Abstract
Background
Viral infection and anti-cardiac immunity are involved in the pathogenesis of dilated cardiomyopathy (DCM). Immunity targeting particular antigens may evoke expansion of reactive T-cell clones.
Material/Methods
Myocardial tissues from explanted hearts were investigated for clonal T-cell-receptor- (TCR-) β rearrangements by an established semi-nested polymerase chain reaction (PCR), followed by high-resolution GeneScan analysis and direct sequencing. From 17 explanted DCM hearts, 3 myocardial samples each were obtained from the right ventricle, the septum, and the left ventricle (total: 9 myocardial samples per case). Six explanted hearts with non-DCM cardiomyopathy entities served as controls.
Results
GeneScan analysis revealed polyclonal TCR-β rearrangements in all controls. In contrast, at least 1 myocardial sample in 9 out of 17 DCM hearts (total: 20 of the 81 DCM specimens) displayed single dominant TCR-β PCR products consistent with the presence of clonal T-cell populations. Direct sequencing of the clonal TCR-β PCR-products disclosed an involvement of Vb 19.01 segments in 14 of the dominant amplificates (70%). Further TCR-Vβ segments involved in clonal TCR-β rearrangements of DCM hearts were Vβ 6-1.01 (n=1), Vβ 6-3.01 (n=2), Vβ 6-5.01 (n=1), Vβ 10-3.02 (n=1), and Vβ 19.03 (n=1).
Conclusions
The detectability of clonal TCR-β rearrangements indicates a pathogenic relevance of this finding in DCM. The predominance of Vβ 19.01 segments suggests that the immune response in DCM patients targets particular epitopes. However, the partly heterogenic TCR-β populations in various myocardial samples from the respective cases support the notion that T-cell immunity may target multiple epitopes in human DCM.
Keywords: dilated cardiomyopathy, myocarditis, pathogenesis, immune system, T-cell receptor, TCR, clonal TCR-rearrangements
Background
Chronic myocardial inflammation can be identified as a specific pathogenic mechanism in a significant proportion of dilated cardiomyopathy (DCM) patients, either with viral persistence (e.g., coxsackievirus, adenovirus, Parvovirus B19/B19V) or in terms of a post-viral anti-cardiac immunity maintained even after viral elimination [1,2]. Immunohistologically detected intramyocardial inflammation has been identified as a feature of myocarditis with prognostic and therapeutic relevance [3–5].
The T-cell receptor (TCR) is a transmembrane heterodimer that recognizes peptides presented by HLA molecules. Adaptive immunity depends on genetic recombination, which produces diverse functional TCR gene rearrangements from a pool of discontinuous segments. The human TCR-β locus consists of 65 V (variable), 2 D (diversity), and 13 J (joining) segments. As a result, 106 different TCR-β chain rearrangements are available in the human naïve T-cell repertoire [6]. Clonal TCR-β rearrangements are detectable in T-cell neoplasms. Chronic activation of the immune system targeting specific antigenic epitopes can also evoke clonal TCR-β expansions in the inflamed tissues [7,8]. A further mechanism that may contribute to the development of clonal TCR-β rearrangements is superantigens (from bacteria or viruses) by cross-linking the lateral side of the MHC class II molecule with the Vβ portion of the TCR, thereby stimulating proliferation of specifically reactive T-cells [9]. Experimental data from both autoimmune [10,11] and coxsackievirus myocarditis [12] have shown restricted TCR-β rearrangements involving V-b 8.2 and 10 by CDR3 (complementary determining region 3) spectratyping in rodents. Depletion of the respective clonotypes by antibodies or DNA vaccines resulted in significant protection from disease development [11,13].
In light of these findings from experimental myocarditis models, we sought to investigate whether clonal TCR-β restrictions are detectable also in human DCM. We furthermore investigated the distribution pattern of clonal TCR-β rearrangements by analyzing multiple samples (3 each) obtained from 3 cardiac regions (9 samples total) from explanted DCM hearts (RV: right ventricle, S: Septum, LV: left ventricle). We hypothesized that if clonal TCR-β rearrangements target particular, but multiple, epitopes, various clonal TCR-β rearrangements with a multifocal pattern might be present in DCM hearts.
