Abstract
We have generated an inducible system to control the timing of transgene expression in zebrafish and chick. An estrogen receptor variant (ERT2) fused to the GAL4 transcriptional activator rapidly and robustly activates transcription within 3 hours of treatment with the drug 4-hydroxy-tamoxifen (4-OHT) in tissue culture and transgenic zebrafish. We have generated a broadly expressed inducible ERT2-GAL4 zebrafish line using the ubiquitin (ubi) enhancer. In addition, use of ERT2-GAL4 in conjunction with tissue-specific enhancers enables the control of transgene expression in both space and time. This spatial restriction and the ability to sustain forced expression are important advantages over the currently used heat-shock promoters. Moreover, in contrast to currently available TET and LexA systems, which require separate constructs with their own unique recognition sequences, ERT2-GAL4 is compatible with the growing stock of UAS lines being generated in the community. We also applied the same inducible system to the chick embryo and find that it is fully functional, suggesting that this strategy is generally applicable.
Keywords: Transgenic, GAL4, Zebrafish, Chick, Tamoxifen
INTRODUCTION
The analysis of protein function is enhanced by the ability to perform gain- and loss-of-function experiments in vivo. The use of zebrafish as a model organism has been making great strides in both regards, particularly in the application of the binary GAL4/UAS forced expression system (Halpern et al., 2008). Transgenic lines expressing the transcriptional activator GAL4 under the control of specific enhancer elements (Sagasti et al., 2005) or driven by random insertional events (‘enhancer traps’) (Davison et al., 2007; Asakawa et al., 2008; Distel et al., 2009) are being created to enable the expression of Upstream Activating Sequence (UAS)-linked transgenes within specific tissues or cell types. In parallel, stable UAS lines are being created to express genes of interest when crossed with these GAL4 driver lines (Bradford et al., 2011). As these developments gather pace, the combinatorial nature of the GAL4/UAS system will greatly expand the experimental repertoire of the zebrafish model organism.
Currently, the pattern and timing of transgenic expression is generally dictated by the inherent activity profile of the enhancer elements being used, or by the genomic environment surrounding the GAL4 transgene in a random insertional event. Although this can have experimental advantages in some contexts, for example to mimic the time and pattern of endogenous gene expression, it is often desirable to control the timing of transgene expression. The heat shock-responsive HSP70 enhancer has enabled temporal control over the initiation of forced transgene expression (Halloran et al., 2000). However, the duration of induced expression is variable, and dependent on the rate of mRNA decay and protein stability of each particular transgene (Geling et al., 2003). Additionally, heat shock affords no tissue specificity without additional engineering of dedicated transgenic lines such as a enhancer or UAS-driven, floxed STOP-transgene constructs (Thummel et al., 2005; Hans et al., 2009) to provide an intersectional expression system. These approaches are complex, and drawbacks include the detrimental effect of heat-shock on primary cilia (Prodromou et al., 2012), and the accelerated development at higher temperatures, a confounding variable when studying a dynamic system or a short-lived biological phenomenon.
We set out to create a flexible conditional expression system amenable to both spatial and temporal control. Importantly, the kinetics of induction needed to be rapid enough to permit analysis during the first few days post-fertilisation, when the embryo is undergoing rapid developmental changes. An additional benefit would be the ability to turn off transgene expression by drug withdrawal (Knopf et al., 2010). A number of different inducible GAL4 systems have been used in Drosophila (RU486) (Osterwalder et al., 2001; Nicholson et al., 2008), Xenopus (RU486) (Chae et al., 2002) and zebrafish (ecdysone hormone) (Esengil et al., 2007). Based on reports of the successful use of 4-hydroxy-tamoxifen (4-OHT) to activate Cre recombinase/estrogen receptor fusions in zebrafish embryos (Hans et al., 2009; Hans et al., 2011; Mosimann and Zon, 2011), we decided to base our system on this approach. We find that fusing the GAL4-VP16 transcriptional activator to the human estrogen receptor (variant ERT2) (Feil et al., 1997) creates a protein with 4-OHT-inducible transcriptional activity, little or no leaky activity, rapid induction kinetics and whose activity is extinguished by drug withdrawal. Importantly, stable transgenics bearing these fusions driven by various enhancers are effective at driving UAS-bearing transgenic lines to give temporally controlled, tissue-specific gene expression. Using the ubiquitin enhancer (Mosimann et al., 2011) as a driver, we have generated a stable zebrafish line that provides ubiquitous conditional GAL4 activity within the first 24 hours post fertilisation (hpf). We find that this inducible system is fully functional in the chick embryo, suggesting that it should be transferable to a broader range of model organisms.
MATERIALS AND METHODS
Fish maintenance
Wild-type and transgenic zebrafish embryos were obtained by natural spawning and raised at 28.5°C, as described previously (Westerfield, 1993). All DNA injections were performed in one-cell stage embryos.
Transgenic constructs
The ERT2 coding sequence (CDS) was amplified by PCR from an intermediate plasmid containing the mutated estrogen ligand-binding domain (G400V/M543A/L544A) (Feil et al., 1997) and cloned in-frame upstream of a GAL4-VP16 fusion CDS, thus creating ERT2-GAL4. This fusion was subsequently cloned into a number of plasmids as follows. EF1α::ERT2-GAL4 was created by cloning the ERT2-GAL4 CDS into a plasmid containing the Xenopus EF1α enhancer/promoter (Johnson and Krieg, 1994) and the mouse Fos minimal promoter (Dorsky et al., 2002). A UAS::tdTomato was also present in this plasmid, but was found to be non-functional. To create HuC::ERT2-GAL4, the HuC enhancer/promoter from HuC::YC2.1 (Higashijima et al., 2003) was cloned upstream of the ERT2-GAL4 CDS. To create cldnb::ERT2-GAL4, a 4.2 kb fragment of the upstream promoter of the zebrafish cldnb locus was PCR amplified and cloned upstream of the ERT2-GAL4 CDS. To create ubi::ERT2-GAL4, the ubiquitin promoter/enhancer was subcloned from pCM206 [a kind gift from Christian Mosimann (University of Zurich, Switzerland) and Leonard Zon (Boston Children’s Hospital, MA, USA)] (Mosimann et al., 2011) upstream of the ERT2-GAL4 CDS. UAS::GFP-dncasp9 was created by first generating a dominant-negative zebrafish caspase9 (Cys298Ala) [based on Rowe et al. (Rowe et al., 2005)], then fusing it downstream of eGFP (from pEGFP-N1, Clontech). The UAS::hoxb8a was generated by fusing a 6× myc tag with a PCR amplified CDS of the zebrafish hoxb8a, and cloned into a 5× UAS plasmid. All the plasmids described have a miniTOL2 backbone (Balciunas et al., 2006) to facilitate genomic integration in zebrafish. Unless otherwise stated, all UAS constructs contain five copies of the UAS sequence. Initial experiments with the cldnb::ERT2-GAL4 line were crosses to a 2×UAS::H2B-citrine that lacked the E1B promoter region E1B [described previously (Distel et al., 2009)]. Maps, plasmids and additional information are available upon request.
Transient and stable transgenic zebrafish
Transgenic embryos were generated by co-injecting 5-20 pg of plasmid DNA with 25 pg of TOL2 transposase mRNA into one-cell stage embryos (Balciunas et al., 2006). Transient transgenics were analyzed over the first few days of development. When more than one plasmid was injected at once, the total amount of DNA was kept below 25 pg in total. Stable transgenics were generated from similarly injected embryos, grown to adulthood and screened for stable integration in their offspring. Some transgenes contained an alpha crystallin::RFP cassette as a genotyping aid to identify carriers (by their red eyes) (Gerety and Wilkinson, 2011). To quantify the degree of ERT2-GAL4 activity present in the absence of 4-OHT in the HuC::ERT2-GAL4 experiments, we collected a 20 image confocal stack spanning the entire thickness of the brains of five untreated embryos (35-40 μm), performed a maximum projection in the Leica software and counted all green cells in these images.
Tissue culture and transfections
HEK 293 cells were grown in standard media and replated into fibronectin-coated multiwell dishes prior to transfection. Up to 1 μg of total plasmid DNA was transfected using FugeneHD according to the manufacturer’s instructions (Roche). Cells were grown overnight (16 hours) in the presence of either 5 μM 4-OHT (H7904; Sigma) or ethanol control, and fixed the next day for analysis. An anti-GAL4 antibody (specific for DNA-binding domain, sc-577, Santa Cruz) was used to detect the expression and localisation of GAL4 protein within cells. Detection of primary antibody was carried out using Alexa Fluor-594 secondary antibodies (1:450, Invitrogen). The nuclear stain DAPI was added with the secondary antibodies for 1-2 hours.
Drug treatments
4-hydroxy-tamoxifen (H7904, Sigma) was dissolved at 12.5 mg/ml in 100% ethanol and stored at -20°C. Subsequent dilutions were made immediately before use, in standard fish water or tissue culture media, to 0.5-5 μM. Controls were treated with an equivalent amount of ethanol diluted in fish water or media. The Bcl2 inhibitor HA14-1 (Sigma-Aldrich, H8787) was dissolved in DMSO at 10 mM and further diluted in fish water immediately prior to application. The dose of 10 μM HA14-1 has been established previously (Gerety and Wilkinson, 2011) as effective at inducing widespread apoptosis within 2 hours. For the apoptosis rescues, embryos from the relevant cross were grown in standard fish water, in their chorions, with 0.625 μM 4-OHT from 50% epiboly until 18 hours post-fertilisation (hpf). Subsequent challenge with 10 μM HA14-1 was carried out in the presence of fresh 0.625 μM 4-OHT to ensure sustained ERT2-GAL4 activity.
qPCR assays
Embryos from three independent clutches of ubi::ERT2-GAL4 fish crossed to UAS::H2B-citrine were treated from 50% epiboly until 18 hpf in 1 μM 4-OHT. Citrine-positive embryos were selected, and half were washed four times in fish water. The other half were transferred into fish water containing fresh 1 μM 4-OHT. Both batches were allowed to grow another 24 hours, until 42 hpf. mRNA was isolated, followed by reverse transcription using oligo-dT primers (SuperscriptIII kit, Invitrogen). qPCR was performed in triplicate on each sample to detect Citrine levels (forward primer, TGACCCTGAAGTTCATCTGC; reverse primer, AAGTCGTGCTGCTTCATGTG) and normalised to ef1alpha (forward primer, CTGGAGGCCAGCTCAAACAT; reverse primer, AAGAGTAGTACCGCTAGCATTAC). Data from washout conditions were plotted relative to sustained exposure (±s.e.m.).
In situ hybridisation and immunohistochemistry
Embryos were grown until the desired stage, then fixed in 4% paraformaldehyde/PBS overnight. Fixed embryos were then stored in 100% methanol, or processed immediately for in situ hybridisation or immunohistochemistry. A probe to detect H2B-citrine was made from a plasmid containing the coding sequence of eGFP. Digoxigenin-UTP labelled riboprobes were synthesized according to the manufacturer’s instructions (Roche). In situ hybridisation and colour development with NTB/BCIP performed as described previously (Xu et al., 1994). Embryos were then re-fixed in paraformaldehyde, cleared in 70% glycerol/PBS, and mounted for photography on a Zeiss Axiovision microscope fitted with a Zeiss Axiocam digital camera.
Primary antibodies used were anti-cleaved caspase 3 (1:250, #9661, Cell Signaling Technology), mouse anti-cMyc (to detect myc-tagged Hoxb8a, clone 9E10, sc40, Santa Cruz), rabbit anti-GFP (to detect Citrine and GFP-dnCasp9, TP401, Torrey Pines Biolabs), mouse anti-HuC/D (clone 16A11, A-21271, Invitrogen) and goat anti-βGal (Biogenesis). Embryos were blocked in PBS+0.1% Tween20 and 5% goat serum. Antibodies were diluted in this blocking solution, applied to embryos and incubated overnight at 4°C. Detection of primary antibodies was carried out using Alexa Fluor-488, -594 or -647 secondary antibodies (1:450, Invitrogen). Fluorescent images were captured using a Leica TCS SP2 confocal microscope, and a Leica M205FA stereoscope. For many experiments, GFP/Citrine fluorescence was detected without staining with anti-GFP.
Manipulation of the chick embryos
In ovo electroporation experiments were performed into the spinal cord of HH stage 11 chick embryos. Whole electroporated embryos were subsequently taken out of the egg and cultured on agarose gel according to the protocol described previously (Chapman et al., 2001). 4-OHT (10 μM diluted in Hank’s Balanced Salt Solution, HBSS; Invitrogen) was introduced into the spinal cord cavity 1 hour after the electroporation, and analysis of induction was performed after an additional 8 hours of culture. At least four embryos were examined for each condition. The electroporation, preparation of chick neural plate explants and immunohistochemistry were performed as described previously (Dessaud et al., 2007), with 1 μM 4-OHT treatment for stated time intervals (n=4 explants for each timepoint and condition). Primary antibodies used were rabbit anti-GFP (Invitrogen) and goat anti-βGal (Biogenesis).
RESULTS
Construction of an estrogen receptor-GAL4-VP16 fusion
As the basis for the inducible GAL4 system, we chose to use a variant of GAL4 in which the DNA-binding domain is fused to two copies of a truncated VP16 activation domain (Asakawa et al., 2008; Ogura et al., 2009). This variant has been shown to be a weaker activator than the fusions containing the full-length VP16 activation domain and thus provides the opportunity for a greater dynamic range in transcriptional activity. ERT2, the modified ligand-binding domain (LBD) of human estrogen receptor α bearing the G400V/M543A/L544A mutations, binds the synthetic antagonist 4-OHT, but is insensitive to 17 β-oestradiol (Feil et al., 1997). This specificity limits the risk of endogenous estrogens activating the fusion protein. Importantly, 4-OHT is not deleterious to zebrafish embryo development at concentrations below 10 μM (Hans et al., 2009). We cloned one copy of the ERT2 domain upstream of GAL4 to generate ERT2-GAL4. For initial testing, this fusion was placed downstream of the Xenopus EF1α enhancer in order to drive expression in a variety of cells and embryonic tissues (Fig. 1, schematic at top).
Fig. 1.
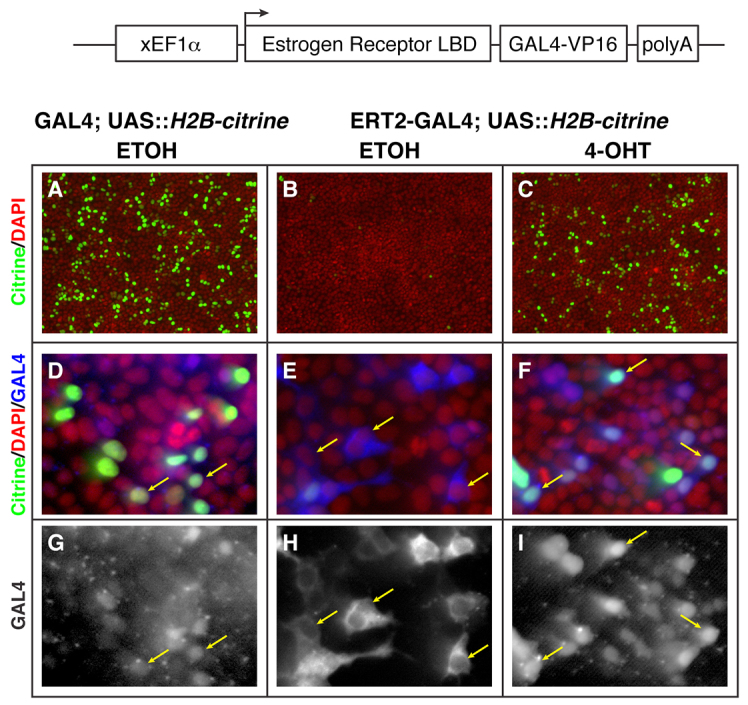
Localisation and activity of GAL4-VP16/estrogen receptor LBD fusion proteins are 4-OHT dependent. (A-I) Forced expression of ERT2-GAL4 (schematised, top) in HEK293 cells shows cytoplasmic localisation of GAL4 in control treatment (E and H, blue and grey, respectively), and nuclear enrichment after treatment with 5 μM 4-OHT for 16 hours (F and I, blue and grey, respectively), when detected with an anti-GAL4 antibody. (D,G) This is in contrast to the constitutive nuclear localisation of GAL4-VP16 alone (D and G, blue and grey, respectively). Nuclei are labelled with DAPI (A-F, red channel). Consistent with the localisation of GAL4 in these samples, expression of Citrine from a co-transfected UAS::H2B-citrine plasmid (A-F, Citrine in green channel) is dependent on nuclear localisation of GAL4, and hence 4-OHT treatment in ERT2-GAL4-expressing cells (compare B and C, green channel). Yellow arrows indicate GAL4 protein localisation.
ERT2-GAL4 fusions are 4-OH-tamoxifen responsive
In the absence of ligand, the estrogen receptor (ER) is bound and held in the cytoplasm by heat-shock proteins (HSPs), effectively inhibiting transcriptional activity. Ligand binding causes HSP/ER dissociation, thereby permitting nuclear translocation and transcriptional activity of the ER (reviewed by Branda and Dymecki, 2004). By fusing the ER ligand binding domain to GAL4, we expected to see a ligand-dependent subcellular localisation of ERT2-GAL4. To test this, we transfected HEK-293 cells with plasmids bearing either EF1α::GAL4 or EF1α::ERT2-GAL4 along with a plasmid containing a UAS::H2B-citrine. These cells were treated overnight (16 hours) with ethanol control (ETOH) or 5 μM 4-OHT. Immunodetection of GAL4 protein revealed that GAL4 alone was constitutively present in the nucleus (Fig. 1D,G, anti-GAL4 in blue and grey, respectively). By contrast, the ERT2-GAL4 protein was present in the cytoplasm and excluded from the nucleus of cells grown in control conditions, whereas in the presence of 4-OHT, there is an accumulation of GAL4 protein in the nucleus (Fig. 1E,H versus 1F,I).
As expected from the subcellular localisation of the GAL4 variants, we found high levels of H2B-Citrine expression in the GAL4-alone condition, and very few positive cells in the ETOH-treated ERT2-GAL4 cultures (Fig. 1A,B). Upon treatment with 5 μM 4-OHT, ERT2-GAL4 transfected cells showed high levels of H2B-Citrine (Fig. 1C). These experiments reveal that the ERT2-GAL4 fusion protein has the anticipated 4-OHT responsive localisation and transcriptional activity.
Stable transgenic expression of ERT2-GAL4 gives rapid 4-OHT dependent induction
We used TOL2-transposase-mediated transgenesis (Kawakami, 2005; Balciunas et al., 2006) to generate a stable zebrafish line carrying an EF1α::ERT2-GAL4 insertion. To determine whether ERT2-GAL4 transactivates UAS-linked transgenes in vivo, we crossed the EF1α::ERT2-GAL4 fish with several UAS effector lines. Embryos from crosses of carriers to UAS::H2B-citrine, UAS::GFP-dominant negative caspase9 (GFP-dnCasp9) and UAS::hoxb8a lines were collected, grown to ∼50% epiboly, and then treated overnight with either ETOH (control) or 0.5 μM 4-OHT. In all cases, we saw no expression of the UAS-linked transgene in controls, and strong expression in 4-OHT-treated embryos (Fig. 2A,B,E,F,I versus 2C,D,G,H,J,K). As has been described previously (Hans et al., 2009), the Xenopus EF1α promoter displays strong widespread expression up to early somite stages, followed by a rapid decrease, with heterogeneous expression maintained in the eye, central nervous system and somites. This is seen in the sustained eye and brain expression of UAS-linked genes (Fig. 2D,H,K).
Fig. 2.
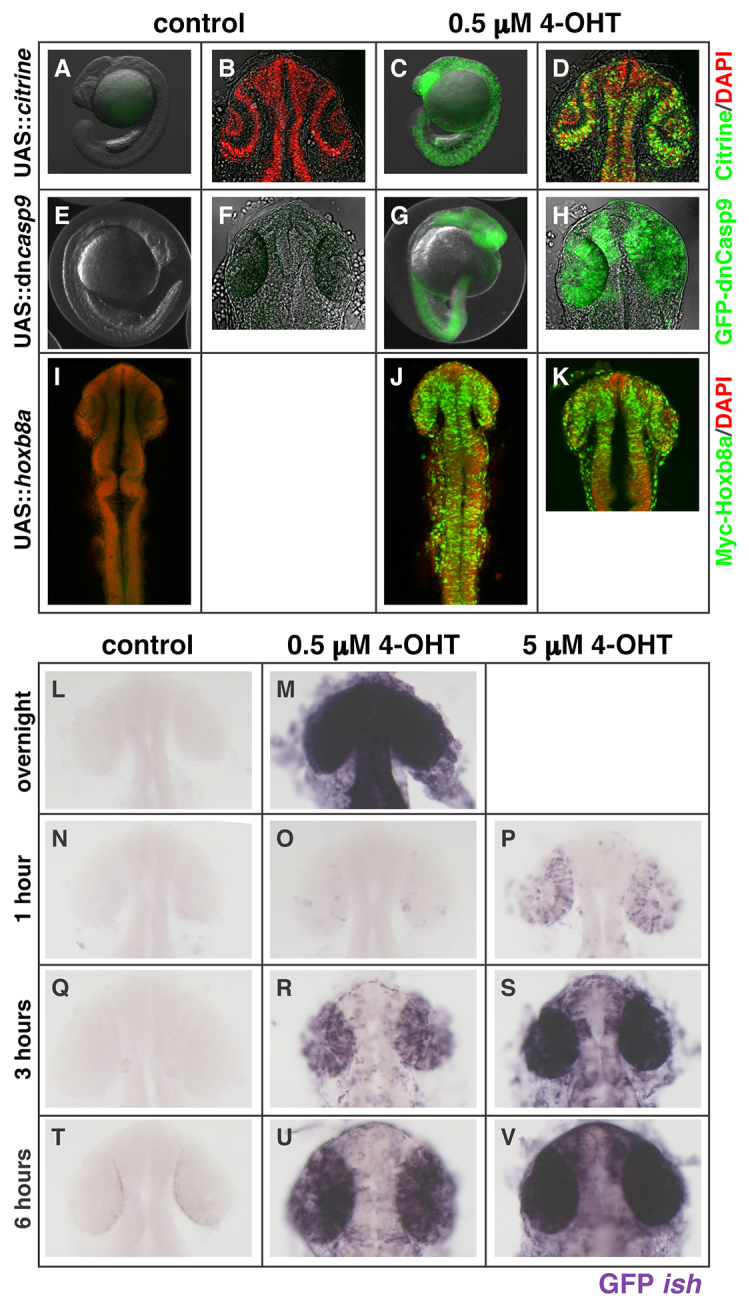
Activity of ERT2-GAL4 in vivo is 4-OHT dependent, rapid and dose sensitive. (A-K) Stable transgenic fish lines expressing ERT2-GAL4 under control of the Xenopus EF1α promoter show robust transactivation of UAS-linked H2B-citrine (C,D), GFP-dnCasp9 (G,H) or Myc-hoxb8a (J,K) when treated overnight with 0.5 μM 4-OHT (all in green channels), compared with controls (A,B,E,F,I, green channel). In some samples, nuclei are labelled with DAPI (B,D,I-K, red channel) and/or brightfield (A-H, grey) to visualize the embryo. (L-V) A time-course of UAS-linked transgene transcription (in situ hybridisation for GFP, purple) driven by ERT2-GAL4 shows 4-OHT dependence (controls in N,Q,T versus 4-OHT treated in O,R,U and P,S,V). The kinetics and intensity of response are dose dependent: embryos treated with 5 μM 4-OHT show more rapid, stronger and less mosaic transcription from the UAS-driven transgene than 0.5 μM 4-OHT-treated embryos (compare P,S,V with O,R,U) over the 6-hour treatment course. Embryos treated overnight with 0.5 μM 4-OHT (control in L, 4-OHT in M) show robust GFP transcription. The light pigmentation around the eye of embryos at 6 hours in T is not in situ signal.
The activation kinetics of any inducible system will determine the temporal resolution of the study. The need for speed is particularly acute when working with early stage zebrafish embryos when rapid developmental changes are taking place. We sought to establish how rapidly after 4-OHT treatment a UAS-linked transgene was activated. In order to visualize directly the transcriptional activity of the ERT2-GAL4, we used in situ hybridisation to detect nascent mRNA synthesis from the UAS-linked transgene. EF1α::ERT2-GAL4 fish were crossed to UAS::GFP-dnCasp9 fish, and the embryos were either grown overnight in fish water, or were treated with ETOH or 0.5 μM 4-OHT from 50% epiboly to 18 hpf, when they were fixed. The untreated embryos were then exposed to either ETOH, 0.5 μM 4-OHT or 5 μM 4-OHT from ∼18 hpf, for either 1, 3 or 6 hours, when they were fixed and processed to detect GFP mRNA.
In agreement with the previously observed GFP fluorescence (Fig. 2G,H), embryos treated overnight with 4-OHT showed strong GFP mRNA expression, whereas no signal was seen in control treated embryos, even after extensive overstaining (Fig. 2M versus 2L). This indicates that, in vivo, the ERT2-GAL4 is inactive in the absence of ligand. Within 1 hour of treatment, GFP mRNA could be detected in a few cells at 0.5 μM 4-OHT, and more strongly and in more numerous cells at 5 μM 4-OHT (Fig. 2N versus 2O,P). There is thus a rapid dose-dependent activation of transcription from the UAS-linked transgene. An important technical note is that these embryos remained in their chorions during treatment. At the 3 and 6 hour time points, double-positive embryos exposed to either concentration of 4-OHT show a progressive increase in transcript accumulation, again in a dose-dependent manner (Fig. 2Q versus 2R,S and 2T versus 2U,V). The 5 μM treatment appeared to plateau after 3 hours, whereas the 0.5 μM-treated embryos had not achieved equivalent levels even at 6 hours (Fig. 2S versus 2U). These data indicate that 4-OHT is highly permeable, fast-acting and capable of generating different levels of transgene induction.
Driving ERT2-Gal4 by tissue-specific enhancers restricts GAL4 activity
In order to generate tissue-restricted GAL4 activity, we cloned the ERT2-GAL4 cassette downstream of tissue-specific enhancers for HuC, an RNA-binding protein expressed in post-mitotic neurons (Higashijima et al., 2003), and ClaudinB (Cldnb), a tight-junction protein expressed in a distinct set of tissues including the skin and migrating lateral line primordium (Haas and Gilmour, 2006). We tested these in a transient transgenesis assay, injected as plasmid DNA along with a UAS::H2B-Citrine plasmid and mRNA for TOL2 transposase (Balciunas et al., 2006). In the absence of 4-OHT, HuC::ERT2-GAL4-injected embryos had a small number of Citrine-positive cells (Fig. 3A, average=28 cells, n=5 embryos). A similar background of Citrine was seen in untreated cldnb::ERT2-GAL4 injected embryos (data not shown). When injected embryos were treated with 0.5 μM 4-OHT, they showed strong enhancer-specific, tissue-restricted expression of Citrine. We confirmed that Citrine expression in the HuC::ERT2-GAL4-injected embryos is neuronally enriched by performing imunohistochemistry for HuC protein (Fig. 3B-E, anti-HuC in red, Citrine in green).
Fig. 3.
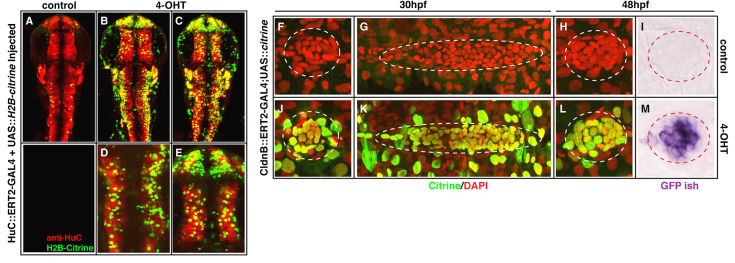
Tissue-specific ERT2-GAL4 produces 4-OHT-dependent, region restricted activity of transgene expression. (A-E) Transient transgenics in which ERT2-GAL4 is under control of the HuC enhancer/promoter show robust neuron-specific expression from co-injected UAS::H2B-citrine when embryos are treated overnight with 0.5 μM 4-OHT, compared with controls (compare B-E versus A, green channel). Staining with anti-HuC/D antibody reveals the endogenous expression pattern of this gene (A-E, red channel). Images in A-C are maximum projections of the entire embryo thickness (36-40 μm). Images in D,E are enlarged maximum projections of a 5.7 μm stack from embryo in C. (F-M) Stable transgenics in which cldnB enhancer/promoter drives ERT2-GAL4 show robust induction and 4-OHT dependence of a stable 5×UAS::H2B-citrine transgene (compare J,K,L versus F,G,H, green channel). When treated with 1 μM 4-OHT from 50% epiboly, embryos express Citrine in the migrating lateral line primordium and deposit neuromasts at 30 hpf (dotted ovals in K and J, respectively, green channel). Persistent expression of the UAS-linked citrine at 48 hpf in neuromasts is detected by in situ hybridisation for citrine transcript (compare M with I, red circled purple signal). Some samples were co-stained with DAPI to reveal nuclei (F-H,J-L, red channel). Additional Citrine-positive cells outside the lateral line structures in J-L reflect the expression of Cldnb in the skin.
Next, we generated a cldnb::ERT2-GAL4 transgenic founder fish (see Materials and methods). When crossed to a stable UAS::H2B-citrine line, the embryos display the expected tissue-restricted Citrine expression in response to 1 μM 4-OHT from 50% epiboly, but not in control conditions. However, Citrine protein expression was highly mosaic, and was only present in a subset of the cells expressing Citrine mRNA (data not shown). We subsequently realised that we had used a transgenic line containing only two UAS repeats, and lacking the E1B promoter described by Distel et al. (Distel et al., 2009). A similar disconnect between UAS-driven transcription and translation has been previously described (Distel et al., 2009). To determine whether this mosaicism was due to a defect in the cldnb::ERT2-GAL4 transgene, or the 2×UAS transgene, we repeated the cross using the H2B-citrine line driven by a 5×UAS and containing the E1B promoter (as used in all other experiments). In response to 1 μM 4-OHT from 50% epiboly, cldnb enhancer activity was evident in the skin, olfactory placode and in the morphologically distinct lateral line primordium, and deposited neuromasts at 28 hpf (Fig. 3J-L versus 3F-H and data not shown) (Kollmar et al., 2001; Haas and Gilmour, 2006). Importantly, the expression of Citrine protein was now near-uniform throughout the target tissues, and correlates strongly with citrine mRNA (Fig. 3J-L; data not shown). The sustained tissue-specific expression of the UAS-linked transgene was also seen after in situ hybridisation with a citrine RNA probe at 48 hpf (Fig. 3I versus 3M). These data indicate that stable lines expressing ERT2-GAL4 fusions conditionally activate UAS transgenes with little or no background, and highlight the importance of continued improvements in transgene architecture (Distel et al., 2009; Goll et al., 2009; Akitake et al., 2011).
Generation of a semi-ubiquitous conditional GAL4 activator strain
As the Xenopus EF1α promoter is not expressed ubiquitously at all time points (Fig. 2), we tested the recently described promoter element of the zebrafish ubiquitin gene, which has been reported to have widespread sustained tissue activity (Mosimann et al., 2011). Transient transgenics from injection of the ubi::ERT2-GAL4 and UAS::H2B-citrine plasmids showed widespread, 4-OHT-inducible GAL4 activity when embryos were treated with 0.75 μM 4-OHT from 50% epiboly to 21 hpf (supplementary material Fig. S1). When founder transgenic ubi::ERT2-GAL4 lines were crossed to a stable UAS::H2B-citrine line, embryos treated under the same conditions showed broad Citrine expression (Fig. 4A,D,G versus 4B,E,H, green channel). Despite a small amount of leaky GAL4 activity in transient transgenics (DNA-injected embryos) in the absence of 4-OHT (supplementary material Fig. S1, control), we find no detectable leakage in stable transgenic lines (Fig. 4A,D,G). In agreement with published data on the zebrafish ubiquitin enhancer, we find that the strength and breadth of Ubi::ERT2-GAL4 activity is insertion dependent: double labelling with the nuclear marker DAPI (Fig. 4, red channel) reveals that, at 21 hpf, expression varies from 97% to 41% of cells expressing Citrine (Fig. 4I versus 4K, 239/246 versus 184/440 cells positive, see Materials and methods) after overnight treatment with 0.75 μM 4-OHT. We are currently raising independent insertions from founders to isolate the line with the broadest ERT2-GAL4 expression.
Fig. 4.
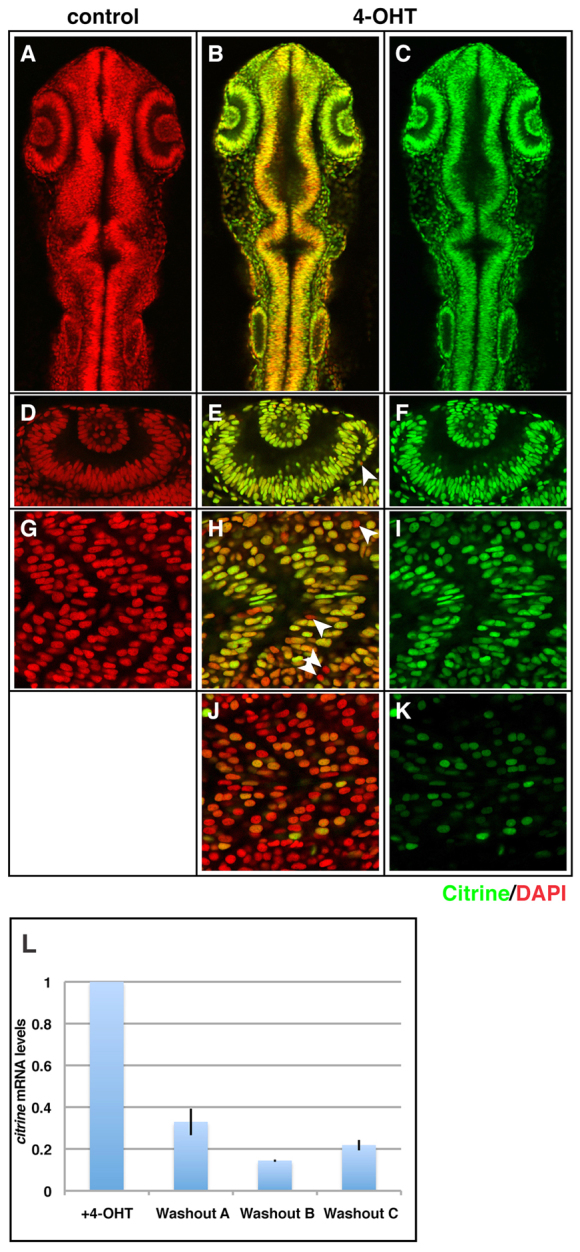
ERT2-GAL4 under the control of the ubiquitin promoter can drive widespread, 4-OHT-dependent activity to 24 hours post-fertilisation. (A-K) Stable ubi::ERT2-GAL4 lines were crossed to a UAS::H2B-citrine line, and embryos were treated overnight with ethanol (A,D,G) or 0.75 μM 4-OHT (B,C,E,F,H-K) from 50% epiboly to 21 hpf. Confocal analysis of single optical planes reveals near-ubiquitous GAL4 activity in a 4-OHT-dependent manner in one line (A,D,G versus B,C,E,F,H,I, Citrine in green channel, DAPI in red channel). Arrowheads indicate Citrine-negative cells. A second independent ubi::ERT2-GAL4 line showed much weaker and more mosaic expression (J,K versus H,I, Citrine in green channel, DAPI in red channel), suggesting insertion site-dependent effects on ubiquitin promoter activity. Representative Citrine-negative cells in E,H are indicated by arrowheads. (L) Upon withdrawal of 4-OHT from Citrine-positive embryos from 18 hpf to 42 hpf (24 hour withdrawal, washouts A, B and C), mRNA levels of citrine drop by 67-85% compared with embryos maintained in 4-OHT (+4-OHT) (n=3 clutches) (compare +4-OHT with washouts A, B and C). qPCR levels are relative to embryos maintained in 4-OHT. Error bars indicate ±s.e.m. (overall signficant difference between +4-OHT and washout conditions, t-test, P<0.002).
ERT2-GAL4 activity can be reversed by drug withdrawal
One of the attractive possibilities of a drug-induced transgene system is the ability to shut it off at will (Knopf et al., 2010). We therefore tested whether ERT2-GAL4 activity was reversible by removal of 4-OHT. The ubi::ERT2-GAL4 line was crossed to a stable UAS::H2B-citrine line, and embryos were treated from 50% epiboly to 18 hpf with 1 μM 4-OHT. Citrine-positive embryos were then selected and divided into two groups. One group was washed in fish water (‘washout’), whereas the other group was moved to fresh 4-OHT (‘+4-OHT’), before both were grown for an additional 24 hours. qPCR analysis of the two conditions showed an average 76% reduction in citrine mRNA (67-85%, Fig. 4L, n=3 clutches). These data indicate that 4-OHT-induced ERT2-GAL4 activity is reversible, thus permitting controlled transgene extinction.
ERT2-GAL4 activity is sufficient to generate gain-of-function phenotypes in vivo
The activity of UAS-linked fluorescent and tagged reporter transgenes in the ERT2-GAL4 system prompted us to examine whether the system gave sufficiently high expression levels to generate gain-of-function phenotypes in vivo. We have previously shown that the drug HA14-1 can rapidly induce apoptosis in the zebrafish embryo, resulting in the activation of caspase 3 (Gerety and Wilkinson, 2011). Conversely, forced expression of a dominant-negative caspase 9 (dnCasp9) protein has been shown to block caspase 3 activation and subsequent apoptosis (Rowe et al., 2005). We therefore attempted to rescue the apoptotic effects of HA14-1 by ERT2-GAL4-induced expression of dnCasp9 in vivo. We crossed carrier ubi::ERT2-GAL4 and UAS::GFP-dnCasp9 lines, treated the resulting embryos with 0.625 μM 4-OHT from 50% epiboly until 18 hpf, then transferred them to fish water containing fresh 4-OHT and 10 μM HA14-1. After 2 hours, the embryos were fixed and processed for detection of cleaved caspase 3. Whereas dnCasp9-negative embryos showed extensive apoptosis in the somites and tail region (100%, n=14), embryos broadly expressing dnCasp9 showed little or no apoptosis (n=12) (Fig. 5K,L versus 5M,N; cleaved caspase 3 in red channel). Importantly, the remaining cleaved caspase 3 present in dnCasp9-expressing embryos was only present in dnCasp9-negative cells (Fig. 5E,J,O). This indicates that the drug-induced ERT2-GAL4 activity is sufficient to drive cell-autonomous rescue of apoptosis by a UAS-linked dominant-negative caspase.
Fig. 5.
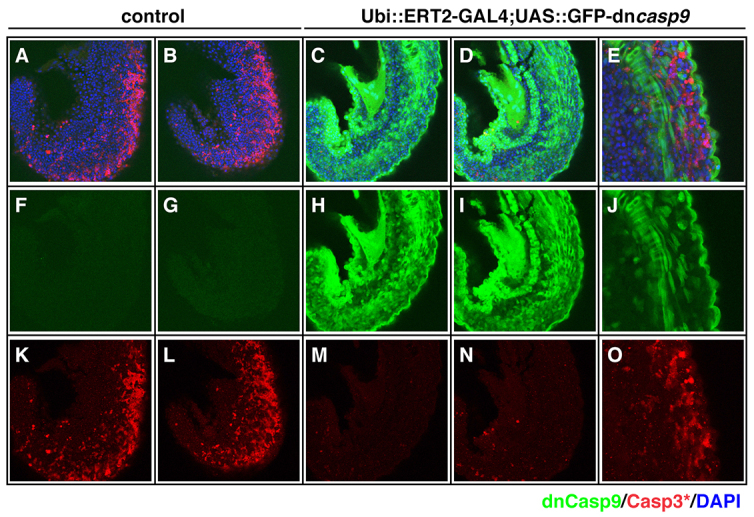
ERT2-GAL4-driven expression is sufficient to generate gain-of-function phenotypes in vivo. (A-O) Embryos from a Ubiquitin::ERT2-GAL4 fish crossed to UAS::GFP-dnCasp9 fish were treated with ethanol (A,B,F,G,K,L) or 0.625 μM 4-OHT (C-E,H-J,M-O) from 50% epiboly to 18 hpf, followed by added treatment with 10 μM HA14-1 for 2 hours. Staining for activated caspase 3 (red channel) and GFP (green channel) reveals a rescue of HA14-1-induced apoptosis by widespread GAL4-induced expression of dominant-negative caspase 9 (compare K,L with M,N, red channel) in embryonic trunk. In embryos with mosaic GAL4 activity (E,J,O, GFP in green channel), the rescue of apoptosis is cell-autonomous to the GFP-dnCasp9-expressing cells (E,J,O, compare non-overlapping green and red). Samples were co-stained with DAPI (A-E, blue channel) to reveal nuclei of all cells. All images are side views of the trunk at 20 hpf.
ERT2-GAL4 fusions function in other vertebrate systems
The use of GAL4 as a transgenic transcriptional driver has expanded dramatically since the popularisation of the VP16 fusion, enabling its use in vertebrates (Köster and Fraser, 2001; Halpern et al., 2008; Distel et al., 2009). Although the use of GAL4 has permeated zebrafish research and has been established in Xenopus, its use in mouse and chick has been sporadic (e.g. Rowitch et al., 1999; Avraham et al., 2009), and drug-regulated GAL4 approaches are even more rare (Pierson et al., 2000). Given the synthetic nature of our conditional system, and that it works in human cells (HEK293), we predicted that it would be functional in other vertebrate systems and decided to test it in the chicken embryo.
The chick is amenable to DNA transfection by electroporation, permitting rapid transgene expression (Itasaki et al., 1999). To test the ERT2-GAL4 system in intact embryos, we electroporated the expression plasmids EF1α::ERT2-GAL4 together with UAS::GFP and a tracer plasmid (CMV::RFP) into the spinal cord of the Hamburger Hamilton stage 11 (HH stage 11) chick embryos (Hamburger and Hamilton, 1992). Whole embryos were subsequently explanted in vitro to allow for controlled dose conditions (Chapman et al., 2001). Although the co-electroporated tracer plasmid shows RFP expression in all conditions (Fig. 6B,E,H, red channel), strong GFP expression occurs only in the presence of 10 μM 4-OHT (Fig. 6F versus 6I, green channel); a small amount of leaky GFP expression was seen in control treated embryos, as well as embryos receiving only UAS::GFP and CMV::RFP (Fig. 6C and 6F, green channel).
Fig. 6.
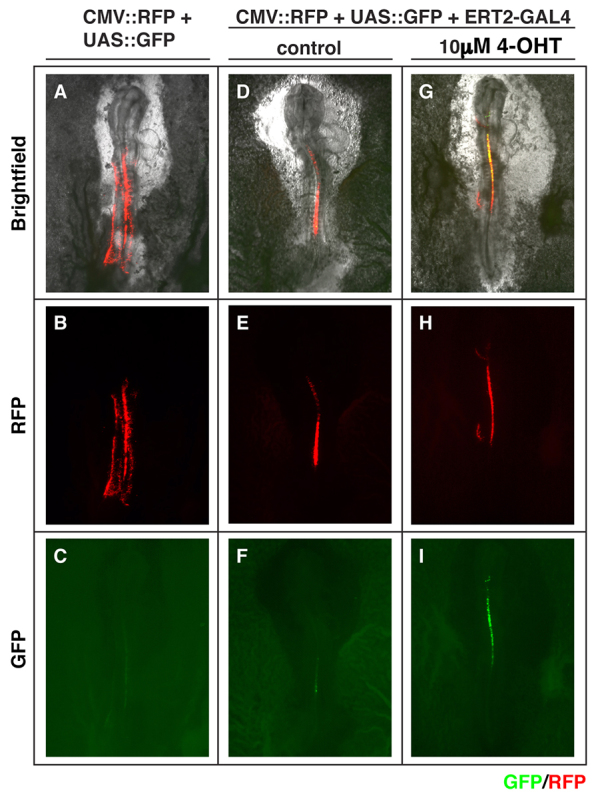
The ERT2-GAL4 system is functional in chicken embryos. (A-I) Plasmids containing 14×UAS::GFP and CMV::RFP were electroporated without (A-C) or with (D-I) EF1α::ERT2-GAL4 into the spinal cord of the HH stage 11 embryos and were cultured in the absence (A-F) or in the presence (G-I) of 10 μM 4-OHT (n=4 embryos per condition). After 8 hours of treatment, strong GFP expression was observed only in the presence of 4-OHT (compare C,F and I). RFP expression in B,E,H confirms effective electroporation in these samples. (A,D,G) The morphology of the explanted whole embryos.
In order to quantitate the 4-OHT-dependent induction, we turned to the chick neural plate explant system, which is widely used in the analysis of neural tube patterning (Dessaud et al., 2007). We used EF1α::ERT2-GAL4 and UAS::H2B-citrine, as well as a tracer plasmid (CMV::β-Galactosidase) to monitor transfection efficiency. These were electroporated, in ovo, into the posterior neural plate of chicken embryos at Hamburger Hamilton stage 9+. Subsequently, explants excised from the neural plate were cultured in the presence of 1 μM 4-OHT for different time intervals. We found Citrine expression in 30% of β-Gal-positive cells within 3 hours of starting 4-OHT treatment, and by 12 hours over 80% of the transfected (β-Gal-positive) cells expressed a saturating level of Citrine (Fig. 7E,K,Q). In the absence of 4-OHT, Citrine expression was observed in fewer than 5% of the cells (Fig. 7B,H,N), suggesting that the induction was specific to the treatment with 4-OHT. Taken together, these data indicate that the ERT2-GAL4 system is amenable to a number of experimental approaches during chick development.
Fig. 7.
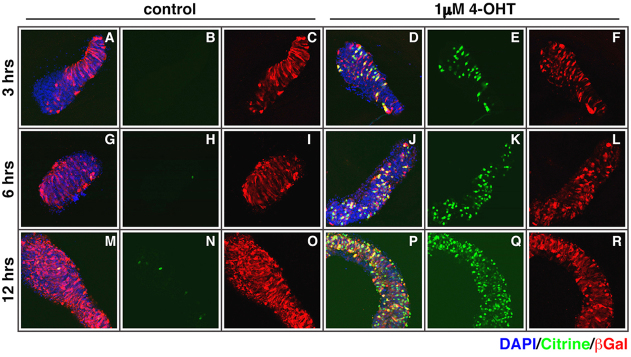
The ERT2-GAL4 system is functional in chick neural plate explants. (A-R) Plasmids containing EF1α::ERT2-GAL4, UAS::H2B-citrine and CMV::βGal were electroporated into the posterior neural plate of chicken embryos at HH stage 9+, and explants were excised and cultured in vitro. We performed a time course of Citrine expression after exposure of the explants to control or 1 μM 4-OHT: after the indicated time interval (3,6,12 hours), the explants were fixed and stained with GFP and βGal antibodies and DAPI (n=4 embryos for each condition and time point). We found a rapid induction of Citrine expression within 3 hours (E versus B) and over 80% of transfected cells expressing Citrine by 12 hours (Q versus N) after the start of treatment with 1 μM 4-OHT. Untreated explants showed fewer than 5% of cells expressing Citrine, indicating the GAL4 activity was 4-OHT dependent. β-Gal expression was used for determining the overall efficiency of the electroporation and DAPI staining (blue channel) was used to reveal all nuclei.
DISCUSSION
The ability to force ectopic and precocious expression of transgenes has made zebrafish a powerful model system for elucidating gene function in development and disease (Esengil and Chen, 2008). By combining two pre-existing technologies, estrogen receptor fusion proteins and the binary GAL4/UAS system, we have created a tool that gives us temporal control of transgene misexpression in zebrafish and chick embryos. We have shown that in vivo the induction is rapid and dose sensitive, and by placing the ERT2-GAL4 fusion under the control of tissue-specific enhancers, we have been able to add spatial control to this system. Drug withdrawal can also be used to extinguish transgene expression. This system complements the existing inducible systems in zebrafish, is compatible with the predominantly UAS-based transgenics available and should provide a means for more refined experimental design. Our results in the chick model system reveal a broader application for both GAL4 and the specific inducible system.
The power of forced expression
Transgenic forced expression provides developmental biologists with a robust method for investigating protein function and developmental processes. The technique of injecting capped mRNA into Xenopus embryos for this purpose was rapidly adopted in the zebrafish field owing to its technical simplicity and speed. This permitted analysis of gene function in vivo, as well as mosaic studies to analyse cellular competition and interactions. Additionally, mosaic transgene expression allows one to distinguish between cell-autonomous and non-autonomous gene effects. However, the inherent limitations of injecting early embryos, namely widespread, uncontrolled and immediate transgene expression subject to dilution and mRNA degradation led to more sophisticated methods for forced expression.
Temporal or spatial control of transgene expression
Heat shock-sensitive promoters enable us to control the onset of transgene expression, bypassing sensitive developmental stages or dissecting out the temporal aspects of particular embryological processes (reviewed by Halloran et al., 2000). However, because these promoters are activated throughout the embryo, they give no control over where transgenes are expressed. Conversely, genomic enhancer elements have been used to drive tissue-specific expression, but the timing of expression is then entirely dependent on the endogenous regulatory mechanisms acting on the element (Köster and Fraser, 2001). Intersectional approaches, where enhancer-driven Cre recombinase removes a stop cassette blocking heat-shock function, or vice versa, can give a degree of dual control (reviewed by Luan and White, 2007), but requires task-specific transgenics (tissue-specific Cre lines, heat-shock Floxed-STOP driven gene of interest, and vice versa) and relies on an excision event that is neither time neutral nor reversible.
The versatility of the binary GAL4/UAS system has revolutionized the way zebrafish biologists perform experiments. A growing catalogue of both GAL4 driver and UAS effector lines is providing new and flexible tools with which to perform experiments (Davison et al., 2007; Distel et al., 2009; Kawakami et al., 2010; Bradford et al., 2011). As has been seen in the Drosophila field, there is a logistical rationale for sticking to one system for the majority of reagents: our approach has sought to take advantage of this by full compatibility with existing UAS effector lines.
Drug-inducible systems
In its most common form, the timing of transgene expression is fully dependent on the enhancer elements used and/or the genomic locus into which a GAL4 enhancer trap has landed. These techniques were followed by the development of a number of ligand-dependent transgene expression systems in zebrafish. The ecdysone receptor-GAL4 fusion (GAL4-EcR) allows GAL4 activity to be temporally controlled by exposure to analogues of the insect hormone (Esengil et al., 2007; Lou et al., 2011). However, we were never able to achieve useful levels of GAL4-EcR protein expression, even when expressed from an identical plasmid context to that of our ERT2-GAL4 (data not shown), suggesting possible protein stability issues.
The mifepristone-inducible LexPR system (Emelyanov and Parinov, 2008) provides a novel and effective way to control activator function, relying on specific LexA binding-site driven transgenes. Finally, the TET-Dual inducible system, which requires tetracycline and a second inducing drug to drive transgenes downstream of the TET-response element (TET-RE) DNA sequence shows promise as a third independent approach (Knopf et al., 2010). However, the LexA and TET-RE strategies lack compatibility with the predominant transgenic methodology, limiting their utility in the context of the hundreds of tissue-specific GAL4 enhancer trap lines and UAS-linked transgenes that have been generated (Asakawa et al., 2008; Halpern et al., 2008; Distel et al., 2009; Ogura et al., 2009; Kawakami et al., 2010; Bradford et al., 2011). Nonetheless, when used in combination, the LexA, TET-Dual inducible and GAL4 systems will expand the sophistication of zebrafish experiments, enabling independent control of multiple transgenes.
During our study, we found a varying degree of mosaicism in GAL4-induced transgene expression. This was probably caused by an older UAS cassette architecture that results in poor transcriptional and translational initiation: using an updated 5×UAS cassette dramatically reduced the incidence of mosaicism. Active work on methylation-induced transgene silencing, and other epigenetic mechanisms in zebrafish (Goll et al., 2009; Akitake et al., 2011) are shedding new light on gene regulation in vivo. These studies and ours highlight the fact that there is still much to learn from the biology for improving transgenic approaches to misexpression.
In their seminal paper, Brand and Perrimon brought a revolution of combinatorial genetic manipulation to the Drosophila community (Brand and Perrimon, 1993). The zebrafish community has embraced this invention, adapting it to suit both the biological peculiarities of vertebrates and their specific research needs. This creative development continues apace, addressing issues of transgene variability, epigenetic silencing, intersectional techniques, permanent cell marking for lineage tracing, mosaic analysis, and more (Goll et al., 2009; Collins et al., 2010). The use of ERT2-GAL4 fusions will contribute to this expanding toolkit of transgenic approaches for application in zebrafish and other vertebrate model organisms.
Supplementary Material
Acknowledgments
We thank Javier Terriente for helpful discussions, and NIMR Biological Services for zebrafish husbandry.
Footnotes
Funding
This work was supported by the Medical Research Council [U117532048]. S.S.G. was supported by a fellowship from the Human Frontier Science Foundation. M.A.B. was supported by Fellowships from the Fondation Fyssen (France) and the European Molecular Biology Organization (EMBO). N.S. is supported by a Marie Curie Fellowship [219939] and the Uehara memorial foundation. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.091520/-/DC1
References
- Akitake C. M., Macurak M., Halpern M. E., Goll M. G. (2011). Transgenerational analysis of transcriptional silencing in zebrafish. Dev. Biol. 352, 191–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K., Suster M. L., Mizusawa K., Nagayoshi S., Kotani T., Urasaki A., Kishimoto Y., Hibi M., Kawakami K. (2008). Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc. Natl. Acad. Sci. USA 105, 1255–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avraham O., Hadas Y., Vald L., Zisman S., Schejter A., Visel A., Klar A. (2009). Transcriptional control of axonal guidance and sorting in dorsal interneurons by the Lim-HD proteins Lhx9 and Lhx1. Neural Dev. 4, 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balciunas D., Wangensteen K. J., Wilber A., Bell J., Geurts A., Sivasubbu S., Wang X., Hackett P. B., Largaespada D. A., McIvor R. S., et al. (2006). Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2, e169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford Y., Conlin T., Dunn N., Fashena D., Frazer K., Howe D. G., Knight J., Mani P., Martin R., Moxon S. A., et al. (2011). ZFIN: enhancements and updates to the Zebrafish Model Organism Database. Nucleic Acids Res. 39, D822–D829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 [DOI] [PubMed] [Google Scholar]
- Branda C. S., Dymecki S. M. (2004). Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev. Cell 6, 7–28 [DOI] [PubMed] [Google Scholar]
- Chae J., Zimmerman L. B., Grainger R. M. (2002). Inducible control of tissue-specific transgene expression in Xenopus tropicalis transgenic lines. Mech. Dev. 117, 235–241 [DOI] [PubMed] [Google Scholar]
- Chapman S. C., Collignon J., Schoenwolf G. C., Lumsden A. (2001). Improved method for chick whole-embryo culture using a filter paper carrier. Dev. Dyn. 220, 284–289 [DOI] [PubMed] [Google Scholar]
- Collins R. T., Linker C., Lewis J. (2010). MAZe: a tool for mosaic analysis of gene function in zebrafish. Nat. Methods 7, 219–223 [DOI] [PubMed] [Google Scholar]
- Davison J. M., Akitake C. M., Goll M. G., Rhee J. M., Gosse N., Baier H., Halpern M. E., Leach S. D., Parsons M. J. (2007). Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol. 304, 811–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessaud E., Yang L. L., Hill K., Cox B., Ulloa F., Ribeiro A., Mynett A., Novitch B. G., Briscoe J. (2007). Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature 450, 717–720 [DOI] [PubMed] [Google Scholar]
- Distel M., Wullimann M. F., Köster R. W. (2009). Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl. Acad. Sci. USA 106, 13365–13370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky R. I., Sheldahl L. C., Moon R. T. (2002). A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev. Biol. 241, 229–237 [DOI] [PubMed] [Google Scholar]
- Emelyanov A., Parinov S. (2008). Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev. Biol. 320, 113–121 [DOI] [PubMed] [Google Scholar]
- Esengil H., Chen J. K. (2008). Gene regulation technologies in zebrafish. Mol. Biosyst. 4, 300–308 [DOI] [PubMed] [Google Scholar]
- Esengil H., Chang V., Mich J. K., Chen J. K. (2007). Small-molecule regulation of zebrafish gene expression. Nat. Chem. Biol. 3, 154–155 [DOI] [PubMed] [Google Scholar]
- Feil R., Wagner J., Metzger D., Chambon P. (1997). Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 237, 752–757 [DOI] [PubMed] [Google Scholar]
- Geling A., Itoh M., Tallafuss A., Chapouton P., Tannhäuser B., Kuwada J. Y., Chitnis A. B., Bally-Cuif L. (2003). bHLH transcription factor Her5 links patterning to regional inhibition of neurogenesis at the midbrain-hindbrain boundary. Development 130, 1591–1604 [DOI] [PubMed] [Google Scholar]
- Gerety S. S., Wilkinson D. G. (2011). Morpholino artifacts provide pitfalls and reveal a novel role for pro-apoptotic genes in hindbrain boundary development. Dev. Biol. 350, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll M. G., Anderson R., Stainier D. Y., Spradling A. C., Halpern M. E. (2009). Transcriptional silencing and reactivation in transgenic zebrafish. Genetics 182, 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas P., Gilmour D. (2006). Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line. Dev. Cell 10, 673–680 [DOI] [PubMed] [Google Scholar]
- Halloran M. C., Sato-Maeda M., Warren J. T., Su F., Lele Z., Krone P. H., Kuwada J. Y., Shoji W. (2000). Laser-induced gene expression in specific cells of transgenic zebrafish. Development 127, 1953–1960 [DOI] [PubMed] [Google Scholar]
- Halpern M. E., Rhee J., Goll M. G., Akitake C. M., Parsons M., Leach S. D. (2008). Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 5, 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V., Hamilton H. L. (1992). A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 195, 231–272 [DOI] [PubMed] [Google Scholar]
- Hans S., Kaslin J., Freudenreich D., Brand M. (2009). Temporally-controlled site-specific recombination in zebrafish. PLoS ONE 4, e4640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S., Freudenreich D., Geffarth M., Kaslin J., Machate A., Brand M. (2011). Generation of a non-leaky heat shock-inducible Cre line for conditional Cre/lox strategies in zebrafish. Dev. Dyn. 240, 108–115 [DOI] [PubMed] [Google Scholar]
- Higashijima S., Masino M. A., Mandel G., Fetcho J. R. (2003). Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J. Neurophysiol. 90, 3986–3997 [DOI] [PubMed] [Google Scholar]
- Itasaki N., Bel-Vialar S., Krumlauf R. (1999). ‘Shocking’ developments in chick embryology: electroporation and in ovo gene expression. Nat. Cell Biol. 1, E203–E207 [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Krieg P. A. (1994). pXeX, a vector for efficient expression of cloned sequences in Xenopus embryos. Gene 147, 223–226 [DOI] [PubMed] [Google Scholar]
- Kawakami K. (2005). Transposon tools and methods in zebrafish. Dev. Dyn. 234, 244–254 [DOI] [PubMed] [Google Scholar]
- Kawakami K., Abe G., Asada T., Asakawa K., Fukuda R., Ito A., Lal P., Mouri N., Muto A., Suster M. L., et al. (2010). zTrap: zebrafish gene trap and enhancer trap database. BMC Dev. Biol. 10, 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf F., Schnabel K., Haase C., Pfeifer K., Anastassiadis K., Weidinger G. (2010). Dually inducible TetON systems for tissue-specific conditional gene expression in zebrafish. Proc. Natl. Acad. Sci. USA 107, 19933–19938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar R., Nakamura S. K., Kappler J. A., Hudspeth A. J. (2001). Expression and phylogeny of claudins in vertebrate primordia. Proc. Natl. Acad. Sci. USA 98, 10196–10201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster R. W., Fraser S. E. (2001). Tracing transgene expression in living zebrafish embryos. Dev. Biol. 233, 329–346 [DOI] [PubMed] [Google Scholar]
- Lou X., Deshwar A. R., Crump J. G., Scott I. C. (2011). Smarcd3b and Gata5 promote a cardiac progenitor fate in the zebrafish embryo. Development 138, 3113–3123 [DOI] [PubMed] [Google Scholar]
- Luan H., White B. H. (2007). Combinatorial methods for refined neuronal gene targeting. Curr. Opin. Neurobiol. 17, 572–580 [DOI] [PubMed] [Google Scholar]
- Mosimann C., Zon L. I. (2011). Advanced zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments. Methods Cell Biol. 104, 173–194 [DOI] [PubMed] [Google Scholar]
- Mosimann C., Kaufman C. K., Li P., Pugach E. K., Tamplin O. J., Zon L. I. (2011). Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development 138, 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson L., Singh G. K., Osterwalder T., Roman G. W., Davis R. L., Keshishian H. (2008). Spatial and temporal control of gene expression in Drosophila using the inducible GeneSwitch GAL4 system. I. Screen for larval nervous system drivers. Genetics 178, 215–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura E., Okuda Y., Kondoh H., Kamachi Y. (2009). Adaptation of GAL4 activators for GAL4 enhancer trapping in zebrafish. Dev. Dyn. 238, 641–655 [DOI] [PubMed] [Google Scholar]
- Osterwalder T., Yoon K. S., White B. H., Keshishian H. (2001). A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl. Acad. Sci. USA 98, 12596–12601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T. M., Wang Y., DeMayo F. J., Matzuk M. M., Tsai S. Y., Omalley B. W. (2000). Regulable expression of inhibin A in wild-type and inhibin alpha null mice. Mol. Endocrinol. 14, 1075–1085 [DOI] [PubMed] [Google Scholar]
- Prodromou N. V., Thompson C. L., Osborn D. P., Cogger K. F., Ashworth R., Knight M. M., Beales P. L., Chapple J. P. (2012). Heat shock induces rapid resorption of primary cilia. J. Cell Sci. 125, 4297–4305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe I., Le Blay K., Du Pasquier D., Palmier K., Levi G., Demeneix B., Coen L. (2005). Apoptosis of tail muscle during amphibian metamorphosis involves a caspase 9-dependent mechanism. Dev. Dyn. 233, 76–87 [DOI] [PubMed] [Google Scholar]
- Rowitch D. H., St.-Jacques B., Lee S. M., Flax J. D., Snyder E. Y., McMahon A. P. (1999). Sonic hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J. Neurosci. 19, 8954–8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagasti A., Guido M. R., Raible D. W., Schier A. F. (2005). Repulsive interactions shape the morphologies and functional arrangement of zebrafish peripheral sensory arbors. Curr. Biol. 15, 804–814 [DOI] [PubMed] [Google Scholar]
- Thummel R., Burket C. T., Brewer J. L., Sarras M. P., Jr, Li L., Perry M., McDermott J. P., Sauer B., Hyde D. R., Godwin A. R. (2005). Cre-mediated site-specific recombination in zebrafish embryos. Dev. Dyn. 233, 1366–1377 [DOI] [PubMed] [Google Scholar]
- Westerfield M. (1993). The Zebrafish Book. Eugene. OR: University of Oregon Press; [Google Scholar]
- Xu Q., Holder N., Patient R., Wilson S. W. (1994). Spatially regulated expression of three receptor tyrosine kinase genes during gastrulation in the zebrafish. Development 120, 287–299 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


