Abstract
MicroRNAs regulate key biological processes and their aberrant expression may lead to cancer. The primary transcript of canonical miRNAs is sequentially cleaved by the RNase III enzymes, Drosha and Dicer, which generate 5′mono-phosphate ends that are important for subsequent miRNA functions. In particular, the recognition of the 5′mono-phosphate of pre-miRNAs by Dicer is important for precise and effective biogenesis of miRNAs. Here we identify a RNA-methyltransferase, BCDIN3D, that O-methylates this 5′mono-phosphate and negatively regulates miRNA maturation. Specifically, we show that BCDIN3D phospho-dimethylates pre-miR-145 both in vitro and in vivo and that phospho-dimethylated pre-miR-145 displays reduced processing by Dicer in vitro. Consistently, BCDIN3D depletion leads to lower pre-miR-145 and concomitantly increased mature miR-145 levels in breast cancer cells, which suppresses their tumorigenic phenotypes. Together, our results uncover a miRNA methylation pathway potentially involved in cancer that antagonizes the Dicer–dependent processing of miR-145 as well as other miRNAs.
Introduction
MicroRNAs (miRNAs) are short single stranded RNA molecules (18–24 nucleotides) that post-transcriptionally interfere with gene expression in a variety of eukaryotes (Ghildiyal and Zamore, 2009). miRNAs target the RNA interference effector complex (RISC) to specific messenger RNAs (mRNAs) through partial base-paring to sequences predominantly found in the 3′ untranslated region (3′UTR). This interaction results in decreased translation of the proteins they encode and/or in the degradation of the mRNAs themselves (Fabian et al., 2010). To date, over 1000 human miRNAs have been identified and each of them potentially regulates many mRNAs (Friedman et al., 2009; Kozomara and Griffiths-Jones, 2011). Consequently, miRNAs have been involved in numerous cellular processes including development, differentiation, proliferation, apoptosis, the stress response and viral defense (Fabian et al., 2010). Importantly, altered expression of miRNAs is a common trait of cancers (Farazi et al., 2011). Indeed, deciphering regulation of miRNA expression may be important not only for diagnostic, but also for therapeutic purposes (Kasinski and Slack, 2011). Due to the way that miRNAs are generated, their expression can be regulated at different levels. The first level is transcriptional as miRNAs are synthesized from larger transcripts by RNA polymerase II or RNA polymerase III complexes. The next level is post-transcriptional, as these primary miRNA precursors (pri-miRNA) undergo at least three steps before the mature single stranded form. The pri-miRNA is first cleaved by Drosha to release a hairpin-loop shaped RNA called pre-miRNA (Lee et al., 2003). The loop of this pre-miRNA is further cleaved by Dicer to generate a miRNA duplex (Chendrimada et al., 2005). The miRNA duplex is dissociated, the passenger strand is discarded, while the guide strand is loaded onto the Argonaute (Ago) protein to form an active RISC complex (Kawamata and Tomari, 2010). Each of these steps is potentially subjected to regulation, as the rate of transcription of a given pri-miRNA does not always correlate with the levels of its mature miRNA (Thomson et al., 2006). One crucial aspect of this process is related to the 5′ terminal end of these RNA molecules. Both RNase III enzymes, Drosha and Dicer, generate 5′ ends that contain a negatively charged mono-phosphate group. This 5′ mono-phosphate is bound by specific positively charged pockets in Dicer and Ago2 and these interactions are necessary for efficient and accurate processing as well as the stability of the mature RISC complex (Frank et al.; Kawamata et al., 2011; Park et al., 2011). Here we unveil an unexpected post-transcriptional modification of the 5′ mono-phosphate group. Specifically, we show that the previously uncharacterized human enzyme BCDIN3D O-methylates both in vitro and in vivo the 5′ terminal mono-phosphate group of the precursor of miR-145, resulting in a complete loss of its negative charge. Consistent with the importance of this charge for interaction with Dicer, methylated pre-miR-145 displays reduced processing by Dicer in vitro. Accordingly, upon BCDIN3D depletion, the association of Dicer with the product of pre-miR-145 processing increases in vivo. As a result, depletion of BCDIN3D in human cells leads to lower levels of pre-miR-145 and a concomitant increase of mature miR-145. Our results also indicate that this is not limited to miR-145, but may be responsible for the regulation of many other miRNAs. Finally, the interest of our findings is further enhanced by the potential involvement of BCDIN3D in cancer, as BCDIN3D depletion in breast cancer cells abolishes their tumorigenic phenotypes. Altogether our results uncover a human enzyme defining a miRNA methylation pathway that antagonizes the Dicer–dependent processing of miRNAs and that has potential as a therapeutic target in cancer.
Results
BCDIN3D is a methyltransferase that targets the 5′ mono-phosphate of nucleic acids
Methylation of DNA and specific residues within histones plays a crucial role in the epigenetic regulation of chromatin-based processes, such as transcription and genomic organization (Goldberg et al., 2007; Klose and Bird, 2006; Kouzarides, 2007; Xhemalce et al., 2011). With the aim of identifying novel epigenetic regulators, we set up a screen for previously uncharacterized methyltransferases that target chromatin. Among our candidates were BCDIN3 and BCDIN3D, the human members of the Bin3 family of putative methyltransferases. Members of this family are found from S. pombe to humans and share homology within their putative S-adenosyl Methionine (SAM) binding motif (Figure 1A). The two paralogs exist from D. melanogaster to humans, while lower eukaryotes possess only one Bin3 family member (Figure 1B).
Figure 1. BCDIN3D is a methyltransferase that targets the 5′ mono-phosphate of nucleic acids.
(A) Alignment of the S-Adenosyl Methionine (SAM) Binding domain of the Bin3 family members from S. pombe, A. thaliana, C. elegans, D. melanogaster, M. musculus and H. sapiens, generated with the CLUSTAL W algorithm. The asterisks indicate the residues that, when mutated to Alanine, abolish the methyltransferase activity of human BCDIN3D (Figure 1F). The arrow indicates human BCDIN3D. (B) Phylogenetic tree based on the alignment shown in (A), generated with the Geneious software. The dashed ellipsoid underlines the BCDIN3D cluster and the arrow indicates human BCDIN3D. (C) In vitro methyltransferase assay with recombinant human BCDIN3 and BCDIN3D, 3H-radioactive SAM as methyl group donor and either histones or nucleosomes as substrate. The reactions were loaded on a 15% SDS-PAGE gel, fixed and subjected to autoradiography. The radioactive band observed in the BCDIN3D/nucleosome lane co-migrates with histone H3. (D) In vitro methyltransferase assay with recombinant BCDIN3D, 3H-SAM and nucleosomes, in which histone H3 is either full length (H3), or is truncated of the 31 first amino-acids (H3ΔNter). The truncation makes H3ΔNter migrate faster (Coomassie staining, right panel), but does not modify the migration of the radioactive band (autoradiography, left panel). (E) 1%g of histones and nucleosomes were loaded on a 15% SDS-PAGE gel and stained with Ethidium Bromide. The band observed in the nucleosome lane corresponds to the 601-DNA used to assemble the nucleosomes. (F) In vitro methyltransferase assay using recombinant human BCDIN3D-WT and BCDIN3D-D72G74A catalytic mutant, 3H-radioactive SAM as methyl group donor and purified 601-DNA as substrate. The 601-DNA is generated by cleavage from a plasmid harbouring multiple tandem repeats of the sequence flanked by EcoRV restriction sites. Prior to the methyltransferase assay with BCDIN3D, the 601-DNA was either CpG methylated using the Ssp1 bacterial methyltransferase and/or treated with Alkaline Phosphatase (AP) that removes the 5′-alpha-phosphate left from the EcoRV digestion. The DNA from the methyltransferase assays with BCDIN3D was purified, loaded on a native 6% PAGE gel, stained with Ethidium Bromide (lower panel), fixed, and subjected to autoradiography (upper panel). (G) Schematic representing the inferred methyltransferase activity of BCDIN3D on a 5′ mono-phosphorylated ribonucleotide. The dashed ellipsoid underlines the 5′ alpha phosphate group.
