Abstract
In Drosophila, CLOCK/CYCLE heterodimer (CLK/CYC) is the primary activator of circadian clock genes that contain the E-box sequence in their promoter regions (hereafter referred to as “E-box clock genes”). Although extensive studies have investigated the feedback regulation of clock genes, little is known regarding other factors acting with CLK/CYC. Here we show that Drosophila C-terminal binding protein (dCtBP), a transcriptional co-factor, is involved in the regulation of the E-box clock genes. In vivo overexpression of dCtBP in clock cells lengthened or abolished circadian locomotor rhythm with up-regulation of a subset of the E-box clock genes, period (per), vrille (vri), and PAR domain protein 1ε (Pdp1ε). Co-expression of dCtBP with CLK in vitro also increased the promoter activity of per, vri, Pdp1ε and cwo depending on the amount of dCtBP expression, whereas no effect was observed without CLK. The activation of these clock genes in vitro was not observed when we used mutated dCtBP which carries amino acid substitutions in NAD+ domain. These results suggest that dCtBP generally acts as a putative co-activator of CLK/CYC through the E-box sequence.
Introduction
Many organisms show circadian rhythms in physiology, metabolism, and behavior. These rhythms are controlled by an endogenous circadian clock [1]. In Drosophila, there are seven transcription factors among core components of the circadian clock. The transcriptional activator CLOCK/CYCLE heterodimer (CLK/CYC) binds to the E-box sequence in the promoter regions of clock genes period (per), timeless (tim), vrille (vri), PAR domain protein 1ε (Pdp1ε), and clockwork orange (cwo) to activate their transcription [1]–[3]. The product proteins of these genes feed back to control their own transcription. Three feedback loops are tightly interlocked to yield the circadian oscillation of clock genes’ products. In one loop, PER/TIM suppresses the function of CLK/CYC to generate the oscillation of their own transcription. In another loop, the transcription of Clk is mediated by VRI and PDP1ε which acts as a suppressor and an activator, respectively. In the other loop, CWO inhibits the transcription of clock genes to bind the E-box sequence. This interlocked feedback loops generate and maintain circadian rhythm in pacemaker cells in the Drosophila head and regulate circadian output pathways that control circadian rhythms in physiology, metabolism, and behavior. Although CLK/CYC is well known as the primary factor regulating the circadian oscillation of transcription of the core clock genes as well as output genes, little is known regarding other factors that act with CLK/CYC. Although NEJIRE (NEJ), a homolog of CBP/p300 [4], has been reported as a co-factor of CLK, conflicting reports have claimed that it acts as a co-activator [5] and co-repressor [6].
Drosophila C-terminal binding protein (dCtBP) [7], [8] is a homolog of human CtBP that binds to the C-terminal region of human adenovirus E1A proteins to negatively modulate an oncogenic transformation [9], [10]. dCtBP was initially reported as a transcriptional co-repressor functioning during embryonic development in Drosophila [11]. dCtBP forms complexes with Knirps, Snail and Hairy, all of which contain a DNA-binding domain, to suppress transcription of their target genes [11], [12]. The consensus sequences P-DLS-K in Knirps and Snail and PLSLV in Hairy have been identified as binding sequences of dCtBP [9], [11], [12]. Although dCtBP is well known to function as a repressor, a recent study reported that dCtBP may also function as an activator in the Wingless signaling pathway [13], [14]. In the adult brain, ubiquitous expression of dCtBP has been reported in virtually all neurons including pacemaker cells [15]. CtBP contains extensive homology with D-2-hydroxy acid dehydrogenases, including the conserved nicotinamide adenine dinucleotide domain (NAD+) and has dehydrogenase activity [16]. In mammal, NAD+ is associated with CLOCK/BMAL1 function through SIRT1 [17], and NAD+ and SIRT1 function as a molecular switch to modulate both expression of clock genes and metabolism [17]. We revealed that dCtBP acts as a putative co-activator of CLK/CYC in the transcription of a subset of the E-box clock genes both in vivo and in vitro and its NAD+ domain is essential for the activation.
