Abstract
Metastatic melanoma is one of the most aggressive forms of cutaneous cancers. Although recent therapeutic advances have prolonged patient survival, the prognosis remains dismal. C-MER proto-oncogene tyrosine kinase (MERTK) is a receptor tyrosine kinase with oncogenic properties that is often overexpressed or activated in various malignancies. Using both protein immunohistochemistry and microarray analyses, we demonstrate that MERTK expression correlates with disease progression. MERTK expression was highest in metastatic melanomas, followed by primary melanomas, while the lowest expression was observed in nevi. Additionally, over half of melanoma cell lines overexpressed MERTK compared with normal human melanocytes; however, overexpression did not correlate with mutations in BRAF or RAS. Stimulation of melanoma cells with the MERTK ligand GAS6 resulted in the activation of several downstream signaling pathways including MAPK/ERK, PI3K/AKT, and JAK/STAT. MERTK inhibition via shRNA reduced MERTK-mediated downstream signaling, reduced colony formation by up to 59%, and diminished tumor volume by 60% in a human melanoma murine xenograft model. Treatment of melanoma cells with UNC1062, a novel MERTK-selective small-molecule tyrosine kinase inhibitor, reduced activation of MERTK-mediated downstream signaling, induced apoptosis in culture, reduced colony formation in soft agar, and inhibited invasion of melanoma cells. This work establishes MERTK as a therapeutic target in melanoma and provides a rationale for the continued development of MERTK-targeted therapies.
Introduction
Although early cutaneous melanoma is usually curable with surgery, distant metastatic melanoma is an aggressive cancer with a median overall survival time of less than 1 year. In 2012, over 75,000 new melanoma diagnoses were expected and over 9,000 deaths were projected (1). Advances in the understanding of distinct melanoma subtypes as well as melanoma immunobiology have resulted in 2 FDA-approved therapies for metastatic melanoma in 2011: vemurafenib, an inhibitor of mutant BRAF — an oncogene present in approximately 50% of melanomas — and ipilimumab, a monoclonal antibody that targets CTLA-4 (2–4). Despite these rather impressive developments, the overall clinical benefit is limited to either small subgroups of patients who may be cured by immunotherapies or to a subset of patients with BRAF-mutant melanoma, most of whom will eventually develop resistance to molecularly targeted therapies (5–9). This implies the need to better understand melanoma biology and identify additional molecular targets that may be amenable to therapeutic manipulation.
Receptor tyrosine kinases (RTKs) are frequently ectopically expressed, overexpressed, or hyperactivated in tumor cells and are therefore attractive targets for cancer therapy. C-MER proto-oncogene tyrosine kinase (MERTK), a member of the TAM (TYRO, AXL, MERTK) family of RTKs, has been characterized as a therapeutic target in hematopoietic malignancies and several solid tumors including lung, prostate, and brain (10–13). As a potent mediator of prosurvival and antiapoptotic signaling pathways, MERTK is an upstream activator of both ERK1/2 and AKT. Additional signaling pathways that lead to antiinflammatory cytokine production as well as enhanced migration and invasion have been identified downstream of MERTK (11, 14, 15). A phosphoproteomic study identified MERTK and other TAM receptors as commonly activated RTKs in melanoma, however no studies have reported on the function of MERTK in melanoma (16). The role of other TAM receptors in melanoma has been described, suggesting that MERTK may also have a significant role in melanoma development and progression. TYRO3 was identified as an overexpressed receptor in melanoma, a regulator of MITF, and a contributor to the proliferative, antiapoptotic, chemoresistant, and tumorigenic phenotypes of melanoma cells (17). In another study, AXL was commonly expressed in NRAS-mutant melanomas lacking MITF expression and contributed to a migratory and invasive phenotype (18). In addition, Sensi et al. found that melanoma cells often secrete GAS6, a ligand of TAM receptors, indicating a mechanism of TAM autocrine signaling in melanoma. Taken together, these observations support the investigation of MERTK as a potential therapeutic target in melanoma.
Here, we report that MERTK expression increases with nevus-to-melanoma disease progression and is frequently overexpressed in melanoma cell lines. We propose an oncogenic role for MERTK in melanoma and demonstrate the suppression of MERTK-mediated signaling, colony formation, and tumorigenesis when MERTK expression is inhibited. Furthermore, pharmacologic targeting of MERTK in melanoma cells using UNC1062, a novel MERTK-selective small-molecule tyrosine kinase inhibitor, inhibited MERTK activation and subsequent signaling downstream of ligand-stimulated MERTK, induced apoptosis, and inhibited colony formation and invasion. These studies establish a potential oncogenic role for MERTK in melanoma and validate MERTK as a novel melanoma therapeutic target.
Results
MERTK expression increases with melanoma progression from nevus to metastatic disease.
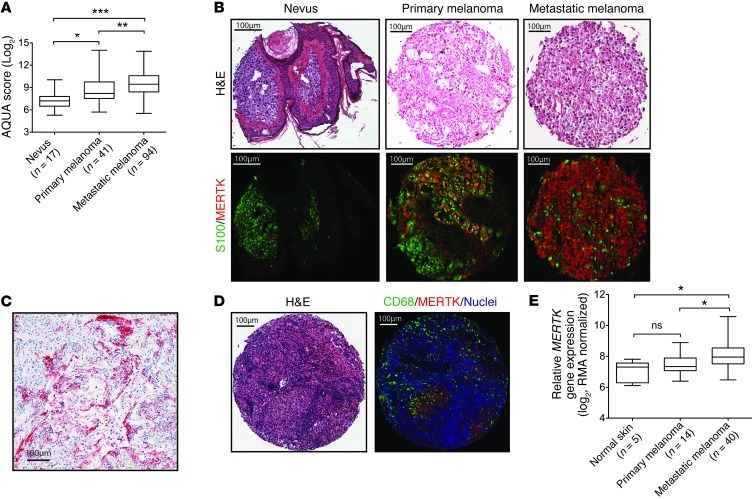
Although MERTK expression has been previously demonstrated in several melanoma cell lines (16), its expression in melanoma tissues has not been previously reported. To investigate the pattern and expression levels of MERTK during nevus-to-melanoma progression, 2 independent tissue microarrays (TMAs) were stained with an antibody against MERTK protein, and immunofluorescence was assessed in S100-positive melanocyte lineage cells. A previously described nevus-to-melanoma TMA (19) and a metastatic melanoma TMA produced at the University of North Carolina (UNC) were used to determine MERTK expression in nevi (n = 17), primary melanomas (n = 41), and metastases (n = 94). As shown in Figure 1A, two-stain immunofluorescence analysis revealed that MERTK protein expression is low in nevi, but is significantly increased in primary melanomas and even more so in metastatic melanomas (trend test via linear regression 0.765, P < 0.0001). Figure 1B shows representative cores from nevi, primary, and metastatic melanoma tumor tissues that were stained with antibodies against MERTK and S100 and counterstained with Hoechst 33258. Immunohistochemical analysis of MERTK in tumor tissues from patients who had undergone craniotomy for melanoma brain metastases showed that MERTK was detectable in melanoma cells in 28% (8 of 29) cases. Figure 1C shows a representative tissue section obtained from a patient who underwent craniotomy for melanoma brain metastases.
Figure 1. MERTK expression in nevus and melanoma tissues.
(A) Box plots of MERTK expression in nevus, primary, and metastatic melanoma human samples. AQUA score of MERTK expression for each of the TMA cores representing nevi, primary, and metastatic melanoma were calculated in S100-positive cells and were normalized using logarithmic transformation. (B) Representative tissue cores from the nevus-to-melanoma TMA. Original magnification, ×20. Core H&E-stained images (top). Merged images (RGB) showing S100 staining pseudocolored with green and MERTK protein pseudocolored with red (bottom). (C) MERTK expression in a representative tissue section from a patient who underwent craniotomy for melanoma brain metastases. Tissues were stained for MERTK using alkaline phosphatase (red) detection and counterstained with hematoxylin. Original magnification, ×40. (D) Representative tissue core from the UNC metastatic melanoma TMA stained with H&E (left) and merged image showing staining with CD68 (pseudocolored with green), MERTK protein (pseudocolored with red), and Hoechst (blue, right). (E) Gene expression analysis of MERTK transcript in patient tissue samples from the GSE7553 microarray dataset (*P < 0.05, Tukey’s multiple comparison test).
Both the staining pattern of the melanoma tissue microarrays and the morphology of MERTK-positive cells indicate that MERTK is not only expressed by melanoma cells, but is also expressed by other cells with monocytoid features (Figure 1D), as we have previously reported (20). To assess whether MERTK is also expressed by macrophages that infiltrate melanoma tumors, the UNC metastatic melanoma TMA was stained with antibodies against MERTK and CD68. Figure 1D shows a representative tissue section from a metastatic melanoma lesion that was stained with MERTK and CD68 antibodies then counterstained with Hoechst 33258. Tissue studio analysis revealed that 53% of CD68+ tumor–infiltrating cells coexpress MERTK (range 0%–94%).
To further validate the MERTK expression trend observed at the protein level, MERTK transcript expression was assessed as a function of melanoma disease progression using a previously published melanoma microarray collection (21). Using microarray data collected from patient tissue samples, MERTK gene expression data were obtained from patient tissue samples derived from normal skin (n = 5), primary melanoma (n = 14), and metastatic melanoma (n = 40) tissue datasets (Figure 1E and Supplemental Table 1). Mean MERTK transcript levels increased with disease progression, and while there was no statistical increase in MERTK transcript expression between normal skin and primary tumors, there was a significant increase in MERTK mRNA expression in metastatic tumors compared with primary tumors (P < 0.05). In addition, MERTK mRNA in metastatic tumors was significantly greater than in normal skin (P < 0.05). Taken together, these data indicate that MERTK expression at both the transcript and protein levels increases with melanoma disease progression and suggest a role for MERTK in melanoma development and progression.
MERTK is overexpressed in melanoma cell lines and can be stimulated to activate MAPK, AKT, and JAK/STAT pathways.
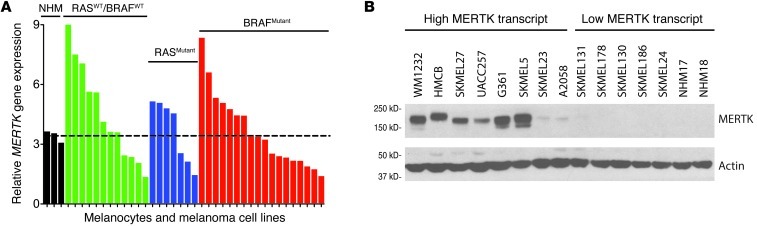
MERTK mRNA transcript levels assessed by microarray were evaluated using 2 cell line datasets. In a cell line collection that underwent microarray analysis at UNC, 55% (20 of 36) of melanoma cell lines had transcript levels greater than those in normal human melanocytes (Figure 2A and Supplemental Table 2). No significant correlation was observed between MERTK expression and oncogenic mutations in RAS or BRAF (RASWT/BRAFWT, RASMutant, BRAFMutant, Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI67816DS1). The finding that approximately 50% of melanoma cell lines overexpress MERTK mRNA was confirmed by an independent analysis of microarray data obtained from melanoma cell lines available through the Cancer Cell Line Encyclopedia (CCLE); 46% (24 of 52) of cell lines overexpressed MERTK compared with normal skin and were independent of molecular subtype (Supplemental Figure 1, B and C and Supplemental Table 3). Based on the mRNA levels observed in the microarray analyses of Figure 2A, a subset of cell lines with either high or low MERTK mRNA were selected for MERTK protein analyses by Western blot. As shown in Figure 2B, most cell lines with high MERTK mRNA levels exhibited similarly high MERTK protein levels, and all cell lines with low MERTK mRNA exhibited low MERTK protein levels, including 2 isolates of normal human melanocytes (NHM17 and NHM18). These results validate the microarray data and support the notion that MERTK mRNA transcript levels correlate with protein abundance.
Figure 2. Melanoma cell lines frequently express MERTK irrespective of oncogenic mutational status.
(A) Microarray analysis of MERTK transcript levels in normal human melanocytes (NHM) and melanoma cell lines. Melanoma cell lines are grouped by oncogenic mutation status. Dotted line represents the average NHM MERTK transcript level. A complete list of melanoma cell lines analyzed is provided in Supplemental Table 2. (B) MERTK protein expression in melanoma cell lines. MERTK protein was detected in lysates from the indicated cell lines by Western blot. Actin was detected as a loading control.
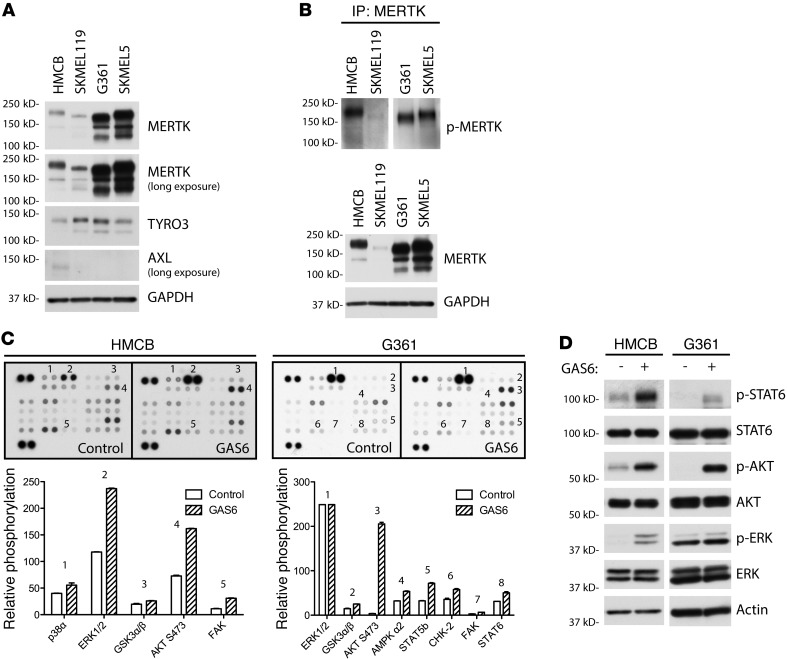
To study the role of MERTK in melanoma, 4 melanoma cell lines were selected that express MERTK and are representative of the most frequent melanoma molecular subtypes; G361 and SKMEL5 melanoma cells harbor the BRAFV600E activating mutation, SKMEL119 harbors the NRASQ61R mutation, and HMCB expresses wild-type BRAF and RAS proteins. Studying the role of MERTK in the context of BRAF and NRAS mutations is important from a clinical perspective, since approximately 50% and 20% of melanomas contain BRAF- and NRAS-activating mutations, respectively (22). To determine the expression of TAM-family RTKs, MERTK, TYRO3, and AXL protein expression was determined by Western blot analysis. As shown in Figure 3A, all 4 melanoma cell lines expressed TYRO3 and MERTK; only 1 cell line (HMCB) weakly expressed AXL. The multiple MERTK species observed are likely due to posttranslational modifications, most notably, glycosylation. MERTK has been described to be both heavily and differentially glycosylated through Asn-linked glycosylation (23, 24). Treatment of melanoma cell lysates with PNGase F resulted in 1 predominant lower molecular weight band (Supplemental Figure 2). To determine whether MERTK is active in these cells, phospho-MERTK levels in the 4 cell lines were identified. Immunoprecipitates were analyzed by Western blot using an antibody directed against the triphosphorylated MERTK activation loop (Figure 3B). All cell lines contained phosphorylated MERTK, suggesting that MERTK is an active receptor in these cells.
Figure 3. Melanoma cell lines express TAM receptors and can be activated to promote antiapoptotic and prosurvival signaling.
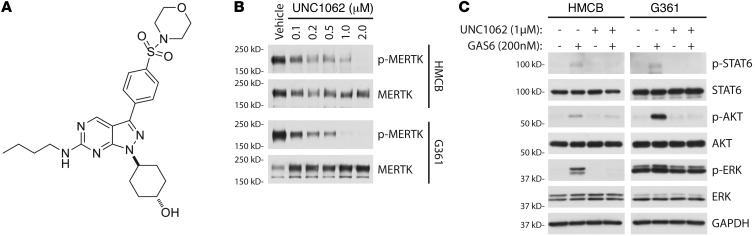
(A) TAM receptor expression in 4 metastatic melanoma cell lines. Lysates from HMCB, SKMEL119, G361, and SKMEL5 were analyzed by Western blot for MERTK, TYRO3, and AXL expression. GAPDH was detected as a loading control. (B) MERTK phosphorylation in melanoma cell lines. Subconfluent cultures were treated with pervanadate. MERTK was immunoprecipitated (IP) from whole-cell lysates then visualized by Western blot for phosphorylated MERTK. As a control, total MERTK protein was detected in whole-cell lysates corresponding to the protein load applied to the immunoprecipitation. (C) GAS6 activation of signaling pathways in melanoma cell lines. Phosphokinase array suggests p38, ERK1/2, GSK3α/β, AKT, AMPK, STAT5, CHK-2, FAK, and STAT6 as signaling molecules downstream of MERTK. Relative phosphorylation between control-treated and GAS6-treated cells is shown graphically for a subset of signaling molecules probed. (D) Western blot of phospho- and total STAT6, AKT, and ERK1/2. Cell cultures were stimulated with GAS6 and whole-cell lysates were evaluated by Western blot analysis. Actin was detected as a loading control.
To determine signaling pathways that are activated downstream of MERTK, a phosphokinase array was employed as an exploratory tool to examine alterations in kinase signaling in response to GAS6 stimulation. In HMCB cells, p38, ERK1/2, GSK3α/β, AKT, and FAK kinase were phosphorylated upon GAS6 treatment relative to control-treated cells (Figure 3C, left). Similarly, GSK3α/β, AKT, and FAK were activated in G361 cells (Figure 3C, right). In addition, AMPKα2, STAT5B, CHK2, and STAT6 were also stimulated in G361 cells relative to the control-treated cells. To confirm the phosphokinase array results, a subset of these signaling proteins was examined by Western blot. STAT6, ERK1/2, and AKT were indeed phosphorylated in both HMCB and G361 cell lines in response to GAS6 stimulation (Figure 3D). Similar trends were observed in SKMEL119 and SKMEL5 cells (Supplemental Figure 3). Overall, these data indicate that MERTK is frequently overexpressed in melanoma cells and that MERTK activation can regulate MAPK/ERK, PI3K/AKT, and JAK/STAT pathways.
MERTK silencing by shRNA inhibits oncogenic properties in melanoma cells.
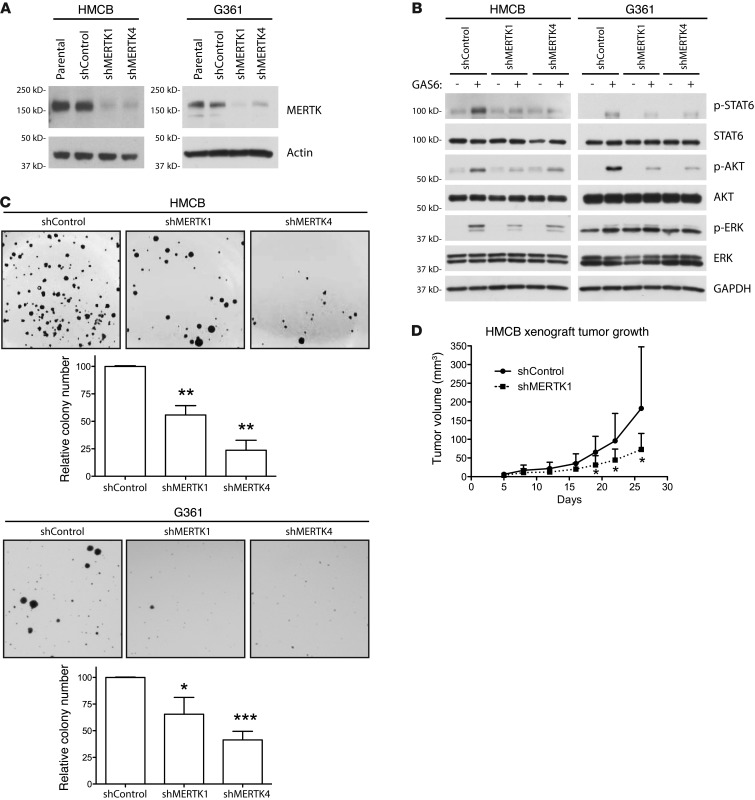
To further investigate the role of MERTK in oncogenic signaling and characterize the effects of long-term MERTK inhibition, we established MERTK knockdown melanoma cell lines. HMCB and G361 cells were transduced with one of two independent shRNAs targeting MERTK (shMERTK1 and shMERTK4) or an shRNA targeting an irrelevant gene (shControl) to produce stable MERTK knockdown derivative cell lines. As shown in Figure 4A, MERTK protein expression was reduced in cell lines expressing shMERTK constructs. This reduction in MERTK protein expression reduced GAS6-mediated downstream signaling through antiapoptotic and prosurvival signaling pathways including STAT6, AKT, and ERK1/2 (Figure 4B). The residual activation of these signaling proteins in GAS6-treated knockdown cells could be due to signaling mediated through other cell-surface TAM receptors or to residual MERTK expression. These results demonstrate that STAT6, AKT, and ERK1/2 phosphorylation in these melanoma cells is mediated by GAS6 activation of MERTK and that inhibition of MERTK with shRNA can attenuate survival and proliferation signaling.
Figure 4. MERTK inhibition by shRNA diminishes MERTK-mediated downstream signaling, colony-forming potential, and tumorigenesis.
(A) MERTK protein expression in cell lines stably expressing shRNA against MERTK. Whole-cell lysates from indicated cell lines were analyzed by Western blot for MERTK protein expression. Actin was detected as a loading control. (B) GAS6-mediated signaling in MERTK knockdown cell lines. shControl and MERTK knockdown cell lines were treated with GAS6 and downstream signaling was evaluated by Western blot. (C) Anchorage-independent colony-forming assays. Cell lines were grown in soft agar for 21 days (HMCB) or 14 days (G361) then stained and counted. Images from a representative experiment are shown. Mean values and standard errors were derived from at least 3 independent experiments. Statistically significant differences compared with shControl cells were determined using a Student’s 2-tailed t test (*P < 0.05; **P < 0.01; ***P < 0.001). (D) Tumor growth in HMCB xenograft mouse model. HMCB shControl and shMERTK1 cells were injected ectopically in mice. Tumor volume was monitored on the indicated days via caliper measurement, and the average tumor volume with standard deviation is plotted against the days after xenograft (*P < 0.05, n = 6).
To characterize the functional consequences of MERTK-mediated prosurvival and antiapoptotic signaling, the long-term effects of MERTK knockdown with shRNA were investigated. Using a soft agar assay, the role of MERTK in anchorage-independent growth of melanoma cells was evaluated. Both HMCB and G361 cell lines transduced with shMERTK constructs developed significantly fewer colonies in soft agar compared with shControl cell lines (Figure 4C). For HMCB, colony numbers decreased by 44% for shMERTK1 (P < 0.01) and by 76% for shMERTK4 (P < 0.01) compared with shControl (Figure 4C, left). Similarly, G361 colony numbers decreased by 35% for shMERTK1 (P < 0.05) and by 59% for shMERTK4 (P < 0.001) compared with shControl (Figure 4C, right). To determine whether MERTK inhibition via shRNA knockdown could mitigate melanoma tumorigenic potential in vivo, HMCB cells were injected subcutaneously into the flanks of SCID mice. At 26 days after implantation, HMCB shMERTK1 xenograft tumors had 60% smaller tumor volumes (73 mm3 mean tumor volume) compared with shControl tumors (183 mm3 mean tumor volume, P < 0.05) (Figure 4D). These in vitro and in vivo data indicate that MERTK plays important roles in the oncogenic/tumorigenic melanoma phenotype and suggest that MERTK is a therapeutic target in melanoma.
A novel MERTK tyrosine kinase inhibitor, UNC1062, inhibits MERTK-mediated signaling, promotes apoptosis, and inhibits colony formation in melanoma cells.
While activating mutations in BRAF and NRAS occur in melanoma at rates of 41% and 18%, respectively (25), lower mutation frequency or gene amplifications in other signaling molecules, such as RTKs, can also contribute to melanoma pathogenesis (26–28). UNC1062 was developed as a MERTK-selective tyrosine kinase inhibitor (Figure 5A). Its structure is based on a previously published pyrazolopyrimidine scaffold (29), and it has an improved affinity and specificity profile compared with its parent compound, UNC569 (Liu and Wang, unpublished observation). UNC1062 potently inhibits MERTK kinase activity in vitro (MERTK IC50 = 1.1 nM, Morrison Ki = 0.33 nM) and exhibits specificity within the TAM family (TYRO3 IC50 = 60 nM, AXL IC50 = 85 nM). Treatment of HMCB and G361 cells with increasing concentrations of UNC1062 resulted in a potent dose-dependent reduction in MERTK phosphorylation (Figure 5B).
Figure 5. UNC1062 is a novel MERTK inhibitor that reduces MERTK-mediated antiapoptotic and prosurvival downstream signaling.
(A) Chemical structure of UNC1062. (B) Inhibition of phospho-MERTK with UNC1062. Subconfluent cultures were treated with indicated doses of UNC1062 for 90 minutes followed by pervanadate treatment. MERTK was immunoprecipitated from whole-cell lysates then visualized by Western blot for phosphorylated MERTK and total MERTK. (C) Downstream signaling inhibition after administration of UNC1062. Cultures were serum starved then treated with UNC1062 for 90 minutes. After treatment with UNC1062, cells were stimulated with GAS6 and lysates were analyzed by Western blot for phospho- and total STAT6, AKT, and ERK1/2. GAPDH was detected as a loading control.
To assess the impact of pharmacological MERTK inhibition on downstream signaling, MERTK-mediated signaling was evaluated in the presence of UNC1062. HMCB and G361 cells were pretreated with 1 μM UNC1062 or vehicle for 90 minutes prior to stimulation with GAS6 or vehicle only. As shown in Figure 5C, cells treated with vehicle exhibited MERTK-mediated activation of STAT6, AKT, and ERK1/2. In contrast, treatment with UNC1062 greatly diminished activation of these signaling molecules.
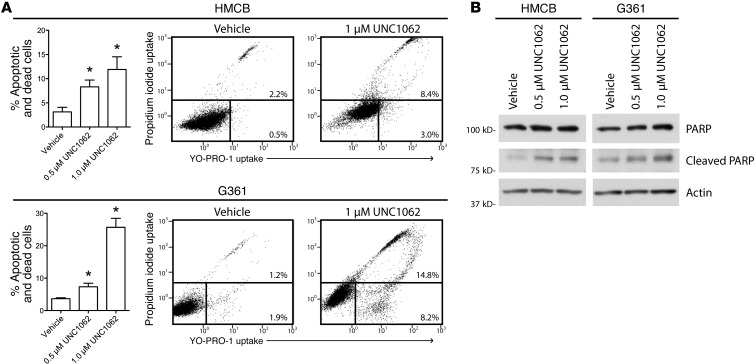
The consequence of inhibiting MERTK-mediated antiapoptotic and prosurvival signaling pathways was investigated by monitoring cell death. Induction of apoptosis in response to treatment with UNC1062 was measured by flow cytometric analysis of cells stained with YO-PRO-1 iodide and propidium iodide (PI), dyes that are selectively taken up by apoptotic (YO-PRO-1+) and/or dead (YO-PRO-1+, PI+) cells. HMCB cell death increased 9%, while G361 cell death increased 22% after treatment with UNC1062 compared with vehicle-treated cells (Figure 6A). Cell death following treatment with UNC1062 was confirmed by Western blot analysis showing increased PARP cleavage in both melanoma cell lines (Figure 6B). These data suggest that pharmacologic inhibition of MERTK can reduce oncogenic signaling and promote apoptosis in melanoma cells regardless of BRAF mutation status.
Figure 6. MERTK inhibition with UNC1062 increases apoptosis in melanoma cell lines.
(A) YO-PRO-1/PI staining and flow cytometric analysis. Cells were treated with the indicated doses of UNC1062 for 48 hours and then stained with YO-PRO-1 iodide (YO-PRO-1) and PI. Stained cells were analyzed on a flow cytometer to determine uptake of YO-PRO-1 and PI in apoptotic and dead cells. Mean values and standard errors were determined from 3 independent experiments. Statistically significant differences relative to vehicle only were determined using a Student’s 2-tailed t test (*P < 0.05). Representative scatter plots and percentages of early apoptotic (bottom right region) and dead (upper region) cells are shown for control- and UNC1062-treated cells. (B) PARP cleavage. Cells were treated with the indicated dose of UNC1062 for 24 hours and lysates were analyzed via Western blot.
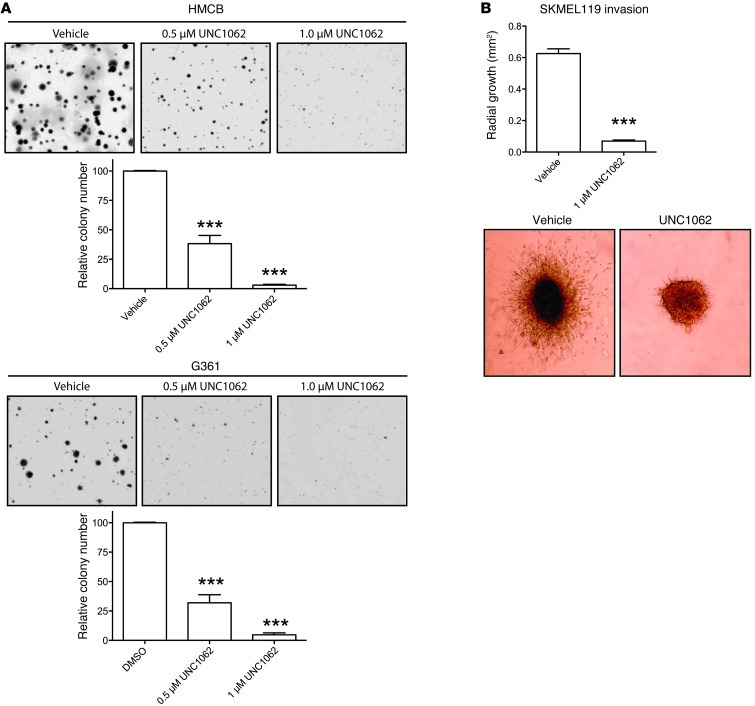
To determine whether pharmacologic MERTK inhibition can abrogate melanoma cell oncogenic properties, the effect of treatment with UNC1062 on colony formation was determined. For these studies, HMCB and G361 soft agar cultures were treated with UNC1062 or vehicle only. As shown in Figure 7A, treatment with 0.5 μM or 1.0 μM UNC1062 resulted in a statistically significant reduction in colony formation relative to vehicle-treated controls in both HMCB cells (62% and 97%, respectively) and G361 cells (68% and 95%, respectively).
Figure 7. MERTK inhibition reduces colony-forming potential and migratory and invasive capabilities.
(A) Anchorage-independent colony-forming assays. HMCB and G361 were grown in soft agar with the indicated doses of UNC1062. Images from a representative experiment are shown. Mean values and standard errors were derived from at least 3 independent experiments. Statistically significant differences were determined using a Student’s 2-tailed t test (***P < 0.001). (B) Invasion of SKMEL119. Cells were grown into spheroids and the spheroids were transplanted into collagen matrix and evaluated for invasion after 96 hours. Images from a representative experiment are shown. Original magnification, ×40. Bar graphs show means and standard errors derived from 6 spheroids (Student’s 2-tailed t test, ***P < 0.001).
MERTK inhibition reduces migration and invasion of melanoma cells.
To assess the impact of MERTK inhibition on migration and invasion, the SKMEL119 cell line, which has substantial growth and invasive capacity, was used. SKMEL119 cells express elevated MERTK levels that are markedly diminished by the expression of shMERTK4 (Supplemental Figure 4A). Time-lapsed video microscopy was used to measure cell migration of shControl and shMERTK4-expressing SKMEL119 cells across a fibronectin-coated surface. A statistically significant 30% decrease in the velocity of individual cell migration was observed in cells expressing shMERTK4 relative to shControl cells (Supplemental Figure 4B). Additionally, using a 3D collagen matrix invasion assay (30), spheroids of SKMEL119 treated with UNC1062 exhibited an 89% reduction in invasion (P < 0.001, Figure 7B). These results demonstrate that MERTK inhibition by either MERTK knockdown or treatment with UNC1062 reduces migration and invasion of melanoma cells.
Discussion
Despite the recent FDA approval of vemurafenib and ipilimumab, treatment of patients with metastatic melanoma remains challenging due to the primary resistance to chemotherapies and molecularly targeted therapies, the development of acquired resistance to BRAF inhibitors, or to the limited benefit of immunotherapies to a small subgroup of patients. Recent advances in next-generation sequencing of melanoma have confirmed earlier observations that only a handful of genes bear mutations or copy number alterations at a frequency of greater than 20% (31). Therefore, it is critical to identify other important potential targets in melanoma development and progression that are amenable to pharmacological inhibition. The studies presented here add melanoma to the growing list of malignancies in which MERTK is aberrantly expressed. We believe this work has led to several novel insights. First, MERTK expression is significantly elevated in distant metastatic tumors compared with primary melanomas. Second, MERTK is overexpressed in approximately half of melanoma cell lines, irrespective of BRAF and NRAS status, and is an active receptor. Third, targeting MERTK suppresses prosurvival pathways such as STAT6, AKT, and ERK1/2. Fourth, targeting MERTK suppresses colony-forming potential and migration. And fifth, targeting MERTK in vivo retards tumor growth in a human melanoma xenograft model.
The finding that MERTK expression is highest in distant metastatic melanomas compared with primary melanomas and the roles of MERTK in colony formation, migration, and invasion suggest that MERTK plays a role in the progression of primary melanomas and the development of distant metastases. Similar to the observations in this report, the migratory nature of glioblastoma cells could be reduced by MERTK inhibition with either shRNA knockdown or a MERTK monoclonal antibody (15), suggesting that increased MERTK expression may contribute to outgrowth of the metastatic tumor.
In agreement with a previous report demonstrating phosphorylation of TAM family receptors in several melanoma cell lines (16), the data presented here confirm that MERTK can be phosphorylated in melanoma cells and further show that MERTK is functionally important for several oncogenic signaling pathways and phenotypes. Specifically, we report here on MERTK-mediated signaling through the MAPK/ERK, PI3K/AKT, and JAK/STAT signaling pathways. The mechanism of MERTK activation in melanoma cells is not clear, but Sensi et al. have previously described melanoma cell expression and secretion of GAS6, the common ligand for all members of the TAM family of proteins, suggesting a method of autocrine and/or paracrine activation of MERTK (18). Since expression of MERTK by melanoma cells increases during progression from primary to metastatic melanoma, it would be interesting to determine whether corresponding increases in GAS6 levels occur in serum from patients with metastatic melanoma, implicating serum GAS6 levels as a potential early marker of melanoma progression, as in other cancers (32).
MAPK/ERK and PI3K/AKT are 2 of the most frequently dysregulated pathways in melanoma (31). These 2 pathways not only play a role in melanoma development and progression, but are also involved in primary and secondary resistance to BRAF inhibitors (5–8). The observation that MERTK signals via both pathways, as well as through others whose roles in melanoma biology are currently unclear (e.g., STAT6), not only highlights the complex regulation of these pathways by membrane receptors, such as MERTK, but may also provide a therapeutic advantage, since targeting MERTK may disrupt signaling in multiple pathways. These observations and the data presented here suggest that MERTK-targeted therapies could potentially be considered for patients, irrespective of BRAF and NRAS status and/or prior treatment with BRAF inhibitors.
MERTK is expressed at high levels in melanoma-infiltrating CD68+ cells, and the role of MERTK in this context warrants further investigation in view of early observations that MERTK knockout mice frequently exhibit autoimmune phenomena due to their inability to engulf and efficiently clear apoptotic cells (20). Since the development of autoimmunity has been associated with clinical benefit in melanoma (33), it is tempting to speculate that increased expression of MERTK in melanoma-infiltrating macrophages increases the efficiency of macrophages to clear apoptotic melanoma cells and thereby limits the time for antigens released by dying cells to stimulate the immune system to develop an effective antitumor response. With respect to this hypothesis, targeting MERTK may potentially have a dual antitumor role; first by directly inhibiting migration, invasion, and growth of tumor cells, and also by an indirect immunomodulatory role.
To our knowledge, this study is the first to characterize roles for MERTK in melanoma and provide proof-of-principle studies toward establishing MERTK as a therapeutic target in melanoma by using the MERTK-specific small-molecule inhibitor, UNC1062. The increased expression of MERTK in a large (greater than 30%) number of metastatic melanomas, its potent effect in migration, invasion, and colony formation that is unrelated to the presence of BRAF and NRAS mutations, its signaling via multiple intracellular pathways that are known to be oncogenic, its potential roles in other host cells, and the development of highly specific small-molecule inhibitors provide the rationale and the means for continued development of a MERTK-targeted therapeutic agent for the treatment of malignant melanoma.
Methods
Melanoma tissue microarrays and other tissues.
The previously described nevus-to-melanoma progression TMA was stained to assess the expression of MERTK in nevi, primary, and metastatic melanomas (19). The University of North Carolina (UNC69A1) metastatic melanoma TMA was created from specimens obtained from the UNC Hospital surgical pathology archive under the protocol approved by the UNC Institutional Review Board (08-0242). TMA was stained to assess the expression of MERTK in mononuclear as well as melanoma cells in melanomas metastatic to brain (n = 11), lung (n = 8), liver (n = 4), and stomach (n = 1). The UNC69A1 TMA was constructed from representative tumor cores (0.6-mm) spotted in triplicate, and 4-micron-thick sections were used for further analysis. Formalin-fixed paraffin-embedded tumor blocks from patients who underwent craniotomy for melanoma brain metastases at the University of Pittsburgh Medical Center (UPMC) were retrieved under the University of Pittsburgh Cancer Institute’s (UPCI) IRB-approved protocol 10-005 and 5-micron sections were available for further analysis.
Immunofluorescence and immunohistochemistry.
Dual immunofluorescent stains of TMA slides with antibody pairs of either MERTK-S100 or MERTK-CD68 were carried out using the Bond fully automated slide-staining system (Leica Microsystems). Slides were de-paraffinized in Bond dewax solution (AR9222) and hydrated in Bond wash solution (AR9590). Antigen retrieval for all antibodies was performed for 30 minutes at 100°C in Bond epitope retrieval solution 1 (AR9961, pH 6.0). Primary antibodies were applied sequentially and slides were incubated with the MERTK antibody (Epitomics Y323, 1:1,000), followed by Bond polymer (DS9800) and the tyramide Cy5 amplification step to visualize MERTK (PerkinElmer). After completion of MERTK staining, the primary antibodies against S100 (Dako) or CD68 (Leica Microsystems) were applied. S100 was detected with Alexa555-labeled goat anti-rabbit secondary antibody (Invitrogen) and CD68 with goat anti–mouse-HRP (EnVision+, Dako), followed by tyramide Cy3 (PerkinElmer). Nuclei were stained with Hoechst 33258 (Invitrogen). The stained slides were mounted with ProLong Gold antifade reagent (Molecular Probes, Inc.). For immunohistochemistry slides were incubated with MERTK antibody (Epitomics, 1:500) followed by Bond Polymer Refine Red Detection (DS9390), including hematoxylin counterstain.
Imaging analyses.
AQUA analysis software, version 2.2 (HistoRx) was used to assess MERTK expression in S100-positive cells. Briefly, high-resolution acquisition (×20 objective) of TMA immunofluorescence (IF) slides in the DAPI (Hoechst 33258), Cy3 (S100), and Cy5 (MERTK) channels was performed using a PM-2000 (HistoRx), as described (34). The AQUA algorithm was used to score the expression of MERTK in an S100 mask, a melanocyte lineage marker. Monochromatic and merged high-resolution (512 × 512 pixel, 1-μm resolution) images for each spot were generated. Tissue Studio analysis (Definiens Architect XD v2.0.4, Tissue Studio v3.5) was used to assess the percentage of MERTK expression in melanoma-infiltrating CD68+ cells. Briefly, a TMA IF slide was scanned in Aperio FL (Aperio Technologies) through DAPI (Hoechst 33258), Cy3 (CD68), and Cy5 (MERTK) channels using a ×20 objective. The Tissue Studio image analysis solution was then applied using the DAPI channel to define the cell nucleus and the Cy5 channel to define the cell membrane. The ratio of cells that coexpress DAPI, Cy3, and Cy5 divided by the cells that coexpress DAPI and Cy3 was used to calculate the percentage of MERTK+CD68+ melanoma-infiltrating cells.
Gene expression analysis.
Raw microarray gene expression profiles were obtained from the NCBI Gene Expression Omnibus (GEO) (35) for GSE36133 (36) and GSE7553 (21) datasets. Robust multiarray average (RMA) (37) was performed on these Affymetrix datasets to extract signal intensities and perform data normalization using the Affymetrix Power Tools program. The GSE36133 dataset was analyzed independently and the 5 normal skin samples were normalized with the 61 melanoma cell lines from GSE7553 using RMA. Mutation data for the 61 melanoma cell lines were assessed by examination of the CCLE project website (36) for each cell line; data for duplicate cell lines were averaged and cell lines without mutation data were excluded from further analysis. We also obtained the normalized data for GSE40047 (38), profiled by Agilent-014850 Whole Human Genome Microarray 4x44K G4112F. Melanoma cell lines or normal melanocyte lines were labeled as Cy5, and reference RNA was labeled as Cy3. The normalized/processed data obtained from the GEO for this dataset were expressed as –log10(Cy3/Cy5). The arrays were scanned and log2 ratios were generated for each gene. The data were median normalized by subtracting out the median log-ratio for each array so that each normalized array had a median log-ratio of 0. Values for the gene of interest were determined and a constant was added to each gene value to adjust the lowest value of each array to 0. Each value was then used as an exponent to 2 (i.e., 2x) to transform the values into fold change from the log2 value generated from the arrays. This set the lowest value in each dataset to 1, and all of the other values represented the fold change difference from the lowest value. Mutational status for this dataset was obtained from the GEO. MERTK expression was extracted and plotted across cell lines for comparison.
Cell culture and reagents.
HMCB was propagated in RPMI medium supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml). G361, SKMEL5, and SKMEL119 were cultured in DMEM and supplemented as described above. HMCB, G361, SKMEL5, and A2058 identities were verified by short tandem repeat (STR) analysis and matched ATCC product information. At this time there is no STR profile publicly available for SKMEL119, WM1232, SKMEL27, SKMEL23, UACC257, SKMEL24, SKMEL130, SKMEL131, SKMEL23, SKMEL178, or SKMEL186; however, the STR profiles are singular and do not match any in the ATCC or DSMZ collections. Pervanadate solution containing 10 nM sodium orthovanadate and 0.15% H2O2 was prepared. UNC1062 inhibitor was synthesized based on a published patent procedure by Wang et al. (39), and stock solutions were prepared in DMSO at 3 mM.
Western blot and signaling assays.
Whole-cell lysates were prepared in lysis buffer containing 50 mM HEPES pH 7.5, 150 mM NaCl, 10 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM Na3VO4, 0.1 mM Na2MoO4, 1 mM NaF, and protease inhibitors (Roche). Equal protein amounts were separated on 8% Tris-glycine SDS-PAGE then transferred to PVDF or nitrocellulose membranes. Antibodies were applied according to the manufacturer’s specifications (Supplemental Table 4). For GAS6 stimulation studies, cells (4 × 105) were incubated overnight and then starved for 3 hours in serum-free medium prior to activation with either 200 nM GAS6 (885-GS; R&D Systems) or control buffer for 10 minutes. UNC1062 signaling inhibition studies were conducted in a similar manner but included the addition of drug after 2 hours of serum starvation and treatment for 90 minutes prior to activation with GAS6. Due to the short lifetime of MERTK phosphorylation in vitro, a pervanadate treatment was used only to observe levels of phospho-MERTK. For phospho-MERTK assays, subconfluent cultures were treated with pervanadate solution for 3 minutes, MERTK was immunoprecipitated from whole-cell lysates (MAB8912; R&D Systems) then visualized with Western blot using p-MERTK antibody (PhosphoSolutions) (11). PNGase F was conducted according to the manufacturer’s instructions (P0704; New England BioLabs). All Western blots are a representative example of at least 3 independent experiments. The phosphokinase array was performed once according to the manufacturer’s instructions (ARY003; R&D Systems).
Stable shRNA cell line production.
Constitutive shRNA cell lines (HMCB, SKMEL119, G361 shMERTK1, and shMERTK4) were developed as previously described (11). Control cell lines were produced in a similar manner with either an shRNA against the human ERBB4 gene (HMCB and SKMEL119) or GFP (G361).
Colony formation assays.
Cells were plated in approximately 0.3% agar over a higher percentage agar base layer. Agar was overlaid with medium (RPMI for HMCB and McCoys for G361). Colonies were grown for 14 to 21 days prior to staining with 1 mg/ml nitrotetrazolium blue (Sigma-Aldrich) and subsequent counting. Medium and UNC1062 were renewed twice weekly. Medium was renewed after 7 days for shRNA experiments.
Mouse xenograft models.
HMCB shControl and shMERTK1 cells (3 × 106 in 100 μl) were injected subcutaneously into the flanks of 8- to 9-week-old female SCID mice (n = 6 per group). Tumor growth was monitored twice weekly using calipers, and tumor volume was calculated using a modified ellipsoid model (π/6 × length × width × depth).
Apoptosis assays.
Cells (1 × 105) were plated and incubated overnight prior to treatment with UNC1062 or an equivalent concentration of DMSO. After 48 hours of treatment, cells were lifted with 0.02% EDTA in PBS and combined with cell culture supernatant. Cells were collected by centrifugation and resuspended in PBS containing 0.5 μM YO-PRO-1 (Y3603; Invitrogen) and 0.5 μg/ml propidium iodide (PI) (P3566; Invitrogen). YO-PRO-1 and PI uptake was analyzed using a Beckman Coulter Gallios flow cytometer and CXP and/or Kaluza software. Cells were plated and treated in the same manner for PARP cleavage assays, but cells were lysed after collection.
Migration assays.
SKMEL119 cells were plated on MatTek dishes coated with 10 μg/ml of fibronectin in PBS and allowed to attach overnight. Cells were subsequently equilibrated in a VivaView FL microscope (Olympus) live-cell imaging chamber at 37°C for 1 hour followed by time-lapse imaging of 25 viewing fields from each dish. Images were taken of each field every 40 minutes for 16 hours (original magnification, ×200) and were converted to stacks using ImageJ software (NIH). Single-cell velocity was measured with ImageJ software (NIH) using the Manual Tracking plug-in. Cells that divided, migrated out of field of view, or touched other cells more than transiently were excluded from the analysis.
Invasion assays.
SKMEL119 spheroids were grown by plating cells (5 × 103) in DMEM supplemented with 30% FBS in a 96-well plate coated with 1.5% noble agar. After 72 hours, spheroids were harvested then implanted into a gel of bovine collagen I containing 1× EMEM supplemented with 10% FBS and DMSO or UNC1062 in 24-well plates. DMEM containing either DMSO or drug was layered over the solidified collagen. Spheroids were allowed to invade the collagen for 96 hours before images were taken. Relative units of invasive growth were determined using area measurement tools in Adobe Reader 9 Pro and subtracting initial spheroid size. Experiments were performed 3 times, with 6 spheroids/invasions analyzed from duplicate wells for each condition in each experiment.
Statistics.
For the analysis of TMA data, R package and GraphPad Prism 5 (GraphPad) software were used. The MERTK TMA AQUA scores were log transformed for normality. A 1-way ANOVA was performed to compare the association between nevi, primary melanomas, and metastatic melanomas. Linear regression was fitted to MERTK expression to assess the significance of the trend in MERTK expression across different stages of nevus-to-melanoma progression.
Data from all in vitro experiments are reported as the mean ± SEM unless otherwise noted. Data were analyzed and presented using GraphPad Prism 5 software (GraphPad). Significance was determined using a Student’s 2-tailed t test unless otherwise noted.
Results were considered significant at P values less than 0.05 and are labeled with a single asterisk. In addition, P values less than 0.01 and less than 0.001 are designated with double and triple asterisks, respectively.
Study approval.
The UNC metastatic melanoma TMA (UNC69A1) was developed under the UNC’s IRB-approved protocol 08-0242 (Chapel Hill, North Carolina, USA). Melanoma brain metastasis samples were collected at the UPMC and were retrieved under the UPCI’s IRB-approved protocol 10-005 (Pittsburgh, Pennsylvania, USA). Animal experiments were performed under an approved UNC Institutional Animal Care and Use Committee protocol. (Chapel Hill, North Carolina, USA).
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Cancer Institute (contract no. HHSN261200800001E), American Cancer Society (RGS082910121B, to D.K. Graham), National Institutes of Health (RO1CA137078, to D.K. Graham; CA115888 and ES014635, to J. Shields), and the American Brain Tumor Basic Research Fellowship in honor of Mark Linder (to M.J. Sambade). The UNC Translational Pathology Laboratory is supported in part by grants from the National Cancer Institute (3P30CA016086) and the UNC University Cancer Research Fund (UCRF). The authors wish to thank Randall Wong of the University of Colorado DERC Molecular Biology Core (NIH P30 DK57516) for cell line authentication; Christine Childs of the University of Colorado Cancer Center Flow Cytometry Shared Resource for technical assistance; and Charlene Santos of the UNC-CH LCCC Animal Studies Facility for conducting animal experiments using human melanoma xenografts.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Citation for this article:J Clin Invest. 2013;123(5):2257–2267. doi:10.1172/JCI67816.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24(26):4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 5.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johannessen CM, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468(7326):968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Atefi M et al. Reversing melanoma cross-resistance to BRAF and MEK inhibitors by co-targeting the AKT/mTOR pathway. PLoS One. 2011612e28973. 10.1371/journal.pone.0028973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poulikakos PI, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature. 2011;480(7377):387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham DK, et al. Ectopic expression of the proto-oncogene Mer in pediatric T-cell acute lymphoblastic leukemia. Clin Cancer Res. 2006;12(9):2662–2669. doi: 10.1158/1078-0432.CCR-05-2208. [DOI] [PubMed] [Google Scholar]
- 11.Linger RMA, et al. Oncogene. Mer or Axl receptor tyrosine kinase inhibition promotes apoptosis, blocks growth and enhances chemosensitivity of human non-small cell lung cancer [published online ahead of print August 13, 2012]. doi: 10.1038/onc.2012.355 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y-M, Robinson DR, Kung H-J. Signal pathways in up-regulation of chemokines by tyrosine kinase MER/NYK in prostate cancer cells. Cancer Res. 2004;64(20):7311–7320. doi: 10.1158/0008-5472.CAN-04-0972. [DOI] [PubMed] [Google Scholar]
- 13.Keating AK, et al. Inhibition of Mer and Axl receptor tyrosine kinases in astrocytoma cells leads to increased apoptosis and improved chemosensitivity. Mol Cancer Ther. 2010;9(5):1298–1307. doi: 10.1158/1535-7163.MCT-09-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linger RMA, Keating AK, Earp HS, Graham DK. TAM receptor tyrosine kinases: biologic functions, signaling, and potential therapeutic targeting in human cancer. Adv Cancer Res. 2008;100:35–83. doi: 10.1016/S0065-230X(08)00002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers AEJ, et al. Mer receptor tyrosine kinase inhibition impedes glioblastoma multiforme migration and alters cellular morphology. Oncogene. 2011;31(38):4171–4181. doi: 10.1038/onc.2011.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tworkoski K, et al. Phosphoproteomic screen identifies potential therapeutic targets in melanoma. Mol Cancer Res. 2011;9(6):801–812. doi: 10.1158/1541-7786.MCR-10-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu S, et al. A genomic screen identifies TYRO3 as a MITF regulator in melanoma. Proc Natl Acad Sci USA. 2009;106(40):17025–17030. doi: 10.1073/pnas.0909292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sensi M, et al. Human cutaneous melanomas lacking MITF and melanocyte differentiation antigens express a functional Axl receptor kinase. J Invest Dermatol. 2011;131(12):2448–2457. doi: 10.1038/jid.2011.218. [DOI] [PubMed] [Google Scholar]
- 19.Nazarian RM, Prieto VG, Elder DE, Duncan LM. Melanoma biomarker expression in melanocytic tumor progression: a tissue microarray study. J Cutan Pathol. 2010;37:41–47. doi: 10.1111/j.1600-0560.2010.01505.x. [DOI] [PubMed] [Google Scholar]
- 20.Scott RS, et al. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature. 2001;411(6834):207–211. doi: 10.1038/35075603. [DOI] [PubMed] [Google Scholar]
- 21.Riker AI, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Medical Genomics. 2008;1(1):13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wellbrock C, et al. V599EB-RAF is an oncogene in melanocytes. Cancer Res. 2004;64(7):2338–2342. doi: 10.1158/0008-5472.CAN-03-3433. [DOI] [PubMed] [Google Scholar]
- 23.Migdall-Wilson J, et al. Prolonged exposure to a Mer ligand in leukemia: Gas6 favors expression of a partial Mer glycoform and reveals a novel role for Mer in the nucleus. PLoS One. 2012;7(2):e31635. doi: 10.1371/journal.pone.0031635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sather S, et al. A soluble form of the Mer receptor tyrosine kinase inhibits macrophage clearance of apoptotic cells and platelet aggregation. Blood. 2007;109(3):1026–1033. doi: 10.1182/blood-2006-05-021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee J-H, Choi J-W, Kim Y-S. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol. 2011;164(4):776–784. doi: 10.1111/j.1365-2133.2010.10185.x. [DOI] [PubMed] [Google Scholar]
- 26.Gray-Schopfer V, Wellbrock C, Marais R.Melanoma biology and new targeted therapy. Natur e. 20074457130851–857. 10.1038/nature05661 [DOI] [PubMed] [Google Scholar]
- 27.Miller AJ, Mihm MC. Melanoma. N Engl J Med. 2006;355(1):51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 28.Dutton-Regester K, Hayward NK. Reviewing the somatic genetics of melanoma: from current to future analytical approaches. Pigment Cell Melanoma Res. 2012;25(2):144–154. doi: 10.1111/j.1755-148X.2012.00975.x. [DOI] [PubMed] [Google Scholar]
- 29.Liu J, et al. Discovery of small molecule Mer kinase inhibitors for the treatment of pediatric acute lymphoblastic leukemia. ACS Med Chem Lett. 2012;3(2):129–134. doi: 10.1021/ml200239k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambade MJ, et al. Melanoma cells show a heterogeneous range of sensitivity to ionizing radiation and are radiosensitized by inhibition of B-RAF with PLX-4032. Radiother Oncol. 2011;98(3):394–399. doi: 10.1016/j.radonc.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gustafsson A, et al. Differential expression of Axl and Gas6 in renal cell carcinoma reflecting tumor advancement and survival. Clin Cancer Res. 2009;15(14):4742–4749. doi: 10.1158/1078-0432.CCR-08-2514. [DOI] [PubMed] [Google Scholar]
- 33.Gogas H, et al. Prognostic significance of autoimmunity during treatment of melanoma with interferon. N Engl J Med. 2006;354(7):709–718. doi: 10.1056/NEJMoa053007. [DOI] [PubMed] [Google Scholar]
- 34.Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8(11):1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- 35. Barrett T et al. NCBI GEO: archive for functional genomics data sets —10 years on. Nucleic Acids Res. 201139Database issueD1005–D1010. 10.1093/nar/gkq1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 38.Carson C, et al. A prognostic signature of defective p53-dependent G1 checkpoint function in melanoma cell lines. Pigment Cell Melanoma Res. 2012;25(4):514–526. doi: 10.1111/j.1755-148X.2012.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang X, Liu J, Yang C, Zhang W, Frye S, Kireev D, inventors; The University of North Carolina at Chapel Hill, assignee. Pyrazolopyrimidine compounds for the treatment of cancer. US patent WO/2011/146313. November 24, 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.