Material and Methods
Patients
Normal tonsillar tissues obtained at autopsy were used as polyclonal controls of the TRC-b PCR method, whereas the T-cell line MOLT4 served as control for TCR-β clonality. From 17 explanted DCM hearts (1 female; age: 48.9±13.1 years; LVEF: 19.7±4.9%), each 3 myocardial samples (ca. 50 mg tissue) were obtained from random locations of the right ventricle (RV; RV 1–3), the septum (S), and the left ventricle (LV) at explantation (9 total myocardial samples per case). Coronary artery disease was excluded in all these patients by the lack of significant coronary stenosis in coronary angiography, and further possible secondary causes (e.g., valvular, hypertensive or systemic diseases) were excluded. Patients subjected to immunomodulatory treatment such as immunosuppression or antiviral interferon treatment were excluded from this study. Single LV myocardial samples from 6 explanted hearts with non-DCM cardiomyopathy entities (1 female; age: 53±10 years; LVEF: 19±4%; ischemic cardiomyopathy: n=3; hypertrophic non-obstructive cardiomyopathy: n=1; transposition of the great vessels and ventricular septum defect: n=2) served as controls. The cardiomyopathy patients gave informed consent for research projects of their explanted hearts. These investigations were approved by the local ethics committee at the Charité – Universitätsmedizin Berlin in the framework of the Sonderforschungsbereich TR19 in compliance with the Helsinki Declaration.
PCR for the detection of clonal TCR-β rearrangements
The PCR for the detection of TCR-β rearrangements was performed as described by Assaf et al. [14]. In brief, DNA was extracted from snap-frozen myocardial tissues using the QIAGEN DNA extraction kit (QIAGEN, Hilden, Germany) according the manufacturer’s instructions. Two hundred ng of genomic DNA from each sample was subjected to a semi-nested PCR with 2 separate reactions involving the same Vβ consensus primer (Vβ pan: 5’-CTCGAATTCT(T/G)T(A/T) (C/T)TGGTA(C/T)C (G/A)(T/A)CA-3’; 200 ng) and 2 Jβ different primer sets (200 ng each set) consisting of 6 (Jβ1 family; JβFS1A) and 7 (Jβ2 family; JβFS2A) Jβ family-specific primers, respectively, employing high-quality high-performance liquid chromatographic purified oligonucleotides. Negative controls (samples without DNA) were included after each sample. Thirty cycles were carried out with a primer annealing temperature of 60°C (40 sec) for the initial 5 cycles and 57°C (40 sec) for the remaining 25 cycles. For re-amplification, an aliquot (1%) of the first 2 reactions was used as a template in 2 additional separate PCRs, comprising 40 cycles each with the same annealing temperature profile as described above. The same Vβ primer (200 ng) was used in combination with 2 nested family-specific Jβ primer mixes (JβFS1 and JβFS2; 200 ng each set). The conditions for denaturation (96°C, 15 sec) and primer extension (72°C, 40 sec) remained constant through all cycles of the first and second PCR, whereas the concentration of MgCl2 was 2.5 mmol/L in the first and 1.5 mmol/L in the second amplification. All reactions were carried out in a final volume of 100 μL with 0.8 mmol/L of dNTPs (200 μmol/L each) and 2.5 units Taq polymerase (Perkin Elmer, Weiterstadt, Germany) in a thermal cycler (TC9600, Perkin Elmer, Weiterstadt, Germany).
High-resolution GeneScan analysis of the PCR products was performed with 5-carboxyflourescein-labeled Vβ primers. Aliquots (2 μL) of PCR products were mixed with loading buffer (2 μL formamide, 0.5 μL EDTA), and 0.5 μL of the internal size standard (Genescan-500) were included for precise determination of the length of the amplificates. After denaturation for 2 min at 90°C, the products were separated on a sequencing gel and analyzed by automatic fluorescence quantification and size determination, using the computer program GENESCAN 672 (ABI 373A, Applied Biosystems, Darmstadt, Germany).
In cases with clonal TCR-β rearrangements, the re-amplification was repeated with unlabeled Vβ primers to generate unlabeled PCR products, which, after purification, can be used for direct fluorescence dye terminator cycle sequencing. The sequencing reactions were analyzed on a 377A DNA sequencer (Applied Biosystems, Darmstadt, Germany) after removal of the unincorporated fluorescence dye. Each sequencing reaction was carried out in both directions using the primers Vβpan and JβFS1 or JβFS2, respectively. The sequences were identified by comparison to published sequences in the international ImMunoGeneTics database (IMGT; website: http://www.imgt.org/) [15].
Statistical analysis
Statistical analysis was performed using JMP Statistical Discovery Software 4.02 (SAS Institute, Inc., Cary, NC, USA). Qualitative data were compared using the chi-square test. A probability value of p<0.05 was considered statistically significant.
Results
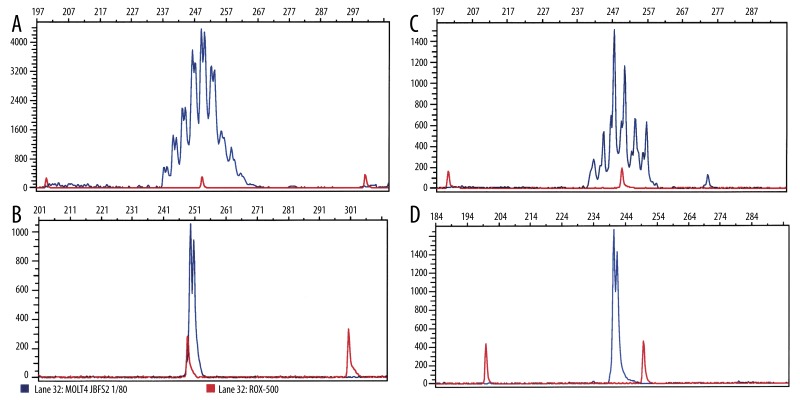
The analysis of DNA extracted from tonsils gave rise to polyclonal TCR-β rearrangements (Gaussian-like distribution of the PCR amplificates), with a size range from 234 to 261 bp as revealed by high-resolution GeneScan analysis (Figure 1A). In contrast, TCR-β PCR employing DNA extracted from a T-cell line (MOLT-4) resulted in a single PCR product, characteristic for the presence of a single TCR-β rearrangement (Figure 1B). In control myocardial tissues (non-DCM hearts), the GeneScan analysis of the TCR-β PCR products revealed a Gaussian-like size distribution consistent with the presence of polyclonal T-cell populations (Figure 1C). In contrast, clonal TCR-β rearrangements were detectable in at least 1 myocardial sample in 9 out of 17 DCM cases (3 each from RV, LV, and S) of the DCM hearts (Figure 1D). The demographic data of this subset of DCM patients were not statistically different compared to the remaining DCM patients without detectable clonal TCR-β rearrangements (female: 0; age: 51.8±11.3 years; LVEF: 18.9±5.2%). A total of 20 of the 81 specimens displayed single dominant TCR-β PCR products consistent with the presence of clonal T-cell populations. There was a non-significant tendency for a higher detection rate of T-cell clonality in the samples from the LV compared with the remaining regions of the DCM hearts (RV: n=4, S: n=4, LV: n=6). Interestingly, the sizes of the dominant PCR products were not identical within the same case or among the respective cases.
Figure 1.
Representative GeneScan analysis results of the fluorescence-labeled TCR-β PCR products (blue color). The x-axes represent molecular size (base pairs) and the y-axes fluorescence intensity. (A) Normal tonsillar tissue with polyclonal TCR-β composition. (B) T-cell lymphoma line MOLT4 with clonal TCR-β composition. (C) Ischemic cardiomyopathy with polyclonal TCR-β composition. (D) Dilated cardiomyopathy with clonal TCR-β composition.