Results
dCtBP Affects Circadian Locomotor Rhythm in tim-positive Cells
To screen new clock genes, we used the EP lines [18], which carries the Upstream Activation Sequence (UAS) insertion in the promoter region of a target gene. The EP lines were crossed with tim(UAS)-Gal4 as a driver [19]. Because tim is expressed in virtually all clock-related cells [20], a target gene downstream of UAS can be activated by GAL4 in these tissues. This allowed us to screen for gene candidates which contribute to the circadian system, regardless of the tissue specificity of the target gene expression. We found EP3352 strain carrying the UAS insertion in the promoter region of dCtBP altered circadian locomotor rhythm when it was crossed with tim(UAS)-Gal4. About 80% of tim(UAS)-Gal4;EP3352 flies became arrhythmic; the remaining flies demonstrated lengthening of the circadian period to over 26 h (Table 1). Because homozygous EP3352 flies were semi-lethal, the UAS insertion in the promoter region of dCtBP might affect the expression of the dCtBP gene. We newly established two lines of UAS-dCtBP transgenic flies, which enabled us to investigate the effect of dCtBP overexpression. tim(UAS)-Gal4/+; UAS-dCtBP-1/+ flies demonstrated a period length of approximately 25.5 h, significantly longer than those of the corresponding parental strains (t test, P<0.05). The other overexpression flies, tim(UAS)-Gal4/UAS-dCtBP-2, became totally arrhythmic (Figure 1 and Table 1). To further check the effect of dCtBP overexpression in limited clock cells, we used pdf-Gal4 in which GAL4 is expressed in a subset of pacemaker neurons [21]. Both pdf-Gal4/Y;+;UAS-dCtBP-1/+ and pdf-Gal4/Y;UAS-dCtBP-2/+ flies showed a significantly longer period than corresponding each parental strains (t test, P<0.05).
Table 1. Free-running periods of dCtBP-overexpressing and knockdown flies.
| lines | Period(mean ± SEM) | NR | NA |
| tim(UAS)-Gal4/+ | 24.06±0.06 | 32 | 0 |
| pdf-Gal4/Y | 24.17±0.07 | 27 | 1 |
| EP3352/+ | 24.00±0.08 | 8 | 0 |
| UAS-dCtBP-1/+ | 23.95±0.04 | 21 | 2 |
| UAS-dCtBP-2 | 23.88±0.06 | 30 | 0 |
| tim(UAS)-Gal4;EP3352 | 26.30±0.38a,b | 4 | 15 |
| tim(UAS)-Gal4/+;UAS-dCtBP-1/+ | 25.51±0.24a,b | 11 | 2 |
| tim(UAS)-Gal4/UAS-dCtBP-2 | – | 0 | 43 |
| pdf-Gal4/Y;+;UAS-dCtBP-1/+ | 25.23±0.21a,b | 10 | 2 |
| pdf-Gal4/Y;UAS-dCtBP-IR2/+ | 25.32±0.27a,b | 12 | 4 |
| UAS-dCtBP-IR1/+ | 24.11±0.06 | 29 | 0 |
| UAS-dCtBP-IR2/+ | 24.02±0.06 | 34 | 2 |
| tim(UAS)-Gal4/+;UAS-dCtBP-IR1/+ | 24.53±0.05a,b | 63 | 6 |
| tim(UAS)-Gal4/+;UAS-dCtBP-IR2/+ | 24.45±0.04a,b | 58 | 1 |
| pdf-Gal4/Y;+;UAS-dCtBP-IR1/+ | 24.46±0.11a,b | 14 | 0 |
| pdf-Gal4/Y;UAS-dCtBP-IR2/+ | 24.46±0.11a,b | 40 | 1 |
NR: Number of rhythmic flies recorded.
NA: Number of arrhythmic flies recorded.
significantly different from the period of the flies carrying the tim(UAS)-Gal4 as a control (t test, P<0.05).
significantly different from the period of the flies carrying the UAS sequence as a control (t test, P<0.05).
Figure 1. The actograms of dCtBP-knockdown and -overexpressing flies.
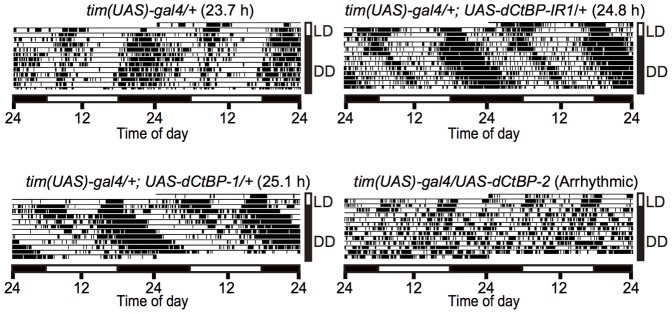
Typical locomotor activity in the control (upper left), dCtBP-knockdown flies (upper right), and dCtBP-overexpressing flies (lower panels). The number in parentheses represents the free-running period of the corresponding flies. Adult flies were entrained to a 12-h light:12-h dark cycle (LD) for 3 days, and then kept in constant darkness (DD). Horizontal bars in white and black indicate times of light and dark, respectively, in LD. Vertical bar in white: LD; vertical bar in black: DD.