See also Figure S1.
We assessed the activity of purified candidate proteins using in vitro methyltransferase assays with purified histones and nucleosomes as substrates and 3H-radioactive SAM as the methyl group donor. In our assays, BCDIN3D but not BCDIN3, showed a specific activity in the form of a radioactive band migrating at the level of histone H3 (around 17 kDa) in a SDS-PAGE gel only in the presence of nucleosomes (Figure 1C). In order to test if BCDIN3D was targeting the tail of histone H3, we repeated the methyltransferase assay with nucleosomes where histone H3 carried a truncation of its 31 N-terminal amino acids (H3ΔNter). As shown in Figure 1D, the radioactive band did not disappear nor changed its migration in the H3ΔNter nucleosomes, suggesting that the methylated product was not histone H3. As the only material difference between histones and nucleosomes was the DNA-601 used to assemble the nucleosomes (Huynh et al., 2005), it was possible that the methylation product was the DNA-601 itself. Staining of the SDS-PAGE gel with Ethidium Bromide showed that the 601-DNA does migrate in an SDS PAGE gel forming a band around 17 KDa (Figure 1E). Moreover, BCDIN3D was able to methylate the 601-DNA in the absence of any histone (Figure 1F). However, BCDIN3D did not target the canonical Cytosines within CpG sites, because the methylation product was still observed when the 601-DNA was pre-methylated to saturation with the bacterial SspI enzyme that methylates these sites (Figure 1F). Rather, BCDIN3D targeted the 5′ mono-phosphate generated by the EcoRV digestion used during the 601-DNA purification procedure, as the methylation product was no longer observed when the 601-DNA was pretreated with alkaline phosphatase (Figure 1F). Importantly, the methyltransferase activity was intrinsic to BCDIN3D as point mutations in its SAM binding domain abolished its observed activity in our assay (Figure 1A, F). While we were performing these experiments, the homolog of BCDIN3D, BCDIN3, was reported to stabilize the nuclear non-coding RNA 7SK by methylating its 5′ tri-phosphate (Jeronimo et al., 2007; Shuman, 2007). In addition, we established that BCDIN3D is localized in the cytoplasm (Figure S1A-C). Finally, the activity that we observed on the 601-DNA was weak, i.e. the methylation reactions were at the limit of detection by liquid scintillation and we needed to expose the radioactive gels on film for 2-4 weeks in order to detect a significant signal. Based on these observations, we hypothesized that the bona fide target(s) of BCDIN3D may be RNA(s) that are mono-phosphorylated at their 5′ end(s) as shown in Figure 1G.
BCDIN3D affects the levels of precursor and mature forms of miR-145
We considered that micro RNAs (miRNA) were good candidates for being BCDIN3D targets. Indeed, while most primary precursors of miRNAs (pri-miRNA) are transcribed by RNA polymerase II and are 7-methyl-guanosine-capped, both the precursor miRNAs (pre-miRNA) generated by Drosha and the mature miRNAs generated by Dicer have 5′ mono-phosphate ends (Sarnow et al., 2006).
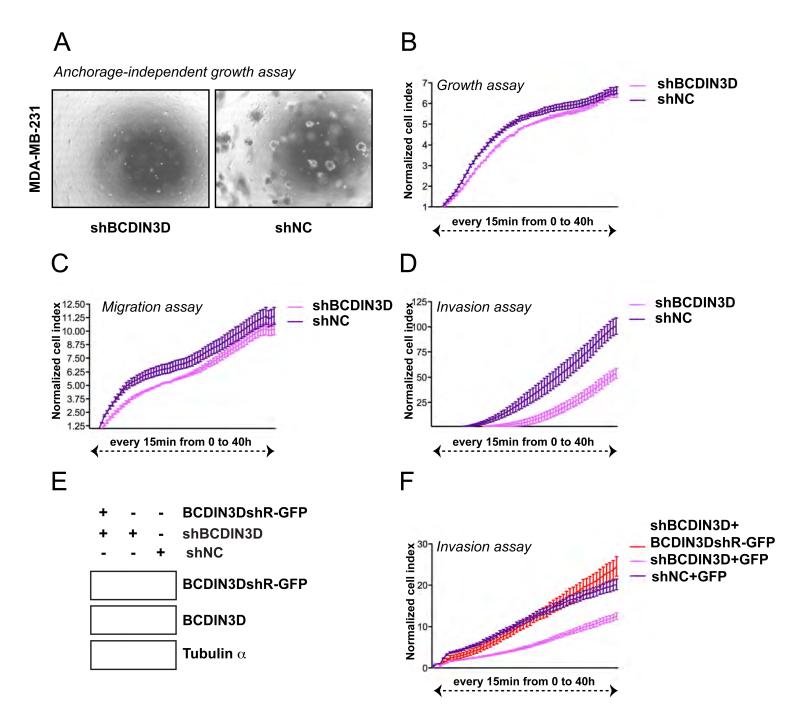
We first sought to determine a cellular system in which to identify the targets of BCDIN3D methylation. Because of global mRNA expression data linking BCDIN3D to breast cancer (Liu et al., 2007), we analyzed the role of BCDIN3D in a number of cellular assays relevant to this cancer. Cells from the triple negative breast cancer cell line MDA-MB-231 have the ability to grow in anchorage independent conditions, a hallmark of cell transformation, and to penetrate through a basement membrane matrix, a key property of cellular invasion. Stable shRNA mediated depletion of BCDIN3D in these cells reduced their ability to form colonies in soft agar medium (anchorage independent growth assay, Figure 2A). Moreover, depletion of BCDIN3D using this shRNA and another siRNA targeting an independent sequence of BCDIN3D (Figure S3A) significantly decreased the invasiveness of the MDA-MB-231 cells (invasion assay, Figure 2D and Figure S2C-D) without greatly affecting their growth and migration abilities (growth assay, Figure 2B; MTT assay, Figure S2A and migration assay, Figure 2C and Figure S2B). Importantly, re-introduction of an shResistant BCDIN3D-GFP protein into the cells expressing shBCDIN3D at levels similar to the endogenous BCDIN3D protein (Figure 2E), fully rescued the invasion defect of these cells (Figure 2F), clearly demonstrating that the observed effects are specific.
