Abstract
Background
Plant cell culture represents an alternative source for producing high-value secondary metabolites including paclitaxel (Taxol®), which is mainly produced in Taxus and has been widely used in cancer chemotherapy. The phytohormone methyl jasmonate (MeJA) can significantly increase the production of paclitaxel, which is induced in plants as a secondary metabolite possibly in defense against herbivores and pathogens. In cell culture, MeJA also elicits the accumulation of paclitaxel; however, the mechanism is still largely unknown.
Methodology/Principal Findings
To obtain insight into the global regulation mechanism of MeJA in the steady state of paclitaxel production (7 days after MeJA addition), especially on paclitaxel biosynthesis, we sequenced the transcriptomes of MeJA-treated and untreated Taxus × media cells and obtained ∼ 32.5 M high quality reads, from which 40,348 unique sequences were obtained by de novo assembly. Expression level analysis indicated that a large number of genes were associated with transcriptional regulation, DNA and histone modification, and MeJA signaling network. All the 29 known genes involved in the biosynthesis of terpenoid backbone and paclitaxel were found with 18 genes showing increased transcript abundance following elicitation of MeJA. The significantly up-regulated changes of 9 genes in paclitaxel biosynthesis were validated by qRT-PCR assays. According to the expression changes and the previously proposed enzyme functions, multiple candidates for the unknown steps in paclitaxel biosynthesis were identified. We also found some genes putatively involved in the transport and degradation of paclitaxel. Potential target prediction of miRNAs indicated that miRNAs may play an important role in the gene expression regulation following the elicitation of MeJA.
Conclusions/Significance
Our results shed new light on the global regulation mechanism by which MeJA regulates the physiology of Taxus cells and is helpful to understand how MeJA elicits other plant species besides Taxus.
Introduction
Taxus, also known as yew, is a genus of gymnosperm and contains at least 14 species [1]. Taxus can produce a number of chemicals with pharmaceutical properties. Among them, paclitaxel (Taxol®) was reported to be a very promising anticancer drug in 1964 [2] and now is used widely in chemotherapy treatment of lung, ovarian, and breast cancer [3]. Paclitaxel was originally extracted from the bark of Taxus brevifolia but yields are low; approximately 0.01% of the dry bark weight [4]. With growing demand for the paclitaxel for medical use, harvesting from bark cannot meet the demand and also raises serious ecological concerns.
Great efforts have been taken to increase the production of paclitaxel by finding alternative sources and methods of synthesis including Taxus needles [4], [5] fungal sources [4], [5], developing semisynthesis method from related taxanes [6], and using plant cell cultures [7]. Due to the high demand, a combined method of production is mostly used in which an abundant natural intermediate metabolite compound is isolated from natural sources (yew bark and needle) or by plant cell culture, and then using semi-synthesis methods to obtain paclitaxel.
Plant cell culture has the potential for producing large quantities of paclitaxel but currently suffers from low yields. Some compounds are known to stimulate the production of paclitaxel when added to the medium. Among them, methyl jasmonate (MeJA) has been found to significantly elicit the production of paclitaxel in the cultured cells of T. × media, T. canadensis, T. cuspidata etc. [8], [9], [10] with the greatest accumulation of paclitaxel, 36.0 mg/L, observed when the cultured cells were elicited with 200 µM MeJA [9]. However, the addition of MeJA in Taxus cell culture also retards cell growth [10] and therefore reduces indirectly the yield of paclitaxel in culture. A detailed knowledge of the biosynthesis of paclitaxel and its regulation by MeJA is required before directed bioengineering methods could be implemented to increase paclitaxel yields in cell culture.
Next generation sequencing technologies have been proved to be rapid and cost-effective means to analyze the genome and transcriptome in non-model species [11], [12], [13]. Improvement in de novo assembly of high-throughput sequencing data and relative accurate estimation of gene expression levels makes this approach also powerful in quantifying gene expression [14], [15], [16]. This technology has been applied to attempt to understand varying aspects of paclitaxel synthesis in Taxus [17], [18], [19]. Hao et al [17] assembled a de novo transcriptome of T. mairie and sequenced three different tissues (root, stem and leaves) using a tag-based digital gene expression system, and found a number of genes involved in tissue specific functions. Wu et al. [18] sequenced the transcriptome of T. cuspidata needles using 454 pyrosequencing. The early response of elicitation with MeJA has been studied by Li et al [19] who found 13,469 differentially expressed genes in T. chinensis, whereas Lenka et al [20] used PCR-based suppression subtractive hybridization (SSH) to find 300 differentially expressed genes between non-elicited and MeJA elicited T. cuspidate cell suspension.
To get a comprehensive understanding of the global regulation mechanism of MeJA on paclitaxel biosynthesis in steady state of paclitaxel production, we sequenced the transcriptomes of Taxus × media cells after 7 days without or with the elicitation of MeJA (Figure S1) when the maximum paclitaxel accumulation is observed [9], [10]. Comparing the gene expression profiles of these two cells revealed that 18 out of all 29 known genes in terpenoid backbone and paclitaxel biosynthesis had higher transcript levels by addition of MeJA. In addition to the increased transcript abundance of specific transcription factors and chromatin modification proteins, we found decreased transcript levels of genes involved in cell cycle, DNA replication, meiosis, and mismatch repair, which provides insights into the mechanism by which MeJA both inhibits cell growth while significantly inducing the production of paclitaxel.
Results
Illumina Sequencing and de novo Assembly of Sequence Reads in Taxus × media
Illumina high-throughput second generation sequencing was carried out to obtain the transcriptomes of Taxus × media cell cultures under nonelicited and elicited conditions. The sequencing produced 77,395,444 reads with each 75 base and with the total nucleotides (nt) of 5,804,658,300 nt (5.8 Gb): i) non-elicited T. × media transcriptome produced 37,323,590 reads (2.8 Gb); ii) T. × media elicited with MeJA produced 40,071,854 reads (3.0 Gb) nt. After removing the adaptor sequences, empty reads and low quality sequences (reads with more than 10% Q20 bases), we obtained 16,340,898 and 16,153,572 high quality reads from the nonelicited and elicited cultures, respectively. The sequence data have been submitted to NCBI Sequence Read Archive (SRA) database under the accession number SRX156706 and SRX156707.
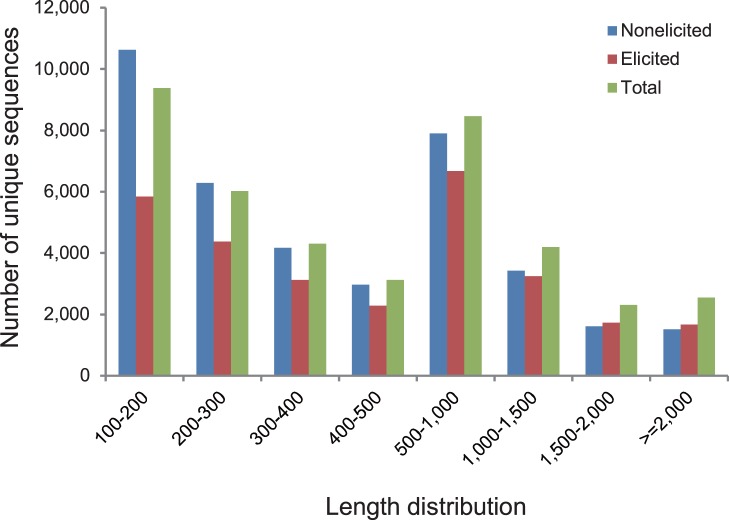
The assembly was independently carried out for each sample using ABySS [14]. From the nonelicited culture, a total of 38,506 contigs with length not less than 100 nt were generated; the N50 size and the mean size are 976 and 581 nt, respectively. Figure 1 shows the length distribution of contigs ranging from 100 nt to more than 2000 nt. From the elicited culture, 28,959 contigs have the N50 size of 1,142 nt and the mean size of 689 nt. We obtained a total of 40,348 unique sequences by further assembly from the two datasets with the N50 size of 1,193 nt and the mean size of 684 nt, of which 1368 probably are complete cDNAs.
Figure 1. Length distribution of contigs from the nonelicited and MeJA elicited cultures, and the integrated data from both cultures (Total) assembled by using ABySS and CAP3.
The y-axis indicates the number of unique sequences with certain length. The x-axis indicates the length range of unique sequences.
Functional Prediction of Unique Sequences by BLAST Search Against GenBank nr Protein Databases
To gain a preliminary insight into the functions of the unique sequences obtained, we performed BLASTX search against GenBank non-redundant (nr) using E-value 10−5 as a cutoff value. 21,652 out of 40,348 unique sequences (53.7%) were detected to have homologs. We found 35 and 74 unique sequences with the relatively high abundance of more than 1000 transcripts per million (relative abundance) in the nonelicited and elicited cells, respectively (Table S1), among which ribosomal protein S2 and gibberellin 3-beta-dioxygenase 4-like protein are highly expressed in both datasets. Compared with the nonelicited culture, the elicited culture contains unique sequences encoding the enzymes in pathogenesis and ethylene biosynthesis such as ACC oxidase. These abundantly expressed genes in cultured cells are distinct from those in needles, which include genes encoding ribulose bisphosphate carboxylase (RuBisCO) and other proteins that involved in photosynthesis [18]. Interestingly, we note multiple sequences with unknown annotation. After domain search against PROSITE database (http://prosite.expasy.org), only one (Contig29843) was found containing putative domains. It would be intriguing to investigate what functions other sequences perform, given the high levels that these genes possess.
COG and GO Classification and KEGG Pathway Enrichment of Unique Sequences
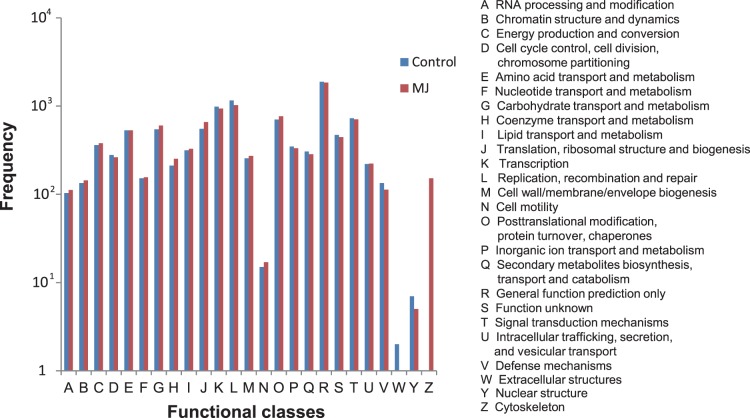
To facilitate the functional classification of the unique sequences, we employed their homologs’ functional classification information in COG database, which emphasizes a classification of genes in an evolutionary view using complete genome sequences [21]. According to COG database, all the unique sequences were classified into 25 different functional classes, which were represented by A to Z (Figure 2). About 19% and 23% unique sequences from the non-elicited cells and the elicited cells respectively were found homologs in COG database. The unique sequence frequency in each functional class varies from 0 to 1,886 in nonelicited cells and from 0 to 1,842 in the elicited cells. Compared with the nonelicited cells, the elicited cells showed more unique sequences in RNA processing and modification (Figure 2A, 5%), amino acid transport and metabolism (Figure 2E, 3%), and secondary metabolite biosynthesis, transport and catabolism (Figure 2Q, 9%) but have less expressed genes in other functional classes (from 1% to 24%).
Figure 2. Histogram presentation of clusters of orthologous groups (COG) classification in nonelicited and MeJA elicited culture.
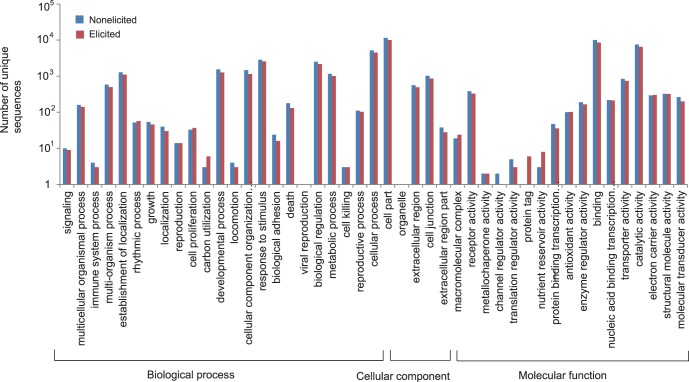
Figure 3. Histogram of Gene ontology (GO) classification of unique sequences in nonelicited and MeJA elicited cells.
Results are categorized into three GO ontologies, biological process, cellular component, and molecular function. The classes without any unique sequences both in the nonelicited and elicited cells were removed from the figure.
After obtaining putative gene functions assigned by homology searches, we predicted the biological processes (26% and 30% unique sequences in non-elicited and elicited cells), cellular components (27% and 41% unique sequences in non-elicited and elicited cells) and molecular functions (30% and 46% unique sequences in non-elicited and elicited cells) that the proteins belong to, by association with GO information using the obtained annotation from GenBank nr database (Figure 3). The results showed that most proteins encoded by these unique sequences take part in the biological processes including response to stimulus, biological regulation, developmental process and metabolic process. As for the cellular component domain, a majority of products of unique sequences lie in cell part, cell junction, and extracellular region. For the molecular function domains, genes involved in binding and catalytic activity are dominant. Notably, almost all the classes have more unique sequences in the nonelicited cells than in the elicited cells (Figure 3).
To understand the metabolism pathways in nonelicited and elicited T. × media cells, we performed BLASTX search against KEGG database to map the unique sequences to canonical pathways. A total of 2,739 and 2,605 unique sequences in the elicited (9%) and non-elicited (7%) cells refer to 137 and 135 KEGG pathways, respectively (Table S2). Interestingly, the largest unique pathways in the two datasets include spliceosome (5%), ribosome (4%), protein processing in endoplasmic reticulum (3%), and purine metabolism (3%) (Table S2). Compared with the nonelicited cells, the elicited cells have slightly more unique sequences in the pathways including glycolysis/gluconeogenesis, DNA replication, mismatch repair, diterpenoid biosynthesis, etc. (Table S2).
Comparison of the Expression Levels of Genes between Nonelicited and Elicited T. × media Cells
To investigate the response of elicitation with MeJA in T. × media cultured cells, we estimated the expression levels of each unique sequence in the two cultures by using RSEM software, which can quantify transcript abundance from RNA-seq data without a reference genome [15], and then performed statistical analysis implemented in EdgeR [16] using the mapped read numbers for each unique sequences calculated by RSEM. About 17% unique sequences (6,748) showed at least two fold changes (adjusted p-value ≤0.05) (Table S3). We found 282 unique sequences with no less than 4 fold changes (adjusted p-value ≤0.05), which are from 93 pathways (Table S4). As some unique genes are involved in more than one pathway, the total number of unique sequences in the 93 pathways reached to 708.
We investigated the expression changes in the view of pathways by calculating the genes with four fold changes. Among the 93 pathways, 26 showed increased transcript abundance, 17 of them displayed decreased transcript abundance, and other pathways had unique sequences with either increased or decreased in transcript abundance. Pathways that showed increased transcript abundance with at least two unique sequences include diterpenoid biosynthesis and terpenoid backbone biosynthesis. Interestingly, two pathways, biosynthesis of terpenoids and biosynthesis of alkaloids derived from terpenoid and polyketide, have at least 14 unique sequences with distinct changes and all but one had increased transcript abundance. Pathways which possessed decreased transcript abundance include for example cell cycle, base excision repair, and citrate cycle (TCA cycle). In addition, all the unique sequences in the pathways of DNA replication, mismatch repair, purine metabolism, homologous recombination, nucleotide excision repair had lower transcript abundance except for one unique sequence (Table S4).
MeJA Responsive Genes Involved in the Regulation of Gene Expression
We examined the genes that regulate gene expression, including transcription factors and genes involved in DNA and histone modification. A total 411 unique sequences were identified as transcription factors, including multiple members from bZIP and bHLH transcription factor classes (Table S5). Among them, 46 unique sequences showed at least 4 fold changes (adjusted p-value <0.05). The expression levels of 21 out of the 46 transcription factors showed higher transcript abundance, including members of bHLH, bZIP, TRY, GRAS, WRKY, PLATZ, MYB, and AP2/ERF domain-containing transcription factors; the remaining 25 transcription factors displayed lower transcript abundance, including GRAS, WRKY, E2F, PLATZ, ICE1, DP, TRY, GT-2, and ethylene-responsive transcription factors (Table S5).
We also found 23 unique sequences encoding DNA methyltransferase, 13 encoding methyl-CpG-binding domain-containing proteins, 44 of histone methyltransferase, 16 of histone demethylase, 12 of histone acetyltransferase, 27 of histone deacetylase, and 2 for histone ubiquitination (Table S6). Interestingly, the expression levels of 5 DNA methyltransferases, 2 histone methyltransferases, and 2 histone deacetylases possessed lower transcript abundance. However, 1 histone demethylase and 2 histone deacetylases, displayed at least 3 times increased transcript abundance (adjusted p-value≤0.05).
The Response of Genes in the Biosynthesis of Jasmonic Acid and Ethylene
Because the biosynthesis of JA is potentially important to the signal transduction of MeJA, we investigated the expression changes of genes in the two pathways of JA synthesis. Two of the seven genes in the biosynthesis of jasmonic acid, lipoxygenase (LOX) and allene oxide synthase (AOS), showed increased transcript abundance (Table S7). Similarly, methyl jasmonate esterase (MJE, Contig11373), catalyzing the conversion of MeJA to JA, also showed 5 fold increases in transcript abundance (Table S7). Two of the three genes involved in ethylene biosynthesis [22] were identified in our study, ACC synthase and ACC oxidase (Table S7). However, only ACC oxidase displayed distinct changes in expression levels. The expression levels of one unique sequence (Contig16235) which possessed 95% identity with the reported ACC oxidase in T. cuspidata [20] showed 4 fold increase in transcript abundance.
Genes in the Main Chain of Terpenoid Backbone Biosynthesis and Paclitaxel Biosynthesis, Transport and Degradation and their Transcript Changes
Because the paclitaxel biosynthesis pathway is incomplete in the KEGG database, to find the genes responsible for the biosynthesis of the main chain of terpenoid backbone, we manually identified all genes involved in these two pathways by reciprocal BLAST search against the transcriptome using previous reported enzymes as queries (Table S8). The main chain of terpenoid backbone biosynthesis includes the mevalonate (MVA) pathway and the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway and requires 17 genes in total. Thirty-six unique sequences were obtained by manual BLAST search and all the 17 genes in the main chain of terpenoid backbone biosynthesis were found (Table 1 and Table S9). Interestingly, all the 10 unique sequences from eight genes had increases in transcript abundance. The largest increase in transcript abundance observed in the pathway were from 1-deoxy-D-xylulose-10-phosphate synthase (DXS, 35 and 15 fold), geranylgeranyl diphosphate synthase (GGPPS, 17 fold) (Table 1). Two unique sequences of 4-hydroxy-3-methylbut-2-enyl diphosphate reductase (IDS) show similar transcript abundance changes, 7 and 5 times, respectively, and as they encode different regions of IDS protein it is likely that they are from the same IDS gene. Other genes showing increased transcript abundance in the pathway include 4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS), 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase (CMK), 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (MCS), 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR), and acetyl-CoA acetyltransferase (AACT) (Table1 and Table S9).
Table 1. Genes in main chain of terpenoid backbone biosynthesis and paclitaxel biosynthesis with changes in transcript abundance.
| Sequence_ID | Annotation | Abbreviation | Subject IDs | Identities | Fold change |
| Terpenoid backbone biosynthesis | |||||
| 237932 | 1-deoxy-D-xylulose-10-phosphate synthase | DXS | AAS89342.1 | 99% | 36.1 |
| Contig30899 | Geranylgeranyl diphosphate synthase | GGPPS | AAD16018.1 | 99% | 17.9 |
| Contig29438 | 1-deoxy-D-xylulose-9-phosphate synthase | DXS | AAS89342.1 | 98% | 15.8 |
| Contig22315 | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase | IDS | ABU44490.1 | 99% | 6.8 |
| Contig33460 | 1-deoxy-D-xylulose-5-phosphate reductoisomerase | DXR | AAT47184.1 | 99% | 5.8 |
| Contig28325 | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase | MCS | ABB88956.1 | 94% | 5.3 |
| Contig25640 | 4-hydroxy-3-methylbut-3-enyl diphosphate reductase | IDS | ABU44490.1 | 99% | 5.2 |
| Contig26664 | 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase | CMK | AAZ80384.1 | 71% | 4.9 |
| Contig33194 | 4-hydroxy-3-methylbut-2-enyl diphosphate synthase | HDS | ABB78087.1 | 87% | 4.8 |
| Contig33656 | acetyl-CoA acetyltransferase | AACT | NP_851150.1 | 81% | 3.7 |
| Paclitaxel biosynthesis | |||||
| Contig33713 | taxane 2α-O-benzoyltransferase | TBT | Q9FPW3.1 | 96% | n.d. |
| Contig29447 | taxane 2α-O-benzoyltransferase | TBT | Q9FPW3.1 | 93% | 47.4 |
| Contig28471 | 3′-N-debenzoyl-2′-deoxytaxol N-benzoyltransferase | DBTNBT | Q8LL69.1 | 99% | 32.1 |
| Contig32749 | Taxane 7-beta hydroxylase | T7OH | ACR78247.1 | 99% | 31.0 |
| 203604 | 3′-N-debenzoyl-2′-deoxytaxol N-benzoyltransferase | DBTNBT | Q8LL69.1 | 97% | 25.8 |
| Contig19765 | Taxane 5-alpha hydroxylase | T5OH | AAU93341.1 | 100% | 23.6 |
| Contig30864 | taxadien-5α-ol-O-acetyl transferase | TAT | AAU89980.1 | 93% | 22.2 |
| Contig28796 | taxadien-5α-ol-O-acetyl transferase | TAT | AAU89980.1 | 99% | 22.1 |
| Contig20518 | 10-deacetylbaccatin III-10-O-acetyltransferase | DBAT | ABW84241.1 | 100% | 19.2 |
| Contig24572 | 3′-N-debenzoyl-2′-deoxytaxol N-benzoyltransferase | DBTNBT | Q8LL69.1 | 99% | 17.8 |
| 247064 | taxa-4(5),11(12)-diene synthase | TS | AAZ41362.1 | 100% | 17.2 |
| Contig17866 | Taxane 5-alpha hydroxylase | T5OH | AAU93341.1 | 100% | 16.0 |
| 240648 | baccatin III 3-animo-3-phenylpropanoyltransferase | BAPT | ACN62085.1 | 99% | 14.9 |
| Contig26380 | Taxane 5-alpha hydroxylase | T5OH | Q6WG30.2 | 100% | 12.2 |
| Contig22452 | 10-deacetylbaccatin III-10-O-acetyltransferase | DBAT | ABW84241.1 | 99% | 12.2 |
| Contig20363 | taxadien-5α-ol-O-acetyl transferase | TAT | AAS49031.1 | 100% | 11.5 |
| Contig21313 | 3′-N-debenzoyl-2′-deoxytaxol N-benzoyltransferase | DBTNBT | AAT73199.1 | 98% | 10.3 |
| Contig26054 | taxa-4(5),11(12)-diene synthase | TS | AAR13861.1 | 96% | 10.3 |
| Contig17837 | taxadien-5α-ol-O-acetyl transferase | TAT | AAS49031.1 | 100% | 10.0 |
| Contig27532 | taxadien-5α-ol-O-acetyl transferase | TAT | Q8S9G6.1 | 99% | 8.5 |
| Contig17061 | 3′-N-debenzoyl-2′-deoxytaxol N-benzoyltransferase | DBTNBT | AAT73199.1 | 99% | 7.8 |
| Contig18229 | taxa-4(5),11(12)-diene synthase | TS | AAZ41362.1 | 99% | 7.6 |
| Contig30915 | Taxane 10-beta hydroxylase | T10OH | Q9AXM6.1 | 99% | 6.4 |
| Contig33190 | baccatin III 3-animo-3-phenylpropanoyltransferase | BAPT | ACN62085.1 | 99% | 6.3 |
For the genes in paclitaxel biosynthesis, only genes with more than 90% identity with reported genes are shown. “n.d.” indicates that the estimated relative abundance is zero in the nonelicited culture.
The paclitaxel biosynthesis pathway is still not fully elucidated and only 12 genes are confirmed (Figure S2). A manual BLAST search obtained 130 unique sequences from 11 of 12 genes, with only phenylalanine aminomutase (PAM) missing (Table S10). We searched the assembly results using an alternative assembly program SoapDenovo [23] and found 5 and 8 short PAM fragments in the nonelicited and elicited cultures, respectively. Among the 11 reported genes, T13OH shows the highest 69% identity with one unique sequence and the other ten genes display at least 90% identities to multiple unique sequences.
Of the 130 unique sequences associated with the paclitaxel biosynthesis pathway, 108 showed increased transcript abundance. Nine of 108 showed extremely low transcript levels in the nonelicited cells and therefore their expression changes could not be determined, which encode taxadien-5α-ol-O-acetyl transferase (TAT), taxane 2α-O-benzoyltransferase (TBT), taxane 10-beta hydroxylase (T10OH), 3′-N-debenzoyl-2′-deoxytaxol N-benzoyltransferase (DBTNBT), and taxane 5-alpha hydroxylase (T5OH). The largest expression changes in the remaining genes were 7 fold transcript abundance in baccatin III 3-animo-3-phenylpropanoyltransferase (BAPT) and 10-deacetylbaccatin III-10-O-acetyltransferase (DBAT), 6 fold in taxane 5-alpha hydroxylase (T5OH), taxane 7-beta hydroxylase (T7OH), and taxa-4(5), 11(12)-diene synthase (TS). The three unique sequences with lower transcript levels encode taxadien-5α-ol-O-acetyl transferase (TAT) and taxane 5-alpha hydroxylase (T5OH) (Table 1 and Table S10).
We also obtained some candidates of the unknown enzymes in paclitaxel biosynthesis among the unique sequences that show increases in transcript accumulation (Table S11 and Figure S2). By searching the annotations proposed in previous studies [24], [25], we found 2 unique sequences for epoxidase, 6 for oxomutase, 9 for CoA ligase, and 1 for oxidase. By blast search, we obtained 27 unique sequences encoding P450s that may function as T1OH, T9OH and T2’OH. Most of these candidates showed more than four-fold changes after elicitation of MeJA.
Some transporters, such as ABC transporters, are thought to be involved in the transport of paclitaxel [26], and 47 unique sequences encoding such transporters were found with distinct changes in expression levels. Among them were one unique sequence with increased transcript abundance and six unique sequences with lower transcript abundance (Table S12). In addition, 60 genes potentially associated with paclitaxel degradation, including diverse esterase and hydrolase show distinct changes (Table S13). However, although 10 unique sequences encoding multidrug and toxic compound extrusion transporters (MATE) were identified, none show significant changes in transcript abundance.
Comparison of MeJA Altered Transcript Levels between T. × media, T. chinensis, and T.cuspidata
By using broad gene functional descriptions our gene expression data was compared to other published studies investigating MeJA gene expressions changes in other Taxus species. Between T. × media and T. chinensis we identified seven genes that may function as transcription factors that were identified as possessing abundantly increased transcript levels in our data but not in the results of Li et. al. [19]; including GATA domain class transcription factor, PLATZ transcription factor, BIM1-like, E2FB-like, FER-Like iron deficiency-induced, HY5-like, and TT8 transcription factors. For genes involved in paclitaxel biosynthesis, only 9 of 12 genes with increased transcript levels after MeJA elicitation were also identified as similarly up-regulated by Li et al. [19] (T2OH, T13OH, and TBT were not identified as up regulated).
Only about 22.7% genes with distinct expression changes in T. cuspidata were found in our study and 70 of these genes showed consistent changes to the elicitation of MeJA in the two cell lines with different sampling times (Table S14). Twenty-two unique sequences with lower transcript level changes in T. × media also had lower transcript levels in T. cuspidata. Among the genes with lower transcript levels in both datasets, HD2 type histone deacetylase is known to be involved in jasmonic acid and ethylene signaling in response to pathogens [27]. Similarly, 26 unique sequences with increased transcript changes in T. × media were also up-regulated in T. cuspidata, which contained 29 genes. These include TBT in paclitaxel biosynthesis [28], 1-aminocyclopropane-1-carboxylate oxidase (ACO) in ethylene biosynthesis [22], 12-oxophytodienoate reductase possible in jasmonate biosynthesis [29], cationic peroxidase in pathogen incompatible interaction [30], Chalcone synthases involve in phenylpropanoid biosynthesis which is inducible by pathogens [31], and taxane 14b-hydroxylase participating in the bifurcation in paclitaxel biosynthesis [32]. Notably several genes between the different studies were identified as having opposite transcript changes in response to elicitation with MeJA: 15 genes (include one of two ACC oxidases) in T. × media with lower transcript levels were up-regulated in T. cuspidata,while 7 genes(e.g., dihydroflavonol-4-reductase in anthocyanin biosynthesis [33]) in T. × media with increased transcript levels were identified as down-regulated in T. cuspidata.
Identification of Potential mRNA Targets by miRNAs
Specific miRNAs have been implicated in the regulation of gene expression of T. chinensis cells elicited with MeJA [33]. To investigate the potential role of miRNAs in the synthesis of paclitaxel in T. × media cells, all unique sequences that were potential targets for MeJA regulated miRNAs were identified [33]. In total 200 unique sequences with assigned functions by homology searches, 106 showed changes in transcript abundance (Table S15). Interestingly, increased transcripts encoding the transcription repressor MYB4, myb-related protein 308, ethylene-responsive transcription factor ERF113, and acyltransferase, are all predicted to be the targets of miR-858, miR837, miR172, miR408, miR169, miR397b, miR172e, and miR396a that have lower transcript numbers in T. chinensis following the elicitation of MeJA [33], whereas sequences with lower transcript accumulation encoding alpha-expansin 15 are potential targets of miR164ab and miR164c, which have higher transcript numbers in T. chinensis after the elicitation of MeJA. These results indicate that these predicted targets may be valid targets of miRNAs (Table S15).
Validation of Differentially Expressed Genes with qRT-PCR Analysis
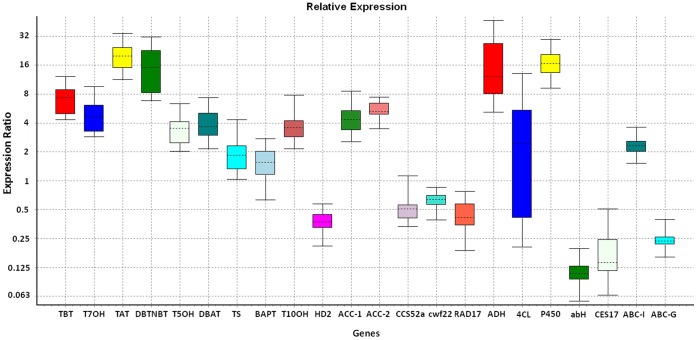
To validate the RNA-seq gene expression results, we selected 22 representative genes encoding 7 groups of enzyme types (Table 2) and designed specific primers for quantitative real-time PCR (Table S16). Consistent with RNA-seq analysis, the relative expression pattern of 20 genes out of 22 tested genes were identified as being significantly up-regulated or down-regulated in the MeJA induced cell cultures as compared to the control (Table 2, Figure 4). Contig787 (cell cycle switch protein CCS52a) has different expression pattern with the RNA-seq analysis. The expression change of 133463 (AMP dependent CoA ligase) is not significant in qRT-PCR assays, while the RNA-seq analysis presumed that it should be up-regulated by MeJA treatment. It is notable that all 9 genes involved in paclitaxel biosynthesis are up-regulated, and 2 candidate genes involved taxol degradation are down-regulated following MeJA treatment with both qRT-PCR and RNA-seq analysis.
Table 2. Comparison of the expression change of the 22 selected candidate genes with RNA-seq and qRT-PCR analysis.
| Putative Function | IDs | Putative Function | Fold change | |
| RNA-seq | qRT-PCR | |||
| Contig31917 | taxane 2α-O-benzoyltransferase, TBT | 17.1 | 6.9 | |
| Contig32749 | taxane 7β-hydroxylase, T7OH | 48.5 | 4.7 | |
| Contig30864 | taxadien-5α-ol-O-acetyl transferase, TAT | 34.3 | 19.2 | |
| Paclitaxel biosynthesis | Contig28471 | 3′-N-debenzoyl-2′-deoxytaxol N-benzoyltransferase, DBTNBT | 52.0 | 14.2 |
| Contig19765 | cytochrome P450 taxadiene 5α-hydroxylase, T5OH | 29.9 | 3.3 | |
| Contig20518 | 10-deacetylbaccatin III-10-O-acetyl transferase, DBAT | 21.1 | 3.9 | |
| 247064 | taxa-4(5),11(12)-diene synthase, TS | 17.1 | 1.8 | |
| 240648 | baccatin III 3-animo-3-phenylpropanoyltransferas, BAPT | 14.9 | 1.5 | |
| Contig30915 | taxane 10β-hydroxylase, T10OH | 5.7 | 3.6 | |
| HD2 type histone deacetylase | 120154 | HD2 type histone deacetylase, HD2 | 0.8 | 0.4 |
| ACC oxidase | Contig16235 | ACC oxidase, ACC-1 | 4.6 | 4.4 |
| 235821 | 1-aminocyclopropane-1-carboxylate oxidase, ACC-2 | 24.3 | 5.4 | |
| Contig787# | cell cycle switch protein CCS52a, CCS52a | 4.9 | 0.5 | |
| Cell cycle | Contig18564 | cell cycle control protein cwf22, cwf22 | 0.4 | 0.6 |
| 114663 | cell cycle checkpoint protein RAD17-like, RAD17 | 0.5 | 0.4 | |
| Candidates in the | Contig24302 | short chain alcohol dehydrogenase, ADH | 14.9 | 14.8 |
| unknown steps of | 133463# | AMP dependent CoA ligase, 4CL | 26.0 | 1.7* |
| paclitaxel biosynthesis | Contig24891 | cytochrome P450 716B2, P450 | 12.1 | 17.0 |
| Taxol degradation | Contig17145 | alpha/beta hydrolase fold-containing protein, abH | 0.1 | 0.1 |
| 84500 | carboxylesterase 17, CES17 | 0.1 | 0.2 | |
| Taxol transport | Contig20317 | ABC transporter I family member 6, chloroplastic, ABC-I | 2.8 | 2.3 |
| 241293 | ABC transporter G family member 3, ABC-G | n.d. | 0.2 | |
“#” indicate different expression models in RNA-seq analysis and qRT-PCR assays. “n.d.” indicates that the expression ratio of the gene in elicited cells to that in nonelicited cells approaches to zero. “*” indicates that the p-value is 0.447.
Figure 4. Expression analysis of 22 representative genes in nonelicited and elicited cells by qRT-PCR.
Y-axis indicates the relative expression distribution of a ratio of elicited cells versus nonelicited cells with actin as the reference gene. X-axis displays 22 selected genes, whose full names were given in Table 2. The boxes indicate 50% of observations near the median represented by the dotted line, and the whiskers show the outer 50% of observations.
Discussion
Transcriptome Re-programming of T. × media Cells After Elicitation with MeJA
Plant cell culture provides an alternative source to produce secondary metabolites including pharmaceuticals, nutraceuticals, and food additives [34], [35]. Different cell lines of Taxus were established but the low yield of paclitaxel has been hindering the commercial application. Phytohormone MeJA not only regulates normal developmental processing but also induces defensive responses to herbivores and pathogens by accumulating secondary metabolites [36]. In an effort towards establishing high paclitaxel producing cell cultures, the plant hormone MeJA was used to elicit the accumulation of paclitaxel. The highest production reaches to 36.0 mg/L after 7 days elicitation with 200 µM MeJA [9], which represents the steady state of paclitaxel biosynthesis and accumulation. As only one biological replicate was sequenced from elicited and nonelicited cells, a limitation of our analysis is that we cannot determine the biological variation of genes we identify as being MeJA responsive. Our list includes false positives as we find the results of 2 out of 22 tested genes are not consistent with the qRT-PCR analysis. Although good consistent changes were obtained for most candidate genes, it is still required to conduct independent validations to accurately measure the expression levels of genes of interest.
By de novo assembly, we obtained 38,506 and 28,959 contigs from the nonelicited and elicited cultures, respectively. Total 40,348 unique sequences were acquired by further assembly from these two datasets. Analysis on the expression levels revealed that the elicitation of MeJA induced the transcriptome re-programming in T. × media cells. We tracked the genes in the signaling network that MeJA triggered as an elicitor [37], [38]. A number of genes were found with transcript abundance changed, which included transcription factors, DNA and histone modification proteins, and the down-stream target genes involved in the biosynthesis of the main chain of terpenoid backbone, paclitaxel, jasmonic acid and ethylene. Among the transcription factors that were identified, Apetala2/ethylene response factor (AP2/ERF)-domain, R2R3 MYB, and MYC transcription factors are MeJA-modulated regulators of secondary metabolite biosynthesis [39], [40], [41]. HY5, a bZIP transcription factor, was recently shown involving in MeJA signaling pathway by interaction with MYC2 transcription factor [42].
Interestingly, we found some other bHLH and bZIP transcription factors, which play central roles in phytochrome signal transduction [43] and are involved in light and stress signaling processes [44], respectively. This implies that supplementation of MeJA may change light signal transduction network. However, the changes of expression levels in the two classes are inconsistent, with some members with increased transcript abundance while others have lower abundance. Considering that bZIP and bHLH transcription factors are large families, different members of these classes probably take part in functionally diverse processes and therefore show diverse changes in expression levels. Similarly, the complex expression changes of WRKY transcription factors following the elicitation of MeJA are consistent with their known diverse functions in biotic and abiotic stresses [45].
Key Regulatory Genes Involved Paclitaxel Biosynthesis, Transport, and Degradation
We found all 17 genes involved in the main chain of terpenoid backbone biosynthesis and 12 genes in paclitaxel biosynthesis, which include 36 and 167 unique sequences in the two biosynthesis pathways respectively. They provide a valuable start point to discovering key regulatory points in the synthesis of paclitaxel. Seven genes in the main chain of terpenoid backbone biosynthesis show changes in transcript abundance. The increased transcript abundance of GGPPS (18 times) was also observed by northern blot analysis in T. × media cells cultured on solid medium [46]. Random sequencing of cDNA clones derived from a MeJA elicited cell culture implicated DXR as being potentially MeJA inducible [47], and our data confirm this, as its transcript levels were 6 times higher.
Eleven of the 12 genes in paclitaxel biosynthesis displayed increased transcript abundance, 6 of which were reported by Lenka et al [20] and 8 by Li et al. [19]. The genes with increased transcript abundance in our data but absent from Li et al.’s data [19] imply that they are specifically increased in the steady state of paclitaxel production but not in the early state of MeJA elicitation. Intriguingly, we also found multiple candidates in the unknown steps of paclitaxel biosynthesis. Further experimental evidence is needed to confirm whether their putative function in paclitaxel biosynthesis. Xylosyltransferase can act as a shunt of paclitaxel biosynthesis by transferring a xylosyl group to the intermediate metabolites of paclitaxel biosynthesis including 10-deacetylpaclitaxel and 10-deacetyl cephalomannine and therefore decrease the intermediates fluxed to paclitaxel. We found 14 unique sequences annotated as xylosyltransferase but none showed changes in transcript abundance to the elicitation of MeJA, implying that the shunt to other chemical compounds is unlikely during elicitation of MeJA.
The transport of paclitaxel is bidirectional as extracellular paclitaxel can be absorbed into the cells and intracellular paclitaxel may be secreted into the extracellular medium [48]. Previous studies implied the presence of a specific transport pathway for the paclitaxel, although the identity of the transporters was unknown [26], [48]. ABC transporters have been previously proposed to be responsible for paclitaxel transport [26] and our data showed that 6 of 45 unique sequences encoding s showed lower transcript abundance. We also found one ABC transporter with higher transcript abundance and the significant expression change was also validated by qRT-PCR assay, which supports their association with the efflux/influx of paclitaxel. Further experiments on these candidates may help to identify the proteins involved in paclitaxel transport and aid in the bioengineering of paclitaxel extraction [48].
After synthesis, a portion of the paclitaxel synthesized is transported into the vacuole for storage [49]. Multidrug and toxic compound extrusion transporter (MATE) has been reported to involve in the vacuolar transport of some secondary metabolites, including alkaloids [50] and flavonoids [51], [52]. We found 10 unique MATE sequences, none of which showed distinct transcript changes. This implies that MATE may not be involved in the transport paclitaxel into the vacuole. Among those esterase and hydrolase genes that potentially associated with paclitaxel degradation, the genes showing lower transcript profiles, such as alpha/beta hydrolase fold-containing protein whose significant decrease in expression level was validated by qRT-PCR assay, are potential targets for enhancing paclitaxel production in Taxus cell culture with gene knock-out or knock-down strategy.
The Response of Genes in Jasmonic Acid and Ethylene Biosynthesis
MeJA cannot activate gene expression until it is converted to jasmonic acid (JA) and further to bioactive JA-Ile derivative [36]. Besides derived from MeJA, JA can also de novo biosynthesized from linolenic acid in chloroplast. The increased transcript abundance of genes in the biosynthesis of JA is consistent with the known self-activation of jasmonic acid biosynthesis from previous studies [53]. The interaction between JA and ethylene signaling pathways affects accumulation of plant secondary metabolites [54] and ethylene, whose accumulation can also enhance the biosynthesis of paclitaxel in T. cuspidata cells together with MeJA [55]. Therefore it is interesting to investigate the gene expression changes of genes in ethylene biosynthesis. Of note, one unique sequences (Contig16235) encoding ACC oxidase with 4 times up-regulation was estimated to have 1,875 transcripts per million, which is one of the genes with highest transcript numbers. The increase of ACC oxidase implies higher accumulation of ethylene, which can also enhance the biosynthesis of paclitaxel in T. cuspidata cells together with MeJA [55].
The Modulation Mechanism of MeJA on the Cell Growth
Paclitaxel is produced as a secondary metabolic product by the host plants to defend pathogens, such as oomycetes and wood decaying fungi [56]. Following the metabolic flux is shunt to various secondary metabolites, the nutrients used for the normal growth of plants or cells were affected and the plants or cells undergo retarded development. However, how such a resource reallocation occurs, and how MeJA mediates the crucial switch from growth to defense is still unclear. The growth repression by MeJA was found to be associated with reduced transcript levels of genes involved in cell cycle, especially the M-phase genes [57]. We found that two genes associated with the cell cycle, DNA-dependent protein kinase and minichromosome maintenance protein, showed four fold decreased transcript abundance, of which minichromosome maintenance protein reportedly displayed up-regulation changes in the fast growing cells [58]. Interestingly, besides the cell cycle progression, our results also revealed that the expression levels of the genes involved in DNA replicates, meiosis, and mismatch repair had decreased transcript abundance, which included DNA replication licensing factor MCM, proliferating cell nuclear antigen, beta-lactamase class C protein, DNA mismatch repair protein, and exonuclease. This implies that MeJA also modulates the growth and development of cells by interfering with the cell cycle, while they mobilize the resources to produce large amount of secondary metabolites for defense.
Expression Change Comparison of Genes in the Early and Steady Elicitation of MeJA
Li et al. [19] investigated the global changes of T. chinensis transcriptome in the early MeJA elicitation events (16h after MeJA addition), and Lenka et al. [20] sampled the cells at 6 hours, 18 hours, and 5 day post-elicitation in T. cuspidata. Therefore, it is interesting to know the change profile of genes along with the treatment of MeJA. Unfortunately, since the original raw RNA-seq data is not available we could only identify genes in this study based on broad functional descriptions to compare transcript changes. Seven genes encoding transcription factors and three genes in paclitaxel biosynthesis were uniquely found with increased transcriptional abundance in our data. The genes with similar and distinct expression models in our data and in T. cuspidata potentially provides us a set of targets for the following metabolic engineering to directly increase paclitaxel production, such as TBT in paclitaxel biosynthesis [28] and the gene expression regulator HD2 type histone deacetylase in epigenetics [27]. Notably, taxane 14b-hydroxylase directs a side-route of paclitaxel pathway which can shunts paclitaxel production, and transcription inhibition to this gene can decrease the product of other compounds [59]. The unique sequence encoding taxane 14b-hydroxylase may provide a good target to indirectly increase paclitaxel production. Further research on these genes specifically their temporal response to MeJA elicitation would provide valuable information about the regulatory mechanism of paclitaxel biosynthesis.
The Regulatory Roles of miRNAs in the Elicitation of Methyl Jasmonate
Qiu et al [33] identified 58 miRNAs from 25 families in T. chinensis cells with 14 miRNAs (7 families) showing MeJA altered expression. The lack of extensive sequence information on the T. chinensis genome precluded further identification of Taxus specific targets in that study. However because of the high conservation of miRNAs among closely related species in plants [60] and the stringent complementarity between plant miRNAs and their mRNA targets, we can use the mRNA sequences from T. × media in this study to identify potential mRNA targets. Two hundred potential targets with known functions were found (Table S15) and multiple pairs of miRNAs and predicted targets show complementary expression patterns (higher and lower transcript abundance), including transcription factors and other regulatory genes possibly involved in the biosynthesis of paclitaxel, such as MYB, ethylene-responsive transcription factors, acyltransferase, and geranylgeranyl transferase type I. Among these targets, Courdavault et al. once proposed that geranylgeranyl transferase type I is involved in early jasmonate signaling transduction pathway [61]. The complementary expression patterns of predicted targets with miRNAs provides further support for the notion that miRNA-mediated gene expression regulation may also involve in the signal network following the elicitation of MeJA, including the biosynthesis of paclitaxel. It may be possible to develop a miRNA-based method for improving paclitaxel yield in the future.
Materials and Methods
Plant Materials
As solid cell culture has simpler and more reproducible generation procedures, but possesses similar growth characteristics as suspension cell culture, solid medium was used to culture all material in this study. The cell line was created from leaf segments of Taxus × media on the B5 medium containing 100 mg/L myo-inositol, 1.25 mg/L nicotinic acid, 1.0 mg/L vitamin B1, 0.5 mg/L vitamin B6, 1.0 g/L lactalbumin hydrolysate, 0.1 mg/L 6-BA, 1.0 mg/L 2,4-D, 8.0 g/L agar and 20.0 g/L sucrose. Subculture was carried out every four weeks on 67-V solid medium supplemented with 2.0 mg/L IAA, 1.5 mg/L 2,4-D, 0.2 mg/L 6-BA, 100 mg/L myo-inositol, 8.0 g/L agar and 30 g/L sucrose. T. × media is a hybrid designation for crosses between English yew (Taxus baccata) and Japanese yew (Taxus cuspidata). These hybrids are noted for high palcitaxel content in their needles and shoots. The elicitor, methyl jasmonate (MeJA) was dissolved in an equal volume ethanol, filter-sterilized and then added to the media before solidification with the final concentration of 200 µM. For the control experiment, an equivalent amount of ethanol was added to the medium. The cells were harvested on 7 days after elicitation for total RNA isolation.
Total RNA Isolation and cDNA Library Construction
The total RNAs were extracted using a modified CTAB method from Chang [62]. Briefly, 1g of fresh cells was ground in liquid nitrogen and the powder was mixed with 100 µL mercaptoethanol and 5 mL extraction solution preheated to 65°C. After a water bath at 65°C for 3 min, the mixtures were centrifuged for 10 min at 12,000 rpm at 4°C. The upper aqueous phase was extracted two times with equal volume of chloroform:isoamyl alcohol (24∶1). Then, 1/4 volume 10 M LiC1 was added to the supernatant. After placement overnight at −20°C, the precipitation was collected by centrifuging for 20 min at 12,000 rpm at 4°C and dissolved in 500 µL SSTE. The solution was further extracted with equal volume of phenol:chloroform:isoamyl alcohol (25∶24:1) and with 2.5 time volume of ethanol. The precipitate was dried and dissolved in water. The quantity and quality of total RNA were evaluated using a Nanodrop ND-1000 (Nanodrop technologies, Wilmington, DE, USA), gel electrophoresis and Agilent 2100 (Agilent Technologies, Palo Alto, Calif.). RNA samples showing good quality in RNA electrophoresis and RNA integrity number more than 8.4 in Agilent 2100 were stored at −80°C until sequencing. The cDNA libraries were constructed by using Illumina’s kit following the manufacturer’s protocol (TruSeq RNA Sample Preparation Kits v2, San Diego, USA).
Sequencing and Data Analysis
The paired-end cDNA library sequencing was performed on an Illumina Solexa 1G Genome Analyzer. Image deconvolution and quality calculation were conducted by using the Illumina GA pipeline 1.3. Adaptor sequences, empty reads and low quality sequences were removed from the raw reads to produce high quality reads. The high quality reads were then assembled by using ABySS (version 3.8) [14]. In brief, all the high quality reads were firstly assembled into contigs using different k-mer values, then the assembly results from diverse k-mer values were assessed based on the total number of contigs with length no less than 100 nt, the N50 value, the median and mean lengths of contigs. The optimal assemblies were selected for the control and MeJA treatment datasets. For the convenience of comparison on gene expression levels, the contigs of the two dataset was assembled into one integrated dataset using CAP3 [63]. We call the resulting integrated sequence fragments as unique sequences in this study.
After removing all the unique sequences with ambiguous or unknown bases, the number of complete cDNAs was estimated as the following: the putative open reading frames of all unique sequences were predicted with OrfPredictor (http://proteomics.ysu.edu/tools/OrfPredictor.html), which were then used as queries to search against Swiss-Prot database with E-value set to 1e-5. The unique cDNAs were considered complete cDNAs if the alignment coverage of the top hit was more than 90%.
Unique sequences were used for BLAST search and annotation against NCBI non-redundant (nr) databases, cluster of orthologous groups of protein (COG) database, kyoto encyclopedia of genes and genomes (KEGG) database, and gene ontolog (GO) database with an cut-off E-value of 10−5, respectively. Functional annotations were performed by sequence similarity against nr database and the annotations of first sequence with highest sequence similarity and clear function annotation were endowed to the corresponding unique sequences. Both functional annotation by GO was analyzed against the GO database, and the pathways annotation was retrieved using the internal KEGG information of hits in GO database. The expression levels of unique sequences in each dataset were calculated by using RSEM software (version 1.1.13) [15]; the resulting mapped reads number for each unique sequences was feed to EdgeR (version 2.2.6) [16] to perform the significance analysis. The normalization factor was calculated using calcNormFactors, the common dispersion across the read numbers mapped to each unique sequences was estimated using estimateCommonDisp, the genewise exact test for difference in the means between the nonelicited and elicited cells of negative-binomially distributed counts was computed using exactTest. The unique sequences with no less than a twofold change in the transcript level and p-value ≤0.05 were considered as significantly differently expressed between the nonelicited cells and elicited cells.
Homolog Search for Identifying Genes Involved in the Main Chain of Terpenoid Backbone Biosynthesis and Paclitaxel Biosynthesis
The genes involved in the main chain of terpenoid backbone biosynthesis and paclitaxel biosynthesis were manually identified by BLAST search against the assembled contigs. The queries were all from Taxus if available and the genes from Arabidopsis thaliana (TAIR, www.arabidopsis.org) were used if they are still unknown in Taxus. All the hits with E-value less than 10−5 in Taxus were then used as queries to search GenBank nr database again and were kept if their subjects also were annotated as the enzymes involved in the main chain of terpenoid backbone biosynthesis and paclitaxel biosynthesis.
Comparison of Genes with Significant Expression Changes After Elicitation with MeJA between T. × media and T. cuspidata
The genes with significant changes in expression levels after elicitation with MeJA were compared between T. × media and T. cuspidata by blast search. All the unique sequences in our data showing two fold expression changes after elicitation with MeJA were used as queries to search against all the genes obtained by SSH [20]. All the subjects from T. cuspidata were kept if their identity values with T. × media and the alignment lengths are not less than 95% and 50 amino acids, respectively.
Prediction of Potential Target mRNAs of miRNAs
The miRNAs found in T. chinensis [33] were used to identify the potential miRNA targets against all the assembly unique sequences with known functions by target-align (http://www.leonxie.com/targetAlign.php) [64], which was developed by ourselves. The sequences that satisfied the following parameters were considered as the potential targets of miRNAs: i) the total mismatched nucleotides and the contiguous mismatched nucleotides in the miRNA:mRNA duplexes were no more than four and two respectively; ii) the mismatched nucleotides from the first site to the ninth site in miRNAs do not exceed one; iii) no mismatch occurs on the tenth and eleventh sites; iv) the score for G:U match was set to 0.5.
Real-time PCR Assays
Total RNA was extracted using Column Plant RNAout kit (TIANDZ, Beijing, China) from the T. × media MeJA elicited cells and nonelicited cells as those prepared for RNA-seq sequencing, according to the manufacturer's protocol. RNA was treated with DNase I (Takara, Dalian, China) to remove residual genomic DNA. The concentration of the isolated RNA and the 260/280-absorbance ratio was measured in triplicates with Nanodrop ND-8000 (Thermo, USA). The quality of RNA samples was confirmed by electrophoresis on a 1.2% agarose. Total RNA was reverse-transcribed to cDNA using PrimeScript RT Master Mix (Perfect Real Time) (Takara, Dalian, China) in a total volume of 20 µl containing 1 µg of total RNA. The reaction was carried out at 37°C for 15 min and 85°C for 5 s. Forward (Fwd) and reverse (Rev) primers were designed using Primer3 [65]. Table S16 shows the primers for the selected genes and the reference gene actin. The real-time PCR assays were conducted in an optional 96-well plate with ABI7500 system (ABI, USA) and a commercial SRBR-Green master mix kit (Takara, Dalian, China). The reactions were performed with 1 µl cDNA in 20 µl reaction mix containing 10 µl 2 × SYBR Premix Ex Taq and 1 µl primers. The conditions were as follows: initial holding at 95°C for 30 s, followed by a two-step program of 95°C for 5 s and 55°C for 34 s for 40 cycles. Two independent biological replicates, three technical replicates for each PCR reaction were performed. Reverse transcriptase negative controls and “no template controls” (without cDNA in PCR) were included. Data analysis was performed with REST 2009 Software [66]. Statistical difference are significant when p<0.05.
Supporting Information
Taxus × media cells cultured on solid medium.
(DOC)
Flow chart of paclitaxel biosynthetic pathway in T. × media .
(DOC)
Unique sequences with over 1000 transcripts/million in nonelicited and elicited cultures.
(XLS)
Comparison of unique sequences in KEGG pathways between nonelicited and elicited cultures.
(XLS)
Unique sequences with no less than two-fold changes in transcript levels (adjusted p-value ≤0.05) following the elicitation of MeJA.
(XLS)
KEGG pathway analysis of unique sequences with distinct changes in expression levels following MeJA elicitation.
(XLS)
Transcription factors and expression levels comparison between nonelicited and elicited cultures.
(XLS)
Genes involved in DNA and histone modification and expression level comparison between nonelicited and elicited cultures.
(XLS)
Genes involved in biosynthesis of jasmonic acid and ethylene and expression level comparison between nonelicited and elicited cultures.
(XLS)
Queries used in the BLAST search to identify genes in the main chain of terpenoid backbone biosynthesis and paclitaxel biosynthesis.
(DOC)
Genes involved in main chain of terpenoid backbone biosynthesis and expression level comparison between nonelicited and elicited cultures.
(XLS)
Genes involved in paclitaxel biosynthesis and expression level comparison between nonelicited and elicited cultures.
(XLS)
Candidates of unknown enzymes in paclitaxel biosynthesis.
(XLS)
Genes potentially involved in paclitaxel transport and expression level comparison between nonelicited and elicited cultures.
(XLS)
Genes potentially involved in paclitaxel degradation and expression level comparison between nonelicited and elicited cultures.
(XLS)
Comparison of genes with changes in transcript abundance after elicitation with MeJA between T. × media and T. cuspidate .
(XLS)
miRNA targets and expression level comparison between nonelicited and elicited cultures.
(XLS)
Primers used for qRT-PCR assays.
(DOC)
Acknowledgments
We thank Prof. Jun-Gui Dai (the Institute of Materia Medica, Chinese Academy of Medical Sciences) for providing Taxus × media cell line and Prof. Jinsong Wu (Kunming Institute of Botany, Chinese Academy of Sciences, Kunming 650201, China) for useful suggestions. Most computation work was performed on the HPC Center, Kunming Institute of Botany, CAS, China.
Funding Statement
This work is supported by a grant from the National Natural Science Foundation of China (31170628) and a startup grant from Kunming Institute of Botany, Chinese Academy of Sciences, to the Group of Molecular Ecology, a grant from Yunnan Recruitment Program of Experts in Sciences (2012HA016), and a grant from Thousand Youth Talents Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hao D, Huang B, Yang L (2008) Phylogenetic relationships of the genus Taxus inferred from chloroplast intergenic spacer and nuclear coding DNA. Biological & pharmaceutical bulletin 31: 260–265. [DOI] [PubMed] [Google Scholar]
- 2. McGuire WP (2001) The story of taxol - Nature and politics in the pursuit of an anti-cancer drug. Science 292: 1073–1074. [Google Scholar]
- 3. Slichenmyer WJ, Vonhoff DD (1991) Taxol - a New and Effective Anticancer Drug. Anti-Cancer Drugs 2: 519–530. [PubMed] [Google Scholar]
- 4. Witherup KM, Look SA, Stasko MW, Ghiorzi TJ, Muschik GM, et al. (1990) Taxus Spp Needles Contain Amounts of Taxol Comparable to the Bark of Taxus-Brevifolia - Analysis and Isolation. Journal of Natural Products 53: 1249–1255. [DOI] [PubMed] [Google Scholar]
- 5. Stierle A, Strobel G, Stierle D (1993) Taxol and Taxane Production by Taxomyces-Andreanae, an Endophytic Fungus of Pacific Yew. Science 260: 214–216. [DOI] [PubMed] [Google Scholar]
- 6.Holton RA, Biediger RJ, Boatman PD (1995) Semisynthesis of taxol and taxotere. Taxol science and applications. Suffness M: CRC Press, Boca Raton. 97–121.
- 7. Tabata H (2004) Paclitaxel production by plant-cell-culture technology. Adv Biochem Eng Biotechnol 87: 1–23. [DOI] [PubMed] [Google Scholar]
- 8. Yukimune Y, Tabata H, Higashi Y, Hara Y (1996) Methyl jasmonate-induced overproduction of paclitaxel and baccatin III in Taxus cell suspension cultures. Nature Biotechnology 14: 1129–1132. [DOI] [PubMed] [Google Scholar]
- 9. Ketchum RE, Gibson DM, Croteau RB, Shuler ML (1999) The kinetics of taxoid accumulation in cell suspension cultures of Taxus following elicitation with methyl jasmonate. Biotechnology and bioengineering 62: 97–105. [DOI] [PubMed] [Google Scholar]
- 10. Ketchum RE, Rithner CD, Qiu D, Kim YS, Williams RM, et al. (2003) Taxus metabolomics: methyl jasmonate preferentially induces production of taxoids oxygenated at C-13 in Taxus x media cell cultures. Phytochemistry 62: 901–909. [DOI] [PubMed] [Google Scholar]
- 11. Morozova O, Marra MA (2008) Applications of next-generation sequencing technologies in functional genomics. Genomics 92: 255–264. [DOI] [PubMed] [Google Scholar]
- 12. Shendure J, Ji H (2008) Next-generation DNA sequencing. Nature Biotechnology 26: 1135–1145. [DOI] [PubMed] [Google Scholar]
- 13. Barrero RA, Chapman B, Yang Y, Moolhuijzen P, Keeble-Gagnere G, et al. (2011) De novo assembly of Euphorbia fischeriana root transcriptome identifies prostratin pathway related genes. BMC genomics 12: 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, et al. (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao DC, Ge GB, Xiao PG, Zhang YY, Yang L (2011) The first insight into the tissue specific Taxus transcriptome via illumina second generation sequencing. Plos One 6. [DOI] [PMC free article] [PubMed]
- 18. Wu Q, Sun C, Luo H, Li Y, Niu Y, et al. (2011) Transcriptome analysis of Taxus cuspidata needles based on 454 pyrosequencing. Planta Med 77: 394–400. [DOI] [PubMed] [Google Scholar]
- 19. Li S, Zhang P, Zhang M, Fu CH, Zhao CF, et al. (2012) Transcriptional profile of Taxus chinensis cells in response to methyl jasmonate. BMC genomics 13: 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lenka SK, Boutaoui N, Paulose B, Vongpaseuth K, Normanly J, et al. (2012) Identification and expression analysis of methyl jasmonate responsive ESTs in paclitaxel producing Taxus cuspidata suspension culture cells. BMC genomics 13: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, et al. (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang KL, Li H, Ecker JR (2002) Ethylene biosynthesis and signaling networks. The Plant cell 14 Suppl: S131–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li R, Fan W, Tian G, Zhu H, He L, et al. (2010) The sequence and de novo assembly of the giant panda genome. Nature 463: 311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onrubia M (2012) A MOLECULAR APPROACH TO TAXOL BIOSYNTHESIS. Barcelona: Universitat Pompeu Fabra. 55–59 p.
- 25. Croteau R, Ketchum RE, Long RM, Kaspera R, Wildung MR (2006) Taxol biosynthesis and molecular genetics. Phytochemistry reviews : proceedings of the Phytochemical Society of Europe 5: 75–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fornale S, Esposti DD, Navia-Osorio A, Cusido RM, Palazon J, et al. (2002) Taxol transport in Taxus baccata cell suspension cultures. Plant Physiology and Biochemistry 40: 81–88. [Google Scholar]
- 27. Zhou C, Zhang L, Duan J, Miki B, Wu K (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. The Plant cell 17: 1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walker K, Croteau R (2000) Taxol biosynthesis: Molecular cloning of a benzoyl-CoA : taxane 2 alpha-O-benzoyltransferase cDNA from Taxus and functional expression in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 97: 13591–13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schaller F, Biesgen C, Mussig C, Altmann T, Weiler EW (2000) 12-oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta 210: 979–984. [DOI] [PubMed] [Google Scholar]
- 30. Young SA, Guo A, Guikema JA, White FF, Leach JE (1995) Rice cationic peroxidase accumulates in xylem vessels during incompatible interactions with Xanthomonas-Oryzae Pv Oryzae. Plant Physiology 107: 1333–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Habereder H, Schroder G, Ebel J (1989) Rapid induction of phenylalanine ammonia-lyase and chalcone synthase messenger-RNAs during fungus infection of soybean (Glycine-max L) roots or elicitor treatment of soybean cell-cultures at the onset of phytoalexin synthesis. Planta 177: 58–65. [DOI] [PubMed] [Google Scholar]
- 32. Ketchum REB, Horiguchi T, Qiu DY, Williams RM, Croteau RB (2007) Administering cultured Taxus cells with early precursors reveals bifurcations in the taxoid biosynthetic pathway. Phytochemistry 68: 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Qiu D, Pan X, Wilson IW, Li F, Liu M, et al. (2009) High throughput sequencing technology reveals that the taxoid elicitor methyl jasmonate regulates microRNA expression in Chinese yew (Taxus chinensis). Gene 436: 37–44. [DOI] [PubMed] [Google Scholar]
- 34. Rao SR, Ravishankar GA (2002) Plant cell cultures: Chemical factories of secondary metabolites. Biotechnology advances 20: 101–153. [DOI] [PubMed] [Google Scholar]
- 35. Smetanska I (2008) Production of secondary metabolites using plant cell cultures. Food Biotechnology 111: 187–228. [DOI] [PubMed] [Google Scholar]
- 36. Katsir L, Chung HS, Koo AJ, Howe GA (2008) Jasmonate signaling: a conserved mechanism of hormone sensing. Current opinion in plant biology 11: 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pauwels L, Inze D, Goossens A (2009) Jasmonate-inducible gene: what does it mean? Trends in Plant Science 14: 87–91. [DOI] [PubMed] [Google Scholar]
- 38. Kazan K, Manners JM (2008) Jasmonate signaling: Toward an integrated view. Plant Physiology 146: 1459–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Galis I, Simek P, Narisawa T, Sasaki M, Horiguchi T, et al. (2006) A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid-conjugate biosynthesis in tobacco. Plant Journal 46: 573–592. [DOI] [PubMed] [Google Scholar]
- 40. De Sutter V, Vanderhaeghen R, Tilleman S, Lammertyn F, Vanhoutte I, et al. (2005) Exploration of jasmonate signalling via automated and standardized transient expression assays in tobacco cells. Plant Journal 44: 1065–1076. [DOI] [PubMed] [Google Scholar]
- 41. Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 Are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Prasad BR, Kumar SV, Nandi A, Chattopadhyay S (2012) Functional interconnections of HY1 with MYC2 and HY5 in Arabidopsis seedling development. BMC plant biology 12: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Duek PD, Fankhauser C (2005) bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci 10: 51–54. [DOI] [PubMed] [Google Scholar]
- 44. Jakoby M, Weisshaar B, Droge-Laser W, Vicente-Carbajosa J, Tiedemann J, et al. (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111. [DOI] [PubMed] [Google Scholar]
- 45. Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206. [DOI] [PubMed] [Google Scholar]
- 46. Liao ZH, Gong YF, Kai GY, Zu KJ, Chen M, et al. (2005) An intron-free methyl jasmonate inducible geranylgeranyl diphosphate-synthase gene from Taxus media and its functional identification in yeast. Molecular Biology 39: 11–17. [PubMed] [Google Scholar]
- 47. Jennewein S, Wildung MR, Chau M, Walker K, Croteau R (2004) Random sequencing of an induced Taxus cell cDNA library for identification of clones involved in Taxol biosynthesis. Proceedings of the National Academy of Sciences of the United States of America 101: 9149–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Naill MC, Kolewe ME, Roberts SC (2012) Paclitaxel uptake and transport in Taxus cell suspension cultures. Biochemical Engineering Journal 63: 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Exposito O, Syklowska-Baranek K, Moyano E, Onrubia M, Bonfill M, et al. (2010) Metabolic Responses of Taxus media Transformed Cell Cultures to the Addition of Methyl Jasmonate. Biotechnology Progress 26: 1145–1153. [DOI] [PubMed] [Google Scholar]
- 50. Morita M, Shitan N, Sawada K, Van Montagu MCE, Inze D, et al. (2009) Vacuolar transport of nicotine is mediated by a multidrug and toxic compound extrusion (MATE) transporter in Nicotiana tabacum. Proceedings of the National Academy of Sciences of the United States of America 106: 2447–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhao J, Huhman D, Shadle G, He XZ, Sumner LW, et al. (2011) MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula . Plant Cell 23: 1536–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao J, Dixon RA (2009) MATE transporters facilitate vacuolar uptake of epicatechin 3 '-o-glucoside for proanthocyanidin bosynthesis in Medicago truncatula and Arabidopsis . Plant Cell 21: 2323–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, et al. (2001) Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA research : an international journal for rapid publication of reports on genes and genomes 8: 153–161. [DOI] [PubMed] [Google Scholar]
- 54. Zhao J, Davis LC, Verpoorte R (2005) Elicitor signal transduction leading to production of plant secondary metabolites. Biotechnology advances 23: 283–333. [DOI] [PubMed] [Google Scholar]
- 55. Mirjalili N, Linden JC (1996) Methyl jasmonate induced production of taxol in suspension cultures of Taxus cuspidata: Ethylene interaction and induction models. Biotechnology Progress 12: 110–118. [DOI] [PubMed] [Google Scholar]
- 56. Wagner LJ, Flores HE (1994) Effect of taxol and related-compounds on growth of plant-pathogenic fungi. Phytopathology 84: 1173–1178. [Google Scholar]
- 57. Swiatek A, Lenjou M, Van Bockstaele D, Inze D, Van Onckelen H (2002) Differential effect of jasmonic acid and abscisic acid on cell cycle progression in tobacco BY-2 cells. Plant Physiology 128: 201–211. [PMC free article] [PubMed] [Google Scholar]
- 58. Shultz RW, Lee TJ, Allen GC, Thompson WF, Hanley-Bowdoin L (2009) Dynamic localization of the DNA replication proteins MCM5 and MCM7 in plants. Plant Physiology 150: 658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li FL, Ma XJ, Hu XL, Hoffman A, Dai JG, et al. (2011) Antisense-induced suppression of taxoid 14 beta-hydroxylase gene expression in transgenic Taxus x media cells. African Journal of Biotechnology 10: 8720–8728. [Google Scholar]
- 60. Sun GL (2012) MicroRNAs and their diverse functions in plants. Plant Molecular Biology 80: 17–36. [DOI] [PubMed] [Google Scholar]
- 61. Courdavault V, Burlat V, St-Pierre B, Giglioli-Guivarc'h N (2009) Proteins prenylated by type I protein geranylgeranyltransferase act positively on the jasmonate signalling pathway triggering the biosynthesis of monoterpene indole alkaloids in Catharanthus roseus. Plant Cell Reports 28: 83–93. [DOI] [PubMed] [Google Scholar]
- 62. Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees Plant Molecular Biology Reporter. 11: 113–116. [Google Scholar]
- 63. Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xie F, Zhang B (2010) Target-align: a tool for plant microRNA target identification. Bioinformatics 26: 3002–3003. [DOI] [PubMed] [Google Scholar]
- 65. Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386. [DOI] [PubMed] [Google Scholar]
- 66. Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic acids research 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taxus × media cells cultured on solid medium.
(DOC)
Flow chart of paclitaxel biosynthetic pathway in T. × media .
(DOC)
Unique sequences with over 1000 transcripts/million in nonelicited and elicited cultures.
(XLS)
Comparison of unique sequences in KEGG pathways between nonelicited and elicited cultures.
(XLS)
Unique sequences with no less than two-fold changes in transcript levels (adjusted p-value ≤0.05) following the elicitation of MeJA.
(XLS)
KEGG pathway analysis of unique sequences with distinct changes in expression levels following MeJA elicitation.
(XLS)
Transcription factors and expression levels comparison between nonelicited and elicited cultures.
(XLS)
Genes involved in DNA and histone modification and expression level comparison between nonelicited and elicited cultures.
(XLS)
Genes involved in biosynthesis of jasmonic acid and ethylene and expression level comparison between nonelicited and elicited cultures.
(XLS)
Queries used in the BLAST search to identify genes in the main chain of terpenoid backbone biosynthesis and paclitaxel biosynthesis.
(DOC)
Genes involved in main chain of terpenoid backbone biosynthesis and expression level comparison between nonelicited and elicited cultures.
(XLS)
Genes involved in paclitaxel biosynthesis and expression level comparison between nonelicited and elicited cultures.
(XLS)
Candidates of unknown enzymes in paclitaxel biosynthesis.
(XLS)
Genes potentially involved in paclitaxel transport and expression level comparison between nonelicited and elicited cultures.
(XLS)
Genes potentially involved in paclitaxel degradation and expression level comparison between nonelicited and elicited cultures.
(XLS)
Comparison of genes with changes in transcript abundance after elicitation with MeJA between T. × media and T. cuspidate .
(XLS)
miRNA targets and expression level comparison between nonelicited and elicited cultures.
(XLS)
Primers used for qRT-PCR assays.
(DOC)