medium (5% FBS, 1% penicillin/streptomycin and 1% HEPES in RPMI) 12.
Transfer the cell suspension through a cell strainer (100 μm mesh size, nylon) into a sterile 50-ml centrifuge tube in order to remove bone and leftover tissue. After several wash steps and a lysis step 12, count the viable cells using trypan blue and a hemocytometer.
Resuspend bone marrow cells at a concentration of 400,000 cells ml-1in culture medium (RPMI supplemented with 10% FBS, 1% penicillin/streptomycin, 1% HEPES and 1% L-glutamine) containing 75 ng ml-1 of the mouse granulocyte macrophage-colony stimulating factor (mGM-CSF) obtained from supernatants of 293FT HEK cells transfected with a mouse GMCSF plasmid. Transfer 10 ml of cell suspension (4 x 106 cells) in Petri dishes (100 mm x 20 mm) and incubate in a CO2 incubator (37 °C, 5% CO2). On day 3, replenish old medium with 10 ml fresh medium (keeping end concentration of mGM-CSF at 75 ng ml-1). On day 6, remove 10 ml of old medium and replenish with 10 ml fresh medium (mGM-CSF end concentration = 75 ng ml-1). On day 8, remove 10 ml of old medium and replenish with 10 ml fresh medium (mGM-CSF end concentration = 150 ng ml-1).
On day 10, mature the differentiated DC with 1 μg ml-1 lipopolysaccharide (LPS) for 24 hr.
2. Labeling of Dendritic Cells with 19F-rich Particles
During the 24 hr maturation step (1.5), label the DC with 1 mM fluorine-rich perfluoro-15-crown-5-ether (PFCE) particles (500-560 nm) also for 24 hr. Prepare large fluorine-labeled particles by emulsifying PFCE in Pluronic F-68 (total volume 2.5 ml) using a cell-disrupting titanium sonotrode and employing a continuous pulse program (amplitude 100%, power = 250 W) for 60 sec.
Following the above incubation steps, harvest the mature and fluorine-labeled DC using a tissue culture cell scraper, transfer into a 50-ml centrifuge tube and centrifuge at 500 × g for 5 min at room temperature.
To wash off any leftover particles and dead cells, leave harvested cells to adhere on poly-L-lysine dishes. To prepare for this step, coat Petri dishes with poly-L-lysine (1 mg ml-1) at 37 °C for 1 hr, thereafter wash off the unbound poly-L-lysine with PBS.
Incubate above harvested cells on the poly-L-lysine-coated plates in serum-free medium and allow cells to adhere (1 hr at 37 °C, 5% CO2).
Wash off the medium to discard non-adherent cells, remaining particles and dead cells and wash three times with pre-warmed (37 °C) PBS.
Harvest adherent cells by trypsin-EDTA treatment (0.25% trypsin-EDTA, 3 min incubation at 37 °C).
Following another wash centrifugation step, resuspend the required amount of mature DC in serum-free sterile PBS.
To determine whether the cells have been successfully labeled, it is recommended to check the physicochemical characteristics of the cells using flow cytometry (FACS). The sideward scatter (SSC) should reveal an increased granularity in these cells, as previously described 11. It should be also kept in mind that fibroblasts might contaminate the DC cultures and could also take up the 19F particles in parallel. Therefore the DC purity should be assessed (by measuring the percentage of CD11c+CD11b+ cells using FACS) prior to in vivo application.
3. In Vivo Application of 19F-labeled Dendritic Cells
Administer DC (106 - 107) in a volume of 50 - 100 μl intracutaneously into the hind limbs of a C57BL/6 mouse by positioning the mouse in a holder and carefully inserting a 26 ½ G needle into the upper layers of the skin prior to application (use 0.5 ml tuberculin syringes with permanently-attached needles to minimize dead volume). To ensure an appropriate intracutaneous application, it is important that the inserted part of the needle be partially visible beneath the skin. If the needle is not visible any more, the needle has been inserted too deep into the skin layers. For the untrained individual, the intracutaneous application could be performed under anesthesia.
Image the limbs by in vivo fluorine (19F) / proton (1H) MRI (see next section) 21 hr following injection.
