Abstract
The discovery of small non-coding microRNAs has revealed novel mechanisms of post-translational regulation of gene expression, the implications of which are still incompletely understood. We focused on microRNA 21 (miR-21), which is expressed in cardiac valve endothelium during development, in order to better understand its mechanistic role in cardiac valve development. Using a combination of in vivo gene knockdown in zebrafish and in vitro assays in human cells, we show that miR-21 is necessary for proper development of the atrioventricular valve (AV). We identify pdcd4b as a relevant in vivo target of miR-21 and show that protection of pdcd4b from miR-21 binding results in failure of AV development. In vitro experiments using human pulmonic valve endothelial cells demonstrate that miR-21 overexpression augments endothelial cell migration. PDCD4 knockdown alone was sufficient to enhance endothelial cell migration. These results demonstrate that miR-21 plays a necessary role in cardiac valvulogenesis, in large part due to an obligatory downregulation of PDCD4.
Keywords: Cardiac valves, microRNA, Endothelium
INTRODUCTION
Congenital heart disease is estimated to cause death in ∼1% of newborns and to cause more than 5% of all pre-term mortality, making it the most lethal of all birth defects (Hoffman, 1995). Among congenital cardiac defects, valvular abnormalities and endocardial cushion defects are the most common. Despite its clear developmental and medical importance, there is still a limited understanding of the molecular events governing the complex process of cardiac endocardial cushion formation and valvulogenesis.
The primitive heart is a tube of primary cardiomyocytes lined by a layer of endocardial cells. During cardiac looping, endothelial cells are activated - in part by secreted proteins including transforming growth factor beta (TGFβ) (Potts et al., 1992). Activated endothelial cells downregulate cell-cell contacts and migrate into the cardiac jelly with an accompanying change in morphology. In addition to the TGFβ superfamily, other fundamental pathways involved in the formation of endocardial cushions and cardiac valves include Wnt/β-catenin (Hurlstone et al., 2003), calcium/calcineurin-NFAT (de la Pompa et al., 1998), VEGF (Lee et al., 2006) and Notch (Timmerman et al., 2004), all of which have been implicated in early steps of valvulogenesis. Each pathway is complex, with many different receptors or ligands, and exhibits dynamic temporal regulation throughout development. There is evidence of substantial crosstalk between many of these pathways, which form an incompletely understood signaling network that yields the final three-dimensional valvular structure.
MicroRNAs (miRNAs) are a class of small non-coding RNAs that regulate gene expression at the post-translational level. Each miRNA is thought to regulate multiple genes in diverse biological processes. miRNAs regulate gene expression by binding to complementary sequences in the target mRNA. miRNA binding frequently results in inhibition of translation, which is sometimes associated with degradation of the mRNA target. Given the large number of miRNAs discovered to date and their known and predicted targets, their role in post-translational regulation of gene expression may rival transcriptional regulation. In the heart, several miRNAs have been identified that are important for cardiac development (Zhao et al., 2007), regulation of ion channel expression (Harada et al., 2012), hypertrophy (van Rooij et al., 2006; Carè et al., 2007; Tatsuguchi et al., 2007), myosin isoform switching (van Rooij et al., 2007) and cardiac fibrosis (Thum et al., 2008).
Expression patterns for over 100 miRNAs in zebrafish have been characterized, revealing some striking patterns of tissue expression (Wienholds et al., 2005). Within the heart, microRNA 21 (miR-21) appears to be restricted to valvular endothelium, although it is also expressed in the brain. miR-21 is conserved between zebrafish and humans, with two base changes in the mature form between these species. miR-21 expression has been shown in several neoplasms and is thought to play a role in tumor invasion (Si et al., 2007). Targets identified to date include mRNAs encoding the tumor suppressor programmed cell death 4 (PDCD4) (Frankel et al., 2008; Lu et al., 2008), the PTEN tumor suppressor (Meng et al., 2007) and tropomyosin 1 (Zhu et al., 2007). Knockdown of miR-21 and overexpression of PDCD4 in cancer cells both lead to a reduction in invasion and metastasis (Asangani et al., 2008). Biogenesis of miR-21 has been shown to be regulated by TGFβ superfamily members at the DROSHA microprocessing step (Davis et al., 2008).
miR-21 has also been implicated in the response to several forms of cardiac stress (van Rooij et al., 2006; Tatsuguchi et al., 2007). miR-21 is upregulated in cardiac fibroblasts in the failing heart, where it represses the expression of Sprouty homolog 1 (Spry1), a negative regulator of the extracellular signal-regulated kinase/mitogen-activated protein (ERK-MAP) kinase signaling pathway (Thum et al., 2008). Given its localized expression in prevalvular endothelium and its role in tumor invasion and metastasis, we sought to define the role of miR-21 in cardiac valvulogenesis, specifically in the endothelial migration steps.
MATERIALS AND METHODS
Zebrafish lines
Experiments were performed in wild-type zebrafish (Top Longfin) transgenic strains Tg(cmlc2:dsRed) (gift of Randall Peterson, Massachusetts General Hospital, Boston, BMA, USA) and Tg(Tie2:EGFP)s849. Zebrafish were maintained using standard methods.
Morpholino injection
Morpholino oligonucleotides (MOs) (Gene Tools) were injected into single-cell embryos at the indicated doses. MO sequences (5′-3′) were: dre-miR-21-1 MO (multi-blocker), TGTAACAGCCAACACCAGTCTGATAAGCTAT; dremir-21-2 MO (multi-blocker), ACTCAAAGCCAACACCAGTCTGATAAGCTAC; miR21-2 Guide Dicer, CAAAGCCAACACCAGTCTGATAAGC; Mismatch 21-2, CAAAGaCAgCAaCAGgCTGAgAAGC; PDCD4 TP, GGCACTGTTAGAAGTATCAGCCTAT; and PDCD4 exon 3 splice acceptor, GCCTCTGTGGATGACGACAGCATTT.
Imaging
Images were obtained on a Zeiss LSM Pascal 5 confocal microscope, a Zeiss Axioplan microscope and Photron FASTCAM camera for videos and a Leica Wild M10 with Windows Spot Basic v4.0.8 software for low-magnification images and in situ hybridization results.
In situ hybridization
Between 10 and 20 fixed embryos were subjected to in situ hybridization as previously described (Milan et al., 2006), with the modification that miRNA in situ hybridization embryos were equilibrated in 1-methylimidazole buffer after fixation and cross-linked with 0.16 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (Sigma) (Pena et al., 2009). The probe was a DIG-labeled LNA probe for dre-miR-21 (Exiqon). The probes were as described: bmp4 (Nikaido et al., 1997), notch1b (Westin and Lardelli, 1997), tbx2b (Gross and Dowling, 2005), versican (Thisse and Thisse, 1999), has2 (Bakkers et al., 2004) and spp1 (Peal et al., 2009). The pdcd4b probe was prepared from a cloned pdcd4b template (Open Biosystems, accession number BC065341) by linearizing the vector with EcoRI and transcription with T7 RNA polymerase. The nppa probe template was amplified from a partial cDNA clone (Open Biosystems Accession Number BC124592) using the following PCR primers (5′-3′): TTTGGCAGCAGACGGATGTA and TAATACGACTCACTATAGGGCGTATTGCAGCTAACCTTTT. The probe was synthesized with T7 RNA polymerase. Fixation, plastic embedding, sectioning and Hematoxylin and Eosin staining were performed as previously described (Milan et al., 2006).