The sequence analysis of the clonal TCR-β PCR-products (n=20) from the 9 explanted DCM hearts showed an involvement of Vβ19.01 segments in 14 of the dominant amplificates (70%). Further TCR-Vβ segments involved in clonal TCR-β rearrangements of DCM hearts were Vβ6-1.01 (n=1), Vβ6-3.01 (n=2), Vβ6-5.01 (n=1), Vβ10-3.02 (n=1), and Vβ19.03 (n=1). Clonal TCR-β rearrangements were present in only 1 of the 9 samples from the different cardiac regions in 3 DCM hearts (patient code 05: Vβ6-3.01, 07: Vβ10-3.02, and 20: Vβ6-1.01), and the remaining 6/9 DCM cases demonstrated clonally rearranged TCR-β genes in at least 2 of the 9 different samples investigated from each individual. An involvement of the same Vb-segment was detected in multiple (at least 2) samples in 5/6 (83%) of the DCM patients with Vβ19.01 TCR-Vβ rearrangement (patients 02, 13, 14, 16 and 17), and different Vβ segments were found in the remaining identified clones. Only 1 out of 6 cases with a clonal Vβ19.01 rearrangement (code no. 21) demonstrated no further clonal Vβ19.01 composition in further samples. The results of sequence analysis of clonal TCR-β PCR-products identified by GeneScan analysis in these explanted DCM hearts are summarized in Table 1.
Table 1.
TCR clones analyzed in the different samples from explanted DCM hearts.
| Code | RV 1 | RV 2 | RV 3 | S 1 | S 2 | S 3 | LV 1 | LV 2 | LV 3 |
|---|---|---|---|---|---|---|---|---|---|
| Vβ19.01 | Vβ19.01 | Vβ19.01 | Vβ6-3.01 | Vβ19.01 | |||||
| 02 | J1-2.01 | J1-2.01 | J1-2.01 | Polyclonal | Polyclonal | Polyclonal | J1-1.01 | J1-2.01 | Polyclonal |
| D2.01 | D2.01 | D2.01 | D1.01 | D2.01 | |||||
|
| |||||||||
| Vβ6-3.01 | |||||||||
| 05 | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | J1-1.01 |
| D1.01 | |||||||||
|
| |||||||||
| Vβ10-3.02 | |||||||||
| 07 | J1-2.01 | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal |
| D1.01 | |||||||||
|
| |||||||||
| Vβ19.01 | V19.01 | ||||||||
| 13 | Polyclonal | Polyclonal | Polyclonal | J1-2.01 | Polyclonal | Polyclonal | Polyclonal | J1-2.01 | Polyclonal |
| D2.01 | D2.01 | ||||||||
|
| |||||||||
| Vβ19.01 | Vβ19.01 | Vβ19.01 | |||||||
| 14 | Polyclonal | J1-2.01 | J1-2.01 | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | J1-2.01 |
| D2.01 | D2.01 | D2.01 | |||||||
|
| |||||||||
| Vβ19.01 | Vβ19.01 | ||||||||
| 16 | Polyclonal | Polyclonal | Polyclonal | Polyclonal | J1-5.01 | J1-2.01 | Polyclonal | Polyclonal | Polyclonal |
| D2.01 | D1.01 | ||||||||
|
| |||||||||
| Vβ19.03 | V19.01 | Vβ19.01 | |||||||
| 17 | Polyclonal | J2-5.01 | Polyclonal | Polyclonal | Polyclonal | J2-7.01 | J2-7.01 | Polyclonal | Polyclonal |
| D2.01 | D1.01 | D1.01 | |||||||
|
| |||||||||
| Vβ6-1.01 | |||||||||
| 20 | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | Polyclonal | J1-6.01 | Polyclonal |
| D1.01 | |||||||||
|
| |||||||||
| Vβ6-5.01 | Vβ19.01 | ||||||||
| 21 | Polyclonal | Polyclonal | Polyclonal | J2-2.01 | J1-2.01 | Polyclonal | Polyclonal | Polyclonal | Polyclonal |
| D1.01 | D1.01 | ||||||||
The table depicts the distribution of the analyzed TCR clones (VDJ-regions) in the various samples (RV – right ventricle; S – septum; LV – left ventricle) from explanted DCM hearts.
Discussion
One important finding of our investigation is that the presence of clonal TCR-β rearrangements is a disease-associated phenomenon in 9 out of 17 DCM patients, but not in other non-DCM cardiomyopathies. These first data on the application of this analysis system of clonal TCR-β rearrangements are limited by the fact that only single myocardial samples of the control hearts were investigated, but multiple samples (3 each) were analyzed from 3 cardiac regions (9 samples total) from explanted DCM hearts (RV: right ventricle, S: Septum, LV: left ventricle). Therefore, sampling bias may have contributed to the obtained results. However, the presence of disease-specific TCR- dominances was also confirmed by different methodological approaches in further investigations [16,17]. One major goal of this study was to determine the applicability of the methodology published by Assaf et al. [14], followed by high-resolution GeneScan and direct sequencing. Secondly, we sought to determine the distribution of detectable clonal TCR-β rearrangements in DCM hearts. Analysis of 9 different tissue samples (3 each from the RV, S, and LV) revealed that these clonal T-cell populations were not identically distributed within the respective DCM hearts, albeit with a clear predominance of Vβ 19.01 segments. The clear predominance of the Vβ19.01 segment suggests that a common antigen may evoke and maintain such a selective clonal TCR-β expansion in the majority of DCM cases. Clonal TCR-β rearrangements with further TCR-β segments (Vβ6-1.01, Vβ6-3.01, Vβ6-5.01, Vβ10-3.02 and Vβ19.03) were identified less frequently. However, we were not able to confirm findings by Luppi et al on the restricted usage of Vβ 3, 7, and 13.1 families [17]. Firstly, this discrepancy may result from different patient cohorts. Whereas Luppi et al analyzed 3 children with fulminant myocarditis, we analyzed adult DCM patients with a chronic history of symptoms at end-stage heart failure. Furthermore, methodological differences are also important to consider. The CDR3 spectratyping used by Luppi et al strongly depends on the PCR results from 24 different TCR-β PCRs obtained from healthy controls (ie, peripheral blood lymphocytes), and restricted TCR-Vβ usage is interpreted as the relative increase of distinct PCR products in impaired tissues. In contrast, a determination of clonal TCR-β rearrangements is possible by use of the TCR-β PCR system. This family-specific PCR with multiple primers, however, may be hampered by pseudoclonality issues [14,18]. Nonetheless, this TCR-β PCR approach can discriminate between polyclonal and oligoclonal TCR-β compositions without the need of interpretations based on a control population.
Interestingly, whereas the same Vb-segment was involved in multiple (at least 2) samples in 5/6 (83%) of the DCM patients with Vβ19.01 TCR-Vβ rearrangements, different Vβ segments were found with respect to the remaining identified clones (Vβ6-1.01, Vβ6-3.01, Vβ6-5.01, Vβ10-3.02, Vβ19.03). These data are consistent with clonal expansion of T-cells at multiple intramyocardial foci, and rather incompatible with a diffuse infiltration of the whole DCM heart with a single T-cell population. Our data may be furthermore compatible with the typically non-homogeneously distributed T-lymphocytic clusters suggestive of focal expansion and myocytolysis in DCM [3,19,20]. We therefore conclude that clonal TCR-β rearrangements may have a multifocal pattern in DCM. Different TCR-Vβ skewing in different cardiac regions were also confirmed by CDR3 spectratyping in 2 out of 3 pediatric patients with acute myocarditis [17]. The clear predominance of the Vβ19.01 segment suggests that a common antigen may evoke and maintain such a selective clonal TCR-β expansion in the majority of DCM cases. Clonal TCR-β rearrangements with further TCR-β segments (Vβ6-1.01, Vβ6-3.01, Vβ6-5.01, Vβ10-3.02 and Vβ19.03) were identified less frequently. The multifocal presence of different T-cell clones might be a decisively important issue for the design of TCR-based immunotherapies in DCM, since such approaches would need to target not just one, but several, different clonal T-cell populations simultaneously. In line with these considerations, Matsumoto et al reported that only the combined deletion of both TCR-Vβ 8.2 and TCR-Vβ 10 clones proved efficacious in experimental autoimmune myocarditis, while the approach of a single TCR-Vβ deletion did not improve disease severity [11].
These insights indicate that multiple antigens may be responsible for clonal expansion of reactive T-cells, and warrant further investigations on the respective epitopes. Clonal expansion of T-lymphocytes in DCM may occur in response to specific stimulation by particular epitopes. Potential candidates may be either viral proteins in the framework of antiviral immunity [21,22], or myocardial antigens such as in molecular mimicry [23]. Given the insights from both viral [12] and autoimmune myocarditis [10,24], both options may also hold true for the appearance of clonal T-cell populations in human DCM. Viral proteins might also evoke clonal T-cell expansions by means of superantigens [9,17]. Importantly, investigations on coxsackievirus B4 did not confirm superantigen activity [25]. The focal nature of viral infection and cell-to-cell viral spreading [21,26] might be a further factor contributing to the observed multifocal clonal TCR-β rearrangements in DCM. Regarding autoimmunity, it is important to consider that deletion of self-reactive T-cells in the thymus is not complete, and a minority of self-reactive T-cells may also surpass peripheral selection mechanisms. These potentially self-reactive T-cells can be activated when encountering autoantigens, which are usually not accessible to or are ignored by circulating T-cells [27]. With the ensuing tissue destruction in viral myocarditis, cryptic myocardial antigens can be presented to the immune system, which may evoke local recruitment and clonal expansion of self-reactive T-cells targeting these epitopes [1,2].
An association of findings of skewed CDR3 spectratyping with enteroviral protein (VP-1) expression has been reported in myocarditis patients [17]. These conclusions were drawn by associations to VP-1 (enteroviral protein-1) immunoreactivity, which was reported in all cardiac samples (100%) [17]. However, PCR failed to confirm enteroviral genomes in these tissues. The presence of enteroviral genomes assessed by PCR, however, is reported in about 10–20% of myocarditis and DCM patients [1,28]. Sensitivity and, especially, specificity of VP-1 immunostaining may be an important issue to solve this discrepancy. Further cardiotropic viruses such as adenovirus and Parvovirus B19 (B19V) have been identified even more frequently in myocarditis and DCM patients in recent investigations [4,29]. These viral genomes were not investigated in the aforementioned study by Luppi et al. [17]. On the other hand, the debate on the pathogenic and prognostic significance of the presence of B19V genomes in myocardial tissues is controversial [4,30]. It may be deduced that the antigen-specific T-cells reactive to these proteins might be different from the T-cell clones targeting enteroviral VP-1 [22], with relevance also for DCM patients with multiple viral infections [31].
Conclusions
Viral infection and anti-cardiac immunity are involved in the pathogenesis and prognosis of dilated cardiomyopathy (DCM). The disease-associated presence of clonal TCR- rearrangements, as investigated by semi-nested PCR, GeneScan, and direct sequencing, indicates a pathogenic relevance of this finding in DCM, since it was not detected in non-DCM cardiomyopathies. The predominance of Vβ 19.01 segments suggests that the immune response in DCM patients targets particular epitopes. This finding is consistent with immunity targeting particular antigens involved in DCM, which leads to intramyocardial expansion of reactive T-cell clones. However, the partly heterogenic TCR-β populations in various myocardial samples from the respective cases support the notion that T-cell immunity may target multiple epitopes in human DCM. Further analyses are warranted to investigate the prognostic relevance of clonal TCR-β rearrangements, and the impact of immunomodulatory treatment strategies in DCM patients. These insights might also be interesting for the analysis of T-cell-mediated cardiac allograft rejection.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft through the SFB TR19 (Sonderforschungsbereich Transregio 19/SFB TR19; TPB2 to MH and MN; and TPC6 to RH). The authors declare that they have no competing interests.
Abbreviations
- B19V
Parvovirus B19
- CDR3
complementary determining region 3
- DCM
dilated cardiomyopathy
- LV
left ventricle
- RV
right ventricle
- S
septum
- TCR
T-cell receptor
- VP-1
enteroviral protein-1
Footnotes
Source of support: Departmental sources
References
- 1.Liu PP, Mason JW. Advances in the understanding of myocarditis. Circulation. 2001;104(9):1076–82. doi: 10.1161/hc3401.095198. [DOI] [PubMed] [Google Scholar]
- 2.Esfandiarei M, McManus BM. Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol. 2008;3:127–55. doi: 10.1146/annurev.pathmechdis.3.121806.151534. [DOI] [PubMed] [Google Scholar]
- 3.Noutsias M, Seeberg B, Schultheiss HP, Kühl U. Expression of cell adhesion molecules in dilated cardiomyopathy: evidence for endothelial activation in inflammatory cardiomyopathy. Circulation. 1999;99(16):2124–31. doi: 10.1161/01.cir.99.16.2124. [DOI] [PubMed] [Google Scholar]
- 4.Kindermann I, Kindermann M, Kandolf R, et al. Predictors of outcome in patients with suspected myocarditis. Circulation. 2008;118(6):639–48. doi: 10.1161/CIRCULATIONAHA.108.769489. [DOI] [PubMed] [Google Scholar]
- 5.Frustaci A, Russo MA, Chimenti C. Randomized study on the efficacy of immunosuppressive therapy in patients with virus-negative inflammatory cardiomyopathy: the TIMIC study. Eur Heart J. 2009;30(16):1995–2002. doi: 10.1093/eurheartj/ehp249. [DOI] [PubMed] [Google Scholar]
- 6.Arstila TP, Casrouge A, Baron V, et al. A direct estimate of the human alphabeta T cell receptor diversity. Science. 1999;286(5441):958–61. doi: 10.1126/science.286.5441.958. [DOI] [PubMed] [Google Scholar]
- 7.Hofbauer M, Wiesener S, Babbe H, et al. Clonal tracking of autoaggressive T cells in polymyositis by combining laser microdissection, single-cell PCR, and CDR3-spectratype analysis. Proc Natl Acad Sci USA. 2003;100(7):4090–95. doi: 10.1073/pnas.0236183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muraro PA, Wandinger KP, Bielekova B, et al. Molecular tracking of antigen-specific T cell clones in neurological immune-mediated disorders. Brain. 2003;126(Pt 1):20–31. doi: 10.1093/brain/awg021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lafon M, Lafage M, Martinez-Arends A, et al. Evidence for a viral superantigen in humans. Nature. 1992;358(6386):507–10. doi: 10.1038/358507a0. [DOI] [PubMed] [Google Scholar]
- 10.Hanawa H, Inomata T, Sekikawa H, et al. Analysis of heart-infiltrating T-cell clonotypes in experimental autoimmune myocarditis in rats. Circ Res. 1996;78(1):118–25. doi: 10.1161/01.res.78.1.118. [DOI] [PubMed] [Google Scholar]
- 11.Matsumoto Y, Jee Y, Sugisaki M. Successful TCR-based immunotherapy for autoimmune myocarditis with DNA vaccines after rapid identification of pathogenic TCR. J Immunol. 2000;164(4):2248–54. doi: 10.4049/jimmunol.164.4.2248. [DOI] [PubMed] [Google Scholar]
- 12.Seko Y, Yagita H, Okumura K, Yazaki Y. T-cell receptor V beta gene expression in infiltrating cells in murine hearts with acute myocarditis caused by coxsackievirus B3. Circulation. 1994;89(5):2170–75. doi: 10.1161/01.cir.89.5.2170. [DOI] [PubMed] [Google Scholar]
- 13.Hanawa H, Kodama M, Inomata T, et al. Anti-alpha beta T cell receptor antibody prevents the progression of experimental autoimmune myocarditis. Clin Exp Immunol. 1994;96(3):470–75. doi: 10.1111/j.1365-2249.1994.tb06053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assaf C, Hummel M, Dippel E, et al. High detection rate of T-cell receptor beta chain rearrangements in T-cell lymphoproliferations by family specific polymerase chain reaction in combination with the GeneScan technique and DNA sequencing. Blood. 2000;96(2):640–46. [PubMed] [Google Scholar]
- 15.Ruiz M, Giudicelli V, Ginestoux C, et al. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 2000;28(1):219–21. doi: 10.1093/nar/28.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seko Y, Ishiyama S, Nishikawa T, et al. Restricted usage of T cell receptor V alpha-V beta genes in infiltrating cells in the hearts of patients with acute myocarditis and dilated cardiomyopathy. J Clin Invest. 1995;96(2):1035–41. doi: 10.1172/JCI118089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luppi P, Rudert WA, Zanone MM, et al. Idiopathic dilated cardiomyopathy: a superantigen-driven autoimmune disease. Circulation. 1998;98(8):777–85. doi: 10.1161/01.cir.98.8.777. [DOI] [PubMed] [Google Scholar]
- 18.van Dongen JJ, Langerak AW, Bruggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 19.Noutsias M, Pauschinger M, Schultheiss HP, Kuhl U. Cytotoxic perforin+ and TIA-1+ infiltrates are associated with cell adhesion molecule expression in dilated cardiomyopathy. Eur J Heart Fail. 2003;5(4):469–79. doi: 10.1016/s1388-9842(03)00037-0. [DOI] [PubMed] [Google Scholar]
- 20.Noutsias M, Pauschinger M, Schultheiss H, Kühl U. Phenotypic characterization of infiltrates in dilated cardiomyopathy – diagnostic significance of T-lymphocytes and macrophages in inflammatory cardiomyopathy. Med Sci Monit. 2002;8(7):CR478–87. [PubMed] [Google Scholar]
- 21.Li Y, Bourlet T, Andreoletti L, et al. Enteroviral capsid protein VP1 is present in myocardial tissues from some patients with myocarditis or dilated cardiomyopathy. Circulation. 2000;101(3):231–34. doi: 10.1161/01.cir.101.3.231. [DOI] [PubMed] [Google Scholar]
- 22.Streitz M, Noutsias M, Volkmer R, et al. NS1 specific CD8+ T-cells with effector function and TRBV11 dominance in a patient with parvovirus B19 associated inflammatory cardiomyopathy. PLoS One. 2008;3(6):e2361. doi: 10.1371/journal.pone.0002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gauntt CJ, Arizpe HM, Higdon AL, et al. Molecular mimicry, anti-coxsackievirus B3 neutralizing monoclonal antibodies, and myocarditis. J Immunol. 1995;154(6):2983–95. [PubMed] [Google Scholar]
- 24.Matsumoto Y. Characterization of T cell receptor (TCR) of organ-specific autoimmune disease-inducing T cells and TCR-based immunotherapy with DNA vaccines. J Neuroimmunol. 2000;110(1–2):1–12. doi: 10.1016/s0165-5728(00)00346-5. [DOI] [PubMed] [Google Scholar]
- 25.Varela-Calvino R, Sgarbi G, Wedderburn LR, et al. T cell activation by coxsackievirus B4 antigens in type 1 diabetes mellitus: evidence for selective TCR Vbeta usage without superantigenic activity. J Immunol. 2001;167(6):3513–20. doi: 10.4049/jimmunol.167.6.3513. [DOI] [PubMed] [Google Scholar]
- 26.Klingel K, Rieger P, Mall G, et al. Visualization of enteroviral replication in myocardial tissue by ultrastructural in situ hybridization: identification of target cells and cytopathic effects. Lab Invest. 1998;78(10):1227–37. [PubMed] [Google Scholar]
- 27.Kamradt T, Mitchison NA. Tolerance and autoimmunity. N Engl J Med. 2001;344(9):655–64. doi: 10.1056/NEJM200103013440907. [DOI] [PubMed] [Google Scholar]
- 28.Bowles NE, Ni J, Kearney DL, et al. Detection of viruses in myocardial tissues by polymerase chain reaction. evidence of adenovirus as a common cause of myocarditis in children and adults. J Am Coll Cardiol. 2003;42(3):466–72. doi: 10.1016/s0735-1097(03)00648-x. [DOI] [PubMed] [Google Scholar]
- 29.Kühl U, Pauschinger M, Noutsias M, et al. High prevalence of viral genomes and multiple viral infections in the myocardium of adults with “idiopathic” left ventricular dysfunction. Circulation. 2005;111(7):887–93. doi: 10.1161/01.CIR.0000155616.07901.35. [DOI] [PubMed] [Google Scholar]
- 30.Kuethe F, Lindner J, Matschke K, et al. Prevalence of parvovirus B19 and human bocavirus DNA in the heart of patients with no evidence of dilated cardiomyopathy or myocarditis. Clin Infect Dis. 2009;49(11):1660–66. doi: 10.1086/648074. [DOI] [PubMed] [Google Scholar]
- 31.Noutsias M, Rohde M, Goldner K, et al. Expression of functional T-cell markers and T-cell receptor Vbeta repertoire in endomyocardial biopsies from patients presenting with acute myocarditis and dilated cardiomyopathy. Eur J Heart Fail. 2011;13(6):611–18. doi: 10.1093/eurjhf/hfr014. [DOI] [PubMed] [Google Scholar]