4. In Vivo 19F/1H Magnetic Resonance Imaging
The detailed instructions for setting up the MR scans refer to a Bruker MR scanner using the control software Paravison (version 5.1). Names referring to vendor-specific functions and items have been highlighted in italics. For other MR scanners these steps may have to be adjusted according to the manufacturers' guidelines.
Arrange access to a dedicated small animal MR scanner. For adequate image quality, a system with a magnetic field strength of 9.4 Tesla or more and tailored radio frequency coils (e.g. 1H/19F dual-tunable volume RF coil, 35 mm inner diameter, 50 mm length; Rapid Biomed, Würzburg, Germany) are recommended.
Prior to MRI, prepare the setup of the MRI scanner for anesthesia and physiological monitoring. Ensure that the breathing mask of the animal bed/holder is connected to the isoflurane system and that the temperature probe and breathing pad are connected to a remote animal monitoring system (e.g. Model 1025, SA Instruments Inc., New York, USA). The latter serves to monitor the animal's vital parameters such as body temperature and respiration.
Anesthetize mice by inhalation narcosis using a mouse chamber connected to an isoflurane system. Adjust the flow rate for air and O2 at 0.2 L min-1 and 0.1 L min-1, respectively and 3% isoflurane (adjusted from a vaporizer) for about 2 min until the required level of anesthesia is reached (no response following toe pinch). Optimal flow rates may vary depending on the setup being used. Be aware that high flow rates might lead to dehydration. Careful monitoring of the animals' conditions is required at all times as mentioned above.
Position the mouse prone on the mouse bed/holder of the small animal MR scanner (Figure 1).
While keeping the flow rate for air and O2 constant, adjust the isoflurane vaporizer to 0.8 - 1.5% until an optimal breathing pattern is reached. A respiratory rate of 50 - 70 breaths per min is recommended.
Keep the body temperature at 36-37 °C during the experiment by employing a warm water (or alternatively warm air) circulation system.
Keep the eyes of the mouse moist with sterile eye lubricating ointment during the measurement.
After securing the animal on the imaging holder and the connections to the monitoring system, move the holder to the center of the 1H/19F RF coil (that is positioned at the isocenter of the animal scanner magnet).
Set the position of the mouse such that the knee lies within the isocenter of the magnet, either with an automated method (e.g. using a positioning laser combined with a motor-driven animal holder) or by manually calculating the distance the holder needs to be moved into the magnet to reach the isocenter.
For confirming the correct positioning of the animal in the scanner and later planning of the image slice geometry (i.e. size, position and rotation in space), acquire scout images in three standard orientations (axial, coronal, sagittal) using fast and low resolution image acquisition methods (e.g. TriPilot protocol, which uses a FLASH pulse sequence).
Tune the RF coil to the 1H resonance frequency (e.g. 400.1 MHz for 9.4 T) and to the 19F resonance frequency (e.g. 376.3 MHz for 9.4 T) and match the characteristic impedance of the coil to 50 Ohm using the tuning monitor of the animal MR scanner. Tuning and matching is necessary to achieve the optimum conditions for transmission and signal reception.
Perform the necessary automatic system settings including shimming to fine-tune the homogeneity of the magnetic field (e.g. ADJ_SHIM), system frequency adjustments to tune the RF to the Lamor frequency of the mouse (e.g. ADJ_SF) and reference gain to adjust the RF amplitude (e.g. ADJ_REFG). All these settings are important to make the static and variable magnetics fields (B0, B1) in the region of interest as homogeneous as possible.
Acquire a second set of scout images (as above) along the three orthogonal orientations, after having adjusted the field of view (FOV) such that both lower limbs are visible from pelvis to foot (LxWxH = 50x25x25 mm). These images serve to plan the orientations of the final 3D 1H/19F scans.
Set up a TurboRARE 3D protocol for 1H-scan: TR/TE = 1,500/53 msec, FOV = 50x25x25 mm, Matrix = 400x200x200, RARE Factor = 16 (scan time approx. 60 min). Adjust FOV with the help of the scout images, start the scan (traffic-light).
Load a single pulse FID-sequence with a TR of at least 1,000 msec. Open Edit Scan and set nucleus to 19F. Use manual gain settings by deselecting the automatic reference gain in the Edit Method of the Toolbox.