Neuron-specific knockdown of dCtBP also affected circadian locomotor rhythm, although the effect was relatively smaller than that of dCtBP overexpression (Table 1 and Figure 1). The periods of knockdown flies tested were slightly but significantly longer than the corresponding parental strains– (t test, P<0.05) regardless of the GAL4 driver. About 10% of flies demonstrated arrhythmicity in tim(UAS)-Gal4/+;dCtBP-IR1/+ flies.
Overexpression of dCtBP Increases the Expression Levels of a Subset of E-box Clock Genes
The daily expression profile of dCtBP in the fly head was measured by quantitative PCR analyses (Q-PCR). The expression level of dCtBP in the driver line as a control showed rhythmicity with a low amplitude (Figure 2A). The statistical analysis with Tukey’s test reveals that it peaks at the end of night phase, which is close to that of Clk [22], [23] (Figure 2A). The expression level of dCtBP was also determined at ZT1 and ZT13 in the tim(UAS)-Gal4/UAS-dCtBP-2 flies, which showed arrhythmicity. The former corresponds to the trough phase of E-box clock genes expression, while the latter corresponds to the peak phase. The dCtBP expression level was 17-times higher than that of controls at both phases (Figure 2B).
Figure 2. Temporal dCtBP expression in control and dCtBP-overexpressing flies.
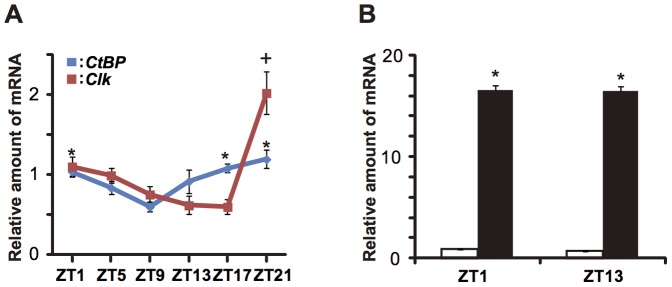
A: Temporal expression profile of dCtBP (blue) and Clk (red) in the head of adult control flies measured by quantitative PCR assay (Q-PCR). ZT1 and ZT13 correspond to 1 h from the onset of light-on and -off conditions in LD, respectively. dCtBP expression reveals a circadian rhythm peaking at the end of night phase. Cross indicates significant difference with trough level of Clk at ZT17 (Tukey’s test, P<0.05). Asterisks indicate a significant difference with the trough level of dCtBP at ZT9 (Tukey’s test, P<0.05). RNAs were sampled three times at each point, and error bars represent S.E.M. B: The expression level of dCtBP at ZT1 and ZT13 in control flies (white) and dCtBP-overexpressing flies (black). dCtBP expression was higher in dCtBP-overexpressing flies than control flies at each phase (*: t test, P<0.05). RNAs were sampled three times at each point, and error bars represent S.E.M. (n = 3).
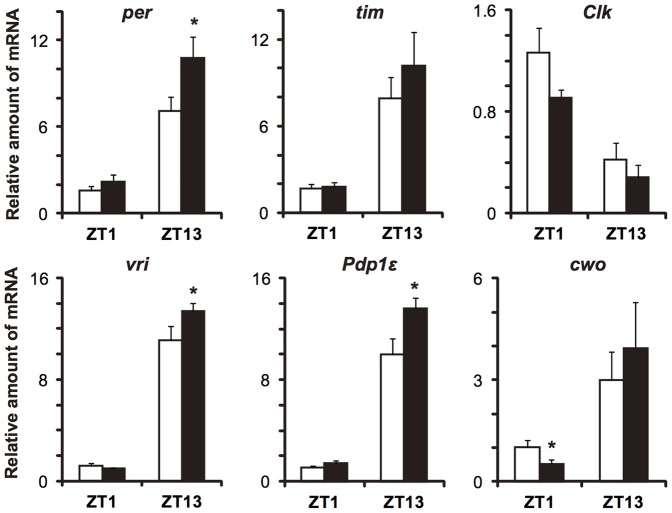
Next, the expression levels of known clock genes were measured in this arrhythmic dCtBP overexpression flies at ZT1 and ZT13. In the case of Clk, whose expression is not controlled through an E-box [24], expression oscillated in antiphase to E-box clock genes. The levels of per, vri, and Pdp1ε increased at their peak phase, whereas that of cwo decreased at the trough phase (Figure 3). The expression level of tim showed no significant change at both phases (Figure 3).
