Abstract
The BCR-ABL T315I mutation causes resistance to imatinib, nilotinib and dasatinib in chronic myeloid leukemia. Forty BCR-ABL positive patients with imatinib resistance were analyzed for T315I mutated clones after six months on nilotinib or dasatinib treatment by quantitative allele-specific ligation polymerase chain reaction with a sensitivity of 0.05%. Ligation polymerase chain reaction revealed 10 patients with more than 10−5 BCR-ABLT315I%/GUS (high levels), none of whom achieved major molecular response after 12 months, and a further 8 patients with 10−5 or below BCR-ABLT315I%/GUS (low levels) who all achieved major molecular response (P<0.001). A second independent group showed molecular response in one of 12 patients with high levels and 5 of 8 patients with low levels (P=0.018). Combining the groups resulted in a sensitivity and specificity of 92.9% and 87.5%, respectively. We conclude that the quantitative level of mutant T315I allele is predictive of major molecular response at 12 months on second-line nilotinib or dasatinib treatment.
www.clinicaltrials.gov: CT00109707, NCT00384228, CA180013, CA180005 CA180006.
Introduction
Resistance to imatinib (IM) is still one of the major problems in the treatment of chronic myeloid leukemia (CML). Several mechanisms have been reported to cause resistance but BCR-ABL kinase domain mutations remain the most common.1–3 Nilotinib (NI) and dasatinib (DA) are active against the majority of mutations causing IM resistance but some mutations confer clinical resistance against NI (Y253H, E255K/V, F359V/C)4 or DA (V299L, T315A, F317L/I/V/C)5 or both (T315I). Hence, mutational screening is recommended as standard of care in patients with suboptimal response or failure either of IM or of second-line NI or DA.6 Sanger sequencing and denaturing high-performance liquid chromatography (D-HPLC) are considered to be suitable techniques for identifying BCR-ABL kinase domain mutations7 and can detect mutated clones with a sensitivity of 10–25%2 and 1–10%,8 respectively. We and others have established mutation specific techniques with higher sensitivities.9–12 Some of these techniques can also provide a quantitative measure of the mutated clones11,12 and have been shown to identify mutations (here referred to as ‘low-level mutations’) overlooked by routine screening techniques.13 Low-level mutations are detectable in advanced disease states but not during first chronic phase CML and do not affect progression-free survival of patients treated with IM.9 In contrast, resistant mutations detected in patients after IM failure either by Sanger sequencing or at a higher sensitivity (0.2%) by mass spectrometry are associated with a low rate of complete cytogenetic responses (CCR),14 suggesting a high prognostic value of low-level mutations detected by more sensitive techniques. Furthermore, the number of low-level mutations at IM failure appears to be associated with a lower rate of CCR and major molecular response (MMR) and a higher incidence of new resistant mutations on NI or DA second line.15 In this study, we have further investigated the prognostic significance of low-level mutations by studying the association between the quantitative level of mutated T315I BCR-ABL six months after the start of second-line NI or DA therapy on the one hand and the subsequent achievement of MMR at 12 months (MMR12) on the other. Furthermore, we sought to define a quantitative cutoff value at which low-level mutations are most informative.