Start measurement without recording data to display an MR signal in real time by using GSP (go setup). If the 19F spectral signal within the acquisition window is too low, add more averages in Edit Method and/or modulate the pulse attenuator (TX0 or SP0 slider) until a signal is clearly visible. Adjust the basic frequency in order to center the 19F spectral peak at 0 Hz in the acquisition window. Apply basic frequency and press Stop. Note that the isoflurane used as anesthesia may generate a background signal. At 9.4 Tesla MRI the chemical shift between the 19F-containing particles and the isoflurane is about 1,800 Hz. Ensure that the excitation bandwidth is smaller than 3,600 Hz to avoid exciting the isoflurane together with the 19F-labeled cells or particles. The basic frequency should be set correctly on the signal generated by the 19F-labeled cells. Alternatively, another method of anesthesia could be used as the experimenter sees fit.
Clone (duplicate) the TurboRARE 3D protocol from the previous 1H scan. Go to Edit Method and deselect the automatic reference gain (as above). Set 19F nucleus from Edit Scan. Change the matrix to 128x64x64 and the RARE-Factor to at least 40 (up to 64). For a TR/TE 1,000/6 msec and 64 averages, the scan time is approximately 70 min. Set the receiver gain to 101 (toolbox) and start the scan without any further adjustments using GOP (go pipeline).
When the scans are finished retract the mouse holder from the MR scanner. Disconnect the mouse carefully from the holder. If the mouse is not sacrificed for ex vivo analysis (e.g. histology or spectroscopy, see below) directly after the MR measurements, closely monitor until it has completely recovered from anesthesia. Body temperature regulation is affected by the anesthesia, so during the recovery process, put the mouse in a separate cage that is placed on a warm temperature-regulated pad. Once the mouse has completely recovered from anesthesia, return it to its holding cage and to the animal room.
To overlay the 19F images to 1H images, use the View Menu option on the same software (Paravison). The Underlay function can be found under Correlate. Save the underlay, load the new image and highlight the 19F layer by defining a new color from the Look up table. To discriminate between the 19F and 1H scans, alter the threshold for the chosen color (cut Look up table) such that the 19F signal will have the chosen color and the background 1H image will remain in grayscale. Other post-processing programs (e.g. ImageJ) are available to create the 1H/19F image overlays.
Should an in vivo quantification of the 19F signal be necessary, follow a simple quantitative protocol as previously described 15,16. In brief, quantification can be done by placing a reference tube with a known concentration of PFCE-Emulsion within the FOV next to the mouse. After acquiring the MR images, quantify the intensities of the 19F signals using region-of-interest (ROI) analysis and compare to the in vitro calibration curve as shown in Figure 2.
5. Lymph Node Magnetic Resonance Spectroscopy (MRS)
After sacrificing the mouse, remove the popliteal lymph node and place in a small 5 mm NMR tube and add 100 μl of 2% PFA.
Install a 19F spectroscopy coil (e.g. a toroidal coil) which has an opening of at least 5 mm for holding the NMR tube containing the lymph node.
Place the NMR tube inside the coil and move the coil into the isocenter of the magnet.
Tune the RF coil to the 19F resonance frequency (e.g. 376.3 MHz for 9.4 T) and match the characteristic impedance of the coil to 50 Ohm using the tuning monitor of the animal MR scanner. Tuning and matching is necessary to achieve the optimum conditions for transmission and signal reception.
Set all shim parameters within the Shimming Toolbox to 0 and press apply. Usually it is not necessary to apply shimming methods since the volume of the sample is relatively small. If a sufficient 19F signal is available, automated methods may be applied for shimming, system frequency and receiver gain as described above.
Load a single pulse FID sequence with a TR of at least 1,000 msec. Open Edit Scan and set the nucleus to 19F. Use manual gain settings by deselecting the automatic reference gain in the Edit Method of the Toolbox. Set the number of averages to 1.
Start the sequence using GSP. If the 19F spectral signal within the acquisition window is too low, add more averages in Edit Method and/or modulate the pulse attenuator (TX0 or SP0 slider) until a signal is clearly visible. Adjust the basic frequency so that the 19F spectral peak is at the center of the acquisition window at 0 Hz and apply basic frequency. Adjust the pulse attenuator again to maximize the signal, to reach 90 ° excitation. When signal is at maximum press Stop.