Figure 3. Expression levels of core clock genes in dCtBP-overexpressing flies.
Relative mRNA levels of the indicated genes at the peak and trough phases were measured using a quantitative PCR assay (Q-PCR). Expression levels of per, vri, and Pdp1ε were higher in the dCtBP overexpression flies (black) than in control (white) at the peak phase. dCtBP overexpression decreased the expression levels of cwo at the trough phase. Asterisks indicate a significant difference from control values (t test, P<0.05). RNAs were sampled three times at each point, and error bars represent S.E.M.
Then in order to investigate whether the effect of dCtBP overexpression can be observed in output genes, we quantified the expression level of takeout (to) [25] whose expression shows circadian rhythm [26], [27]. We compared the expression level of to in three groups of flies, tim(UAS)-Gal4, UAS-dCtBP-2 and tim(UAS)-Gal4/UAS-dCtBP-2. dCtBP overexpression significantly increased to expression both at the peak and trough phases in tim(UAS)-Gal4/UAS-dCtBP-2 flies as compared to those in the parental lines (t test, P<0.05). It seemed that the expression of takeout (to) maintain rhythmicity even in the arrhythmic flies (Figure 4).
Figure 4. Expression level of an output gene, takeout, in dCtBP-overexpressing flies.
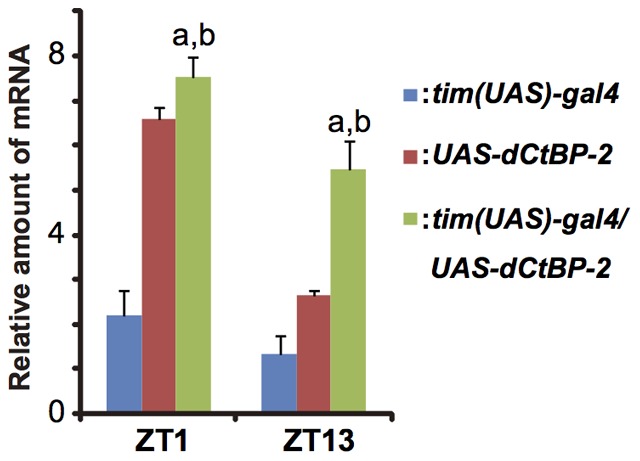
Relative mRNA levels of takeout were measured at ZT1 and ZT13 using a quantitative PCR assay (Q-PCR). The blue, red and green bars represent the tim(UAS)-Gal4, UAS-dCtBP-2 and dCtBP overexpression flies, respectively. The expression level in dCtBP overexpression flies was significantly different from that in tim(UAS)-Gal4 (a: t test, P<0.05) and that of UAS-dCtBP-2 (b: t test, P<0.05) at both phases. RNAs were sampled three times at each point and error bars represent S.E.M.
These results suggest that dCtBP overexpression affects clock-related gene expression. In general, dCtBP overexpression activates the expression of E-box clock genes at the peak phase although there are some exceptions as we observed in tim and cwo. Interestingly, circadian expression rhythm seemed to persist in all clock-related genes we tested, although dCtBP overexpression flies became arrhythmic at the behavioral level.
dCtBP Protein is a Putative Co-activator of CLK/CYC
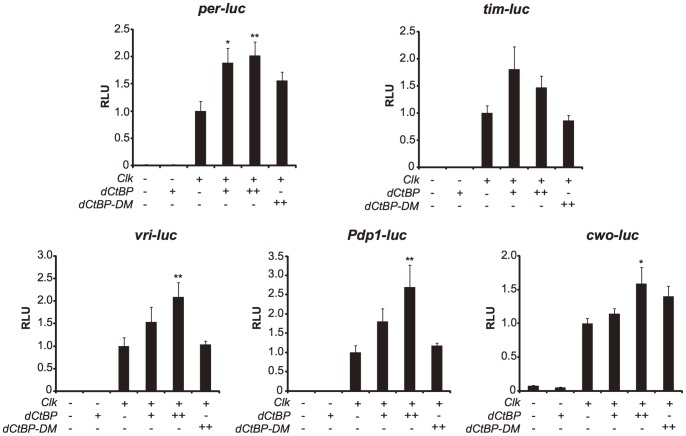
The luciferase assay in cultured Drosophila S2 cells was used to determine whether the gene-specific induction by dCtBP could be observed in vitro. First, we investigated whether dCtBP was able to regulate the E-box clock genes without CLK. S2 cells are reported not to express CLK [22]. Regulation by dCtBP was monitored by promoter-luc, in which firefly luciferase cDNA was linked to the promoter region including the E-box sequence of clock genes. None of those promoters were regulated by dCtBP without CLK (Figure 5). When we further co-transfected the plasmid to express CLK, per-luc, vri-luc, Pdp1-luc and cwo-luc were activated. This activation effect tended to correlate with the amount of dCtBP expression plasmid (Figure 5). However, we could not observe a significant increase of tim-luc under the expression of dCtBP with CLK (Figure 5). Thus, except for the case of cwo-luc, these results obtained in vitro are principally consistent with the results of dCtBP overexpression in vivo.