Figure 2. BCDIN3D depletion suppresses tumorigenic phenotypes in vitro.
(A) Stable depletion of BCDIN3D in the MDA-MB-231 breast cancer cells abolishes anchorage independent growth. Microscopic image of MDA-MB-231 [shBCDIN3D] and MDA-MB-231 [shNC] cells growing in soft agar after 4 weeks incubation (see Extended Experimental Procedures for further detail). (B-D) Stable BCDIN3D depletion substantially impairs invasiveness, but not growth and migration, of MDA-MB-231 cells. MDA-MB-231 [shBCDIN3D] and MDA-MB-231 [shNC] cells were analyzed with the xCELLigence system, which allows to assess cellular growth (B, Growth assay), migration through a 8 %M pore size membrane (C, Migration assay) or invasion through a 8 %M pore size membrane coated with Matrigel (D, Invasion assay) in parallel and in Real-Time. The cells were trypsinized and loaded in quadruplets onto an E-plate (B, Growth assay), CIM-plate (C, Migration assay) or CIM-Plate coated with 1/40 dilution of Matrigel (D, Invasion assay). The cell index was measured every 15 min for 48 h. The results of the assay are represented as normalized cell index. Error bars represent Standard Error of the Mean (SEM) values. (E) MDA-MB-231 [shBCDIN3D+BCDIN3DshR(esistant)-GFP] express BCDIN3DshR-GFP at levels similar to endogenous BCDIN3D protein. MDA-MB-231 [shBCDIN3D+BCDIN3DshR-GFP], MDA-MB-231 [shBCDIN3D+GFP] and MDA-MB-231 [shNC+GFP] cell lines were generated as indicated in the Extended Experimental Procedures. Whole cell extracts were analyzed by Western Blot with anti-BCDIN3D antibody and the loading control alpha-tubulin. The two panels showing the endogenous and BCDIN3DshR-GFP are from the same exposure of the same blot. (F) Expression of BCDIN3DshR-GFP in MDA-MB-231 [shBCDIN3D] cell lines suppresses their invasion defect. The MDA-MB-231 [shBCDIN3D+BCDIN3DshR-GFP], MDA-MB-231 [shBCDIN3D+GFP] and MDA-MB-231 [shNC+GFP] cell lines were analyzed as in (B-D) and the invasion assays are shown. Error bars represent SEM values.
See also Figure S2.
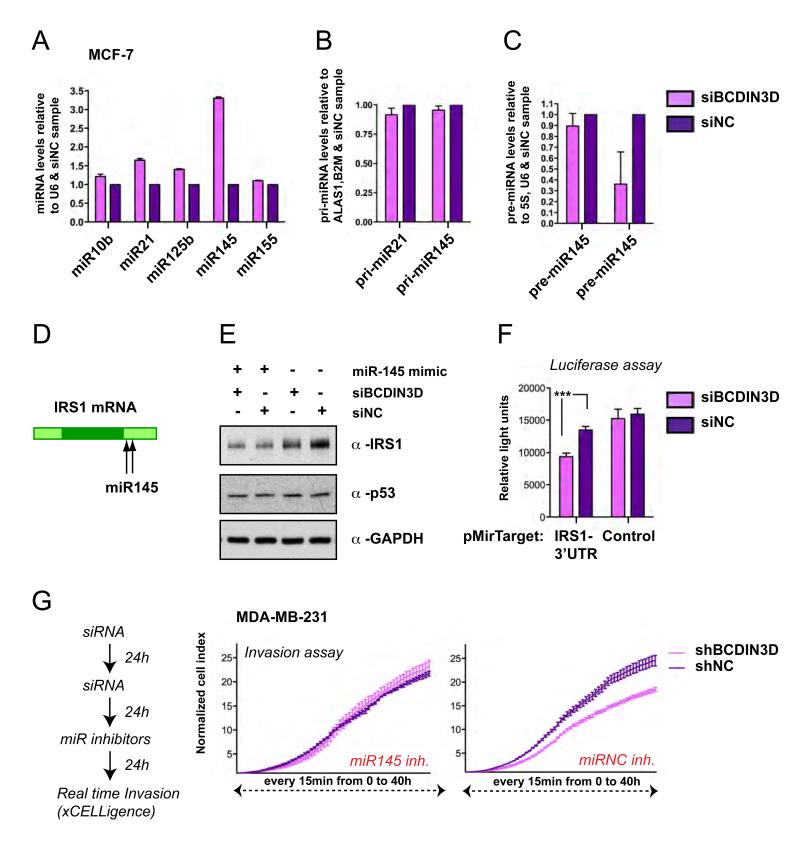
Having confirmed the relevance of BCDIN3D in breast cancer cells, we depleted BCDIN3D in the MCF-7 breast cancer cell line and monitored its effect on the levels of five mature miRNAs (miR-10b, miR-21, miR-125b, miR-145 and miR-155) that are known to be consistently deregulated in breast cancer (Iorio et al., 2005). As shown in Figure 3A, MCF-7 cells transfected with an siRNA targeting the first exon of BCDIN3D (siBCDIN3D, depletion efficiency shown in Figure S1B and Figure S3B) displayed significantly increased levels of mature miR-145 compared to MCF-7 cells transfected with non-targeting negative control siRNAs (siNC). A similar increase in mature miR-145 levels was observed when MCF-7 cells were transfected with an independent pool of siRNAs targeting the second exon of BCDIN3D (Figures S3A-S3C). Moreover, a milder but reproducible increase of mature miR-145 levels was observed in MDA-MB-231 cells transfected with siBCDIN3D compared to siNC (Figure S3D) or the MDA-MB-231 cells stably expressing shBCDIN3D compared to shNC or shBCDIN3D rescued with the BCDIN3DshR-GFP construct (Figure S3E). Importantly, the results obtained by quantitative reverse transcription and PCR (qRT-PCR), were confirmed by Northern Blot with a probe against miR-145 in the non-transformed but immortalized BJ+TERT cells that express higher levels of miR-145 than MCF-7 cells (Figure S3F-S3G). Interestingly, the levels of pri-miR-145 were not increased upon BCDIN3D depletion (Figure 3B), while the levels of pre-miR-145 were significantly decreased (Figure 3C). This suggested that the up-regulation of miR-145 was not due to a transcriptional effect but rather to a downstream effect on either the processing of miR-145 and/or its stability.
Figure 3. siRNA mediated depletion of BCDIN3D changes the levels of precursor and mature forms of miR-145.