Set the averages in Edit Method between 64 and 256 (or more if necessary, depending on the signal) and start the acquisition using GOP. The detected signal correlates linearly with the number of averages. During quantification keep in mind that the detected signal needs to be normalized to one average. Furthermore, a calibration standard of a known concentration or a calibration curve is necessary to calculate the number of cells from the normalized 19F signal (Figure 2).
Once the scan is finished, remove the sample, place the next sample and proceed with step 5.3.
Representative Results
Eighteen to twenty-one hours following intracutaneous application, 19F-labeled dendritic cells (DC) migrate into the draining popliteal lymph node. The movement of DC via the lymphatic ducts into the draining politeal lymph node can be appreciated by overlaying the 1H anatomical images with the 19F DC images (Figure 2A). We have previously reported on the migration of these cells in vivo, as well as the impact of 19F-particle size on DC immunobiology, including uptake efficiency 11. In order to quantify the extent of DC migration into the lymph nodes, we extract the draining lymph nodes and perform 19F MRS (Figure 2B). When we compare the 19F signal obtained from each lymph node with the 19F signal obtained from different numbers of DC labeled with the same PFCE particles (calibration curve, Figure 2C) we can deduce the number of 19F-labeled DC that reach the draining lymph node. In the representative experiment shown in Figures 2A and 2B, we can deduce that 3.6 x 105 antigen-loaded DC reached the right lymph node while 7.5 x 104 DC that were not loaded with antigen reached the right lymph node.
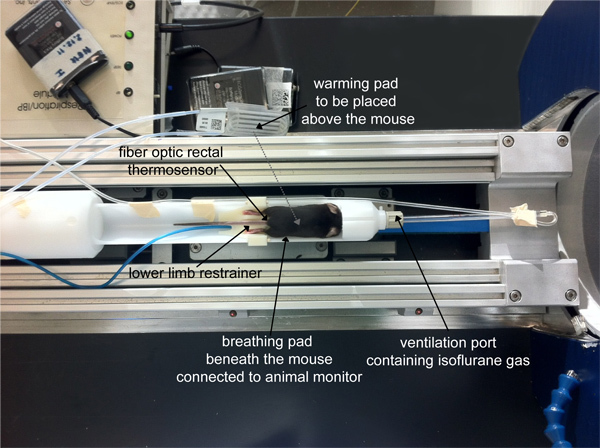 Figure 1. Position of the mouse on the mouse holder of the small animal MR scanner.
Figure 1. Position of the mouse on the mouse holder of the small animal MR scanner.
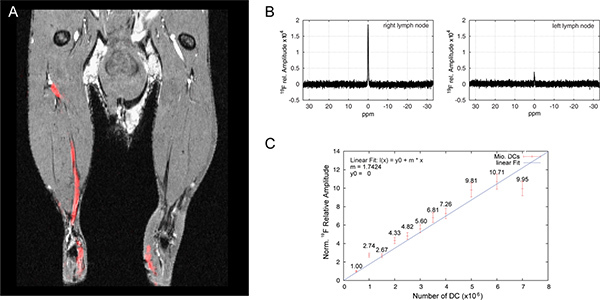 Figure 2. Quantification of 19F-labeled DC migration in vivo using 19F MRS. (A) DC were labeled with 1 mM 560 nm PFCE particles, loaded with (right) or without (left) antigen and administered intracutaneously in hind limb. Whole chicken ovalbumin (OVA) was employed as antigen; OVA was incubated with the DC together with the 19F particles. Dendritic cells are shown as 19F MR signal (red), whereas lymph nodes and lymphatic ducts are shown in the 1H anatomical MR image (grayscale). Images were acquired with a 19F/1H dual-tunable volume birdcage resonator. (B) Lymph nodes from mouse shown in A were extracted and placed in an NMR tube. The 19F signal was measured by 19F MRS (see Protocol Text) and the amplitude was calculated by performing a fast Fourier transformation (FFT) of the acquired free induction decay (FID). (C) Over a period of 24 hr, different amounts of DC were labeled with 1 mM 560 nm PFCE particles and 19F signal was measured by 19F MRS as in B.Click here to view larger figure.