Figure 5. dCtBP regulates transcription of known clock genes with CLK/CYC.
Relative luciferase activities of per-luc, tim-luc, vri-luc, Pdp1-luc, and cwo-luc in the presence of 0 (–) or 100 (+) ng pAc5.1-dCtBP alone, or 0 (–), 100 (+), 400 (++) ng pAc5.1-dCtBP (dCtBP), or 400 (++) ng pAc5.1-dCtBP-G183A/G186A (dCtBP -DM) in conjunction with 100 ng pAct-Clk are represented. The luciferase activity was normalized by the activity of Renilla luciferase as a control reporter, and then the activity was normalized by the activity of pAct-Clk alone. RLU means relative luminescence unit. dCtBP regulates the promoter activity of core clock genes. The difference between values without Clk was calculated by t test. The difference between the values with Clk was calculated by the Tukey’s test, and asterisks indicate significant differences between two values (*P<0.05 and **P<0.01). These experiments were performed independently three times (or four in some cases) and error bars represent S.E.M.
Next, in order to investigate whether these activations are regulated via the nicotinamide adenine dinucleotide domain (NAD+) -dependence of dCtBP, we supplied CLK with the mutated dCtBP which carries two amino acid substitutions in NAD+ binding region (dCtBP-G183A/G186A) [28]. The expression level of all E-box clock genes we tested with the mutated dCtBP was not significantly different from the value without an intact dCtBP.
Discussion
We propose that dCtBP affects the expression of E-box clock genes. The most obvious evidence is that dCtBP acts as a co-activator of CLK as observed in per, vri and Pdp1ε expression in vivo and in vitro. The regulation mechanism is associated with CLK, because our results in vitro suggest that dCtBP have no effect without CLK (Figure 2A). dCtBP may bind to CLK/CYC through an unidentified domain because we could not find a consensus sequence [9], [11], [12] in CLK and CYC to bind with dCtBP (data not shown). Alternatively, more plausible possibility is that an unknown factor acts as a bridge between dCtBP and CLK/CYC. One candidate to act as such a mediator might be NEJIRE (NEJ), which has been reported to directly bind to CLK and function as its co-factor [5], [6]. In mammals CtBP is postulated to antagonaize CBP/p300 [29] which is a homolog of NEJ [4].
We found that the activation in E-box clock genes did not occur with the mutated dCtBP having amino acid substitutions in NAD+ binding domain [16], [28] (Figure 5). The mutated dCtBP might become unstable so that the protein no longer activates those genes [30]. Alternatively, our result suggests that this domain is important for the activation of those genes. In mammal, NAD+ is reported to modulate the rhythmic expression of clock genes downstream of CLOCK/BMAL1, which is a counterpart of CLK/CYC in Drosophila, through Sirt1 [17]. Although it is unknown whether NAD+ contributes to Drosophila circadian clock, the metabolic regulation of the circadian oscillator via the NAD-dependence is probably conserved between mammal and fly.
The expression patterns of all core clock genes seemed to maintain rhythmicity, even in dCtBP overexpressing flies that demonstrated arrhythmicity at the behavioral level (Figure 4). The up-regulation of core clock genes by dCtBP overexpression may induce arrhythmicity in the output pathway both at the molecular and behavioral levels. Pdp1ε is a leading candidate responsible for this loss of rhythmicity because it is known to function not only as a core clock gene but also as a regulator of output genes including to [25], [27], [31]. However, our results reveal that the expression levels of both Pdp1ε and to increased with remaining its rhythmicity even in behaviorally arrhythmic dCtBP overexpressing flies. Thus the responsible output genes that control locomotor rhythmicity may be more strongly affected by the increased level of Pdp1ε and lost rhythmicity. Alternatively, the dCtBP may directly regulate the expression of such output genes and arrhythmicity of dCtBP expression caused by overexpression induced arrhythmicity of expression in those genes.