Design and Methods
Patients
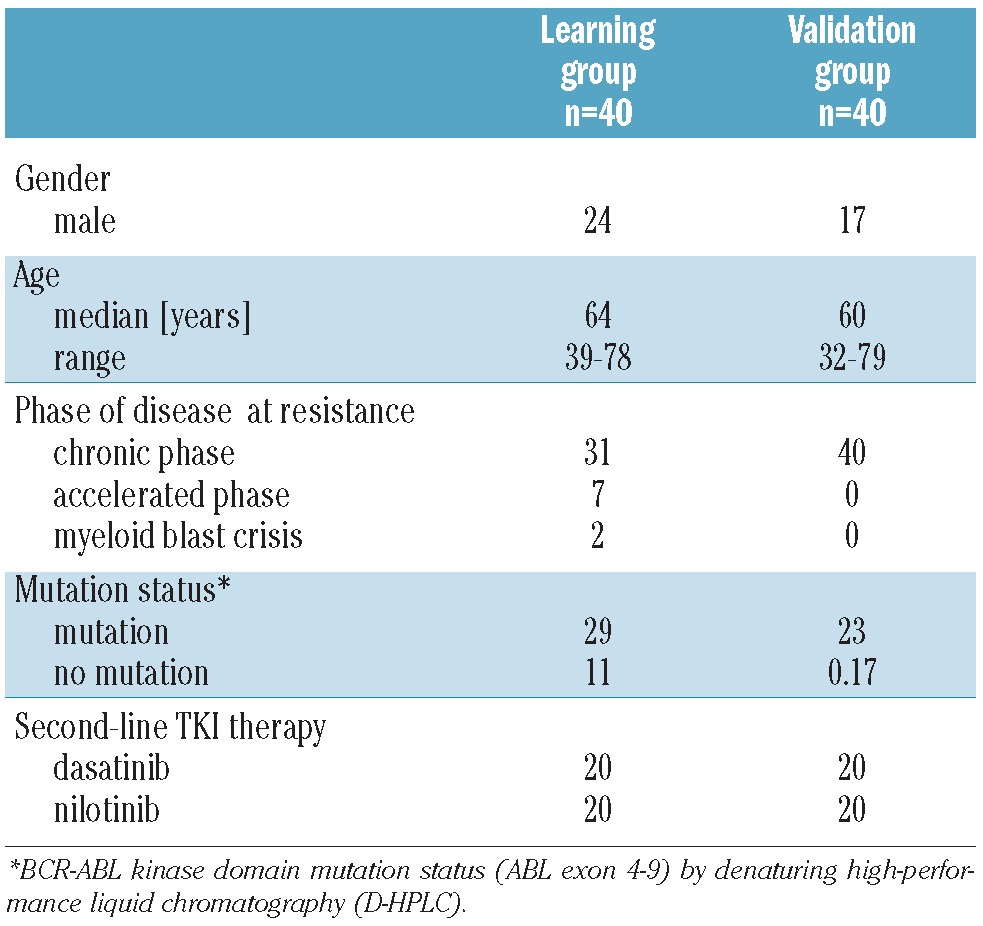
Two independent groups of patients were analyzed. The ‘learning’ group of 40 randomly selected CML patients was used to establish the cut-off level of mutant T315I allele at six months that was most closely associated with MMR12. These patients had already been studied by our group in a comparison study of different techniques for mutation analysis.13 To confirm the findings from the ‘learning’ group, another 40 CML patients were assigned to an independent ‘validation’ group based on the availability of cDNA after six months on NI or DA. All patients in both groups gave written informed consent, had previously received between 400 and 800 mg IM/day, and were receiving second-line NI 800 mg/day or DA 140 mg/day. The underlying clinical trials were conducted in accordance with the Declaration of Helsinki of 1975 as revised in 2000, and approved by the ethics committees of the institutions involved. Patients’ characteristics for both groups prior to the start of second-line tyrosine kinase inhibitor therapy with NI or DA are given in Table 1.
Table 1.
Patients’ characteristics prior to start of second-line tyrosine kinase inhibitor therapy.
Molecular analysis
Samples were analyzed by quantitative reverse-transcriptase polymerase chain reaction (RT-PCR) for BCR-ABL, ABL and β-glucuronidase (GUS) and denaturing high-performance liquid chromatography (D-HPLC) at baseline and after three, six, nine and 12 months of treatment with NI or DA. L-PCR was performed blinded after six months. RNA extraction and cDNA synthesis were performed as previously described.16 Quantitative RT-PCR for BCR-ABL, total ABL and GUS transcripts was performed using the LightCycler™ technology (Roche Diagnostics, Mannheim, Germany).8 D-HPLC was performed on a Transgenomic Wave™ System Model 3500 HT (Transgenomic, Omaha, NE, USA) as previously described.8 We used 10 μL of stored cDNA for L-PCR analysis.12 The dynamic range of the L-PCR approach extends from 100% to between 0.1% to 0.05% mutant (3–3.5 log). The comparative Ct method was used to calculate the percentage of T315I mutated BCR-ABL in the positive samples with additional normalization by GUS expression directly according to the equation: % mutant allele = 2 −(ct BCR-ABLT315I – ct BCR-ABLtotal) × 100.
Statistical analysis
Of the 40 patients who were randomly selected for the learning group, all patients with a positive T315I mutated BCR-ABL allele/GUS level according to L-PCR were qualified for finding the optimal cut-off level in order to identify two groups with maximally different proportions of a later MMR12. The maximum difference was identified by the cut-off level resulting in the highest value for Fisher’s statistical test. However, to obtain a statistically and clinical relevant result, restrictions had to be taken into account: 1) the smaller of the two groups should contain at least 20% of the patients; 2) P value of Fisher’s test was multiplied by the number of different T315I mutated BCR-ABL allele/GUS levels meeting the first restriction. Thus, Bonferroni’s adjustment for the multiple testing of all candidates for optimal cut-off level was performed. In the case of a statistically significant result, the identified cut-off level was tested in an independent validation sample. Level of significance was 0.05. Sensitivity and specificity were calculated. The general suitability of the T315I mutated BCR-ABL allele/GUS level for diagnosis of future MMR12 was described by a receiver operating characteristic (ROC) curve and the area under the curve (AUC) was calculated. All calculations were performed with SAS software version 9.1.3 (SAS Institute, Cary, NC, USA).
Results and Discussion
The T315I mutation causes resistance to IM, NI and DA in CML and is, therefore, an ideal model to study the prognostic potential of low-level mutations, i.e. mutations below the detection limit of Sanger sequencing and D-HPLC. Of the initial learning group of 40 CML patients with IM resistance, 12 achieved MMR12 on either NI (n=5) or DA (n=7). BCR-ABL amplification of the 6-month samples (required for further L-PCR analysis) was successful in 35 patients (88%) by L-PCR; 8 of these achieved MMR12. Eighteen of the 35 patients (52%) were positive for the T315I mutation by L-PCR with a median of 3.71×10−4 % (range 3.91×10−7 – 0.26%) T315I mutated BCR-ABL allele/GUS. Of 10 patients (29%) with a quantitative level over 10−5 BCR-ABLT315I%/GUS, none achieved MMR12 (Table 2). Three of these patients were also positive for T315I by DPLC at six months and had BCR-ABLT315I%/GUS values by L-PCR of 0.23 (#5), 2.6×10−5 (#15) and 0.26 (#35), respectively. Compared to DHPLC, the L-PCR technique, therefore, identified an additional 7 patients who did not achieve MMR by 12 months.
Table 2.
Prediction of major molecular response after 12 months on nilotinib or dasatinib therapy by quantification of the BCR-ABLT315I mutation.
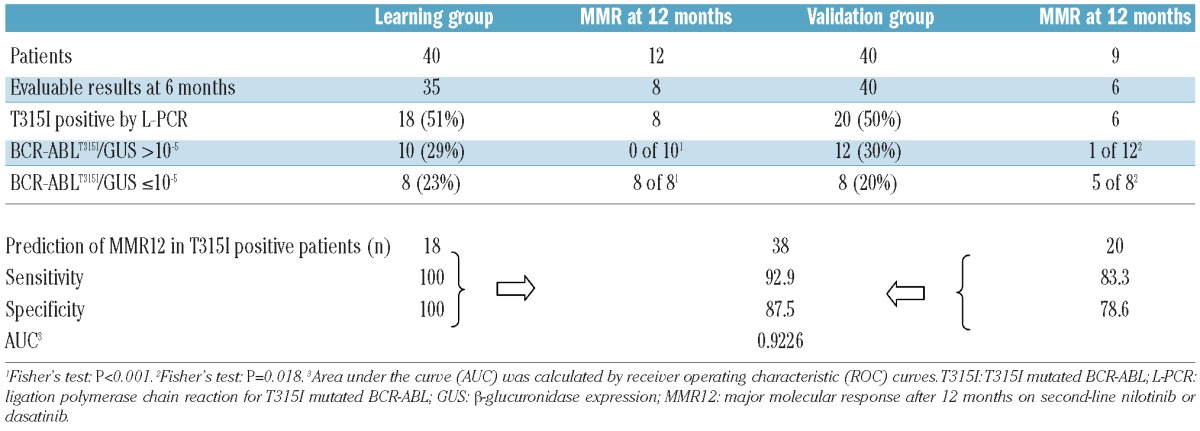
In contrast, all 8 patients (23%) with BCR-ABLT315I%/GUS of 10−5 or below did achieve MMR12.
Thus, the cut-off level of 10−5 BCR-ABLT315I%/GUS by L-PCR precisely separates the T315I positive patients in the learning group according to their subsequent achievement of MMR12 (P<0.001) with a sensitivity and specificity of 100%.
An independent validation group of a further 40 CML patients with IM resistance was analyzed in order to validate the prognostic cut-off level previously identified in the learning group. Nine patients achieved MMR12 on NI (n=4) or DA (n=5). BCR-ABL amplification of the 6-month samples was successful in 40 patients by L-PCR; 6 of those achieved MMR12. Twenty patients (50%) had mutated T315 with a median of 7.27×10−5% (range 8.21×10−8–1.73×10−3) mutated T315I BCR-ABL/GUS. Of these, 12 patients showed BCR-ABLT315I%/GUS over 10−5 with one of the 12 (BCR-ABLT315I%/GUS = 3.16×10−4) achieving MMR12. Of the 8 patients with low levels of mutated T315I (BCR-ABLT315I%/GUS ≤10−5), 5 achieved MMR12 while 3 did not. Thus, the cut-off level of 10−5 maintained significance in the validation group (P=0.018). The sensitivity and specificity to predict MMR12 in the validation group was 83.3% and 78.6%, respectively. The independent validation group, therefore, confirmed the high sensitivity and specificity of the chosen cut-off level (10−5 BCR-ABLT315I%/GUS) as a predictor of MMR12. Combining the results for all T315I positive patients from both groups obtained by L-PCR generated a sensitivity of 92.9% and a specificity of 87.5%. Additional receiver operating curve (ROC) calculations confirmed the high association of the 6-month cut-off value with MMR12 (AUC 0.9226).
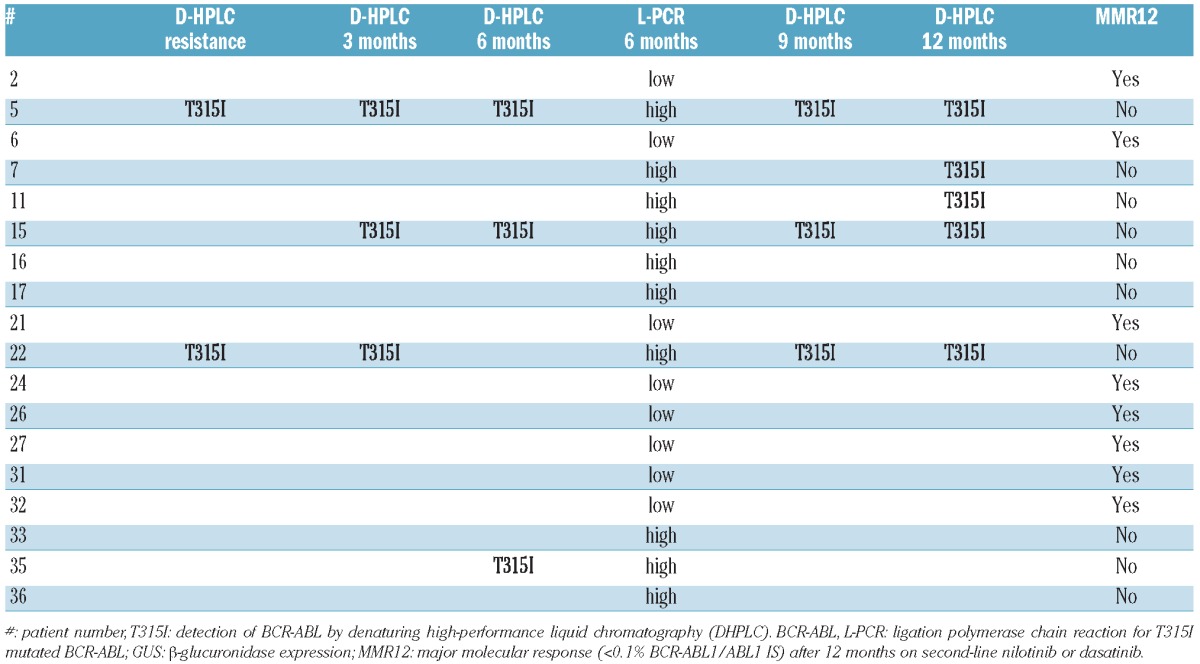
By D-HPLC, a T315I mutation was detected at baseline in 2 patients (#5, 22), at three months in 3 (#5, 15, 22), at six months in 3 (#5,15,35), at nine months in 3 (#5,15,22), and at 12 months in 5 (#5,7,11,15,22) after second-line tyrosine kinase inhibitor treatment (Table 3). None of the patients who were T315I positive by D-HPLC achieved MMR12 and all had BCR-ABLT315I%GUS over 10−5 at six months by L-PCR irrespective of the time of T315I detection by D-HPLC, confirming close qualitative agreement between the two methods. Once positive for T315I by D-HPLC all patients remained positive in further samples with the exception of #22 (6 months) and #35 (9 and 12 months). In all 3 negative patients at six months by D-HLPC (#7, #11, #22), L-PCR revealed a T315I level slightly below the detection limit of the D-HPLC technique. In addition to the 6 patients for whom D-HPLC detected a T315I mutation at least once within the 12-month observation period, L-PCR at six months identified 4 additional patients carrying T315I mutations (#16,17,33,36) that remained undetected by D-HPLC. Having demonstrated the biological and clinical significance of BCR-ABL kinase domain mutations below the detection limit of Sanger sequencing (‘low-level mutations’),14,15 we have now used T315I mutated clones as an example to establish a cut-off level of mutant allele with prognostic significance and validated the cut-off value in an independent group of patients. Our results have several implications for mutation analysis in CML.
Table 3.
Association of BCR-ABLT315I/GUS >10−5 (high) and BCR-ABLT315I/GUS ≤10−5 (low) by ligation-PCR with detection of the T315I mutation by denaturing high-performance liquid chromatography (DHPLC) and achievement of MMR12. Only positive patients by DHPLC and/or L-PCR are shown.
The level of mutated allele appears to be a valuable indicator of whether a mutated cell clone will undergo long-term expansion over time or not. Parker et al. used mass spectrometry (sensitivity 0.2% mutated allele) to show a clear association with response. In our studies, a 0.1% cut-off level corresponded best with response, although the additional normalization with GUS led to an even better correlation (data not shown). In conclusion, we confirm the prognostic relevance of low-level mutations detected by techniques with a sensitivity of 0.1–0.2% mutated allele, although further GUS normalization is recommended if using a quantitative technique. In this respect, it should be noted that the complexity of the molecular techniques involved currently limits the practical use of our findings in the routine monitoring of CML patients.
Vice versa, we have demonstrated that qualitative techniques with sensitivities below this cut-off level may identify additional mutated clones without prognostic consequences. This may explain why Willis et al.9 found no negative impact on progression-free survival in patients for whom low-level mutations were detected at diagnosis by qualitative ASO-PCR with a sensitivity of at least 0.001% mutated clone. The prognostic significance of low-level mutations at CML diagnosis should, therefore, be re-evaluated from a quantitative perspective. Mutations below our cut-off level should only be considered clinically significant if levels rise over time.
A strong association was found between patients with T315I mutations above our cut-off level by L-PCR and T315I mutations detected by D-HPLC. However, L-PCR identified 2 patients six months earlier than did D-HPLC and a further 4 patients with a high-level T315I mutation by L-PCR and no MMR12, who were not identified at all by D-HPLC. Given the evidence for poor response rates in patients with low-level mutations,14 the ability of L-PCR to identify mutations above the clinically significant cut-off level but below the detection level of D-HPLC increases the value of mutation analysis in the optimization of CML therapy.
In summary, sensitive techniques increase the number of patients in whom BCR-ABL mutations can be detected. However, only mutations above a certain cut-off level seem to be of prognostic significance. The cut-off level for the T315I mutation lies below the detection limit of routine screening techniques. Therefore, we recommend that relevant mutations should be quantified in clinical trials to determine mutation specific cut-off levels that have a significant influence on prognosis of the outcome of a given TKI treatment in CML.
Footnotes
Funding
This work was supported in part by the BMBF-grant on Medical Systems biology “HaematoSys” (BMBF-FKZ 0315452).
Authorship and Disclosures
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paqette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–80 [DOI] [PubMed] [Google Scholar]
- 2.Hochhaus A, Kreil S, Corbin AS, La Rosée P, Müller MC, Lahaye T, et al. Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia. 2002; 16(11):2190–6 [DOI] [PubMed] [Google Scholar]
- 3.Hochhaus A, La Rosée P. Imatinib therapy in chronic myelogenous leukemia: strategies to avoid and overcome resistance. Leukemia. 2004;18(8):1321–31 [DOI] [PubMed] [Google Scholar]
- 4.Hughes T, Saglio G, Branford S, Soverini S, Kim DW, Muller MC, et al. Impact of baseline BCR-ABL mutations on response to nilotinib in patients with chronic myeloid leukemia in chronic phase. J Clin Oncol. 2009;27(25):4204–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller MC, Cortes JE, Kim DW, Druker BJ, Erben P, Pasquini R, et al. Dasatinib treatment of chronic-phase chronic myeloid leukemia: analysis of responses according to preexisting BCR-ABL mutations. Blood. 2009;114(24):4944–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baccarani M, Cortes J, Pane F, Niederwieser D, Saglio G, Apperley J, et al. Chronic myeloid leukemia: an update of concepts and management recommendations of European LeukemiaNet. J Clin Oncol. 2009; 27(35):6041–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soverini S, Hochhaus A, Nicolini FE, Gruber F, Lange T, Saglio G, et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: recommendations from an expert panel on behalf of European LeukemiaNet. Blood. 2011; 118(5):1208–15 [DOI] [PubMed] [Google Scholar]
- 8.Ernst T, Erben P, Muller MC, Paschka P, Schenk T, Hoffmann J, et al. Dynamics of BCR-ABL mutated clones prior to hematologic or cytogenetic resistance to imatinib. Haematologica. 2008;93(2):186–92 [DOI] [PubMed] [Google Scholar]
- 9.Willis SG, Lange T, Demehri S, Otto S, Crossmann L, Niederwieser D, et al. High-sensitivity detection of BCR-ABL kinase domain mutations in imatinib-naive patients: correlation with clonal cytogenetic evolution but not response to therapy. Blood. 2005;106(6):2128–37 [DOI] [PubMed] [Google Scholar]
- 10.Oehler VG, Qin J, Ramakrishnan R, Facer G, Ananthnarayan S, Cummings C, et al. Absolute quantitative detection of ABL tyrosine kinase domain point mutations in chronic myeloid leukemia using a novel nanofluidic platform and mutation-specific PCR. Leukemia. 2009;23(2):396–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruber FX, Lamark T, Anonli A, Sovershaev MA, Olsen M, Gedde-Dahl T, et al. Selecting and deselecting imatinib-resistant clones: observations made by longitudinal, quantitative monitoring of mutated BCR-ABL. Leukemia. 2005;19(12):2159–65 [DOI] [PubMed] [Google Scholar]
- 12.Pelz-Ackermann O, Cross M, Pfeifer H, Deininger M, Wang SY, Al-Ali HK, et al. Highly sensitive and quantitative detection of BCR-ABL kinase domain mutations by ligation PCR. Leukemia. 2008;22(12):2288–91 [DOI] [PubMed] [Google Scholar]
- 13.Ernst T, Gruber FX, Pelz-Ackermann O, Maier J, Pfirrmann M, Muller MC, et al. BCR-ABL kinase domain mutations on second line dasatinib or nilotinib therapy in chronic myeloid leukemia patients after imatinib failure: a cooperative evaluation of different detection methods. Haematologica. 2009;94(9):1227–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker WT, Lawrence RM, Ho M, Irwin DL, Scott HS, Hughes TP, et al. Sensitive detection of BCR-ABL1 mutations in patients with chronic myeloid leukemia after imatinib resistance is predictive of outcome during subsequent therapy. J Clin Oncol. 2011; 29(32):4250–9 [DOI] [PubMed] [Google Scholar]
- 15.Parker WT, Ho M, Scott HS, Hughes TP, Branford S. Poor response to second-line kinase inhibitors in CML patients with multiple low-level mutations, irrespective of their resistance profile. Blood. 2012; 119(10):2234–8 [DOI] [PubMed] [Google Scholar]
- 16.Cross NC, Melo JV, Feng L, Goldman JM. An optimized multiplex polymerase chain reaction (PCR) for detection of BCR-ABL fusion mRNAs in haematological disorders. Leukemia. 1994;8(1):186–9 [PubMed] [Google Scholar]