Figure 2. Quantification of 19F-labeled DC migration in vivo using 19F MRS. (A) DC were labeled with 1 mM 560 nm PFCE particles, loaded with (right) or without (left) antigen and administered intracutaneously in hind limb. Whole chicken ovalbumin (OVA) was employed as antigen; OVA was incubated with the DC together with the 19F particles. Dendritic cells are shown as 19F MR signal (red), whereas lymph nodes and lymphatic ducts are shown in the 1H anatomical MR image (grayscale). Images were acquired with a 19F/1H dual-tunable volume birdcage resonator. (B) Lymph nodes from mouse shown in A were extracted and placed in an NMR tube. The 19F signal was measured by 19F MRS (see Protocol Text) and the amplitude was calculated by performing a fast Fourier transformation (FFT) of the acquired free induction decay (FID). (C) Over a period of 24 hr, different amounts of DC were labeled with 1 mM 560 nm PFCE particles and 19F signal was measured by 19F MRS as in B.Click here to view larger figure.
Discussion
This method of employing 19F/1H MRI to follow the movement of DC into the lymph node gives the opportunity to study the migration patterns of immune cells in vivo. Dendritic cells are excellent examples of rapidly migrating immune cells that are able to maneuver through three-dimensional structures without tightly adhering to specific substrates 17. Although the low spatial resolution (μm range) of the described technique is not comparable with the high resolution (nm range) that can be achieved with multiphoton microscopy, with this technique it is still possible to study the nature of cell migration in vivo and over a longer period of time. Furthermore, microscopy has a limited depth penetration and a limited field of view, making it currently unsuitable for imaging large areas within a living organism.
Due to the noninvasiveness of the technique, it is possible to monitor the migration of immune cells for several days without sacrificing the mouse after investigation. Another advantage of the technique is the possibility to overlay the 19F images that capture the migrating cells with anatomical 1H scans, thereby allowing an accurate localization of the cells within the living organism. This technique will enable us to study molecular mechanisms underlying DC migration. Unlike typical in vitro migration assays, this method enables the study of cell migration in a physiological 3D environment.
The potential application of 19F in MRS and MRI have long been recognized 7,18. The high gyromagnetic ratio, high spin and natural isotopic abundance makes 19F an ideal candidate for MR imaging 7. In addition, the similarity between 19F and 1H (regarding their NMR properties) is a prerequisite for a meaningful overlay between the anatomical 1H MRI with the 19F MRI of labeled cells. Indeed one advantage of 1H/19F MRI over other 1H/X nuclei MRI is that the same RF resonator can be used for both 19F and 1H nuclei. In most applications used for 19F/1H MRI, a volume resonator is employed that can be tuned to both 19F and 1H. Due to the almost identical B1 field for both channels, a transfer of power settings from the 1H channel (that is easier to measure) to the 19F channel is therefore possible.
One important factor to be considered when labeling cells with particles of any size (from ultra-small to large micro-sized particles) is the potential of biological manipulation and toxicity. We have recently shown that by increasing the size of the labeling particles, we could promote the maturation status of DC 11. A possible explanation for our finding is the preponderance for particles larger than 500 nm to be taken up by phagocytosis rather than endocytosis 19; the former uptake mechanism defining the nature of DC. Other physical characteristics (e.g. particle shape and surface topology) could also alter the biological function of the labeled cells and should also be carefully considered 20. For any given experimental setup that requires labeling of DC with particles, it is thus highly recommended that typical cell biology assays are performed in parallel to the experiment to determine/exclude any influence of the particles on the biological properties of these cells. Several assays can be performed, depending on the experimental study in question. To exclude an influence on DC maturation, measurements of cell surface marker (e.g. CD80, CD86) expression by FACS is recommended. To exclude an influence on antigen uptake and presentation, phagocytosis experiments and T cell priming experiments, respectively, are recommended. In the case of migration experiments, the expression of chemokine (CC) receptors (such as CCR7) and in vitro migration assays are recommended.
Although 19F/1H MRI holds promise for studying cell migration particularly for clinical purposes, there are currently a number of limitations. These include (i) a spatial resolution that precludes direct visualization of individual cells, (ii) insufficient 19F sensitivity (detection limit of several 105 cells within one ROI), (iii) increased scan durations as a result of increased averaging to compensate for the previous limitation, (iv) a limited range of 19F RF resonators on the market and (v) possible undesirable background 19F signal by other fluorine-containing elements such as isofluorane and polytetrafluoroethylene (PTFE) within MR hardware components (e.g. capacitors and connecting cables).
Apart from factors such as magnetic field strength and gradient strengths, one main determinant that dictates the level of spatial resolution is the sensitivity of the radio frequency (RF) resonator used 21. MRI resolution is closely related to the signal-to-noise ratio (SNR) 21 and upon reducing voxel size to amplify spatial resolution, a loss in SNR is to be expected. One way to increase spatial resolution without compromising signal sensitivity is to employ cryogenically-cooled coils that boost SNR by reducing thermal noise 22. With the aid of a 1H head coil that uses a cryogenic system, we could recently visualize cellular infiltrates in the experimental autoimmune encephalomyelitis after using an in plane resolution of (35x35) μm2 23. The application of this cryogenically-cooled technology for 19F/1H MRI would be an opportunity to overcome some of the MRI limitations associated with cell signal detection and resolution.
One important issue for in vivo tracking of cells by MRI is the quantification of these cells in a particular location within a living organism. For this it is possible to perform 19F MRS (see 5.1-5.9). By employing calibration curves that are obtained from 19F MRS measurements of different numbers of 19F-labeled DC, it is possible to deduce the number of cells reaching the lymph node. The 19F MRS measurements of the lymph nodes can be compared to the 19F MRS DC calibration curve. Furthermore, by comparing the MRS data from the DC cell calibration curves with the pure PFCE, we can deduce that it is possible to detect approximately 1013 19F spins per cell following labeling. According to MR principles, the lowest detection limit is 1018 spins per voxel 24. Thus, we can assume that we are able to detect a minimum number of 105 DC per voxel using a 9.4 T MRI.
In summary, the protocol that we describe here is beneficial for in vivo investigations studying cell migration during pathophysiological processes. The noninvasiveness of the technique enables longitudinal studies without the necessity of sacrificing large numbers of animals; the ability to overlay 19F images from the labeled cells on 1H anatomical images promotes optimal and highly selective tracking of the migrating cells; and 19F MRS provides an opportunity to estimate the number of cells localizing to specific anatomical areas in vivo.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This study was funded by the Deutsche Forschungsgemeinschaft to S.W. (DFG WA 2804) and a university grant to S.W. from the Experimental and Clinical Research Center, a cooperation of the Max Delbrück Center for Molecular Medicine and Charité Medical Faculty in Berlin. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank Mr. Robert Westphal for technical support during his internship in our laboratory.
References
- Yeh TC, Zhang W, Ildstad ST, Ho C. In vivo dynamic MRI tracking of rat T-cells labeled with superparamagnetic iron-oxide particles. Magn. Reson. Med. 1995;33(2):200–208. doi: 10.1002/mrm.1910330209. [DOI] [PubMed] [Google Scholar]
- Weissleder R, Cheng HC, Bogdanova A, Bogdanov A. Magnetically labeled cells can be detected by MR imaging. J. Magn. Reson. Imaging. 1997;7(1):258–263. doi: 10.1002/jmri.1880070140. [DOI] [PubMed] [Google Scholar]
- Franklin RJ, Blaschuk KL, Bearchell MC, Prestoz LL, Setzu A, et al. Magnetic resonance imaging of transplanted oligodendrocyte precursors in the rat brain. NeuroReport. 1999;10(18):3961–3965. doi: 10.1097/00001756-199912160-00043. [DOI] [PubMed] [Google Scholar]
- de Vries IJ, Lesterhuis WJ, Barentsz JO, Verdijk P, van Krieken JH, et al. Magnetic resonance tracking of dendritic cells in melanoma patients for monitoring of cellular therapy. Nat. Biotechnol. 2005;23(11):1407–1413. doi: 10.1038/nbt1154. [DOI] [PubMed] [Google Scholar]