Both overexpression and knockdown of dCtBP caused to lengthen circadian period. This is inconsistent with the general idea that an opposite effect on period could be induced by the excess and less product of the clock-related gene. Although we do not have a definitive explanation of this inconsistency at the present, it might be valuable to point out that recent reports reveal that dCtBP has dual roles as an activator and repressor of Wnt target genes [13], [14]. However, no reports to date have indicated an association between Wnt signaling and circadian gene expression in Drosophila. In addition, because the Wnt signaling pathway does not function in the S2 cells we used [32], we have not been able to obtain any supporting evidence at molecular level. The further extensive study is needed to determine whether dCtBP has dual roles as an activator and an repressor in Drosophila circadian clock. Given that CtBP in mammal is supposed to antagonize to CBP/p300, which is the counterpart of NEJ [29], our results may give a hint to dissolve the problem that there are conflicting reports that NEJ acts as a co-activator [5] and co-repressor [6] of CLK in Drosophila. Our study sheds new light on the regulation mechanism of the E-box clock genes by CLK/CYC and its co-factors.
Experimental Procedures
Fly Strains
tim(UAS)-Gal4 strain [19] was used as the driver to knock down and overexpress dCtBP. UAS-IR lines [33] were established at the National Institute of Genetics. Knockdown flies were obtained by mating females of the driver line to males in each of the UAS-IR lines. The EP3352 line [18] was obtained from the Harvard Stock Center. UAS-dCtBP transgenic lines were established by injection of UAS-dCtBP plasmid into w1118 embryos (BestGene). dCtBP-overexpressing flies were obtained by mating the driver females to EP3352 males or UAS-dCtBP transgenic males.
Recording of Locomotor Activity Rhythm
Flies were kept on standard glucose-cornmeal medium under 12-h light:12-h dark cycles (LD) at 25°C. We measured the locomotor activity of the adult flies using Drosophila activity monitors (Trikinetics Inc.) for 3 days in LD cycles, then over 10 days in constant darkness (DD). A single fly was introduced into a measuring glass tube containing agar gel with 100 mg/ml glucose. The periods were calculated with a χ2 periodogram [34] programmed using the Matlab R2007b software (MathWorks Inc.).
Q-PCR to Analyze Temporal Expression Levels of Clock Genes
The tim(UAS)-Gal4 strain [19] was used as control. Control and dCtBP-overexpressing flies entrained for at least 3 days under LD were sampled three times at each point. Total RNA was isolated from 100 heads at each time point as described elsewhere [35]. cDNA was synthesized from 5 µg total RNA using Ready-To-Go T-Primed First-Strand Kit (Amersham) according to the standard protocol. Q-PCR was performed using Applied Biosystems 7300 and Power SYBR Green PCR Master Mix (Applied Biosystems). PCR reactions were performed with samples containing 1× Power SYBR Green PCR Master Mix (Applied Biosystems), 5 µM primers, and 1 µL synthesized cDNA in a 20 µL volume using the following amplification procedure: 10 min at 95°C, then 40 cycles of 15 s at 95°C, 30 s at 60°C, and 1 min at 72°C. Gapdh2 expression levels were quantified and used as the internal control. We purified total RNA at each time point. Each RNA was used as a template to synthesize cDNA. We repeated these steps and obtained three different cDNAs at each point. One time-series of cDNAs were analyzed by Q-PCR at once with the primer sets in Table S1. The data finally obtained were calculated with the 2−ΔΔCt Method [36] using the following equation, ΔΔCt = (Ct target – Ct Gapdh2) ZT x – (Ct target – Ct Gapdh2) ZT1. We confirmed that all primer sets we used didn’t yield any non-specific amplification by a melting curve analysis using the products of Q-PCR.
Construction of Expression Plasmids
The coding sequence of dCtBP (see http://flybase.org/reports/FBgn0020496.html) was cloned into a pAc5.1B-V5/His plasmid (Invitrogen) by the SA-cloning method [37] using the sets of primers in Table S2.
To construct the pAc5.1-dCtBP-G183A/G186A plasmid, mutagenesis of pAc5.1-dCtBP was performed by site-directed mutagenesis PCR method using PCR with primers 5′- CTGGTGGGACTGGCCCGCATTGCTAGCGCCGTGGCCCTG-3′ and 5′- CAGGGCCACGGCGCTAGCAATGCGGGCCAGTCCCACCAG-3′.