Rescue experiments
miR-21 RNA used for rescue experiments was prepared by first employing PCR amplification from the pre-miR-21-2 expression vector generated for luciferase assays (see below). PCR primers contained a T7 RNA polymerase site, allowing mRNA to be produced from the PCR product using the T7 mMessage mMachine Kit (AM1344, Ambion). miR-21-2 morphants were rescued by co-injection of 1 ng/μl miR-21 RNA with the miR-21-2 MO. An AVC defect was identified by the combination of: (1) absence of a constriction between the atrial and ventricular chambers; (2) presence of blood regurgitation during ventricular systole; and (3) absence of a looped heart. Pericardial edema and pooling of blood in the absence of these features were not considered AVC defects. For Pdcd4 knockdown rescue of the miR-21 and PDCD4 TP morphants, 10 ng PDCD4 splice acceptor MO was co-injected with either 4 ng miR-21-2 MO or 10 ng PDCD4 TP MO, respectively. All embryos were scored at 72 hpf.
miRNA northern blot
RNA samples were prepared from 100 72-hpf embryos using Trizol (Invitrogen) according to the manufacturer’s instructions. Then, 20 μg of total RNA per lane was run on a 15% TBE-urea gel, transferred to Hybond-N+ membrane (Amersham, GE Healthcare) and probed with the miR-21 LNA probe (Exiqon), incubated with anti-DIG-alkaline phosphatase Fab antibody (Roche) and visualized with CDP Star detection (Roche) according to the manufacturer’s directions. The blot was washed and reprobed with 32P end-labeled oligonucleotide to tRNA-valine (5′-TGGTGTTTCCGCCCGGTTT-3′) and imaged after stringent washing.
qPCR of zebrafish miRNA
Quantitative (q) PCR of zebrafish mature miR-21 was performed using a TaqMan Small RNA Assay System according to the manufacturer’s instructions. Briefly, total RNA from 48-hpf zebrafish embryos was purified using Trizol and cDNA was prepared using RNA assay kits for miR-21 and miR-92a (4440886 and 4427975, Applied Biosystems) and the TaqMan MicroRNA Reverse Transcription Kit (4366596, Applied Biosystems). Primers used for qPCR were supplied with each kit, and consisted of stem-looped oligonucleotides that contained the sequence of the mature miR-21 mRNA. qPCR was then performed in triplicate using an ABI Fast 7500 instrument. Results are displayed as mean ± s.d. and compared using a two-tailed Student’s t-test (P<0.05).
Luciferase assays
Luciferase reporter constructs were generated by PCR amplification of candidate gene target 3′ UTRs followed by cloning into pMIR-REPORT (Ambion). The miR-21 binding site was deleted from the PDCD4 clone using Quikchange II (Invitrogen). Sequence-verified clones were transfected into HEK293 cells using Lipofectamine 2000 (Invitrogen), with pMIR-REPORT β-gal control vector as a transfection efficiency control. Mutually primed synthesis was employed to generate a pre-miR 21-2 expression vector driven by the CMV promoter. Forty-eight-hour lysates were assessed for luciferase and β-gal activity using standard assays with three to eight replicates.
GFP western blots
GFP fusion constructs were generated by PCR amplification of the 3′ UTR of pdcd4b and cloning the product into pcDNA3.1/NT-GFP-TOPO (Invitrogen). Capped mRNA (mMessage mMachine, Ambion) was injected into single-cell embryos at 500 ng/μl with or without pre-miR-21 (Ambion) at 50 μM and with or without the miR-21 MO at 100 μM. Ten to twenty embryos were collected at 30 hpf, manually deyolked, suspended in 2× Laemmli buffer, homogenized through a 23 g needle, and heated to 100°C for 2 minutes. SDS-PAGE was followed by western blotting using a mouse anti-GFP primary antibody [GFP(B-2), sc-9996, Santa Cruz Biotech], an HRP-labeled goat anti-mouse secondary antibody (sc-2005, Santa Cruz Biotech) and detection by chemiluminescence (32209, ECL Pierce). The membrane was stripped and reprobed with a mouse anti-human α-tubulin antibody (clone DM1A, Millipore).
PDCD4 western blots
Zebrafish embryos injected with the miR-21-2 MO and uninjected controls were collected at 48 hpf and prepared as above. Western blotting was performed using a rabbit anti-PDCD4 (human) primary antibody (ab51495m, Abcam) and an HRP-labeled anti-rabbit secondary antibody and detection by chemiluminescence (ECL). The membrane was stripped and reprobed with the mouse anti-human α-tubulin antibody. Western blotting of PDCD4 from HPVECs was performed by first collecting cells in Trizol 24 hours after transfection, and purifying the protein according to the manufacturer’s instructions. Blots were probed with mouse anti-PDCD4 antibody (sc-130545, Santa Cruz Biotech), an HRP-labeled goat anti-mouse secondary antibody (sc-2005, Santa Cruz Biotech) and detection by chemiluminescence (ECL). The membrane was stripped and reprobed with the mouse anti-human α-tubulin antibody.
qPCR analysis of mouse cardiac cushion cells
Primary cells from murine AV cushions (E12.5) were isolated and cultured for 3 weeks. These cells were transiently transfected with vector (control) or floxed antigomiR-21 construct. Transfected cells were grown for 72 hours. Both long and short RNAs were isolated using an RNA purification kit from Norgen Biotek. An Mm_miR-21_2 miScript Primer Assay Kit (MS00011487, Qiagen) was used for qRT-PCR with a mature miR-21 primer (5′-TAGCTTATCAGACTGATGTTGA-3′).
Migration assay
Cultivation of HPVECs was as previously described (Paruchuri et al., 2006). miR-21 overexpression was performed by transfection of the miR-21 miRNA precursor (pre-miR miRNA PM12979, Ambion) into HPVECs according to the manufacturer’s instructions. Lentiviral transduction (Mission shRNA lentiviral TRCN0000059079, Sigma) was performed according to the manufacturer’s directions at a MOI of one. TGFβ2 treatment was as described (Paruchuri et al., 2006). Migration assays were performed 6 days after miR-21 transfection and 5 days after TGFβ2 (R&D Systems) treatment, or 6 days after TGFβ2 treatment and 5 days after lentiviral transduction using 6.5 mm Transwells (Costar 8.0 μm pore polycarbonate inserts) as previously described (Paruchuri et al., 2006), with 10% fetal bovine serum as chemoattractant. Cells were fixed in ice-cold acetone and stained with 2% Crystal Violet (Sigma). Three high-magnification fields were imaged and cells counted offline. All conditions were performed in triplicate.
qPCR analysis of HPVECs
HPVECs were transfected and treated with TGFβ2 as described above. Cells were collected in Trizol and total RNA was purified according to the manufacturer’s instructions. cDNA was prepared using the Superscript III Kit (Invitrogen), and PDCD4 and the normalization gene β-actin were measured using SYBR Green (Applied Biosystems) and an ABI Fast 7500 instrument. PDCD4 primers were: 5′ primer, CCGGACATTAATCTGGATGTCCC; 3′ primer, GACGACCTCCATCTCCTTCGC.
RESULTS
miR-21 is required for normal atrioventricular valve valvulogenesis
In zebrafish embryos, the earliest detectable cardiac expression of miR-21 occurs at 48 hours post-fertilization (hpf) and is localized to the atrioventricular valve (AV) ring endocardium (Fig. 1A). By 72 hpf, expression in the AV ring is stronger, and expression in the cardiac outflow tract is observed (Fig. 1B) along with expression in the developing central nervous system.
Fig. 1.