- Liu W, Frank JA. Detection and quantification of magnetically labeled cells by cellular MRI. Eur. J. Radiol. 2009;70(2):258–264. doi: 10.1016/j.ejrad.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens ET, Flores R, Xu H, Morel PA. In vivo imaging platform for tracking immunotherapeutic cells. Nat. Biotechnol. 2005;23(8):983–987. doi: 10.1038/nbt1121. [DOI] [PubMed] [Google Scholar]
- Holland GN, Bottomley PA, Hinshaw WS. F-19 Magnetic-Resonance Imaging. Journal of Magnetic Resonance. 1977;28(1):133–136. [Google Scholar]
- Partlow KC, Chen J, Brant JA, Neubauer AM, Meyerrose TE, et al. 19F magnetic resonance imaging for stem/progenitor cell tracking with multiple unique perfluorocarbon nanobeacons. FASEB J. 2007;21(8):1647–1654. doi: 10.1096/fj.06-6505com. [DOI] [PubMed] [Google Scholar]
- Ruiz-Cabello J, Walczak P, Kedziorek DA, Chacko VP, Schmieder AH, et al. In vivo "hot spot" MR imaging of neural stem cells using fluorinated nanoparticles. Magn. Reson. Med. 2008;60(6):1506–1511. doi: 10.1002/mrm.21783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas M, Morel PA, Ernst LA, Laidlaw DH, Ahrens ET. Fluorine-19 MRI for visualization and quantification of cell migration in a diabetes model. Magn. Reson. Med. 2007;58(4):725–734. doi: 10.1002/mrm.21352. [DOI] [PubMed] [Google Scholar]
- Waiczies H, Lepore S, Janitzek N, Hagen U, Seifert F, et al. Perfluorocarbon particle size influences magnetic resonance signal and immunological properties of dendritic cells. PLoS ONE. 2011;6(7):e21981. doi: 10.1371/journal.pone.0021981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu MP, Sen D, Cahalan MD, Parker I. Generation of Bone Marrow Derived Murine Dendritic Cells for Use in 2-photon Imaging. J. Vis. Exp. 2008. p. e773. [DOI] [PMC free article] [PubMed]
- Inaba K, Inaba M, Romani N, Aya H, Deguchi M, et al. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. Journal of Experimental Medicine. 1992;176(6):1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzavecchia A, Sallusto F. Ralph M. Steinman. 1943-2011. Cell. 1943;147(6):1216–1217. doi: 10.1016/j.cell.2011.11.040. [DOI] [PubMed] [Google Scholar]
- Srinivas M, Turner MS, Janjic JM, Morel PA, Laidlaw DH, et al. In vivo cytometry of antigen-specific t cells using 19F. MRI. Magn. Reson. Med. 2009;62(3):747–753. doi: 10.1002/mrm.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas M, Heerschap A, Ahrens ET, Figdor CG, de Vries IJ. (19)F MRI for quantitative in vivo cell tracking. Trends Biotechnol. 2010;28(7):363–370. doi: 10.1016/j.tibtech.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammermann T, Bader BL, Monkley SJ, Worbs T, Wedlich-Soldner R, et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 2008;453(7191):51–55. doi: 10.1038/nature06887. [DOI] [PubMed] [Google Scholar]
- Liu MS, Long DM. Perfluoroctylbromide as a diagnostic contrast medium in gastroenterography. Radiology. 1977;122(1):71–76. doi: 10.1148/122.1.71. [DOI] [PubMed] [Google Scholar]
- Kwiatkowska K, Sobota A. Signaling pathways in phagocytosis. Bioessays. 1999;21(5):422–431. doi: 10.1002/(SICI)1521-1878(199905)21:5<422::AID-BIES9>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Mitragotri S, Lahann J. Physical approaches to biomaterial design. Nat. Mater. 2009;8(1):15–23. doi: 10.1038/nmat2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoult DI, Richards RE. The signal-to-noise ratio of the nuclear magnetic resonance experiment. J. Magn. Reson. 1976;24(1):71–85. doi: 10.1016/j.jmr.2011.09.018. [DOI] [PubMed] [Google Scholar]
- Kovacs H, Moskau D, Spraul M. Cryogenically cooled probes - a leap in NMR technology. Prog. Nucl. Magn. Reson. Spectrosc. 2005;46(2-3):131–155. [Google Scholar]
- Waiczies H, Millward JM, Lepore S, Infante-Duarte C, Pohlmann A, et al. Identification of Cellular Infiltrates during Early Stages of Brain Inflammation with Magnetic Resonance Microscopy. PLoS ONE. 2012;7(3):e32796. doi: 10.1371/journal.pone.0032796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haacke EM. Magnetic resonance imaging physical principles and sequence design. John Wiley & Sons; 1999. [Google Scholar]