To construct the UAS-dCtBP plasmid for transgenic flies, dCtBP was amplified by PCR using head cDNA in w1118 as a template with primers 5′-AGCGAAATGGACAAAAATCTG-3′ and 5′-CTACGGCGCCTCCGTTGACT-3′ and cloned into pCR2.1 vector (Invitrogen). To construct the UAS-dCtBP plasmid, the dCtBP PCR fragment in pCR2.1 was doubly digested by SpeI and XbaI (New England Biolabs), purified, and cloned into the XbaI site of the pUAST plasmid [38].
Luciferase Assay in Drosophila Cultured Cells
Cultured Drosophila S2 cells were plated in 24-well tissue culture plates with Shields and Sang M3 insect medium (Sigma-Aldrich) supplemented with 12.5% fetal bovine serum (Biowest) and antibiotics (12.5 U/mL penicillin, 12.5 mg/mL streptomycin; Invitrogen) and transfected by a standard method [22] using Effectene Transfection Reagent (QIAGEN) with 100 ng of each promoter-luc [39], [40] in the presence of 0 or 100 ng pAc5.1- dCtBP alone or 0, 100, or 400 ng pAc5.1- dCtBP in conjunction with 100 ng pAct-Clk. As a positive control for the luciferase assay, cells were transfected with 610 ng pAc5.1B empty vector (Invitrogen) with 10 ng pAc5.1-Rluc. Each Luciferase activity was measured 48 h after transfection as described elsewhere [39], [40]. The mean values were calculated from data obtained by three (or four in some cases) independent experiments.
Supporting Information
(DOC)
(DOC)
Acknowledgments
UAS-IR transgenic strains and the EP3352 line were obtained from Genetic Strains Research Center, National Institute of Genetics and the Harvard Stock Center, respectively. We thank Justin Blau and Jeffrey C. Hall for providing tim(UAS)-Gal4, and pdf-Gal4, respectively. We also thank Steve A. Kay for pAct-Clk, per-luc, and tim-luc plasmids. We are grateful to Kenji Tomioka for comments on this manuscript, and Kiyo Kimura, Makiko Haruta, and Kyoko Sakamoto for technical assistance.
Funding Statement
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to TT and AM, and the Japan Society for the Promotion of Science (JSPS) to TQI. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rosbash M (2009) The implications of multiple circadian clock origins. PLoS Biol 7: e1000062 doi:10.1371/journal.pbio.1000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hardin PE (2006) Essential and expendable features of the circadian timekeeping mechanism. Curr Opin Neurobiol 16: 686–692. [DOI] [PubMed] [Google Scholar]
- 3. Tomioka K, Matsumoto A (2010) A comparative view of insect circadian clock systems. Cell Mol Life Sci 67: 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Akimaru H, Chen Y, Dai O, Hou DX, Nonaka M, et al. (1997) Drosophila CBP is a co-activator of cubitus interrupts in hedgehog signaling. Nature 386: 735–738. [DOI] [PubMed] [Google Scholar]
- 5. Hung HC, Maurer C, Kay SA, Weber F (2007) Circadian transcription depends on limiting amount of the transcription co-activator nejire/CBP. J Biol Chem 282: 31349–31357. [DOI] [PubMed] [Google Scholar]
- 6. Lim C, Lee J, Choi C, Kim J, Doh E, et al. (2007) Functional role of CREB-binding protein in the circadian clock system of Drosophila melanogaster. Mol Cell Biol 27: 4876–4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davis RL, Turner DL (2001) Vertebrate Hairy Enhancer of split regulated proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene 20: 8342–8357. [DOI] [PubMed] [Google Scholar]
- 8. Chinnadurai G (2007) Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol 39: 1593–1607. [DOI] [PubMed] [Google Scholar]
- 9. Nibu Y, Zhang H, Levine M (1998) Interaction of short-range repressors with Drosophila CtBP in the embryo. Science 280: 101–104. [DOI] [PubMed] [Google Scholar]
- 10. Schaeper U, Subramanian T, Lim L, Boyd JM, Chinnadurai G (1998) Interaction between a cellular protein that binds to the C-terminal region of adenovirus E1A (CtBP) and a novel cellular protein is disrupted by E1A through a conserved PLDLS motif. J Biol Chem 275: 8549–8552. [DOI] [PubMed] [Google Scholar]
- 11. Poortinga G, Watanabe M, Parkhurst SM (1998) Drosophila CtBP: A Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J 17 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nibu Y, Zhang H, Bajor E, Barolo S, Smaii S, et al. (1998) dCtBP mediates transcriptional repression by Knirps, Krüppel and Snail in the Drosophila embryo. EMBO J 17: 7009–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fang M, Li J, Blauwkamp T, Bhambhani C, Campbell N, et al. (2006) C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J 25: 2735–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bhambhani C, Chang JL, Akey DL, Cadigan KM (2011) The oligomeric state of CtBP determines its role as a transcriptional co-activator and co-repressor of Wingless targets. EMBO J 30: 2031–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, et al. (2010) Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Pros Natl Acad Sci U S A 103: 13497–13502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, et al. (2002) Transcription corepressor CtBP is an NAD+-regulated dehydrogenase. Mol Cell 10: 857–869. [DOI] [PubMed] [Google Scholar]