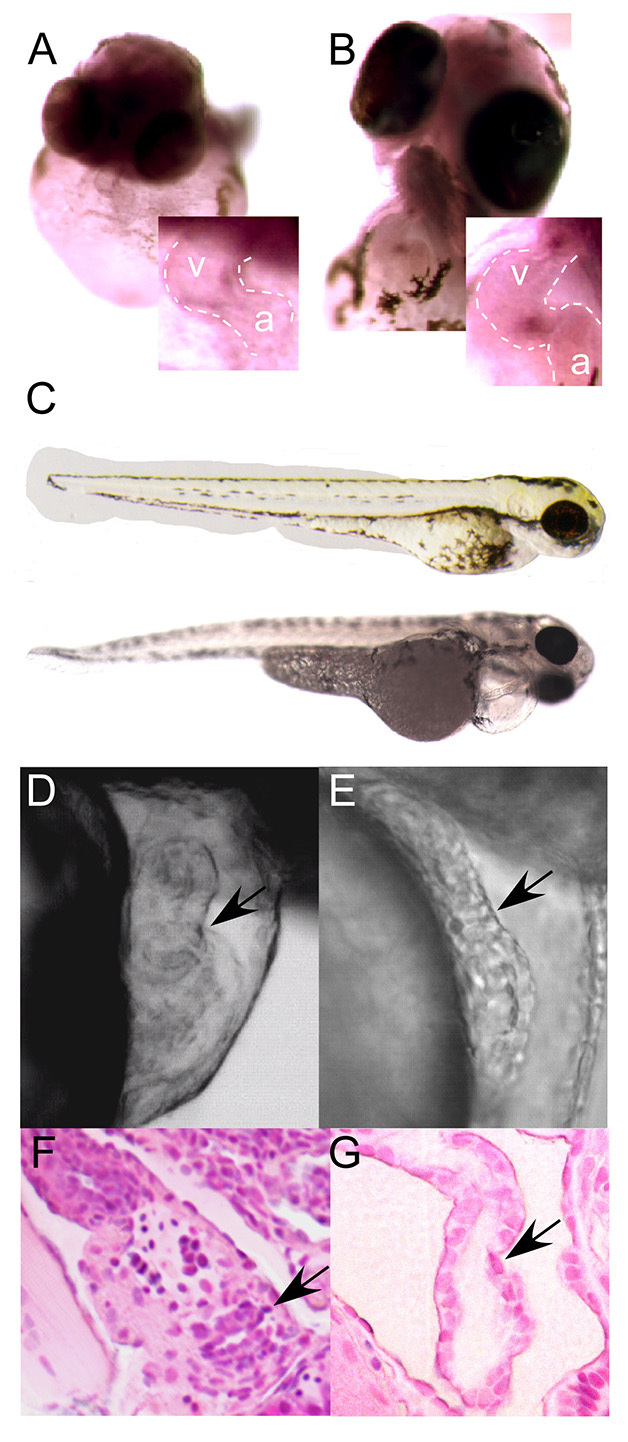
MicroRNA 21 expression and knockdown. (A,B) In situ hybridization with mature miR-21 locked nucleic acid (LNA) probe at 48 (A) and 72 (B) hpf demonstrates expression in zebrafish atrioventricular canal (AVC) at 48 hours (A, inset) and staining of atrioventricular valve (AV) and outflow tract (B, inset). (C) miR-21 knockdown does not perturb the normal body plan or axis at 72 hpf. Control, top; morphant, bottom. (D,E) Mismatch MO control-injected embryos show normal AV ring constriction (D, arrow) at 48 hpf, whereas miR-21 knockdown results in loss of the normal AV constriction (E, arrow). There is also failure of normal cardiac looping, as well as pericardial edema. (F,G) Hematoxylin and Eosin stained sections at day 5 post-fertilization demonstrate normal AV in controls (F, arrow) but failure of AV development in miR-21 knockdown embryos (G, arrow). In all images, the atrial chamber (a) is below and to the right and the ventricle (v) is above and to the left.
There are two genomic copies of mir21 in zebrafish, mir21-1 and mir21-2, each giving rise to the identical mature miRNA (miR-21-1 and miR-21-2). Use of antisense morpholino oligonucleotides (MOs) that extend beyond the mature miRNA sequence allows selective targeting of each miR-21 isoform. Injection into zebrafish embryos of an antisense MO targeting miR-21-1 at doses up to 40 ng did not result in any consistent cardiac phenotype. By contrast, injection of 4 ng of a multi-blocker antisense MO that overlaps both the Drosha and Dicer processing sites in pre-miR-21-2 resulted in failure of development of the AV ring (Fig. 1D,E) with consequent atrioventricular regurgitation (supplementary material Movies 1, 2) and failure of cardiac looping in 80% (104/130) of embryos. Although there were some morphological differences between the miR-21 morphant and the control, the general body plan and axis were unperturbed (Fig. 1C) and later time points revealed progressive pericardial edema. Owing to concerns that developmental delay might be responsible for the failure of valve formation, we studied the hearts at later developmental time points. Even as late as 5 days post-fertilization, when the valve leaflets are normally well formed (Fig. 1F; supplementary material Movie 1), there was no evidence of AV ring constriction, endocardial cushions or leaflet formation in miR-21 MO-injected embryos (Fig. 1G; supplementary material Movie 2). Results were confirmed by injection of a second MO at 2 ng that targeted only the miR-21-2 Dicer processing site and yielded an identical phenotype with 68% penetrance (72/106).
Antisense MO constructs interfere with miRNA function by inhibiting the biosynthesis of the mature miRNA. When this happens, the longer pre-miRNA can be detected by northern blot. Injection of the MO targeting miR-21-2 resulted in detectable immature miR-21, whereas an MO targeting miR-21-1 resulted in only a faint upper band, and a mismatch control MO had no effect (supplementary material Fig. S1A). To confirm that the MO actually prevented formation of the mature form of miR-21, we performed qPCR. miR-92a was used as a normalization control, as it has been shown to be expressed in a constant and highly abundant manner in most cell types (Liang et al., 2007; Bargaje et al., 2010). We observed a significant reduction of mature miR-21 in miR-21-2 MO-injected embryos compared with uninjected controls (supplementary material Fig. S1B). The mismatch control MO had no effect on valvulogenesis, and co-injection of mature miR-21 reduced the effect of the miR-21 MO, with only 45% of embryos showing cardiac valvular defects (supplementary material Fig. S2A).
miR-21 is essential for endocardial and myocardial patterning
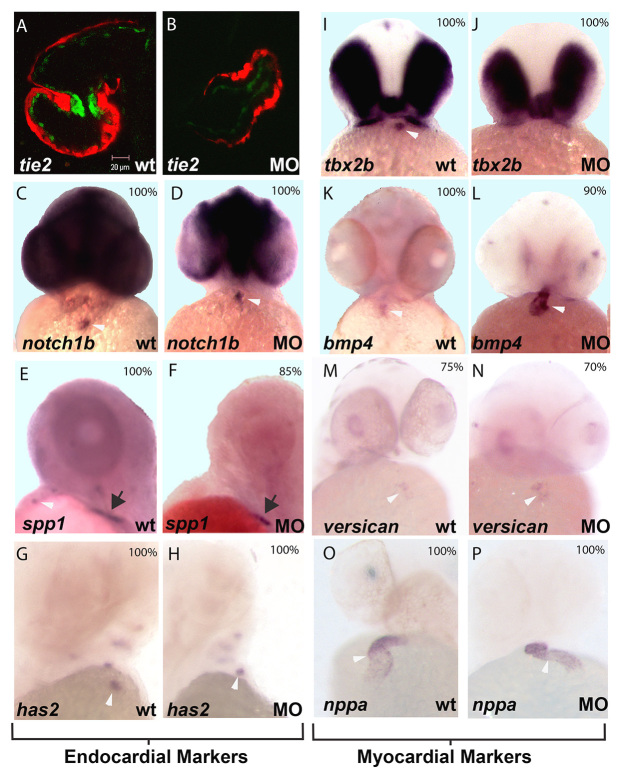
In zebrafish, one of the earliest markers of cardiac AV formation is upregulation of green fluorescent protein (GFP) expression in pre-valve endothelium in Tg(Tie2:EGFP)s849 (Motoike et al., 2000). This transgenic line has been useful in understanding the genetic and epigenetic regulation of cardiac valvulogenesis in the atrioventricular canal (AVC) mutant jekyll (in ugdh) and the contractile mutant silent heart (in tnnt2a) (Walsh and Stainier, 2001; Bartman et al., 2004). The GFP marker is normally upregulated at 48 hpf in the AV ring endothelium (Fig. 2A). Knockdown of miR-21 with the miR-21 multi-blocker MO in Tg(Tie2:EGFP)s849 fish led to loss of endothelial upregulation of the GFP marker in 18/20 embryos, as shown by confocal microscopy at 72 hpf (Fig. 2B). A 5 bp mismatch control MO did not alter Tie2:EGFP expression. These data suggest that miR-21 acts at an early step in valvulogenesis by disrupting endocardial development.
Fig. 2.
miR-21 knockdown affects both endothelial and myocardial AV ring markers. (A,B) Confocal microscopy of control transgenic embryos (cMLC2:dsRed/Tie2:EGFP) show GFP expression (green) in pre-valve AV ring endothelium at 48 hpf (A), whereas miR-21 knockdown embryos do not (B). (C-P) In situ images are oriented with atrium at bottom and ventricle at top. The probe is indicated bottom left and the experimental condition [wild-type control (wt) versus miR-21 MO injected (MO)] bottom right. The percentage of embryos that display the representative in situ pattern is indicated top right (n=10-20). (C-H) Endocardial markers; (I-P) myocardial markers. White arrowheads indicate AV ring staining, where present. Black arrows (E,F) highlight spp1 expression in the fin bud. The majority of the AV ring markers are unchanged upon knockdown of miR-21. However, the myocardial marker tbx2b is absent from the AV ring after miR-21 knockdown (I,J), whereas bmp4 is expanded throughout the ventricle (L), as compared with control embryos (K).
We further examined endocardial markers by in situ hybridization to determine whether signaling pathways known to be important for endocardial development are interrupted (Fig. 2C-H). No change was observed in the expression patterns for notch1b or has2. However, spp1 expression in both the AVC and outflow tract was absent with miR-21 knockdown, whereas expression in the early fin bud persisted (Fig. 2E,F). Absence of spp1 expression was rescued by co-injection of mature miR-21 with the MO (supplementary material Fig. S2C). spp1 is the zebrafish homolog of osteopontin, a gene known to be important in cell migration events in higher vertebrates (Katagiri et al., 1999; Alonso et al., 2007; Saika et al., 2007).
AVC formation and subsequent valve development have been shown to depend on signals exchanged between the endocardium and myocardium (Milan et al., 2006; Laux et al., 2011). To determine whether miR-21 knockdown also affects the myocardial layer, we performed in situ hybridization for the myocardial markers tbx2b, bmp4, versican and nppa (Fig. 2I-P). There was no change in the myocardial expression patterns of nppa or versican in the miR-21 morphants compared with controls (Fig. 2M-P). However, AV ring expression of tbx2b was absent in miR-21 morphant hearts but persisted in the retina (Fig. 2I,J). We also found that the normal restriction of bmp4 expression to the AV (Fig. 2K) is perturbed in knockdown embryos, which display more diffuse expression throughout the ventricle (Fig. 2L). The mechanism for this expansion of bmp4 expression into the ventricle is unknown, although ventricular expansion of bmp4 has been reported previously in other heart valve phenotypes in zebrafish (Walsh and Stainier, 2001; Peal et al., 2009). Thus, the effects of miR-21 knockdown are not limited to the endothelium, but also disrupt patterning of the myocardial cells in the AV ring.
Rescue experiments using mature miR-21 co-injected with the miR-21 MO rescued not only the cardiac morphological defects observed in knockdowns, but also restored the wild-type expression patterns of tbx2b and bmp4 (supplementary material Fig. S2D,E).
Zebrafish pdcd4 is a target of miR-21
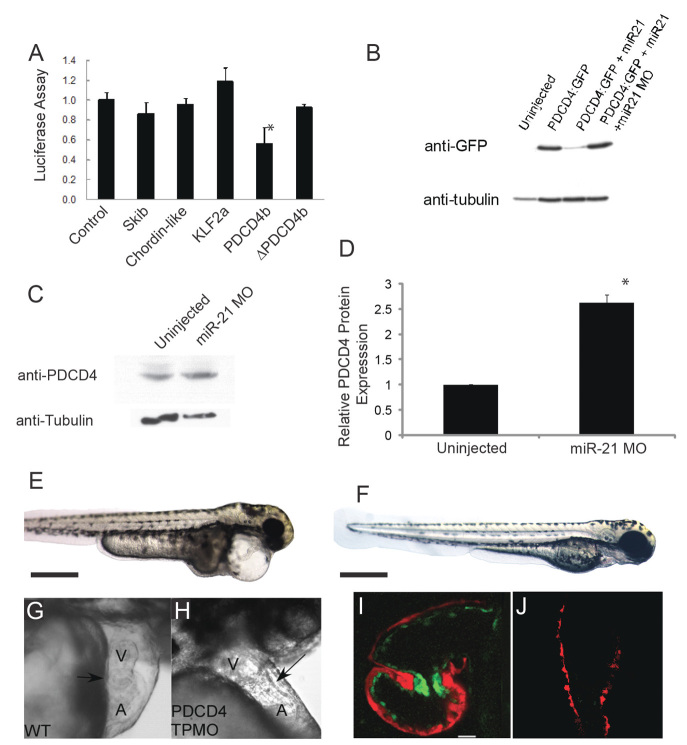
Given the limited degree of sequence homology required for miRNA targeting (Lewis et al., 2003), many algorithms have been developed to predict miRNA targets. Review of targets predicted using miRBase (http://www.mirbase.org) and miRanda (http://www.microrna.org/) identified over 150 candidate targets, of which four were initially chosen (klf2, skib, chordin-like 2 and pdcd4b) based on either biological plausibility or a high degree of conservation across species. Candidate gene 3′ UTR sequences were cloned downstream of the firefly luciferase gene and transfected with and without primiR-21 into HEK293 cells. β-galactosidase served as a transfection efficiency control. We found that the 3′ UTR of pdcd4b rendered the luciferase reporter susceptible to knockdown by cotransfected miR-21 (Fig. 3A). Site-directed mutagenesis to remove the putative miR-21 binding site from the pdcd4b 3′ UTR eliminated the effect of cotransfected miR-21. These results establish that zebrafish pdcd4b is a target for miR-21 (Asangani et al., 2008; Frankel et al., 2008).
Fig. 3.
pdcd4b is a target of miR-21 during zebrafish valve development. (A) Luciferase assays of candidate miR-21 targets. ΔPDCD4b refers to the pdcd4b 3′ UTR with the miR-21 binding site deleted. Data are mean ± s.d.; *P<0.05. (B) Western blot of whole zebrafish lysates injected at the single-cell stage with mRNA encoding GFP:Pdcd4b, co-injected with miR-21 or with miR-21 plus an miR-21 MO. Tubulin staining serves as a loading control. (C) Representative western blot of endogenous zebrafish Pdcd4 in uninjected and miR-21 morphants. (D) Quantification of zebrafish Pdcd4 western blots demonstrating a significant increase in Pdcd4 when miR-21 is knocked down. Data are mean ± s.d.; *P<0.01. (E-H) Injection of a PDCD4b target protector MO (TPMO) does not perturb the body plan or axis at 72 hpf (E, compared with control F), but does cause pericardial edema, loss of the normal cardiac looping, and AV constriction (G and H, arrow) at 48 hpf. (I,J) Confocal microscopy of the cMLC2:dsRed/Tie2:GFP line shows loss of upregulation of the Tie2:GFP marker (green) in the AV ring in pdcd4-TPmiR-21-injected fish (I, atrium at top, ventricle at bottom) compared with controls (J) at 48 hpf. A, atrium; V, ventricle.
As an in vivo test of miR-21 targeting of pdcd4b, and to additionally demonstrate the efficacy of the miR-21 MO, we employed an expression construct in which GFP is fused to the pdcd4b 3′ UTR (Carè et al., 2007; Flynt et al., 2007). In vitro synthesized GFP:pdcd4b mRNA was injected into fertilized zebrafish oocytes. If pdcd4b is a target of miR-21, co-injection of mature miR-21 should diminish GFP expression, an effect that should in turn be rescued by additional co-injection of the miR-21 MO. As shown in Fig. 3B, injection of GFP:pdcd4b mRNA resulted in robust GFP expression at 24 hours of development, as demonstrated by western blot. Co-injection of miR-21 caused a marked reduction of GFP expression, and injection of GFP:pdcd4b mRNA, miR-21, and the antisense miR-21 MO together rescued GFP expression. These results demonstrate that the 3′ UTR of pdcd4b functions in vivo as a target of miR-21 and that the antisense MO blocks miR-21 knockdown of GFP:Pdcd4b expression.
In order to determine whether miR-21 affects the endogenous expression of Pdcd4 protein, we used western blotting to demonstrate that when miR-21 is inhibited the protein levels of Pdcd4 more than double (Fig. 3C,D). Thus, a function of miR-21 is to repress production of Pdcd4 protein, presumably through binding the pdcd4b 3′ UTR. As an additional control, we employed in situ hybridization to determine the endogenous expression levels of the pdcd4b transcript. We observe that pdcd4b is expressed in the AVC in a similar manner to other AV markers (supplementary material Fig. S4). Injection of the miR-21 MO resulted in no detectable change in the mRNA expression pattern or intensity, suggesting that miR-21 does not cause degradation of pdcd4b mRNA. This result does not contradict the above data showing that pdcd4b is a miR-21 target, as miRNA-target interactions do not always result in message degradation.
Target protection of pdcd4b
In order to determine whether pdcd4b is an important and functionally relevant target of miR-21 during valvulogenesis, we employed a target protector (TP) MO (PDCD4-TPmiR-21) designed to specifically block miR-21 interaction with pdcd4b by binding to the seed sequence and also neighboring, transcript-specific sequence (Choi et al., 2007). A search of the zebrafish genome database with the Basic Local Alignment Search Tool confirmed that pdcd4b is the only sequence complementary to the target protector MO. Injection of 10 ng PDCD4-TPmiR-21 MO resulted in phenocopy (95%, 126/131) of the miR-21 knockdown with failure of AV constriction (Fig. 3C-F), development of atrioventricular regurgitation (supplementary material Movie 3) and loss of endothelial Tie2 expression assayed in Tg(Tie2:EGFP)s849 embryos (Fig. 3G,H).
As the PDCD4-TPmiR-21 MO presumably functions by allowing pdcd4b mRNA to be translated into protein, we reasoned that the TP morphant phenotype could be rescued by downregulating Pdcd4b using an MO directed at a pdcd4b splice site. Co-injection of the PDCD4-TPmiR-21 MO and the splice site MO resulted in suppression of the TP morphant heart valve defects from 95% to 32% (supplementary material Fig. S2B). To further investigate the importance of pdcd4b as a target of miR-21, we co-injected the miR-21 MO and the PDCD4 splice site MO. Since miR-21 normally reduces Pdcd4b expression, miR-21 knockdown alone should lead to increased Pdcd4b levels. If pdcd4b is the major functional target of miR-21 in AV endothelium, knockdown of Pdcd4b with the splice site MO should rescue the miR-21 phenotype. This was indeed the case, as 47% of the co-injected embryos displayed the AV defects (compared with 80% with miR-21 MO alone) (supplementary material Fig. S2C).
These data provide evidence for a crucial requirement for miR-21-mediated downregulation of the tumor suppressor Pdcd4b during valve development and further suggest that pdcd4b is the major functional target of miR-21 in cardiac valvulogenesis.
Cardiac cushion expression of miR-21 is conserved in mammals
In order to determine the relevance of miR-21 in mammalian systems, we examined cells from mouse endocardial cushions at E12.5. qPCR revealed expression of miR-21 (data not shown). As a control, these cells were either transfected with a miR-21 antagomir or with a control vector. Transfection of the miR-21 antagomir resulted in reduction of miR-21 expression compared with control transfected cells (P<0.05; supplementary material Fig. S3). Thus, as in zebrafish, miR-21 is present in mammalian cardiac cushion endothelial cells.
Cellular assays of miR-21 function
Clonal populations of endothelial cells derived from human valve tissue have been shown to recapitulate many of the hallmark features of classical endothelial to mesenchymal transition (EMT), including endothelial cell migration in vitro (Paruchuri et al., 2006). Treatment of clonal human pulmonary valve endothelial cells (HPVECs) with TGFβ2 promotes cell migration in transwell assays (Paruchuri et al., 2006). We overexpressed the miR-21 precursor primiR-21 in order to determine its function in HPVECs. We hypothesized that if miR-21 is an important effector of TGFβ signaling, then overexpression of miR-21 might recapitulate some of the effects seen with TGFβ treatment. Overexpression of miR-21 in HPVECs resulted in a 2-fold increase in cell migration (P=0.005), similar to the increase seen when HPVECs are treated with TGFβ alone (Fig. 4A). The effect does not appear to be additive, as combined miR-21 transfection and TGFβ treatment results in migration similar to either treatment alone. These results reveal a role for miR-21 in regulating human valve cell migration during EMT, and imply a conservation of function for miR-21 between human and zebrafish valve development.
Fig. 4.
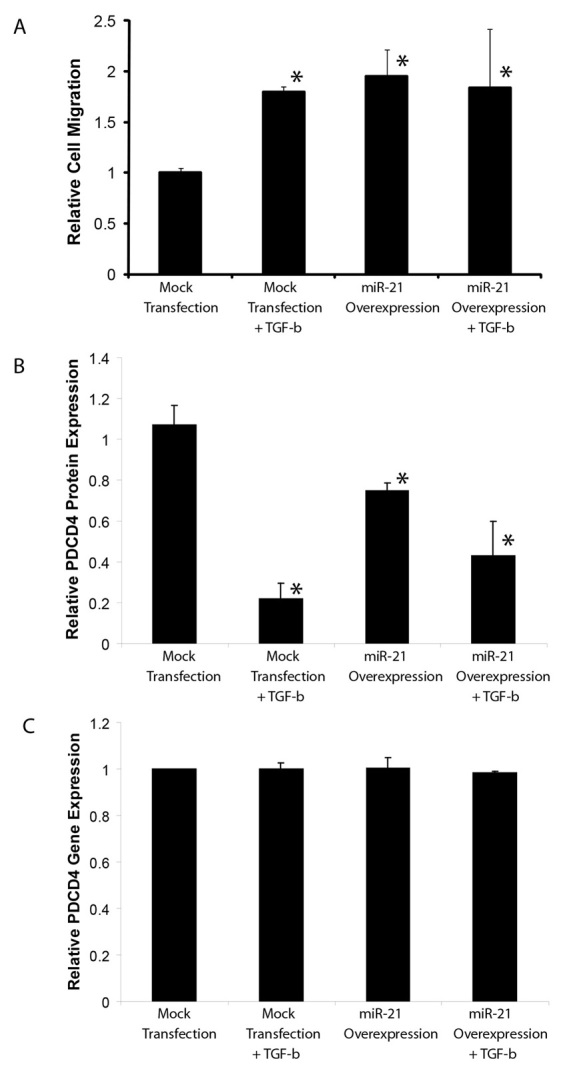
miR-21 knockdown in human pulmonary valve endothelial cells (HPVECs) triggers cell migration. (A) Pre-miR-21 overexpression causes increased HPVEC migration. The relative number of migrating cells (average of three different experiments) shows increased cell migration with TGFβ2 treatment and with miR-21 overexpression. (B) Quantification of western blot showing variation in PDCD4 protein levels under the same conditions as in A. (C) qPCR data demonstrating that PDCD4 mRNA levels do not change under the same conditions as in A. Data are mean ± s.d.; *P<0.05.
Cellular assays of PDCD4B function
Our data suggest that pdcd4b is a key target of miR-21 during zebrafish valvulogenesis. In order to evaluate the relevance of PDCD4 in human valve cells, we evaluated PDCD4 levels in HPVECs in the setting of TGFβ treatment and with miR-21 overexpression. In both of these settings, we observed decreased levels of PDCD4 protein (Fig. 4B). This regulation occurs at a post-transcriptional level, as neither TGFβ treatment nor miR-21 overexpression reduced PDCD4 transcript levels (Fig. 4C).
We next examined whether reduction of PDCD4 expression alone would be sufficient to enhance cell migration. As demonstrated above, treatment of HPVECs with TGFβ resulted in downregulation of PDCD4 (Fig. 5A) compared with untreated cells. As expected, transduction with lentivirus encoding shRNA targeting PDCD4 also resulted in downregulation of the protein, whereas a control lentivirus encoding GFP did not (Fig. 5A). There were no clear changes in cell morphology or proliferation rates. HPVEC migration assays were performed on PDCD4-depleted cells, which revealed a 3-fold increase in cell migration upon lentiviral PDCD4 knockdown compared with untreated controls (P=0.006), whereas treatment with GFP lentivirus resulted in no significant change in cell migration (Fig. 5B-F). The increase in migration with PDCD4 knockdown is similar to that observed during miR-21 overexpression, and is consistent with the hypothesis that miR-21 acts to negatively regulate PDCD4 during the endothelial cell migration events that occur during valvulogenesis.
Fig. 5.
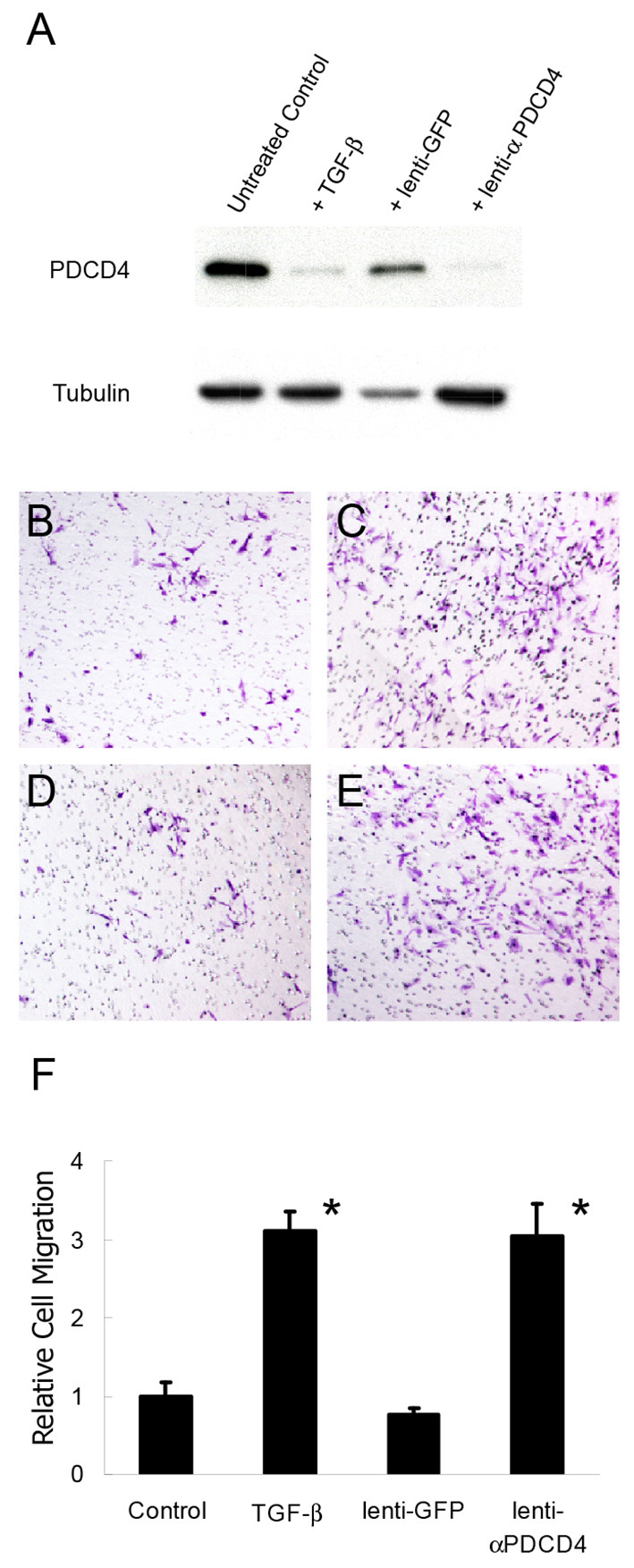
PDCD4 knockdown in HPVECs triggers cell migration. (A) Western blot of HPVEC lysates demonstrating decreased PDCD4 expression resulting from treatment with either TGFβ2 or lentiviral shRNA targeting PDCD4. Lenti-GFP control treatment did not affect PDCD4 levels. Tubulin staining serves as a loading control. (B-E) Migration assays of untreated HPVECs (B) or those treated with TGFβ2 (C), lenti-GFP (D) or lenti-αPDCD4 (E) and stained with Crystal Violet. (F) Relative number of migrating cells (average of three high-magnification fields) shows increased cell migration resulting from TGFβ2 and lenti-αPDCD4 treatments. Data are mean ± s.d.; *P<0.05.
DISCUSSION
Here, we present a model in which miR-21 binds to pdcd4, reducing protein expression and leading to increased endothelial cell migration during valvulogenesis. In zebrafish, knockdown of miR-21 leads to failure of valvulogenesis, and this effect is phenocopied by blocking miR-21 binding to its target pdcd4. Knockdown of miR-21 results in a failure to activate early endothelial valve-forming cells, which might be mediated in part by spp1. The endothelial defects are accompanied by changes in the expression of the myocardial AV ring markers Tbx2b and Bmp4. Interestingly, knockdown of Pdcd4 in the setting of reduced miR-21 partially rescues the cardiac development phenotype, suggesting that pdcd4 is the major functional target of miR-21 during valvulogenesis. In human pulmonic valve endothelial cells, both miR-21 and PDCD4 are regulators of migration.
Early valvulogenesis in mammals is marked by activation of specific endothelial cells in the developing heart tube, which then migrate into the extracellular matrix to form endocardial cushions. If our observations in zebrafish are relevant to mammals, we would expect to find miR-21 in the pre-valve endothelium, i.e. the cardiac cushions. This was indeed the case, as an examination of mouse endocardial cushion cells confirmed expression of miR-21. We next turned to the primary HPVEC cell line, and observed that either upregulation of miR-21 or downregulation of the miR-21 target PDCD4 resulted in enhanced cell migration. miR-21 can therefore be considered a pro-migration factor in pre-valve endothelium, operating at least in part via downregulation of PDCD4. miRNAs are generally thought to provide a mechanism for the post-transcriptional regulation of multiple genes. It is somewhat surprising, therefore, that the miR-21 knockdown (which presumably results in increased expression of multiple miR-21 targets) could be rescued with an MO targeting only pdcd4b. These data suggest that pdcd4b is the primary relevant target of miR-21 during zebrafish valvulogenesis. Our data also suggest that miR-21 acts on pdcd4b mRNA to prevent translation without causing degradation, as neither miR-21 knockdown in the zebrafish model nor overexpression in HPVECs leads to a change in pdcd4b/PDCD4B transcript levels, although both affect protein levels.
Interestingly, PDCD4 is known to be a negative regulator of tumor cell migration in a chick explant model, in part by transcriptional control of the known tumor enhancer gene u-PAR (Asangani et al., 2008). Similarly, knockdown of miR-21 in a colorectal tumor line resulted in an increased level of PDCD4 and a reduction in cell migration, whereas overexpression of miR-21 leads to decreased levels of PDCD4 and an enhancement of cell migration (Asangani et al., 2008). Thus, the functional interaction of miR-21 and PDCD4 to control cell migration appears to be conserved in both development and cancer biology.
In a similar manner, spp1/osteopontin has been shown to control cell migration events during tumor metastasis (Tuck et al., 1999; Tuck et al., 2000). Spp1 is normally repressed by multiple genes, and during metastasis escapes repression to allow cell migration to occur (Furger et al., 2003; Allan et al., 2006; Hedley et al., 2008). Our data indicate that knockdown of miR-21 also affects spp1 expression during valvulogenesis, suggesting that miR-21 and spp1 might work in a common pathway to regulate endothelial cell migration during development.
A previous study has demonstrated that loss of the miR-processing enzyme Dicer leads to cardiac defects (Giraldez et al., 2005). Recent work has demonstrated that knockdown of miR-23 phenocopies the Dicer AVC defect, and that the resulting endocardial cushion expansion is due to upregulation of the enzyme Has2, which leads to a preponderance of the extracellular matrix component gylcosaminoglycan hyaluronic acid (Lagendijk et al., 2011). It is probable that multiple miRNAs are needed during AVC development, at least one of which controls cell migration (miR-21) and one of which controls the extracellular milieu needed for cell migration (miR-23).
Of interest in the field of cardiac development is whether classic EMT occurs during valvulogenesis in zebrafish (Scherz et al., 2008). Although our results do not bear directly on this question, they do add to the growing list of signaling pathways that are conserved in valvulogenesis from mammals to zebrafish, including Notch (Timmerman et al., 2004; Rutenberg et al., 2006), VEGF (Lee et al., 2006), NFAT (Chang et al., 2004), neuregulin (Milan et al., 2006), EGF (Goishi et al., 2003), and Wnt/β-catenin (Hurlstone et al., 2003). These data imply that, although the cellular events might be debated, many of the molecular pathways associated with valvulogenesis are conserved in zebrafish.
Further work to elucidate targets of miR-21 might identify additional downstream TGFβ signaling effectors and regulators of cell identity and migration, and aid in the discovery of candidates for therapeutic intervention. Disease processes such as myocardial fibrosis or even mitral valve prolapse, a condition characterized by excess valve leaflet tissue growth, might be due to pathologic uncontrolled cell migration, raising the possibility that miR-21 could be a useful therapeutic target in these diseases.
Supplementary Material
Acknowledgments
We thank Jill Wylie-Sears for expert technical assistance.
Footnotes
Funding
This work was supported in part by the Fondation Leducq, Paris, France for the Transatlantic MITRAL Network of Excellence to D.S.P, J.B., R.A.N, R.R.M. and D.J.M. [07CVD04]; by the American Heart Association [2261354 to D.J.M. and 11SDG5270006 to R.A.N.]; and by the National Institutes of Health [HL33756 to R.R.M.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.084475/-/DC1
References
- Allan A. L., George R., Vantyghem S. A., Lee M. W., Hodgson N. C., Engel C. J., Holliday R. L., Girvan D. P., Scott L. A., Postenka C. O., et al. (2006). Role of the integrin-binding protein osteopontin in lymphatic metastasis of breast cancer. Am. J. Pathol. 169, 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso S. R., Tracey L., Ortiz P., Pérez-Gómez B., Palacios J., Pollán M., Linares J., Serrano S., Sáez-Castillo A. I., Sánchez L., et al. (2007). A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 67, 3450–3460 [DOI] [PubMed] [Google Scholar]
- Asangani I. A., Rasheed S. A. K., Nikolova D. A., Leupold J. H., Colburn N. H., Post S., Allgayer H. (2008). MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27, 2128–2136 [DOI] [PubMed] [Google Scholar]
- Bakkers J., Kramer C., Pothof J., Quaedvlieg N. E. M., Spaink H. P., Hammerschmidt M. (2004). Has2 is required upstream of Rac1 to govern dorsal migration of lateral cells during zebrafish gastrulation. Development 131, 525–537 [DOI] [PubMed] [Google Scholar]
- Bargaje R., Hariharan M., Scaria V., Pillai B. (2010). Consensus miRNA expression profiles derived from interplatform normalization of microarray data. RNA 16, 16–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartman T., Walsh E. C., Wen K.-K., McKane M., Ren J., Alexander J., Rubenstein P. A., Stainier D. Y. R. (2004). Early myocardial function affects endocardial cushion development in zebrafish. PLoS Biol. 2, E129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carè A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.-L., Segnalini P., Gu Y., Dalton N. D., et al. (2007). MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 13, 613–618 [DOI] [PubMed] [Google Scholar]
- Chang C.-P., Neilson J. R., Bayle J. H., Gestwicki J. E., Kuo A., Stankunas K., Graef I. A., Crabtree G. R. (2004). A field of myocardial-endocardial NFAT signaling underlies heart valve morphogenesis. Cell 118, 649–663 [DOI] [PubMed] [Google Scholar]
- Choi W.-Y., Giraldez A. J., Schier A. F. (2007). Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318, 271–274 [DOI] [PubMed] [Google Scholar]
- Davis B. N., Hilyard A. C., Lagna G., Hata A. (2008). SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pompa J. L., Timmerman L. A., Takimoto H., Yoshida H., Elia A. J., Samper E., Potter J., Wakeham A., Marengere L., Langille B. L., et al. (1998). Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature 392, 182–186 [DOI] [PubMed] [Google Scholar]
- Flynt A. S., Li N., Thatcher E. J., Solnica-Krezel L., Patton J. G. (2007). Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 39, 259–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel L. B., Christoffersen N. R., Jacobsen A., Lindow M., Krogh A., Lund A. H. (2008). Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J. Biol. Chem. 283, 1026–1033 [DOI] [PubMed] [Google Scholar]
- Furger K. A., Allan A. L., Wilson S. M., Hota C., Vantyghem S. A., Postenka C. O., Al-Katib W., Chambers A. F., Tuck A. B. (2003). β(3) integrin expression increases breast carcinoma cell responsiveness to the malignancy-enhancing effects of osteopontin. Mol. Cancer Res. 1, 810–819 [PubMed] [Google Scholar]
- Giraldez A. J., Cinalli R. M., Glasner M. E., Enright A. J., Thomson J. M., Baskerville S., Hammond S. M., Bartel D. P., Schier A. F. (2005). MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833–838 [DOI] [PubMed] [Google Scholar]
- Goishi K., Lee P., Davidson A. J., Nishi E., Zon L. I., Klagsbrun M. (2003). Inhibition of zebrafish epidermal growth factor receptor activity results in cardiovascular defects. Mech. Dev. 120, 811–822 [DOI] [PubMed] [Google Scholar]
- Gross J. M., Dowling J. E. (2005). Tbx2b is essential for neuronal differentiation along the dorsal/ventral axis of the zebrafish retina. Proc. Natl. Acad. Sci. USA 102, 4371–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada M., Luo X., Qi X. Y., Tadevosyan A., Maguy A., Ordog B., Ledoux J., Kato T., Naud P., Voigt N., et al. (2012). Transient receptor potential canonical-3 channel-dependent fibroblast regulation in atrial fibrillation. Circulation 126, 2051–2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley B. D., Welch D. R., Allan A. L., Al-Katib W., Dales D. W., Postenka C. O., Casey G., Macdonald I. C., Chambers A. F. (2008). Downregulation of osteopontin contributes to metastasis suppression by breast cancer metastasis suppressor 1. Int. J. Cancer 123, 526–534 [DOI] [PubMed] [Google Scholar]
- Hoffman J. I. E. (1995). Incidence of congenital heart disease: I. Postnatal incidence. Pediatr. Cardiol. 16, 103–113 [DOI] [PubMed] [Google Scholar]
- Hurlstone A. F. L., Haramis A.-P. G., Wienholds E., Begthel H., Korving J., Van Eeden F., Cuppen E., Zivkovic D., Plasterk R. H. A., Clevers H. (2003). The Wnt/β-catenin pathway regulates cardiac valve formation. Nature 425, 633–637 [DOI] [PubMed] [Google Scholar]
- Katagiri Y. U., Sleeman J., Fujii H., Herrlich P., Hotta H., Tanaka K., Chikuma S., Yagita H., Okumura K., Murakami M., et al. (1999). CD44 variants but not CD44s cooperate with beta1-containing integrins to permit cells to bind to osteopontin independently of arginine-glycine-aspartic acid, thereby stimulating cell motility and chemotaxis. Cancer Res. 59, 219–226 [PubMed] [Google Scholar]
- Lagendijk A. K., Goumans M. J., Burkhard S. B., Bakkers J. (2011). MicroRNA-23 restricts cardiac valve formation by inhibiting Has2 and extracellular hyaluronic acid production. Circ. Res. 109, 649–657 [DOI] [PubMed] [Google Scholar]
- Laux D. W., Febbo J. A., Roman B. L. (2011). Dynamic analysis of BMP-responsive smad activity in live zebrafish embryos. Dev. Dyn. 240, 682–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. M., Cope J. J., Ackermann G. E., Goishi K., Armstrong E. J., Paw B. H., Bischoff J. (2006). Vascular endothelial growth factor receptor signaling is required for cardiac valve formation in zebrafish. Dev. Dyn. 235, 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003). Prediction of mammalian microRNA targets. Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- Liang Y., Ridzon D., Wong L., Chen C. (2007). Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8, 166–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Liu M., Stribinskis V., Klinge C. M., Ramos K. S., Colburn N. H., Li Y. (2008). MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 27, 4373–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S. T., Patel T. (2007). MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan D. J., Giokas A. C., Serluca F. C., Peterson R. T., MacRae C. A. (2006). Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development 133, 1125–1132 [DOI] [PubMed] [Google Scholar]
- Motoike T., Loughna S., Perens E., Roman B. L., Liao W., Chau T. C., Richardson C. D., Kawate T., Kuno J., Weinstein B. M., et al. (2000). Universal GFP reporter for the study of vascular development. Genesis 28, 75–81 [DOI] [PubMed] [Google Scholar]
- Nikaido M., Tada M., Saji T., Ueno N. (1997). Conservation of BMP signaling in zebrafish mesoderm patterning. Mech. Dev. 61, 75–88 [DOI] [PubMed] [Google Scholar]
- Paruchuri S., Yang J.-H., Aikawa E., Melero-Martin J. M., Khan Z. A., Loukogeorgakis S., Schoen F. J., Bischoff J. (2006). Human pulmonary valve progenitor cells exhibit endothelial/mesenchymal plasticity in response to vascular endothelial growth factor-A and transforming growth factor-beta2. Circ. Res. 99, 861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peal D. S., Burns C. G., Macrae C. A., Milan D. (2009). Chondroitin sulfate expression is required for cardiac atrioventricular canal formation. Dev. Dyn. 238, 3103–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena J. T. G., Sohn-Lee C., Rouhanifard S. H., Ludwig J., Hafner M., Mihailovic A., Lim C., Holoch D., Berninger P., Zavolan M., et al. (2009). miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat. Methods 6, 139–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts J. D., Vincent E. B., Runyan R. B., Weeks D. L. (1992). Sense and antisense TGF β 3 mRNA levels correlate with cardiac valve induction. Dev. Dyn. 193, 340–345 [DOI] [PubMed] [Google Scholar]
- Rutenberg J. B., Fischer A., Jia H., Gessler M., Zhong T. P., Mercola M. (2006). Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development 133, 4381–4390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saika S., Shirai K., Yamanaka O., Miyazaki K.-i., Okada Y., Kitano A., Flanders K. C., Kon S., Uede T., Kao W. W.-Y., et al. (2007). Loss of osteopontin perturbs the epithelial-mesenchymal transition in an injured mouse lens epithelium. Lab. Invest. 87, 130–138 [DOI] [PubMed] [Google Scholar]
- Scherz P. J., Huisken J., Sahai-Hernandez P., Stainier D. Y. R. (2008). High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development 135, 1179–1187 [DOI] [PubMed] [Google Scholar]
- Si M. L., Zhu S., Wu H., Lu Z., Wu F., Mo Y. Y. (2007). miR-21-mediated tumor growth. Oncogene 26, 2799–2803 [DOI] [PubMed] [Google Scholar]
- Tatsuguchi M., Seok H. Y., Callis T. E., Thomson J. M., Chen J.-F., Newman M., Rojas M., Hammond S. M., Wang D.-Z. (2007). Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J. Mol. Cell. Cardiol. 42, 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C., Thisse B. (1999). Antivin, a novel and divergent member of the TGFbeta superfamily, negatively regulates mesoderm induction. Development 126, 229–240 [DOI] [PubMed] [Google Scholar]
- Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S., et al. (2008). MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 456, 980–984 [DOI] [PubMed] [Google Scholar]
- Timmerman L. A., Grego-Bessa J., Raya A., Bertrán E., Pérez-Pomares J. M., Díez J., Aranda S., Palomo S., McCormick F., Izpisúa-Belmonte J. C., et al. (2004). Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18, 99–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck A. B., Arsenault D. M., O’Malley F. P., Hota C., Ling M. C., Wilson S. M., Chambers A. F. (1999). Osteopontin induces increased invasiveness and plasminogen activator expression of human mammary epithelial cells. Oncogene 18, 4237–4246 [DOI] [PubMed] [Google Scholar]
- Tuck A. B., Elliott B. E., Hota C., Tremblay E., Chambers A. F. (2000). Osteopontin-induced, integrin-dependent migration of human mammary epithelial cells involves activation of the hepatocyte growth factor receptor (Met). J. Cell. Biochem. 78, 465–475 [DOI] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L. B., Liu N., Williams A. H., McAnally J., Gerard R. D., Richardson J. A., Olson E. N. (2006). A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA 103, 18255–18260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E., Sutherland L. B., Qi X., Richardson J. A., Hill J., Olson E. N. (2007). Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579 [DOI] [PubMed] [Google Scholar]
- Walsh E. C., Stainier D. Y. R. (2001). UDP-glucose dehydrogenase required for cardiac valve formation in zebrafish. Science 293, 1670–1673 [DOI] [PubMed] [Google Scholar]
- Westin J., Lardelli M. (1997). Three novel Notch genes in zebrafish: implications for vertebrate Notch gene evolution and function. Dev. Genes Evol. 207, 51–63 [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W. P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H. R., Kauppinen S., Plasterk R. H. A. (2005). MicroRNA expression in zebrafish embryonic development. Science 309, 310–311 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Ransom J. F., Li A., Vedantham V., von Drehle M., Muth A. N., Tsuchihashi T., McManus M. T., Schwartz R. J., Srivastava D. (2007). Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129, 303–317 [DOI] [PubMed] [Google Scholar]
- Zhu S., Si M.-L., Wu H., Mo Y.-Y. (2007). MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 282, 14328–14336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.