MCF-7 cells were transfected with a siRNA targeted against the first exon of BCDIN3D (siBCDIN3D) or with non targeting siRNAs as a negative control (siNC). (A) The levels of the indicated miRNAs were analyzed by quantitative Reverse Transcription and PCR (qRT-PCR) analysis. The data are normalized to the U6 RNA and to siNC cells. Error bars represent SEM values. (B) Transcript levels of the pri-miR-145 were not significantly altered upon BCDIN3D depletion. The pri-miRNA levels were analyzed by qRT-PCR and normalized to the ALAS1 and B2M mRNAs and to siNC cells. Error bars represent SEM values. (C) The levels of pre-miR-145 are reduced upon BCDIN3D depletion. The pre-miRNA levels were analyzed by qRT-PCR and normalized to the 5S and U6 RNAs and to siNC cells. Error bars represent SEM values. (D) Schematic view of the IRS-1 mRNA and the sequences targeted by miR-145. (E) In a similar manner to miR-145 over-expression, BCDIN3D depletion affects IRS-1 expression at the protein level. These effects are not due to p53 stabilization. MCF-7 cells were co-transfected with miR-145 mimic, siBCDIN3D and siNC as indicated and analyzed by Western Blot with antibodies against IRS-1, p53 and the loading control GAPDH. (F) BCDIN3D depletion significantly reduces the expression of the luciferase reporter gene fused to the 3′UTR of the IRS-1 mRNA. siBCDIN3D and siNC transfected MCF-7 cells were transfected with pMirTarget (Origene) containing or not the 3′-UTR of IRS1 fused to the Luciferase Reporter gene and after 24h, the cells were lysed and the luciferase activity was measured in a luminometer. Shown are results from a typical experiment in quadruplets. Error bars represent SEM values. The transfection efficiency of the plasmids, as assessed by the expression of RFP gene contained in the pMirTarget plasmid, was identical in all samples. (G) miR-145 inhibition rescues the invasion defect of MDA-MB-231 cells transfected with siBCDIN3D. MDA-MB-231 were transfected according to the procedure schematized on the left with siRNAs against BCDIN3D and Negative Control, followed by anti miR-145 inhibitors and negative control. 24 h after the transfection of the miRNA inhibitors, the cells were trypsinized and analyzed as in Figure 2, (B-D) and the invasion assays are shown. Error bars represent SEM values.
See also Figure S3.
The increase in the levels of mature miR-145 seen after BCDIN3D depletion resulted in several expected functional consequences. Indeed, similar to miR-145 over-expression, BCDIN3D depletion decreased the levels of IRS1, a previously established miR-145 target (Shi et al., 2007; Spizzo et al., 2009), at both the protein and RNA levels (Figure 3D-E, and data not shown) and significantly reduced the expression of the luciferase reporter gene fused to the 3′UTR of the IRS1 mRNA (Figure 3F). Moreover, the miR-145 motif was significantly enriched (p=0.0026) in the 3′UTR of genes down-regulated upon BCDIN3D depletion, while the other four miRNAs tested were not (see below). Finally, despite the apparently mild increase of miR-145 levels in MDA-MB-231 cells transfected with siBCDIN3D compared to siNC (Figure S3D), the transfection of miR-145 inhibitors totally or partially rescued the invasion defect of these cells, depending on the density of the matrigel (Figure 3G and data not shown). Altogether, these results strongly suggest that BCDIN3D regulates mature miR-145 levels and that miR-145 is a functional mediator of the phenotypic effects of BCDIN3D depletion.
BCDIN3D dimethylates the 5′-Phosphate group of (pre-)miR-145 in vitro
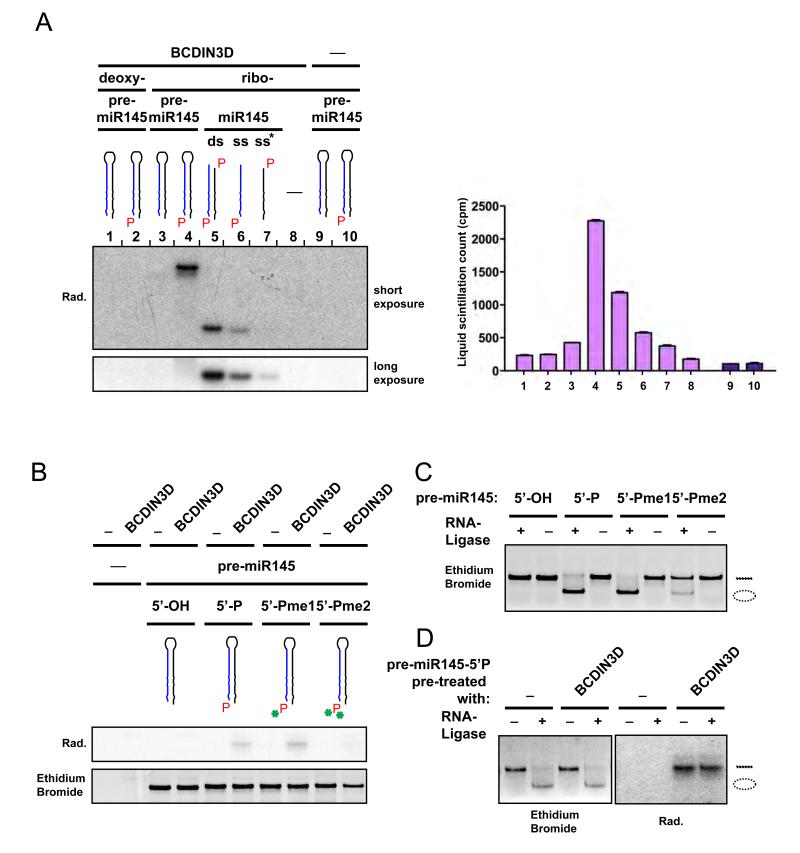
To test if miR-145 is a direct target of BCDIN3D activity, we performed in vitro methyltransferase assays using recombinant BCDIN3D, synthetic forms of miR-145 and 3H-SAM, and analyzed them by autoradiography and liquid scintillation counting. In these assays, BCDIN3D showed a strong activity and a preference for the precursor form of miR-145 (pre-miR-145) compared to either the double stranded miR-145 or the single stranded miR-145 and its passenger miR-145* (Figure 4A). Importantly, BCDIN3D did not methylate the 5′OH unphosphorylated form of neither pre-miR-145, miR-145 or miR-145* (Figure 4A and data not shown), validating the specificity of BCDIN3D activity towards the 5′ mono-phosphate. Finally, BCDIN3D could not methylate the 5′P deoxy-ribonucleotide form of pre-miR-145 confirming that BCDIN3D has indeed a preference for RNA molecules (Figure 4A).
Figure 4. BCDIN3D dimethylates the 5′Phosphate group of (pre-)miR-145 in vitro.
(A) BCDIN3D preferentially methylates pre-miR-145. In vitro methyltransferase assay using recombinant GST-BCDIN3D (1-8) or GST (9-10), 3H-radioactive SAM as methyl group donor and the following synthetic DNA and RNA molecules as substrate (1) deoxy-pre-miR-145 [5′-OH]; (2) deoxy-pre-miR-145 [5′-P]; (3&9) pre-miR-145 [5′-OH]; (4&10) pre-miR-145 [5′-P]; (5) duplex miR-145 [5′-P]; (6) miR-145 [5′P]; (7) miR-145* [5′P]; (8) mock. The nucleic acids were purified and analyzed by autoradiography (left) and liquid scintillation (right). Error bars represent Standard Deviation (SD) values. Full recovery of nucleic acids after purification was verified with a spectrophotometer. (B-D) The product of BCDIN3D methylation is a 5′-Phospho-dimethyl. (B) In vitro methyltransferase assay as in (A) using as substrate synthetic pre-miR-145 molecules that have [5′-OH], [5′-P], [5′-Pme1] or [5′-Pme2] ends (see Figure 1G). The lower panel shows the Staining with ethidium bromide of the gel used for the autoradiography shown in the upper panel. (C) Synthetic pre-miR-145 molecules that have [5′-OH], [5′-P], [5′-Pme1] or [5′-Pme2] ends were ligated with the T4 RNA Ligase 1. The circular RNA molecules migrate faster than the linear molecules. pre-miR [5′-Pme2] is resistant to ligation. (D) Synthetic pre-miR-145 [5′-P] was in vitro methylated as in (A) prior to ligation with T4 RNA Ligase 1. The left panel shows the ethidium bromide staining of the gel used for the autoradiography shown in the right panel. The pre-miR-145 molecules that were methylated by BCDIN3D were also resistant to ligation.
As illustrated in Figure 1G, the 5′ mono-phosphate group contains two negatively charged oxygen moieties [-O−] that can be subject to methylation [-O-CH3]. We therefore sought to determine if BCDIN3D methylates one or both available oxygen moieties. To this purpose, we synthesized pre-miR-145 molecules in which the 5′ mono-phosphate group was either mono- (5′Pme1) or di-methylated (5′Pme2). We first submitted them to in vitro methyltransferase assays with recombinant BCDIN3D and 3H-radioactive SAM. BCDIN3D could still methylate pre-miR-145 [Pme1] but not pre-miR-145 [Pme2] (Figure 4B), strongly suggesting that BCDIN3D can methylate both oxygen moieties of the 5′ mono-phosphate end. In order to determine the ratio of mono- and di-methylated 5′ mono-phosphate products in our in vitro methyltransferase assays, we exploited the enzymatic properties of T4 RNA Ligase 1. This enzyme catalyzes the ligation of a 5′ mono-phosphate nucleic acid donor to a 3′ hydroxyl-terminated nucleic acid acceptor through the formation of a 3′→5′ phosphodiester bond. As expected, the ligation reaction circularized the synthetic pre-miR-145 5′ mono-phosphate RNA molecule resulting in a faster migrating band (Figure 4C). In our conditions, T4 RNA Ligase 1 could still efficiently ligate pre-miR-145 [Pme1] but not pre-miR-145 [Pme2], thus providing us with a tool to discriminate between the mono- and the di-methylated forms of the 5′ mono-phosphate end (Figure 4C). As shown in Figure 4D, pre-miR-145 methylated with BCDIN3D (radioactive band on the right), was fully resistant to ligation with T4 RNA Ligase 1, indicating that the product of BCDIN3D methylation is a 5′Pme2.
Pre-miR-145 is modified by BCDIN3D in vivo
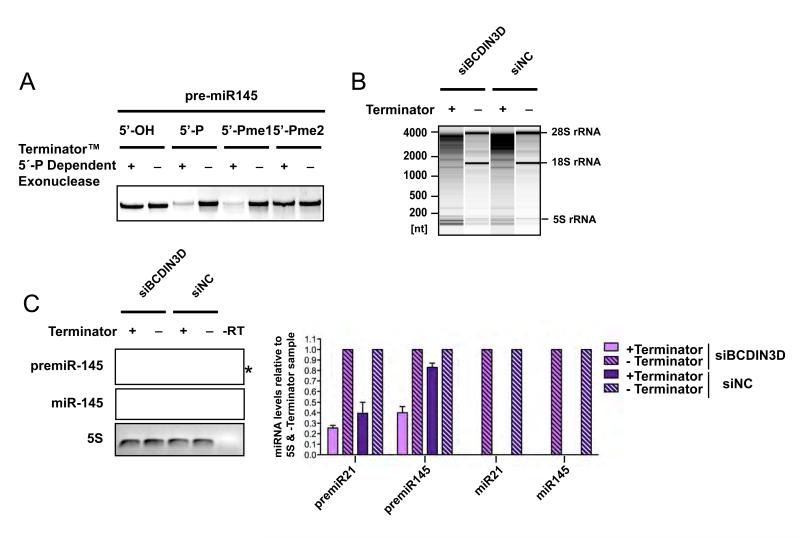
To determine whether BCDIN3D also targets miR-145 and/or its precursor in vivo, we set up an assay that takes advantage of the fact that the 5′Pme2 end is also resistant to treatment with the Terminator™ 5′Phosphate-Dependent Exonuclease that specifically digests RNAs having a 5′ mono-phosphate (Figure 5A). We treated RNA purified from MCF-7 cells transfected with siBCDIN3D and siNC with the Terminator enzyme and analyzed the resulting RNAs by quantitative reverse transcription and PCR with specific primers for pre-miR-145 and miR-145 as well as controls (Figure 5B-C). To verify that the treatment with Terminator™ was effective, we analyzed the RNAs treated with Terminator™ and mock by electrophoresis on an Agilent Total RNA Pico chip. As shown on Figure 5B, Terminator™ fully digested the 28S and 18S rRNAs which have 5′ mono-phosphate ends but left intact the 5S rRNA which has a 5′ triphosphate end. The treatment also fully digested miR-21 which is known to be mono-phosphate (Encode, 2009) (Figure 5C). Under these conditions, a significant proportion of pre-miR-145, but not miR-145, was resistant to digestion with Terminator™ in siNC transfected MCF-7 cells (Figure 5C). Moreover, the Terminator™ resistant fraction of pre-miR-145 was significantly reduced in siBCDIN3D treated MCF-7 cells (Figure 5C). This shows that the 5′end of pre-miR-145 is modified in a BCDIN3D dependent manner in vivo. Together with the in vitro activity of BCDIN3D on pre-miR-145, this suggests that pre-miR-145 is dimethylated at its terminal 5′ mono-phosphate. Interestingly, a smaller portion of pre-miR-21 was also resistant to Terminator™ suggesting that miR-21 is also a BCDIN3D target. This could explain the mild increase of the mature levels of miR-21 in some of our assays.
Figure 5. Pre-miR-145 is also modified in a BCDIN3D dependent manner in vivo.
(A) Synthetic pre-miR-145 molecules that have [5′-OH], [5′-P], [5′-Pme1] or [5′-Pme2] 5′ ends were treated with Terminator 5′-P Dependent exonuclease that is a processive 5′-3′ exonuclease that digests RNAs having a 5′ mono-phosphate. pre-miR [5′-Pme2] is resistant to this treatment. (B) RNA from MCF-7 cells treated or not with Terminator was migrated on an Agilent Total Eukaryotic RNA pico Chip. The treatment was specific as the Terminator fully digested the 18S and 28S rRNA which have 5′mono-phosphates, but did not digest the 5S RNA which has a 5′ tri-phosphate. (C) RNA treated as in (B) was analyzed by Reverse Transcription and PCR with primers specific for pre-miR-21, pre-miR-145, miR-21, miR-145 and 5S. The 5S RNA is known to be 5′ triphosphate and is used to control for equal RNA recovery. The indicated RNAs were analyzed by quantitative RT-PCR and normalized to the 5S RNA and to the [-Terminator] sample (graph on the right). Error bars represent SEM values. To facilitate the visualization of the assay, the PCRs were also analyzed by semi-quantitative PCR (panels on the left). For each primer, the semi-quantitative PCRs were performed in parallel with the quantitative PCRs and were stopped at the same cycle as the quantitative PCR reached the threshold of the less abundant sample. The treatment with Terminator fully digests miR-145 but not pre-miR-145. The asterisk indicates a non-specific band.
See also Figures S4, S5 & S6.
Our global whole RNA analysis in siBCDIN3D and siNC treated MCF-7 cells, also suggested that miR-145 is not the only target of BCDIN3D. Indeed, mRNAs down-regulated in siBCDIN3D cells were very significantly enriched with multiple putative miRNA targeting sequences (Figure S4 and Extended Experimental Procedures). Moreover, we found additional miRNAs whose expression is increased in siBCDIN3D compared to siNC cells through miRNA profiling with the miRCURY LNA PCR platform from Exiqon (Figure S5 and Extended Experimental Procedures). In particular, we determined that the precursor of miR-23b was also modified in a BCDIN3D dependent manner in vivo (Figure S5D) and in vitro (Figure S6), indicating that BCDIN3D robustly modifies other miRNA targets.
BCDIN3D affects miR-145 association with Dicer
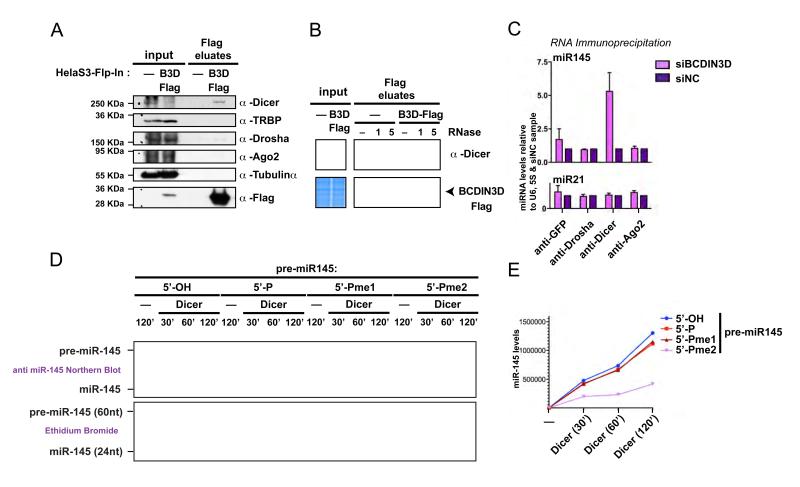
These results prompted us to determine at what step in the RNAi pathway BCDIN3D participates. To this end, we first probed for in vivo interactions between BCDIN3D and proteins involved in the RNAi pathway. We performed anti-Flag co-immunoprecipitations (co-IP) from Hela-S3-Flp-In control and -BCDIN3D-Flag stable cell lines followed by elution with a Flag peptide. This procedure highly reduced non-specific interactions, as shown by the lack of Tubulin alpha in our coIPs, a common contaminant of Flag-affinity purifications (Gregory et al., 2004) (Figure 6A). We reproducibly detected an interaction of BCDIN3D with Dicer but not with other proteins of the RNAi pathway (Figure 6A). Importantly, we could not detect an interaction between BCDIN3D and TRBP, which stably interacts with Dicer and participates in the processing of pre-miRNAs to mature miRNAs (Chendrimada et al., 2005) (Figure 6A). Interestingly, the observed interaction between BCDIN3D and Dicer is disrupted by treatment with RNase A (Figure 6B), suggesting that the interaction may be mediated through RNA(s). The binding of BCDIN3D to Dicer is an additional strong indicator that BCDIN3D has a direct role in the processing of miRNAs.
Figure 6. BCDIN3D interacts with Dicer and its depletion affects miR-145 association with Dicer.
(A) BCDIN3D specifically interacts with Dicer. Flag eluates from isogenic HELA-S3-Flp-In (-) and HELA-S3-Flp-In-BCDIN3D-Flag (BCDIN3D-Flag) stable cell lines co-immunoprecipitated with an anti Flag Antibody were analyzed by Western Blotting with the indicated antibodies (see Extended Experimental Procedures for further detail). (B) The interaction between BCDIN3D and Dicer is sensitive to treatment with RNase A. Flag co-Ips were performed as in (A) in the presence of mock or 1 or 5 %l of RNAse A (see Extended Experimental Procedures for further detail). The co-IPs were analyzed by Coomassie Staining to confirm equal recovery of BCDIN3D (lower panel) and by Western Blot with the Dicer antibody (upper panel). (C) Upon BCDIN3D depletion, the association of Dicer with miR-145, the product of pre-miR-145 processing, is significantly increased. MCF-7 cells transfected with siBCDIN3D or siNC were subjected to RNA-immunoprecipitation with the indicated antibodies (see Extended Experimental Procedures for further detail). RNA from the immunoprecipitates was purified and the levels of miR-21 and miR-145 were analyzed by qRT-PCR. The data are normalized to the 5S and U6 RNAs and to siNC cells. Error bars represent SEM values. (D-E) Synthetic pre-miR-145 molecules that have [5′-OH], [5′-P], [5′-Pme1] or [5′-Pme2] 5′ ends were incubated with human Dicer or Mock at 1mM of MgCl2 for 30, 60 or 120 min as indicated (see Extended Experimental Procedures for further detail). The Dicer processing reactions were loaded on a 15% Urea-PAGE gel and stained with Ethidium Bromide. The gel was photographed with the Chemidoc XRS+ system from Biorad (D, lower panel) and analyzed by Northern Blot with an anti-miR-145 probe (D, upper panel). The miR-145 product was quantified using the Image Lab Software (E).
See also Figure S7.
The observed increase in the levels of miR-145 upon BCDIN3D depletion could be due to either increased processing of miR-145, or to increased stability of the mature miR-145, or both. In order to distinguish between these three possibilities, we determined how depletion of BCDIN3D affected the association of miR-145 with proteins involved with its processing from pri- to pre- to mature miRNA. We performed RNA immunoprecipitations (RIP) with specific antibodies against Drosha, Dicer and Ago2 in MCF-7 cells treated with either siBCDIN3D or siNC. As shown in Figure 6C, depletion of BCDIN3D significantly and specifically increased the association of miR-145 with Dicer. This suggests that depletion of BCDIN3D augments the processing of pre-miR-145 by Dicer into miR-145. Surprisingly, BCDIN3D depletion did not increase miR-145 association with Ago2, although it mildly increased the association of Ago2 with miR-21, another lesser BCDIN3D target. This may be a direct consequence of the fact that miR-145 has a Guanine at position 1. The structure of the MID domain of human Ago2 was recently solved and indicated that a loop within this domain discriminates against Cytosines and Guanines thereby lowering hAgo2 affinity for miRNAs that start with these residues (Frank et al.). Another alternative albeit not exclusive explanation is that while the association of miR-145 with hAgo2 remains apparently unchanged, the increased association rate is actually compensated by a similarly increased dissociation rate.
The 5′ phospho-dimethyl modification reduces the processing of pre-miR-145 by Dicer
All of our results point to the fact that down-regulation of BCDIN3D augments the Dicer dependent processing of pre-miR-145 into miR-145. Firstly, depletion of BCDIN3D leads to a reduction of pre-miR-145 levels and a concomitant increase of mature miR-145 levels (Figure 3A-C). Secondly, upon BCDIN3D depletion, the association of Dicer with the product of pre-miR-145 processing increases (Figure 6C). Finally, we find that BCDIN3D modifies the pre- but not the mature miR-145 in vivo (Figure 5C), even though they have the same 5′end. Indeed, since the mature miR-145 is positioned on the 5p arm of the pre-miR-145 hairpin, the 5′end of the mature miR-145 is not produced by Dicer and is therefore the same as the 5′end of pre-miR-145. Together with the RIP results, this strongly suggests that the 5′ methylated pre-miR-145 molecules are not efficiently processed by Dicer in vivo. To test whether this effect is directly mediated by Dicer, we performed in vitro Dicer processing assays where human recombinant Dicer was incubated with synthetic pre-miR-145 molecules that have [5′-OH], [5′-P], [5′-Pme1] or [5′-Pme2] ends at physiological Mg2+ ions concentration (0.5-1mM) (Gunther, 2006). Under these conditions, the processing of pre-miR-145 [5′-Pme2] by Dicer was impaired compared to the other pre-miR-145 molecules (Figure 6D and Figure S7). The decrease in processing efficiency may result from the loss of the negative charge following methylation (Figure 1G) that in turn may affect the recognition of the 5′mono-phosphate of the pre-miRNA by Dicer (Figure 7). This is supported by recent findings that human Dicer contains a basic motif that anchors the 5′ mono-phosphate terminus (Park et al., 2011).
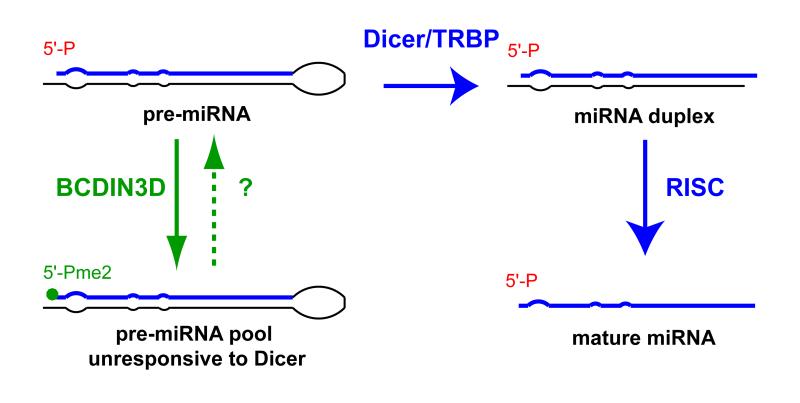
Figure 7.
Model for the mode of action of BCDIN3D.
Discussion
Here we uncover an enzymatic activity mediated by human BCDIN3D, that methylates both oxygen moieties of a phosphate group on nucleic acids. Specifically, we show that BCDIN3D modifies the 5′ mono-phosphate end of miRNAs including pre-miR-145 and pre-miR-23b. Methylation of the 5′ mono-phosphate of pre-miRNAs by BCDIN3D inhibits their processing by Dicer. This inhibition is consistent with the recent finding that human Dicer possesses a basic motif which accommodates the negatively charged 5′ mono-phosphate of pre-miRNAs (Park et al., 2011). Importantly, the interaction between Dicer and the 5′mono-phosphate is necessary for efficient and accurate processing of pre-miRNAs. (Park et al., 2011). Methylation of both oxygen moieties by BCDIN3D neutralizes the negative charges of the 5′ mono-phosphate group, This abrogation of charge may prevent the association of pre-miRNAs with the Dicer basic motif and result in the inhibition of Dicer activity. Together these findings suggest a model whereby direct methylation of pre-miRNAs by BCDIN3D generates a pool of pre-miRNAs that is less responsive to processing by Dicer (Figure 7). The fact that BCDIN3D can associate with Dicer (but not TRBP), may be an indication that BCDIN3D acts on miRNAs while complexed with Dicer. However, it is also possible that the interaction between Dicer and BCDIN3D functions in other aspects of this pathway.
BCDIN3D prefers RNA to DNA, since the deoxy- form of pre-miR-145 is a far inferior substrate compared to its ribonucleotide counterpart (Figure 4A). Given that the 5′ mono-phosphate group is identical between these molecules, other features, such as tri-dimensional structure or the presence of uracil instead of thymine, could determine this specificity. Of course, the possibility remains that BCDIN3D could also target DNA, which may be revealed under specific conditions or biological processes. This notion is supported by an example from C. elegans where Dicer is converted from a ribonuclease to a deoxyribonuclease through the action of a caspase during apoptosis (Nakagawa et al., 2010).
miRNAs are involved in numerous cellular effects and often several miRNAs function in concert to regulate a given biological function. Since BCDIN3D regulates a subset of miRNAs, this enzyme may function in specific biological processes depending on the cellular and/or physiological context. One possible role of BCDIN3D in cancer cells may be to promote cellular invasion (Figure 2). This idea is consistent with the finding that BCDIN3D mRNA is over-expressed in highly tumorigenic breast cancer stem cells and BCDIN3D is included in an “invasiveness” gene signature (Liu et al., 2007). In addition, two established in vivo targets of BCDIN3D, pre-miR-145 and pre-miR23b, are able to suppress the process of cellular invasion (Sachdeva et al., 2009; Shi et al., 2007; Spizzo et al., 2009; Zhang et al., 2011). Collectively, these findings strongly suggest that the negative effect of BCDIN3D depletion on invasion could be due to its effects on several miRNAs. Indeed, a recent genome-wide functional screen showed that 69 out of 900 miRNAs tested were able to inhibit cellular invasion (Zhang et al., 2011). The potential of BCDIN3D to affect many miRNAs simultaneously may allow it to tightly control specific pathways, including cellular invasion. Future work should allow us to establish whether this effect may be explored for therapeutic targeting in breast and possibly other cancers.
Experimental Procedures
The details of all siRNAs, synthetic RNAs, primers and antibodies used in the study are provided in Tables S1, S2 and S3 in the Supplemental Information section. Extended Experimental Procedures are included in the Supplemental Information and/or are available upon request.
Knock down experiments
ON-TARGETplus siRNAs against BCDIN3D and control were purchased from Dharmacon and reverse-transfected in two rounds into MCF7, MDA-MB-231 or BJ+TERT cells using LipofectamineTM RNAiMAX from Invitrogen or HiPerfect from Qiagen. miR-145 mimics and inhibitors were from Ambion. The MDA-MB-231 clones stably expressing shRNAs against BCDIN3D or control were generated by transfecting MDA-MB-231 cells with the pRS vectors containing 29 mer shRNA against BCDIN3D (TR317908, t1368843) and Non-effective scrambled shRNAs (TR30012) from Origene respectively. The shBCDIN3D rescue clones were generated by transfecting tGFP-tagged BCDIN3D carrying a silent C324/C327A mutation.
Cellular assays
For the soft agar colony assays, 3 × 104 cells were plated in 0.35% agar in complete growth medium in 6-well plates and colonies were analyzed after 4-5 weeks. The Real Time growth, migration and invasion assays were performed with the xCELLigence system from Roche following the manufacturer’s instructions. 1/40th dilution of Matrigel was used to coat the upper chamber of the CIM plate for the invasion assays.
RNA analysis
Total RNA and proteins were extracted from the same cells using the RNA/Protein purification kit from Norgen. 20-200 ng of total RNA was used for detection of miRNAs with the mirVana™ qRT-PCR miRNA detection kit and/or the Taqman® MicroRNA Reverse Transcription Kit from Applied Biosystems. Real-time PCR analysis was performed on a StepOne Plus system or Vii7A from Applied Biosystems. The Northern Blots were performed as previously described (Xhemalce and Kouzarides, 2010).
Modified RNAs
Precursor, mature and passenger miR-145 carrying 5′OH, 5′P, 5′Pme1 or 5′Pme2 modifications were custom synthesized by IBA GmbH.
in vitro RNA Methyltransferase assays
These assays were performed in a total volume of 100 %l in 25mM Tris-HCl, pH8, 150mM NaCl, 2mM KCl, 10% glycerol, 1 mM EDTA, 1mM DTT supplemented with EDTA-free Complete Protease Inhibitor cocktail from Roche and 80 U of RNaseOUT from Invitrogen with 1 %g of recombinant BCDIN3D, 4 %l of 20 %M synthetic RNAs and 4 %l of 3H-SAM (PerkinElmer NET155250UC) for 2h at 37°C.
in vitro Dicer processing assays
These assays were performed essentially as described (Park et al., 2011) with 100 pmol of synthetic pre-miRNA, 500 ng of recombinant human Dicer in a total volume of 15 %l in 100mM KCl, 10mM Tris-HCl, pH8, 0.1mM EDTA, 0.5 or 1 mM MgCl2, 0.5mM dTT supplemented with 0.5 U/%l RNaseOUT for the indicated times at 37°C.
Supplementary Material
Supplemental Information. The Supplemental Information includes Extended Experimental Procedures, 3 Supplemental Tables, 7 Supplemental Figures and References.
Acknowledgments
We thank Dr. Till Bartke for the recombinant nucleosomes and the parental Hela-S3-FlpIn cell line, Dr. Kyle M Miller for help with the xCELLigence system, the DNA core facility at the Institute of Cellular and Molecular Biology (ICMB) at the University of Texas at Austin, USA for use of equipment, and Dr. Andrew J Bannister for helpful discussions. BX was funded by a start-up grant from ICMB at the University of Texas at Austin and a Cancer Research UK project grant. The T.K. laboratory was funded by grants from CRUK and the 6th Research Framework Program of the European Union (Epitron, SMARTER and HEROIC).
References
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. Epub 2009 Jan 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–115. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AG02. Nature. 465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. Epub 2008 Oct 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Gunther T. Concentration, compartmentation and metabolic function of intracellular free Mg2+ Magnes Res. 2006;19:225–236. [PubMed] [Google Scholar]
- Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined “30 nm” chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio Mv, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. doi: 810.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y. Making RISC. Trends Biochem Sci. 2010;35:368–376. doi: 10.1016/j.tibs.2010.03.009. Epub 2010 Apr 2013. [DOI] [PubMed] [Google Scholar]
- Kawamata T, Yoda M, Tomari Y. Multilayer checkpoints for microRNA authenticity during RISC assembly. EMBO Rep. 2011;12:944–949. doi: 10.1038/embor.2011.128. doi: 910.1038/embor.2011.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. Epub 2006 Jan 2005. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. Epub 2010 Oct 2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- Nakagawa A, Shi Y, Kage-Nakadai E, Mitani S, Xue D. Caspase-dependent conversion of Dicer ribonuclease into a death-promoting deoxyribonuclease. Science. 2010;328:327–334. doi: 10.1126/science.1182374. Epub 2010 Mar 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. doi:210.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, Elble R, Watabe K, Mo YY. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnow P, Jopling CL, Norman KL, Schutz S, Wehner KA. MicroRNAs: expression, avoidance and subversion by vertebrate viruses. Nat Rev Microbiol. 2006;4:651–659. doi: 10.1038/nrmicro1473. [DOI] [PubMed] [Google Scholar]
- Shi B, Sepp-Lorenzino L, Prisco M, Linsley P, deAngelis T, Baserga R. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem. 2007;282:32582–32590. doi: 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- Shuman S. Transcriptional networking cap-tures the 7SK RNA 5′-gamma-methyltransferase. Mol Cell. 2007;27:517–519. doi: 10.1016/j.molcel.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, Zagatti B, Fabbri M, Veronese A, Liu X, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 2009;17:246–254. doi: 10.1038/cdd.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. Epub 2006 Aug 2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xhemalce B, Dawson MA, Bannister AJ. Histone Modifications. Encyclopedia of Molecular Cell Biology and Molecular Medicine. 2011 [Google Scholar]
- Xhemalce B, Kouzarides T. A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev. 2010;24:647–652. doi: 10.1101/gad.1881710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Hao Y, Yang J, Zhou Y, Li J, Yin S, Sun C, Ma M, Huang Y, Xi JJ. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat Commun. 2011;2:554. doi: 10.1038/ncomms1555. 10.1038/ncomms1555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.