- 17. Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, et al. (2008) The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134: 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rørth P (1996) A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci U S A. 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blau J, Young MW (1999) Cycling vrille expression is required for a functional Drosophila clock. Cell 99: 661–671. [DOI] [PubMed] [Google Scholar]
- 20. Kaneko M, Hall JC (2000) Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol 422: 66–94. [DOI] [PubMed] [Google Scholar]
- 21. Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791–802. [DOI] [PubMed] [Google Scholar]
- 22. Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, et al. (1998) Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280: 1599–1603. [DOI] [PubMed] [Google Scholar]
- 23. Lee C, Bae K, Edery I (1998) The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER-TIM complex. Neuron 21: 857–867. [DOI] [PubMed] [Google Scholar]
- 24. Glossop NR, Lyons LC, Hardin PE (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science 286: 766–768. [DOI] [PubMed] [Google Scholar]
- 25. Benito J, Hoxha V, Lama C, Lazareva AA, Ferveur JF, et al. (2010) The circadian output gene takeout is regulated by Pdpε. Proc Natl Acad Sci U S A. 107: 2544–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sarov-Blat L, So WV, Liu L, Rosbash M (2000) The Drosophila takeout gene is a novel molecular link between circadian rhythm and feeding behavior. Cell 101: 647–656. [DOI] [PubMed] [Google Scholar]
- 27. So WV, Sarov-Blat L, Kotarski CK, McDonald MJ, Allada R, et al. (2000) takeout, a novel Drosophila gene under circadian clock transcription regulation. Mol Cell Biol 20: 6935–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuppuswamy M, Vijayalingam S, Zhao LJ, Zhou Y, Subramanian T, et al. (2007) Role of the PLDLS-binding cleft region of CtBP1 in recruitment of core and auxiliary components of the corepressor complex. Mol Cell Biol 28: 259–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chinnadurai G (2007) Transcriptional regulation by C-terminal binding protein. Int Biochem Cell Biol 39: 1593–1607. [DOI] [PubMed] [Google Scholar]
- 30. Mani-Telang P, Sutrias-Grau M, Williams G, Arnosti DN (2007) Role of NAD binding and catalytic residues in the C-terminal binding protein corepressor. FEBS Lett 581: 5241–5246. [DOI] [PubMed] [Google Scholar]
- 31. Benito J, Zheng H, Hardin PE (2007) PDP1ε functions downstream of the circadian oscillator to mediate behavioral rhythms. J Neurosci 27: 2539–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yanagawa S, Lee JS, Ishimoto A (1998) Identification and characterization of a novel line of Drosophila Schneider S2 cells that respond to wingless signaling. J Biol Chem 273: 32353–32359. [DOI] [PubMed] [Google Scholar]
- 33. Pili-Floury S, Leulier F, Takahashi K, Saigo K, Samain E, et al. (2004) In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against gram-positive bacterial infection in Drosophila adults. J Biol Chem 279: 12848–12853. [DOI] [PubMed] [Google Scholar]
- 34. Sokolove PG, Bushnell WN (1978) The chi square periodogram: its utility for analysis of circadian rhythm. J Theor Biol 72: 131–160. [DOI] [PubMed] [Google Scholar]
- 35. Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, et al. (2002) Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem 277: 14048–14052. [DOI] [PubMed] [Google Scholar]
- 36. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 37. Matsumoto A, Itoh TQ (2011) Self Assembly (SA)-cloning: A rapid construction method for recombinant molecules from multiple fragments. Biotechniques 51: 55–56. [DOI] [PubMed] [Google Scholar]
- 38. Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- 39. Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, et al. (2007) A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev 21: 1687–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Itoh TQ, Tanimura T, Matsumoto A (2011) bHLH-ORANGE family genes regulate the expression of E-box clock genes in Drosophila. Appl Entomol Zool 46: 391–397. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOC)