Abstract
Purpose
Tumor antigen (TA)-specific monoclonal antibodies (mAb) block oncogenic signaling and induce Fcγ receptor (FcγR)-mediated cytotoxicity. However, the role of CD8+ cytotoxic T lymphocyte (CTL) and FcγR in initiating innate and adaptive immune responses in mAb-treated human cancer patients is still emerging.
Experimental Design
FcγRIIIa codon 158 polymorphism was correlated with survival in 107 cetuximab-treated head and neck cancer (HNC) patients. Flow cytometry was performed to quantify EGFR-specific T cells in cetuximab-treated HNC patients. The effect of cetuximab on NK cell, dendritic cell (DC), and T cell activation was measured using IFN-γ release assays and flow cytometry.
Results
FcγR IIIa polymorphism did not predict clinical outcome in cetuximab-treated HNC patients, however elevated circulating EGFR -specific CD8+ 853-861 T cells were found in cetuximab-treated HNC patients (p<0.005). Cetuximab promoted EGFR-specific cellular immunity through the interaction of EGFR+ tumor cells and FcγRIIIa on NK cells, but not on the polymorphism per se. Cetuximab-activated NK cells induced IFN-γ dependent expression of DC maturation markers, antigen presentation machinery (APM) components such as TAP-1/2, and Th1 chemokines through NKG2D/MICA binding. Cetuximab initiated adaptive immune responses via NK-cell induced DC maturation, which enhanced cross-presentation to CTL specific for EGFR as well as another TA, MAGE-3.
Conclusion
Cetuximab-activated NK cells promote DC maturation and CD8+ T cell priming, leading to TA spreading and Th1 cytokine release through ‘NK-DC cross-talk.’ FcγRIIIa polymorphism did not predict clinical response to cetuximab, but was necessary for NK-DC interaction and mAb induced cross-presentation. EGFR-specific T cells in cetuximab treated HNC patients may contribute to clinical response.
Keywords: Cetuximab, Panitumumab, EGFR, ADCC, CTL, cross-presentation, head and neck cancer, immunotherapy
Introduction
Tumor antigen (TA)-specific monoclonal antibodies (mAb) are clinically effective in a variety of malignancies. However, these results are only seen in a subset (~20%) of patients (1). The large majority of treated patients, who are not likely to benefit from this therapy emphasizes the need to characterize the mechanisms underlying the activity of TA-specific mAb, and to optimize the selection of patients to be treated. In the present study we have utilized cetuximab, an epidermal growth factor receptor (EGFR)-specific, chimeric IgG1 isotype mAb that is FDA-approved for colorectal carcinoma (CRC) and head and neck carcinoma (HNC), as a model. Despite overexpression of EGFR in 80-90% of HNC, cetuximab is effective in only in 10-20% of patients (2, 3).
A growing body of experimental and clinical evidence suggests that mAb-induced anti-tumor immunity underlie treated patients’ clinical responses and may play a role in the differential clinical responses. Specifically, the statistically significant correlations of mAb-binding Fcγ receptor (FcγR) polymorphisms with clinical outcome in patients treated with rituximab (4, 5) trastuzumab (6, 7), and cetuximab (8, 9) argue in favor of a role of immune cell activation in the clinical responses which have been described. Recently, trastuzumab emtansine (T-DM1) an Ab-drug conjugate showed significantly prolonged progression free and overall survival in breast cancer patients (10) and such a strategy may be enhanced by immune cell activation. However, the non absolute nature of the correlations in these malignancies (8, 9), the strong influence of k-ras mutation (8) on clinical response to cetuximab in CRC and conflicting information about the beneficial effect of FcγRIIIa genotype in CRC patients (8,9), raise the possibility of additional immunological mechanisms in the clinical benefit from mAb-based immunotherapy. This possibility is supported by the growing evidence in animal model systems (11) and in clinical settings (12) that TA-specific mAb may elicit cellular immunity.
Therefore, in this study we have investigated whether the FcγRIIIa polymorphism on NK cells correlates with the anti-tumor activity of cetuximab using human HNC cells and with the clinical course of the disease in HNC patients. We have selected these patients for our investigations, since in recent studies we and others have shown in in vitro experiments a significant correlation of FcγRIIIa polymorphism with the anti-tumor activity of cetuximab (13). Furthermore, we have tested whether the interaction of cetuximab with FcγRIIIa on NK cells was required to trigger DC maturation and TA-specific cellular immune responses in HNC patients. We demonstrate for the first time that cetuximab-activated NK cells trigger cross-talk and maturation of DC in an FcγR and NK group 2, member D (NKG2D) dependent manner, and this results in TA-specific priming of CTL in cetuximab treated HNC patients. Lastly we have analyzed the mechanism (s) underlying the TA-specific immune response elicited by cetuximab and its potential clinical relevance.
Materials and Methods
Tumor cell lines
The HNC cell lines HLA-A2−EGFR+ PCI-15B, HLA-A2−EGFR+ and MAGE-3+-JHU-029 (14-16), the breast cancer cell line MCF-7 and the lymphoid T2 cell line were grown in IMDM (Sigma, St. Louis, MO) supplemented with 10% FBS (Cellgro, Manassas, VA), 2% L-glutamine and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA) at 37°C in a 5% CO2 , 95% humidity. Adherent tumor cells were detached by warm Trypsin-EDTA (0.25%) solution (Invitrogen, Carlsbad, CA).
Patients and demographics
The cohort of 107 cetuximab treated stage III/IV HNC patients described in Figure 1 combined 60 patients enrolled on two prospective, cetuximab containing clinical trial regimens, UPCI-05-003 and UPCI 05-005 and 47 additional patients treated with cetuximab off protocol, as described in Table 1. The majority of these patients were treated with cetuximab plus cisplatin/paclitaxel/radiotherapy (UPCI 05-003, ref. 17) or cetuximab plus pemetrexed/radiotherapy (UPCI 05-005, ref. 18). Both trial cohorts were single arm phase II trials for locoregionally advanced, previously untreated disease. Patients were assigned to either trial by the treating physician at the time. The remainder of the patients was treated off-trial with cetuximab alone or in conjunction with palliative radiotherapy. EGFR tetramer measurements were performed on protocol patients who were receiving single agent cetuximab during the 6 month cetuximab maintenance phase of UPCI 05-003 (Table 1), or other newly diagnosed HNC patients with stage III-IV disease while receiving cetuximab alone as primary treatment on a newly initiated, prospective phase II trial of single agent cetuximab (UPCI 08-013). The comparison (cetuximab-naïve) HNC cohorts were gender and age-matched, previously cetuximab untreated HNC patients. No patients were excluded as a result of prior treatments or performance status. Blood from cetuximab naïve HNC patients was drawn within the same period after completing therapy without cetuximab.
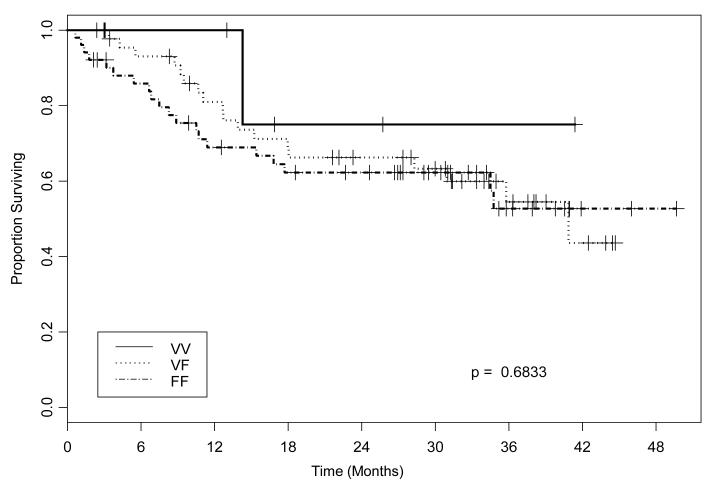
Figure 1.
Kaplan-Meier estimates of disease specific (DSS) survival in cetuximab treated HNC patients. Lack of correlation between FcγRIIIa polymorphisms (based on VV, VF, FF genotype) and survival of cetuximab treated HNC patients. Genomic DNA of HNC patients treated with cetuximab at the University of Pittsburgh Cancer Institute was tested for FcγRIIIa (at codon 158) polymorphisms. HNC patients (n=107) were treated in two prospective cetuximab-based clinical trial regimens or with cetuximab + RT, as described in materials and methods. DSS was measured and a Kaplan-Meier plot was constructed to determine whether the tested FcγR polymorphisms were correlated with clinical outcome. No differences were observed in patients’ DSS based on FcγRIIIa genotype (P=0.683).
Table1.
Demographics of FcγRIIIa genotyped cohort
| Demographics of Cetuximab-treated, FcγRIIIa genotyped cohort | ||||||
|---|---|---|---|---|---|---|
| Regimen | No. of Patients | Tumor Site | Mean Age (yrs) | Males | Females | |
| UPCI 05-0031 | 36 | OC | 3 | 52.9 | 32 | 4 |
| OP | 23 | |||||
| L | 4 | |||||
| HP | 3 | |||||
| Other | 1 | |||||
| Unknown primary | 2 | |||||
| UPCI 05-0052 | 24 | OC | 0 | 58.2 | 20 | 4 |
| OP | 9 | |||||
| L | 5 | |||||
| HP | 4 | |||||
| Other | 4 | |||||
| Unknown primary | 2 | |||||
| cetuximab | 11 | OC | 4 | 65.4 | 10 | 1 |
| OP | 1 | |||||
| L | 1 | |||||
| HP | 0 | |||||
| Other | 5 | |||||
| Unknown primary | 0 | |||||
| cetuximab+RT | 36 | OC | 10 | 62.5 | 30 | 6 |
| OP | 12 | |||||
| L | 3 | |||||
| HP | 2 | |||||
| Other | 8 | |||||
| Unknown primary | 1 | |||||
cetuximab 250 mg/m2 days weekly (after an initial dose of 400 mg/m2), docetaxel 75 mg/m2 day 1, cisplatin 75 mg/m2 day 1, repeated every 21 days × 3 cycles, then radiotherapy to 70 Gy (2 Gy/day) with concurrent cetuximab 250 mg/m2 weekly + cisplatin 30 mg/m2, then maintenance cetuximab for 6 months. Blood was drawn during single-agent cetuximab maintenance.
cetuximab 250 mg/m2 (after an initial dose of 400 mg/m2) weekly + pemetrexed on days 1, 22, and 43 with concurrent radiotherapy (2 Gy/day)
Antibodies and cytokines
The EGFR-specific chimeric IgG1 mAb cetuximab (Erbitux™, BMS Imclone, Princeton, NJ) and the EGFR-specific human IgG2 mAb panitumumab (Vectibix, Amgen, Thousand Oaks, CA) were purchased from the University of Pittsburgh Hillman Cancer Center Pharmacy. The nonspecific control, human isotype IgG1 was purchased from Axxora LLC (San Diego, CA). FITC or PE-Cy7-anti-CD11c mAb (R & D systems), anti-CD80 mAb, anti-CD86 mAb, anti-CD83 mAb, anti-HLA-DR mAb, anti-CCR7 mAb (BD Biosciences Pharmingen) were purchased. Neutralizing anti-FcγRIIIa Ab, and PE- and FITC-conjugated IgG isotypes for flow cytometry were purchased from BD Biosciences. FITC-goat-anti-human Fc specific IgG and FITC-goat anti-mouse IgG were purchased from Invitrogen. The antigen processing machinery (APM) components TAP-1-specific mAb NOB1 and TAP-2 specific mAb NOB2 were developed and characterized as described (19). Anti-CXCL10 Ab, recombinant human GM-CSF, rec. human IFN-α, rec. human IL-1β, rec. human IL-2, rec. human IL-4, rec. human IL-6, rec. human IL-7, PGE2, Poly I:C, and TNF-α were purchased from R&D Systems Inc (Minneapolis, MN). IFN-γ (Interferon-γ) was purchased from InterMune (Brisbane, CA).
Lymphocyte isolation from blood and PBMC
After approval by our Institutional Review Board (UPCI protocol 99-069), informed consent was obtained from each subject. Blood from healthy donors (Western PA blood bank) or HNC patients treated with cetuximab during or within one month of treatment. Lymphocytes were purified by Ficoll-Paque™ PLUS centrifugation (Amersham Biosciences, Uppsala, Sweden) and stored frozen. DC were generated as described previously (19). NK cells and CD8+ T cells were purified using EasySep kits (Stem cell technologies, Vancouver, BC, Canada) and purity was >95% FcγRIIIa +, CD56+, CD3− (13).
PBMC were genotyped at FcγRIIIa (codon 158) using quantitative PCR (Applied Biosystems, Framingham, MA). Genomic DNA was extracted using the DNeasy Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol. Genomic DNA (5 to 50 ng) was added to a 25-μL reaction using 2X Taqman master mix (Applied Biosystems). Plates were analyzed using an ABI prism 7700 (13).
In vitro stimulation of EGFR-specific CD8r+ T cells
Mature DC were incubated with autologous negatively isolated CD8+ T cells for 48 hr at 37°C with rhIL-2 (20 U/ml) and rhIL-7 (5ng/ml). On day 7 and weekly thereafter, lymphocytes were re-stimulated with autologous irradiated DC pulsed with peptide in culture medium supplemented with IL-2 (20 U/ml) and IL-7 (5 ng/ml). The stimulated CD8+ T cells were analyzed for TA specificity in IFN-γ ELISPOT at day 21 and then every 7 days thereafter. We have previously published a novel wild type EFGR853-861 specific tetramer that recognizes EFGR853-861 specific CTL and also shown that this specific tetramer recognizes EFGR853-861 specific CTL clone that was generated by pulsing DC with EFGR853-861 peptide in vitro (14).
CTL lines and ELISPOT assay
IFN-γ ELISPOT assays were performed as described (14). The MAGE-3271–279 (FLWGPRALV) peptide and wild type EGFR853-861 peptide (ITDFGLAKL) were produced by the peptide synthesis facility at the University of Pittsburgh, using F-moc technology (19, 20). The HLA-A2-MAGE-3271–279 tetramer was synthesized by the NIH Tetramer Facility (Emory University, Atlanta, GA). CTL lines were sorted using PE-labeled HLA-A2-EGFR853-861 peptide loaded tetramers (14) and restimulated every 7-10 days. For ELISPOT assays, CD8+ T cells were seeded in triplicate (5×104 ) for bulk CD8+ T cells and 1×103 for CD8+ T cell clones) in multiscreen HTS plates. Synthetic peptides were then added to ELISPOT assays after APCs were seeded. After 18h, spots were enumerated using computer-assisted image analysis software.
Flow cytometry
Lymphocytes were prepared for flow cytometry by washing with PBS (Sigma-Aldrich, St. Louis, MO) and FACS buffer (2% FBS in PBS). For flow cytometric analysis using HLA-A2-peptide tetramers, PE-labeled HLA-A2-EGFR853-861 tetramers were obtained from the Tetramer Facility of the National Institutes of Health (Atlanta, GA). Cells were analyzed on cyan flow cytometer (Beckman Coulter) using Summit v4.3 software. Intracellular IFN-γ staining of CD56+ NK cells was performed by using cytofix/cytoperm fixation/permeabilization kit (BD Bioscience). Briefly, cells were treated with BD GolgiSTOP reagent and stained with FITC-anti-CD56 Ab. After 4 h, cells were washed, fixed, permeabilized and stained with FITC-anti-IFN-γ Ab.
Cytokine and chemokine analysis by Luminex
A standard calibration curve was generated by serial dilutions of recombinant cytokine. Cytokine concentrations in culture supernatants were determined by a multiplexed ELISA (Luminex™) at the UPCI facility. Hu30 Plex kit ( Invitrogen, catalog number LHC 6003) detected IL-1β, IL-1RA, IL-2, IL-2R, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL-12 (p40/p70), IL-13, IL-15, IL-17, TNF-α, IFN-α, IFN-γ, GM-CSF, MIP-1α, MCP-1,MIP-1β, CXCL-10, MIG, Eotaxin, RANTES, VEGF, G-CSF, EGF, FGF-basic and HGF.
T cell chemotaxis assay
Cell migration assays was performed in 5.0μm pore-size 96 well polycarbonate filtration plate (Millipore). Briefly, the lower chambers were filled with cell-free culture supernatant collected from various experimental conditions. T cells (5×104/100μl of fresh media) were plated in the upper well. Following a 4 hr incubation at 37°C, the upper wells containing filtration unit were gently removed and the cells in bottom chamber were counted by hemocytometer. After 4h incubation and counting, Cell numbers were plotted as the % migrated cells.
Statistical and patient follow up analysis
Disease specific survival of HNC patients was defined as the time elapsed from the first treatment with cetuximab until death from HNC. Patients were censored if they were alive at last follow-up or had died but were cancer-free at the time of death. FcγR genotype subclasses were tested for association with disease free survival with the log rank test. Other factors were tested for association with FcγR subclasses including cancer type (primary vs recurrent), disease site and whether the patient was treated on protocol. These results were used to insure the conclusions regarding the influence of FcγR upon disease-specific mortality were not confounded by other potentially influential covariates. Two-tailed unpaired t-test was performed for statistical analysis to compare the significant difference between two groups and p < 0.05 was considered significant. T cell reactivity as measured by the ELISPOT assay was considered positive if the number of spots in test wells was significantly higher than that in background wells when using a one-tailed permutation test for α ≤ 0.05.
Results
Survival of cetuximab treated HNC patients does not correlate with the FcγRIIIa polymorphism
We have previously demonstrated that NK cells expressing an FcγRIIIa V-encoding allele (at codon158) are more potent than NK cells expressing an FcγRIIIa F-encoding allele in ADCC against HNC cells in vitro (13). The clinical significance of our previously published in vitro data (13, 15) was then investigated in a cohort of 107 consecutive HNC patients treated with cetuximab at our institution and genotyped for polymorphic FcγRIIIa position 158 VV, VF or FF. Interestingly, FcγRIIIa genotype was not associated with disease specific survival (DSS) in this large cohort of cetuximab treated HNC patients (Figure 1 and Table 1). The low frequency of the beneficial VV genotype (5%) (13), in light of the higher observed rate of clinical response (~15-20%) (21), raised the likelihood that an additional immune mechanism may play a role in the anti-tumor activity of cetuximab. Since cetuximab can activate NK cells in the presence of EGFR+ tumor cells, and subsequently NK cells may provide a crucial DC maturation signal, we hypothesized that cetuximab-activated NK cells might facilitate the generation of TA-specific CTL.
Enhancement by cetuximab of EGFR-specific CTL in HNC patients
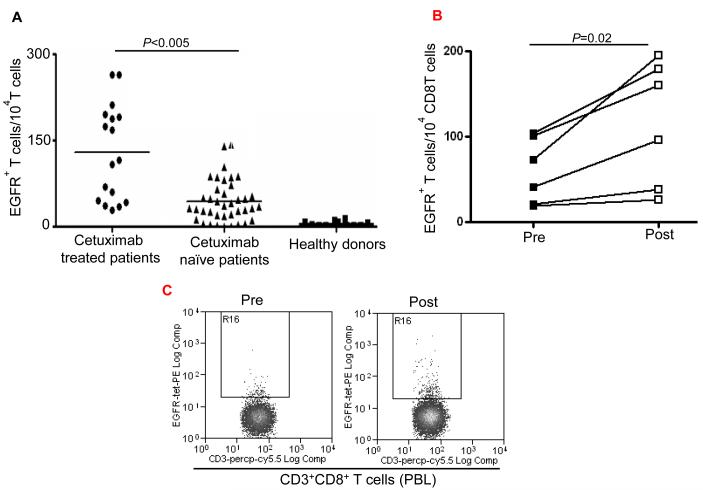
To evaluate whether induction of CD8+ T cells by cetuximab occurred in vivo, we measured the frequency of EGFR853-861-specific CTL in the circulation of cetuximab-treated HNC patients. PBMC were obtained from HLA-A2+ cetuximab-treated (n=17) and HLA-A2+ cetuximab-naïve HNC patients (n=39), and compared to HLA-A2+ healthy donors (n=24). Based on the analysis of CD3+CD8+ T cells stained with an EGFR853-861-specific tetramer (14), a significantly higher frequency of EGFR853-861-specific T cells was found in cetuximab-treated HNC patients than in cetuximab-naïve HNC patients (Figure 2A). As a control, we observed low background of EGFR853-861-specific precursor CTL in healthy HLA-A2+ donors. These data strongly support that the administration of cetuximab to HNC patients leads to EGFR853-861 peptide cross-presentation by DC, triggering expansion of EGFR-specific T cells. Similarly, higher frequency of EGFR853-861-specific T cells was also found in cetuximab-treated HNC patients who received 4 weeks of single agent cetuximab treatment (Figure 2B,C). Taken together, these results indicate that cetuximab alone or in combination with chemotherapy induces expansion of EGFR-specific T cells.
Figure 2.
Enhancement by cetuximab of EGFR853-861-specific CTL in the HNC patients. (A) Higher frequency of EGFR + 853-861 peptide-specific tetramer CD8+T cells in HLA-A2+ cetuximab-treated HNC patients (n=17) compared to HLA-A2+ cetuximab-naïve HNC patients (n=39) was found after EGFR853-861 tetramer staining of CD3+CD8+ T cells by using flow cytometry. The data comparing mean frequencies between cetuximab treated (•) and naïve (▲) patient groups are presented. As a negative control, staining of EGFR853-861 peptide-specific tetramer+ CD3+ CD8+ T cells (from HLA-A2+ healthy donors (n=24) was performed. A two-tailed unpaired t-test was performed for statistical analysis. (B) Expansion of EGFR853-861-specific CTL in single agent cetuximab treated, newly diagnosed HNC patients. Higher frequency of EGFR853-861-specific tetramer+ CD8+T cells in HLA-A2+, single agent cetuximab-treated HNC patients (n=6) after 4 weekly doses using flow cytometry. Mean frequencies were compared from the same patients before ( ■ ) and post ( □ ) cetuximab therapy. (C) A representative dot plot is shown for a single agent cetuximab treated patient (pre and post cetuximab therapy). (D) Enhancement by cetuximab of EGFR cross-presentation to EGFR853- 861-specific CTL by DC that was pre-incubated with cetuximab-activated NK cells. EGFR853-861 peptide-specific CTL were stimulated for 36h at 37°C with DC (from a HLA-A 2.1+ healthy donors) fed with UV-irradiated PCI-15B HNC cells (EGFR+, HLA-2.1−) coated with IgG1 or cetuximab (each at 10μg/ml), with or without autologous NK cells (DC:NK:PCI-15B at 1:1:1 ratio). Cetuximab (IgG1 isotype) binds FcγRIIIa expressed by NK cells avidly. EGFR853-861 peptide-specific CTL activated under the indicated conditions were examined for IFN-γ production by ELISPOT assay. Mature DC generated by 36h incubation with cytokines IL-1β, IL-6, PGE2 and TNF-α were used as a negative control in the assay. Data are representative of three individual experiments. A two-tailed unpaired t-test was performed for statistical analysis. (E) Enhancement by cetuximab of MAGE-3 cross-presentation to MAGE-3271–279-specific CTL by DC that was pre-incubated with cetuximab-activated NK cells. MAGE-3271–279 peptide-specific CTL were stimulated for 36h at 37°C with DC that were prior (from a HLA-A 2.1+ healthy donor) fed with UV-irradiated JHU-029 HNC cells (MAGE-3+, HLA-2.1−) coated with IgG1 or cetuximab (each at 10 μg/ml), with or without autologous NK cells (DC:NK:JHU-029 at 1:1:1 ratio). MAGE-3271–279 peptide-specific CTL activated under the indicated conditions were examined for IFN-γ production by ELISPOT assay. Mature DC generated by 36h incubation with cytokines IL-1β, IL-6, PGE2 and TNF-α were used as a negative control in the assay. We previously showed that an immunogenic EGFR-encoded CTL could be generated by pulsing DC with EGFR853-861 peptide that could stimulate cognate CTL in vitro (14). Total recombinant EGFR protein was not used in this study as a source of TA since this would not mimic FcγRIIIa mediated effects and NK:DC cross talk that cetuximab stimulates. Data are representative of two independent, repeated experiments. A two-tailed unpaired t-test was performed for statistical analysis.
Enhancement by cetuximab of TA cross-presentation to cognate CTL in the presence of NK cells
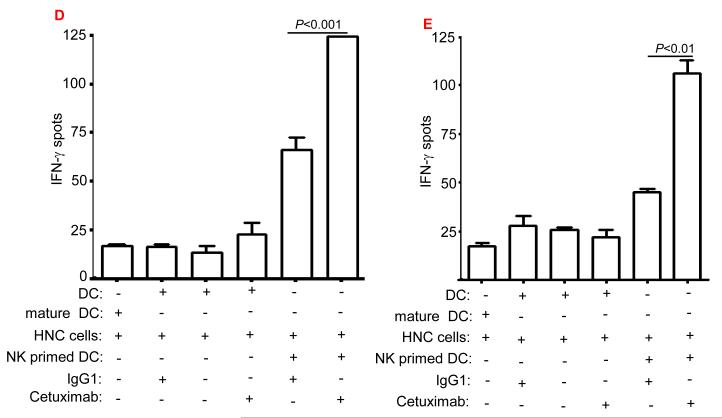
Having observed a higher level of EGFR-specific T cells in cetuximab-treated HNC patients, we investigated in vitro whether cetuximab enhanced EGFR-specific T cell immunity and whether this effect was modulated by NK cells. We studied the effect of cetuximab with or without NK cells on DC cross-presentation using EGFR+, HLA-A2− HNC cells to stimulate cognate CTL (19). First, DC alone (HLA-A2+ donors) were incubated with cetuximab and HLA-A2− PCI-15B HNC cells, which did not show enhanced cross-presentation. We furthermore included FcγRIIIa+ NK cells with DC to evaluate their effect on the cross-presentation capacity of DC. Although addition of NK cells alone showed a 3-fold increase in the cross-presentation by DC, the addition of cetuximab + NK cells resulted in a 5-fold enhancement in the EGFR853-861-specific CTL activation in comparison to IgG1 control mAb + NK cells (p<0.001) . The cross-presentation by NK cell -stimulated, HLA-A2+ DC was quantified using IFN-γ ELISPOT assays and HLA-A2+, EGFR853-861-specific CTL (14). DC incubated with HLA-A2− PCI-15B HNC cells and cetuximab enhanced NK cell dependent cross-presentation to EGFR-specific CTL. A low basal level of cross-presentation was observed with apoptotic PCI-15B HNC cells incubated with an isotype control IgG1 or without NK cells (Figure 2D), or without HNC cells. A positive control included CTL recognition of EGFR853-861 peptide loaded DC.
Enhancement by cetuximab-activated NK cells of non-EGFR (MAGE-3) cross-presentation to MAGE-specific CTL
We selected another HNC cell line JHU-029, which expresses EGFR as well as MAGE-3 (14, 20), in order to investigate whether cross-presentation of MAGE-3 by DC could also be enhanced by cetuximab-activated NK cells. DC incubated with HLA-A2− JHU-029 HNC cells and cetuximab significantly enhanced the cross-presentation to MAGE-3271–279-peptide specific CTL, primarily in the presence of NK cells. Addition of NK cells alone showed a two-fold increase in EGFR cross-presentation by DC. Moreover, addition of cetuximab with NK cells resulted in approximately four-fold enhancement in the MAGE-3271–279-peptide specific CTL activation in comparison to control IgG1 mAb (p<0.01) (Figure 2E). Cytokine matured DC alone were used as a control in the cross-presentation assay. An isotype control IgG1 failed to facilitate enhancement of cross-presentation of EGFR or MAGE-3 antigens in the presence or absence of NK cells. The importance of FcγRIIIa-activated NK cells to promote the activation of EGFR-specific (Figure 2D) as well as non-EGFR antigen (MAGE-3) specific CD8+ T cells, highlights the beneficial effect of enhanced DC maturation and antigen presentation after NK cell activation by cetuximab (Figure 2D and E).
To model cetuximab-dependent cross-priming of EGFR to CTL, we performed in vitro stimulation for two weeks, with DC matured using NK cells activated by PCI-15B HNC cells coated with cetuximab. Consequently, we measured the efficiency of cross-priming of EGFR-specific CTL by tetramer quantification by flow cytometry. DC matured with cetuximab-activated NK cells and PCI-15B HNC cells induced significantly higher levels of EGFR-specific CTL than panitumumab. The latter IgG2 mAb was used as control mAb (supplementary figure 1) due to its reduced FcγRIIIa binding. Taken together, these data strongly support that the administration of cetuximab to HNC patients leads to EGFR853-861 peptide cross-presentation by DC, triggering expansion of EGFR-specific T cells.
Enhancement of DC maturation by cetuximab in the presence of NK cells
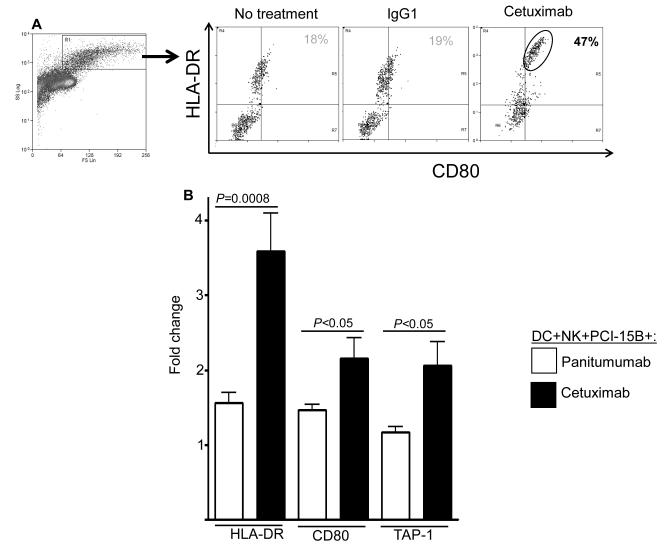
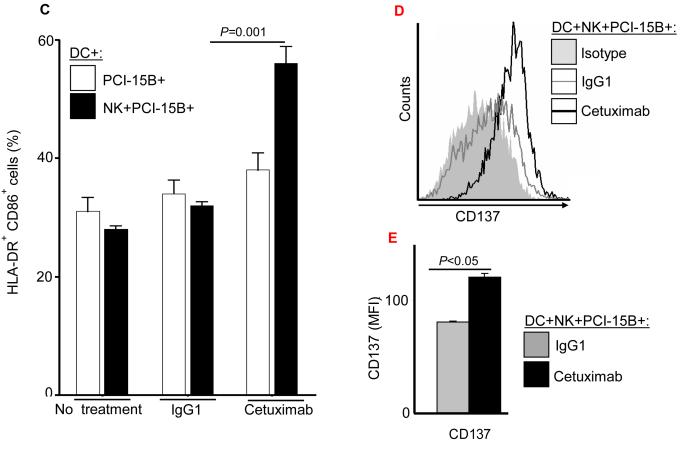
To determine whether enhanced TA cross-presentation by cetuximab was mediated through enhanced DC maturation (19), we analyzed the expression level of surface maturation markers by DC incubated with cetuximab, PCI-15B HNC cells and NK cells. FACS analysis of DC showed significant upregulation of HLA-DR, co-stimulatory molecule CD80,CD83,CD86 (Figure 3A, supplementary figure 2), and APM component TAP-1 (Figure 3B). The latter is highly associated with efficient DC cross-presentation (19). Similarly, increased HLA-DR+/CD86+ expression was observed on the cetuximab-activated, NK cell treated DC (Figure 3C). Enhanced frequencies of HLA-DR+CD83+ DC or HLA-DR+/CD86+ DC were also observed after culture with cetuximab-activated NK cells (supplementary figure 3 A-B). Interestingly, enhanced expression of CD137 by DC was also observed after culture with cetuximab-activated NK cells (Figure 3D-E) (22).
Figure 3.
Enhancement of DC maturation by cetuximab-activated NK cells. (A) Histogram analysis of the upregulation of maturation markers HLA-DR+/CD80+ on DC co-cultured with NK:PCI-15B in the presence of cetuximab. (B) Analysis of upregulation of maturation markers on DC co-cultured with NK:PCI-15B in the presence of cetuximab or panitumumab. Expression level of HLA-DR (represented in MFI, n=13 donors), CD80 (represented in % positive cells, n=8 donors) and TAP-1 (represented in MFI, n=4 donors) on DC co-cultured with NK:PCI-15B (1:1:1 ratio) with no treatment or panitumumab or cetuximab (each 10μg/ml, 48h) were measured. The fold change of DC marker expression were related to levels on control DC co-cultured with NK:PCI-15B alone are shown. A two-tailed unpaired t-test was performed for statistical analysis. (C) Percentages of DC (HLA-DR+ CD86+) that were co-cultured with PCI-15B (1:1 ratio) or with NK:PCI-15B (1:1:1 ratio) with no treatment or with IgG1 or cetuximab (each at 10μg/ml, 48h) were measured. Data are representative of three experiments from three different donors. Enhancement of CD137 expression on DC by cetuximab-activated NK cells. (D-E) Histogram analysis of the upregulation of CD137 expression on DC co-cultured with NK:PCI-15B in the presence of IgG1 or cetuximab (each 10μg/ml, 48h). (B) The quantitative bar diagram showing the significant differences in CD137 expression is also shown. A two-tailed unpaired t-test was performed for statistical analysis.
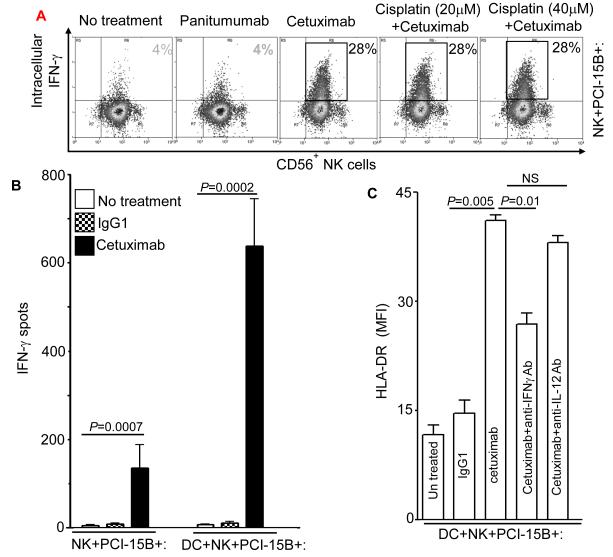
Although differences in lytic activity between NK cells expressing the FcγRIIIa VV genotype versus those expressing the FF genotype were reported (13), both VV and FF expressing, cetuximab-activated NK cells showed a similar effect on the expression of the DC maturation markers, CD80 and TAP-1 (unpublished data). Despite the lack of influence of codon 158 polymorphism on NK cell-induced DC maturation, the absolute dependence of FcγRIIIa binding by NK cells was evident, since the EGFR-specific mAb panitumumab, which binds very poorly to FcγRIIIa due to its IgG2 isotype (23), did not mediate downstream DC maturation (Figure 3B, supplementary figure 2) or NK cell activation (Figure 4A).
Figure 4.
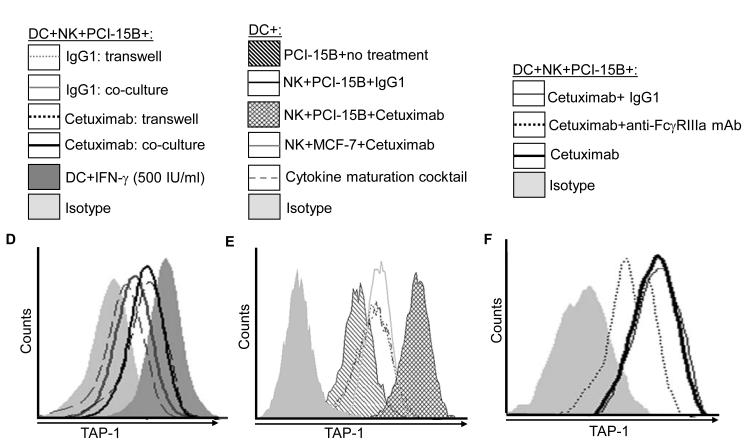
Enhancement by cetuximab of IFN-γ secretion by NK cells. (A) Flow cytometric analysis of intracellular IFN-γ in CD56+ NK cells, which were co-cultured with PCI-15B (1:1 ratio) with no treatment or with panitumumab or cetuximab (each 10μg/ml, 12h) in the presence or absence of cisplatin (20μm, 40μM). Representative figure of two independent experiments are shown. (B) Enhancement by cetuximab of IFN-γ secretion by NK-DC cross-talk in the presence of PCI-15B. The level of IFN-γ in the co-culture of NK:PCI-15B (1:1 ratio) or DC:NK:PCI-15B (1:1:1 ratio) with no treatment or with IgG1 or cetuximab (each at 10μg/ml) was measured after 12h of incubation by ELISPOT assay. Cumulative data of two donors are shown. A two-tailed unpaired t-test was performed for statistical analysis. (C) Importance of IFN-γ released by cetuximab activated NK cells in the enhancement of DC maturation. Percentages of HLA-DR+ DC in DC preparations co-cultured with NK:PCI-15B (at 1:1:1 ratio) with no treatment or with IgG1 or cetuximab (each at 10μg/ml, 48h) were measured by flow cytometry. In parallel, anti-IFN-γ mAb or anti-IL-12p40/70 mAb (each at 10μg/ml) were added along with cetuximab to the co-culture of DC:NK:PCI-15B, and DC maturation markers (HLA-DR+) were analyzed. Data are representative of two experiments from different donors. Enhancement by cetuximab of TAP-1 upregulation by DC in the presence of NK cells. (D-F) Enhancement of DC maturation by soluble factor released from cetuximab-activated NK cells. (D) TAP-1 upregulation in DC co-cultured with cetuximab-activated NK cell is mediated by a soluble factor. Flow cytometric analysis of intracellular TAP-1 expression (and TAP-2 expression, data not shown) was performed for DC incubated with NK:PCI-15B (1:1:1 ratio) with IgG1 or cetuximab (each at 10μg/ml, 48h). DC were physically separated from NK:PCI-15B co-culture using a transwell micropore system. TAP-1 expression in DC was enhanced after co-culture with NK:PCI-15B with cetuximab in both contact dependent and transwell culture conditions. Isotype (gray filled histogram), IgG1 transwell (gray dotted line), IgG1 co-culture (gray line), cetuximab transwell (black dotted line), cetuximab co-culture (black line), IFN-γ (black filled histogram). Data are representative of two experiments from two different donors. (E) Enhancement by cetuximab of TAP-1 upregulation in DC in the presence of NK cells with EGFRhigh HNC cells (PCI-15B), but not with breast cancer cells (MCF-7). By flow cytometry, intracellular TAP-1 expression (and TAP-2 expression, data not shown) was measured in DC that were incubated with NK:PCI-15B (at 1:1:1 ratio) or NK:MCF-7 (1:1:1 ratio) with IgG1 or cetuximab (each at 10μg/ml, 48h). No upregulation of TAP-1 was detected in DC that was incubated with NK:MCF-7 breast cancer cell cells (1:1:1 ratio) in the presence of cetuximab (each at 10μg/ml, 48h). As a control, the level of TAP-1 was measured in DC matured with cytokines (TNF-α, IL-1β, IL-6, PGE2 for 48h). (F) Enhancement by cetuximab of TAP-1 upregulation by DC in the presence of NK cells is FcγRIIIa-dependent. TAP-1 expression was measured in DC incubated with NK:PCI-15B (1:1:1 ratio) in the presence of IgG1 isotype or cetuximab (each at 10μg/ml, 48h). A blocking FcγRIIIa-specific mAb (3G8) or IgG1 isotype control were used to demonstrate dependence of the observed TAP-1 upregulation (in DC) on FcγR expressed by NK cells, but not FcγR expressed by DC. Isotype (filled histogram), cetuximab plus IgG1 (thin line), cetuximab plus anti-FcγRIIIa-specific mAb (dotted line), cetuximab (thick line).
Enhancement by cetuximab of IFN-γ secretion by NK cells
Next, we investigated the cytokine(s) secreted by cetuximab-activated helper NK cells which could be responsible for this DC maturation. Cetuximab treatment of NK cells in the presence of PCI-15B HNC cells was found to induce a high level of IFN-γ secretion by NK cells (Figure 4A). Moreover, cisplatin does not negatively affect cetuximab-induced activation and secretion of IFN-γ by NK cells (Figure 4A). Notably, cetuximab-activated, NK cell treated DC further stimulated these NK cells in a reciprocal fashion, leading to significantly increased secretion of IFN-γ by cetuximab-activated NK cells when incubated with DC. Control IgG1 did not activate NK cells (Figure 4B), consistent with the need for mAb binding of both EGFR and FcγRIIIa. Furthermore, the absence of cetuximab in the NK:DC co-culture (without HNC cells) or DC:PCI-15B co-culture (without NK cells) abrogated the IFN-γ secretion (supplementary figure 4,5). We then determined that DC maturation was mediated through IFN-γ, since an IFN-γ-specific neutralizing Ab blocked the induction of HLA-DR (Figure 4C) on DC by cetuximab-activated NK cells. Blockade of other cytokines, such as IL-12p40/70 showed a slight ability to promote HLA-DR expression on DC, however no significant differences were found on DC maturation (Figure 4C). Similarly, a notable effect of the blocking of IFN-γ was observed on the maturation markers HLA-DR, CD80, HLA-DR+/CD86+ or HLA-DR/CD83+ on DC by cetuximab-activated NK cells (supplementary figure 3 A,B). These changes in the expression of DC maturation markers are specific, since no significant increase in their expression level was detected on DC after incubation with autologous NK cells (supplementary figure 5), PCI-15B cells alone or isotype control IgG1 (supplementary figure 2-3).
Role of FcγRIIIa in the enhancement by cetuximab and NK cells of TAP-1 upregulation in DC
In the experiments described above (Figure 2D), we observed that DC cross-priming of EGFR-specific T cells was enhanced in the presence of cetuximab-activated NK cells. The latter cells had been previously shown to secrete high levels of Th1 cytokines and chemokines (13, 24). Because these cytokines strongly upregulate the expression of certain APM components, such as TAP-1 and TAP-2, which are crucial for TA derived peptide presentation to cognate CTL (19), we investigated whether these APM components were upregulated in DC incubated with cetuximab-activated NK cells. Consistent with this notion, the enhanced level of TAP-1 was observed in DC that were co-cultured with cetuximab-activated NK cells, even if they were physically separated from the NK cells:PCI-15B cells co-culture (in the presence of cetuximab) through a transwell membrane (Figure 4D). The increase in TAP-1 expression in cetuximab-activated, NK cell treated DC was found after incubation with PCI-15B HNC cells (EGFRhigh) but not with MCF-7 breast cancer cells. These results indicate that high EGFR expression on tumor cells is necessary for cetuximab-induced NK cell activation to induce DC maturation (Figure 4E). Also, the upregulation of TAP-1 in DC co-cultured with cetuximab-activated NK cells was abrogated by the blocking FcγRIIIa-specific mAb 3G8, but was not affected by a control isotype IgG1 (Figure 4F). Furthermore, incubation of NK cells with the EGFR-specific, IgG2 isotype mAb panitumumab which binds poorly to FcγRIIIa on NK cells (15, 23, 25), failed to upregulate the expression of co-stimulatory molecules on DC, in accordance with the lack of effect on EGFR cross-priming (supplementary figure 1,2,3). Thus, cetuximab-bound HNC cells activate NK cells through an FcγRIIIa-dependent mechanism, leading to APM component and costimulatory molecule upregulation in DC, manifested by their enhanced ability to mediate TA cross-presentation.
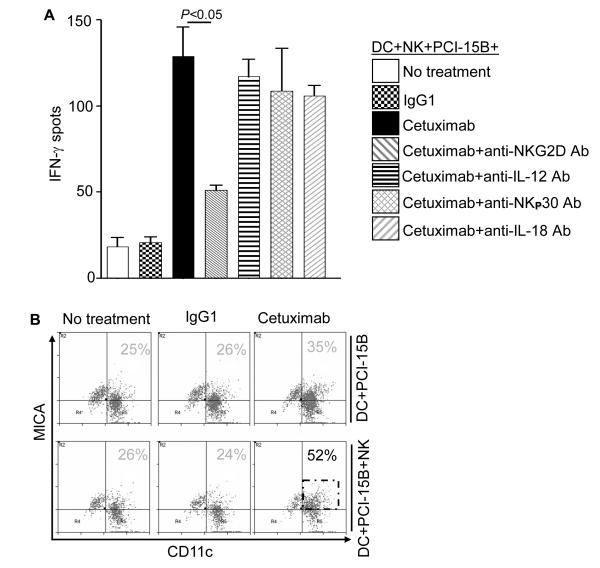
Enhancement by cetuximab of NK-DC cross-talk is NKG2D mediated
In light of the reciprocal activation of NK cells by DC (Figure 4B), we investigated the molecular mechanism underlying the interaction of cetuximab-activated NK cells cultured with mature DC. Specifically blocking mAb for known modulators of NK-DC cross-talk were used to disrupt NK:DC interactions. Interestingly, only a blocking NKG2D-specific mAb abrogated the reciprocal activation of cetuximab-activated NK cells and DC, leading to significantly reduced IFN-γ secretion (Figure 5A). Blockade of other important contributors to NK-DC cross-talk, including NKp30 expressed by NK cells, as well as IL-12 and IL-18, were not responsible for this effect. We observed upregulation of MHC class I chain-related protein A (MICA) on DC but not on HNC cells, suggesting a major role of NKG2D in the reciprocal NK cell activation mediated by cetuximab-induced DC maturation (Figure 5B).
Figure 5.
Enhancement by cetuximab of NK-DC cross-talk is NKG2D mediated. (A) Levels of IFN-γ were measured after co-culture of NK:PCI-15B (1:1 ratio) or DC:NK:PCI-15B (1:1:1 ratio) with no treatment or with IgG1 or cetuximab (each at 10μg/ml) after 24h by ELISPOT assay. Furthermore, the molecular mechanism that may modulate NK-DC cross-talk was assessed by using anti-NKG2D Ab or anti-IL-12 Ab or anti-NKp30 Ab or anti-IL-18 Ab along with cetuximab (each at 10μg/ml) in parallel assays. A two-tailed unpaired t-test was performed for statistical analysis. Cumulative data of two donors are shown. (B) Enhancement by cetuximab of NK-DC cross-talk is dependent on MICA upregulation by DC. The level of NKG2D ligand MICA, on CD11c+ DC in co-cultures of NK:PCI-15B (1:1 ratio) or DC:NK:PCI-15B (1:1:1 ratio) with no treatment or with IgG1 or cetuximab (each at 10μg/ml, 48h) was measured by flow cytometry. A representative histogram is also shown.
Enhancement by cetuximab of Th1 cytokines and chemokines in NK cell-DC co-cultures
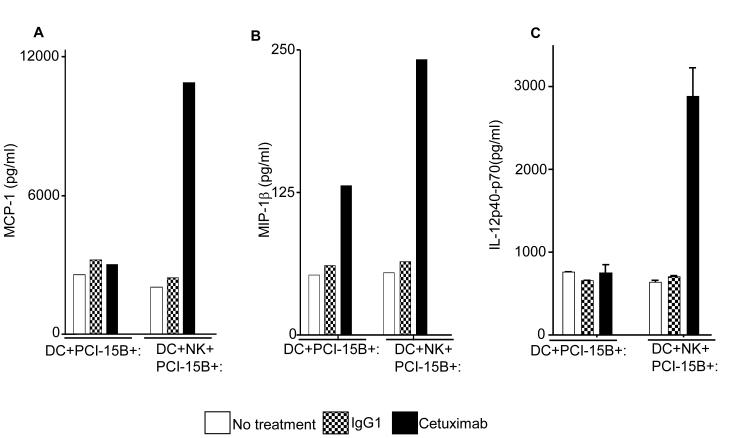
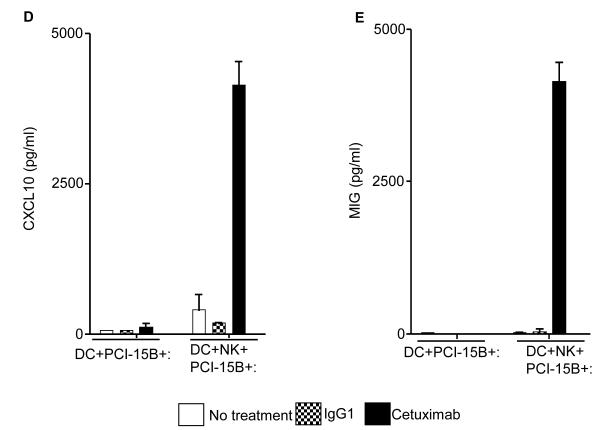
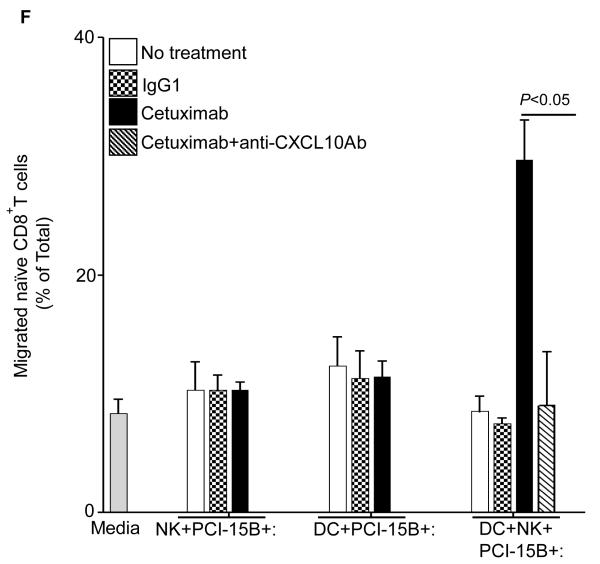
The consequences of NK cell-DC stimulation include functional and phenotypic activation, such as secretion of chemokines and cytokines. We performed multiplex ELISA (Luminex™) analysis to profile the cytokines and chemokines in the supernatants of co-culture (DC:PCI-15B or DC:NK:PCI-15B) untreated or treated with IgG1 or cetuximab. Secretion of the cytokines and chemokines, MCP-1, MIP-1β, IL-12p40/70, (CCR5 ligand), CXCL10 and MIG (CXCR3 ligands) was enhanced only in the cetuximab-activated NK cell-treated, DC (Figures 6A-E). Since cetuximab-activated, NK cell treated DC supernatants contained higher amounts of chemokines responsible for recruiting and activating CD8+ T cells, we confirmed the enhanced migratory ability of naïve CD8+ T cells under the influence of chemotactic factor/s present in supernatant from cetuximab-activated NK cell treated DC. The induced T cell migration was completely abrogated by a CXCL10-specific mAb (Figure 6F).
Figure 6.
Enhancement by cetuximab of Th1 polarizing cytokines in the NK:DC:PCI-15B co-culture. (A-C) Luminex™ analysis for the estimation of level of MCP-1, MIP-1β and IL-12 in the supernatant of DC:PCI-15B (1:1 ratio) or DC:NK:PCI-15B (1:1:1 ratio) co-culture with no treatment or with IgG1 or cetuximab (each at 10μg/ml for 48h). Data are representative of two different donors. (D and E) Enhancement by cetuximab of CXC chemokines in the NK-DC co-culture. The levels of CXC chemokines CXCL10 and MIG were determined in the supernatant of from co-culture of DC:PCI-15B (1:1 ratio) or DC:NK:PCI-15B (1:1:1 ratio) incubated with no treatment or with IgG1 or cetuximab (each at 10μg/ml for 48h). Values are mean ± SEM of two independent experiments from separate donors. (F) Enhancement by cetuximab of the migration of CD8+T cells under the influence of CXCL10 in the supernatant of NK:DC:PCI-15B co-cultures. Migration of CD8+ T cells under the influence of migratory factor(s) present in the fresh media or in the supernatant of NK:PCI-15B (at 1:1 ratio) co-culture, or in the supernatant of DC:PCI-15B (1:1 ratio) co-culture, or in the supernatant of DC:NK:PCI-15B co-culture (1:1:1 ratio), with no treatment or with IgG1 or cetuximab (each at 10μg/ml for 48h) was quantified. After 4h of incubation of CD8+ T cells with co-culture supernatants in the upper chamber of transwell plate, cells were collected from lower chamber and counted. Each condition was plated in triplicate. In parallel to the cetuximab condition anti-CXCL10 Ab (10 μg/ml) was used for the blocking experiments. Data shown are from a representative donor performed in triplicate. Two-tailed unpaired t-test was performed for statistical analysis.
Discussion
While blockade of proliferative and anti-apoptotic pathways in tumor cells plays a major role in the clinical efficacy of therapeutic mAb, such as rituximab, cetuximab and trastuzumab, the role of FcγRIIIa dependent innate immunity (ADCC) induced by these anti-tumor mAbs have generated conflicting findings (8, 9, 26-29). In addition, using murine models, the role of adaptive anti-tumor immune mechanisms in the clinical activity of these mAb has been suggested (30-31), though data from treated cancer patients is generally lacking. Our in vitro data showed that NK cells significantly enhance the anti-tumor effects of cetuximab via interaction with FcγRIIIa (13), which parallel similar results obtained with trastuzumab in breast carcinoma bearing mice (7, 32). Taken together, these results indicated that FcγRIIIa on NK cells could play a critical role in an immuno-naïve environment. However, this effect becomes less marked in ‘immunocompetent’ cancer patients, since we did not find a significant correlation between FcγRIIIa genotype and disease free survival in patients with HNC treated with cetuximab and radiotherapy and chemotherapy (Figure 1), as seen in other cancer patients (26-29). Nevertheless, the data are in agreement with the dominant role of FcγRIIIa on NK cells, although not absolutely dependent on the index of affinity of cetuximab, but rather to initiate proximity of FcγR bearing NK cells and DC with EGFR+ HNC cells and with each other, leading to NK-DC cross-talk mediated through NKG2D-MICA. NK-DC cross-talk is crucial to cetuximab-mediated DC maturation and cross-presentation, which may potentially lead to CTL expansion, and potentially clinical effects, in vivo in cetuximab treated HNC patients. While the sample size of this retrospective analysis was insufficient to correlate T cell frequencies or phenotype with clinical outcome, such a study is ongoing as part of a separate, prospectively treated cohort. Nevertheless, higher frequency of EGFR-specific T cells was also found in prospectively treated HNC patients who received cetuximab alone (without chemotherapy), strongly supporting our conclusions (Figure 2B,C).
Interestingly, that adaptive immune responses might contribute to therapeutic efficacy, based on higher frequency of EGFR-specific CD8+ T cells, must be validated prospectively in a larger cohort cetuximab treated patients (Figure 2) with sufficient follow up (2-3 years) for outcome measurements. This extends recent reports of rituximab and anti-HER2/neu inducing long lasting anti-tumor adaptive immune responses in murine systems, as support the plausible role of adaptive anti-tumor immune responses triggered by anti-tumor specific mAb in treated patients (30, 31, 33). The dependence on IFN-γ secretion by NK cells supported that NK cells provide helper function in DC cross-presentation, extending our previous finding that an immunogenic EGFR-encoded CTL could be generated by pulsing DC with EGFR853-861 peptide that could stimulate cognate CTL in vitro (14). However, total recombinant EGFR protein was not used in this study as a source of TA since this would not mimic FcγRIIIa mediated effects and NK:DC cross talk that cetuximab stimulates.
Although FcγRIIIa polymorphism dependent survival of cancer patients were reported in CRC cohorts (8, 9), adaptive immune responses probably contribute in the cetuximab mediated immune responses, and may better predict clinical outcome of immunotherapy, because NK cells also provide helper function in shaping host adaptive immune responses. Moreover, the low frequency of VV patients often ‘negatively’ influences statistical analysis (26, 29). These data clearly support the paradigm that anti-tumor mAb participate in the long term control of tumor in HNC patients, probably accounting for the clinical activity (30, 31, 33-35).
To analyze the importance of FcγRIIIa, we used the two FDA-approved, EGFR-specific mAb cetuximab and panitumumab which bind the same epitope on EGFR (23, 25), but differ in their isotype and FcγRIIIa affinity, to show that only cetuximab (IgG1) induced NK-DC crosstalk and DC cross-presentation ability, whereas panitumumab (IgG2) does not, despite their equal EGFR binding (Figure 3B, supplementary figure 1,2). It is notable that this IgG2 isotype mAb, despite demonstrated potency in EGFR signaling blockade, has failed multiple prospective HNC trials of clinical efficacy, supporting the proposition that cetuximab imparts greater clinical activity due to the IgG1 isotype-induced immunologic effects.
We have recently shown that cetuximab-naïve HNC patients possess elevated levels of EGFR853-861-specific T cells confirming that EGFR expressed on HNC cells induces a specific immune response in vivo (36). Although antigen-specific CD4+ T cell activation has been noted in a high proportion of breast cancer patients treated with trastuzumab and correlated with clinical responses (7), this is the first demonstration of enhanced TA specific CTL in mAb treated patients (Figure 2), which are crucial to mediating strong anti-tumor immunity in vivo. This effect provides a general yet novel immune mechanism of enhanced anti-tumor activity relevant to clinical response, as well as a potential biomarker to monitor cetuximab effects in mAb-treated cancer patients. Since the frequency of HLA-A2.1 in HNC patients in different regions varies (ranging from 30-50% worldwide), additional HLA-restricted, EGFR-specific T cell epitopes should be identified to expand application of such a biomarker.
Our results indicate that cetuximab:EGFR+ tumor cell complexes originating in the presence of NK cells are provided to DC to generate polyclonal TA (MAGE-3 as well as EGFR) processing and presentation (36-38). In addition, activated-NK cells facilitate DC maturation, as indicated by the concomitant up-regulation of the co-stimulatory molecules CD80, CD86, and CD137 on the DC surface (Figure 3A-E, supplementary figure 2-3). At the same time, cytokines released by NK cells activated through binding of EGFR-cetuximab complexes via FcγRIIIa induce activation markers and TAP-1 (39-42) (Figure 3B, Figure 4A-F), which we have shown to predict cross-priming capacity of DC (19). Our data shows that with the release of cytokines from cetuximab-activated NK cells DC becomes highly responsive to the tumor microenvironment and undergoes maturation along with uptake of dead tumor materials (37). This finding is in strong accordance with the established fact (24, 37) that release of cytokines from cetuximab-activated NK cells may trigger Th1 responses against tumor microenvironment. The ability of activated/mature DC to generate or activate EGFR-specific CTL is an attractive opportunity to subvert immunosuppressive environment, which could be easily achieved with anti-tumor mAb administrations (43). Through this mechanism, therapeutic mAb may enhance antigen cross-presentation by DC to T cells, resulting in augmentation of TA-specific CTL generation (43-45). If this interpretation is correct, cetuximab administration should be probably combined with vaccines targeting EGFR (14) or other TA, as well as adjuvant(s) to specifically stimulate cellular immunity.
In addition to their ability to mediate ADCC, cetuximab-activated NK cells, secrete cytokines, such as IFN-γ MCP-1, MIP-1β that inhibit tumor cell proliferation, enhance antigen presentation, and chemokines such as IP10 and MIG that aid in the chemotaxis of T cells (13, 39). The resulting potent activating bi-directional signaling between NK cells and DC can shape both the innate immune response within inflamed peripheral tissues and the adaptive immune response in secondary lymphoid organs (46) enhancing cross-presentation and priming of T cells, as we observed (Figure 2 and 3). Through direct cellular interactions and secretion of cytokines/chemokines NK cells function as helper cells (40, 47, 48), and enhance DC cross-presentation and T cell induction, with the potential to strengthen TA-specific cellular immune response and to spread this priming to multiple TA, including private antigens. The effects observed can be blocked using a neutralizing IFN-γ mAb consistent with our previous findings regarding optimal APM upregulation in mature DC mediating cross-presentation (19). However, the beginning of DC maturation is clearly dependent on secreted IFN-γ from NK cells whereas IL-12 might contribute for the cross-priming activities and reciprocal NK cell activation (Figure 4B-C, 5 A-B). We have also observed that NKG2D, a potent tumor recognition molecule on NK cells may interplay in shaping the NK-DC cross-talk and reciprocal activities, subsequently NKG2D can also interact with its ligand expressed on tumor cells and or activated DC (Figure 5).
However, the generation and functional activation of NK cells and CTL for elimination of tumor cells might be influenced by the impact of concomitant chemotherapy and/or radiotherapy (8, 22), often administered in conjunction with cetuximab (49). We observed that cetuximab-induced IFN-γ secretion by NK cells activation is not negatively influenced by cisplatin (Figure 4A). Recently, trastuzumab emtansine (T-DM1) an Ab-drug conjugate showed a significantly prolonged progression-free and overall survival in breast cancer patients (10). The induction of T cells might be more relevant and efficacy even further enhanced, when mAb-conjugated cytotoxics can induce immunogenic cell death. Currently we are investigating the combination of cetuximab with NK cell activating molecules and with cytotoxic drugs to further enhance the adaptive immune responses in patients and functional properties of HLA class I or APM components as potential immune escape mechanisms, especially in HNC cells (50). These mechanisms and variables are now being studied in prospective clinical trials using single agent cetuximab, as well as cetuximab in combination with chemoradiotherapy, since in both situations clinical efficacy is observed.
Supplementary Material
Translational Relevance.
The anti-epidermal growth factor receptor (EGFR) mAb cetuximab may act through blocking oncogenic signals and by inducing Fcγ receptor (FcγR) mediated antibody dependent cellular cytotoxicity (ADCC). We show that polymorphism of FcγRIIIa (expressed by NK cells) does not correlate with clinical outcome of cetuximab-treated treated head and neck cancer (HNC) patients. Interestingly, however, cetuximab induces natural killer (NK) cell-dendritic cell (DC) cross-talk, which promotes DC maturation and CD8+ T cell priming, leading to tumor antigen spreading and Th1 cytokine release. The identification of EGFR-specific T cells in cetuximab treated HNC patients now permits a biomarker of response to be correlated with outcome in prospectively collected specimens. The implications of this finding include potential combinatorial vaccines with EGFR-specific mAbs, or in combination with immune checkpoint inhibitors to provide sustained CD8+ T activity after their induction by cetuximab-activated NK-DC priming.
Acknowledgments
Jennifer Ridge-Hetrick, assisted in extracting data from the head and neck cancer database at the University of Pittsburgh Cancer Registry. We acknowledge the core University of Pittsburgh Cancer Institute flow cytometry and Luminex facilities. We appreictae the efforts of Drs. Ethan Argiris, Michael Gibson, and James Ohr for trial coordination and caring for cetuximab treated patients studied here. We thank Ferris lab members for helpful suggestions.
Grant Support This work was supported by National Institute of Health grants R01 DE19727, P50 CA097190, CA110249 and University of Pittsburgh Cancer Institute grant P30CA047904.
Footnotes
Authors’ Contributions Conception and design: R.M. Srivastava, S.C. Lee, S. Ferrone, R.L. Ferris
Acquisition of data: R.M. Srivastava, S.C. Lee, P.A. Andrade-Filho, C.A. Lord, Hyun-bae Jie, H.C. Davidson, A. Lopez-Albaitero, S.P. Gibson
Analysis and or interpretation of data: R.M. Srivastava, S.C. Lee, Hyun-bae Jie, W.E. Gooding, S. Ferrone, R.L. Ferris
Writing and editing of manuscript: R.M. Srivastava, S.C. Lee, S. Ferrone, R.L. Ferris
Study supervision: R.L. Ferris
Conflict of interest: The authors disclose no potential conflicts of interest.
References
- 1.Ferris RL, Jaffee EM, Ferrone S. Tumor antigen-targeted, monoclonal antibody-based immunotherapy: clinical response, cellular immunity, and immunoescape. J Clin Oncol. 2010;28:4390–9. doi: 10.1200/JCO.2009.27.6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371:1695–709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim S, Grandis JR, Rinaldo A, Takes RP, Ferlito A. Emerging perspectives in epidermal growth factor receptor targeting in head and neck cancer. Head Neck. 2008;30:667–74. doi: 10.1002/hed.20859. [DOI] [PubMed] [Google Scholar]
- 4.Weng WK, Levy R. Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J Clin Oncol. 2003;21:3940–7. doi: 10.1200/JCO.2003.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Weng WK, Czerwinski D, Timmerman J, Hsu FJ, Levy R. Clinical outcome of lymphoma patients after idiotype vaccination is correlated with humoral immune response and immunoglobulin G Fc receptor genotype. J Clin Oncol. 2004;22:4717–24. doi: 10.1200/JCO.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Taylor C, Hershman D, Shah N, Suciu-Foca N, Petrylak DP, Taub R, et al. Augmented HER-2 specific immunity during treatment with trastuzumab and chemotherapy. Clin Cancer Res. 2007;13:5133–43. doi: 10.1158/1078-0432.CCR-07-0507. [DOI] [PubMed] [Google Scholar]
- 7.Varchetta S, Gibelli N, Oliviero B, Nardini E, Gennari R, Gatti G, et al. Elements related to heterogeneity of antibody-dependent cell cytotoxicity in patients under trastuzumab therapy for primary operable breast cancer overexpressing Her2. Cancer Res. 2007;67:11991–9. doi: 10.1158/0008-5472.CAN-07-2068. [DOI] [PubMed] [Google Scholar]
- 8.Bibeau F, Lopez-Crapez E, Di Fiore F, Thezenas S, Ychou M, Blanchard F, et al. Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol. 2009;27:1122–9. doi: 10.1200/JCO.2008.18.0463. [DOI] [PubMed] [Google Scholar]
- 9.Zhang W, Gordon M, Schultheis AM, Yang DY, Nagashima F, Azuma M, et al. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J Clin Oncol. 2007;25:3712–8. doi: 10.1200/JCO.2006.08.8021. [DOI] [PubMed] [Google Scholar]
- 10.Verma S, Miles D, Gianni L, Krop IE, Welslau M, Baselga J, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367:1783–91. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim PS, Armstrong TD, Song H, Wolpoe ME, Weiss V, Manning EA, et al. Antibody association with HER-2/neu-targeted vaccine enhances CD8 T cell responses in mice through Fc-mediated activation of DCs. J Clin Invest. 2008;118:1700–11. doi: 10.1172/JCI34333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Disis ML, Wallace DR, Gooley TA, Dang Y, Slota M, Lu H, et al. Concurrent trastuzumab and HER2/neu-specific vaccination in patients with metastatic breast cancer. J Clin Oncol. 2009;27:4685–92. doi: 10.1200/JCO.2008.20.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Albaitero A, Lee SC, Morgan S, Grandis JR, Gooding WE, Ferrone S, et al. Role of polymorphic Fc gamma receptor IIIa and EGFR expression level in cetuximab mediated, NK cell dependent in vitro cytotoxicity of head and neck squamous cell carcinoma cells. Cancer Immunol Immunother. 2009;58:1853–64. doi: 10.1007/s00262-009-0697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrade Filho PA, Lopez-Albaitero A, Gooding W, Ferris RL. Novel immunogenic HLA-A*0201-restricted epidermal growth factor receptor-specific T-cell epitope in head and neck cancer patients. J Immunother. 2010;33:83–91. doi: 10.1097/CJI.0b013e3181b8f421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez-Albaitero A, Ferris RL. Immune activation by epidermal growth factor receptor specific monoclonal antibody therapy for head and neck cancer. Arch Otolaryngol Head Neck Surg. 2007;133:1277–81. doi: 10.1001/archotol.133.12.1277. [DOI] [PubMed] [Google Scholar]
- 16.Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2007;29:163–88. doi: 10.1002/hed.20478. [DOI] [PubMed] [Google Scholar]
- 17.Argiris A, Heron DE, Smith RP, Kim S, Gibson MK, Lai SY, et al. Induction docetaxel, cisplatin, and cetuximab followed by concurrent radiotherapy, cisplatin, and cetuximab and maintenance cetuximab in patients with locally advanced head and neck cancer. J Clin Oncol. 2010;28(36):5294–300. doi: 10.1200/JCO.2010.30.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Argiris A, Karamouzis MV, Smith R, Kotsakis A, Gibson MK, Lai SY, et al. Phase I trial of pemetrexed in combination with cetuximab and concurrent radiotherapy in patients with head and neck cancer. Ann Oncol. 2011;22(11):2482–8. doi: 10.1093/annonc/mdr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Albaitero A, Mailliard R, Hackman T, Andrade Filho PA, Wang X, Gooding W, et al. Maturation pathways of dendritic cells determine TAP1 and TAP2 levels and cross-presenting function. J Immunother. 2009;32:465–73. doi: 10.1097/CJI.0b013e3181a1c24e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Filho PA, Lopez-Albaitero A, Xi L, Gooding W, Godfrey T, Ferris RL. Quantitative expression and immunogenicity of MAGE-3 and -6 in upper aerodigestive tract cancer. Int J Cancer. 2009;125:1912–20. doi: 10.1002/ijc.24590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 22.Kohrt HE, Houot R, Weiskopf K, Goldstein M, Lund P, Scheeren F, et al. Effect of stimulation of natural killer cells with an anti-CD137 mAb on the efficacy of trastuzumab, cetuximab, and rituximab. J Clin Oncol. 2012;30 (suppl; abstr 2514) [Google Scholar]
- 23.Patel D, Guo X, Ng S, Melchior M, Balderes P, Burtrum D, et al. IgG isotype, glycosylation, and EGFR expression determine the induction of antibody-dependent cellular cytotoxicity in vitro by cetuximab. Hum Antibodies. 2010;19:89–99. doi: 10.3233/HAB-2010-0232. [DOI] [PubMed] [Google Scholar]
- 24.Roda JM, Joshi T, Butchar JP, McAlees JW, Lehman A, Tridandapani S, et al. The activation of natural killer cell effector functions by cetuximab-coated, epidermal growth factor receptor positive tumor cells is enhanced by cytokines. Clin Cancer Res. 2007;13:6419–28. doi: 10.1158/1078-0432.CCR-07-0865. [DOI] [PubMed] [Google Scholar]
- 25.Schneider-Merck T, Lammerts van Bueren JJ, Berger S, Rossen K, van Berkel PH, Derer S, et al. Human IgG2 antibodies against epidermal growth factor receptor effectively trigger antibody-dependent cellular cytotoxicity but, in contrast to IgG1, only by cells of myeloid lineage. J Immunol. 2010;184:512–20. doi: 10.4049/jimmunol.0900847. [DOI] [PubMed] [Google Scholar]
- 26.Pennell NM, Bhanji T, Zhang L, Seth A, Sawka CA, Berinstein NL. Lack of prognostic value of FCGR3A-V158F polymorphism in non-Hodgkin’s lymphoma. Haematologica. 2008;93:1265–7. doi: 10.3324/haematol.12638. [DOI] [PubMed] [Google Scholar]
- 27.Farag SS, Flinn IW, Modali R, Lehman TA, Young D, Byrd JC. Fc gamma RIIIa and Fc gamma RIIa polymorphisms do not predict response to rituximab in B-cell chronic lymphocytic leukemia. Blood. 2004;103:1472–4. doi: 10.1182/blood-2003-07-2548. [DOI] [PubMed] [Google Scholar]
- 28.Lin TS, Flinn IW, Modali R, Lehman TA, Webb J, Waymer S, et al. FCGR3A and FCGR2A polymorphisms may not correlate with response to alemtuzumab in chronic lymphocytic leukemia. Blood. 2005;105:289–91. doi: 10.1182/blood-2004-02-0651. [DOI] [PubMed] [Google Scholar]
- 29.Carlotti E, Palumbo GA, Oldani E, Tibullo D, Salmoiraghi S, Rossi A, et al. FcgammaRIIIA and FcgammaRIIA polymorphisms do not predict clinical outcome of follicular non-Hodgkin’s lymphoma patients treated with sequential CHOP and rituximab. Haematologica. 2007;92:1127–30. doi: 10.3324/haematol.11288. [DOI] [PubMed] [Google Scholar]
- 30.Park S, Jiang Z, Mortenson ED, Deng L, Radkevich-Brown O, Yang X, et al. The therapeutic effect of anti-HER2/neu antibody depends on both innate and adaptive immunity. Cancer Cell. 2010;18:160–70. doi: 10.1016/j.ccr.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abes R, Gelize E, Fridman WH, Teillaud JL. Long-lasting antitumor protection by anti-CD20 antibody through cellular immune response. Blood. 2010;116:926–34. doi: 10.1182/blood-2009-10-248609. [DOI] [PubMed] [Google Scholar]
- 32.Musolino A, Naldi N, Bortesi B, Pezzuolo D, Capelletti M, Missale G, et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol. 2008;26:1789–96. doi: 10.1200/JCO.2007.14.8957. [DOI] [PubMed] [Google Scholar]
- 33.Alduaij W, Illidge TM. The future of anti-CD20 monoclonal antibodies: are we making progress? Blood. 2011;117:2993–3001. doi: 10.1182/blood-2010-07-298356. [DOI] [PubMed] [Google Scholar]
- 34.Lee SC, Lopez-Albaitero A, Ferris RL. Immunotherapy of head and neck cancer using tumor antigen-specific monoclonal antibodies. Curr Oncol Rep. 2009;11:156–62. doi: 10.1007/s11912-009-0023-5. [DOI] [PubMed] [Google Scholar]
- 35.Sirianni N, Ha PK, Oelke M, Califano J, Gooding W, Westra W, et al. Effect of human papillomavirus-16 infection on CD8+ T-cell recognition of a wild-type sequence p53264-272 peptide in patients with squamous cell carcinoma of the head and neck. Clin Cancer Res. 2004;10:6929–37. doi: 10.1158/1078-0432.CCR-04-0672. [DOI] [PubMed] [Google Scholar]
- 36.Schuler PJ, Boeckers P, Engers R, Boelke E, Bas M, Greve J, et al. EGFR-specific T cell frequencies correlate with EGFR expression in head and neck squamous cell carcinoma. J Transl Med. 2011;9:168. doi: 10.1186/1479-5876-9-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nouri-Shirazi M, Banchereau J, Bell D, Burkeholder S, Kraus ET, Davoust J, et al. Dendritic cells capture killed tumor cells and present their antigens to elicit tumor-specific immune responses. J Immunol. 2000;165:3797–803. doi: 10.4049/jimmunol.165.7.3797. [DOI] [PubMed] [Google Scholar]
- 38.Rafiq K, Bergtold A, Clynes R. Immune complex-mediated antigen presentation induces tumor immunity. J Clin Invest. 2002;110:71–9. doi: 10.1172/JCI15640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roda JM, Parihar R, Magro C, Nuovo GJ, Tridandapani S, Carson WE., 3rd Natural killer cells produce T cell-recruiting chemokines in response to antibody-coated tumor cells. Cancer Res. 2006;66:517–26. doi: 10.1158/0008-5472.CAN-05-2429. [DOI] [PubMed] [Google Scholar]
- 40.Parihar R, Dierksheide J, Hu Y, Carson WE. IL-12 enhances the natural killer cell cytokine response to Ab-coated tumor cells. J Clin Invest. 2002;110:983–92. doi: 10.1172/JCI15950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalinski P, Giermasz A, Nakamura Y, Basse P, Storkus WJ, Kirkwood JM, et al. Helper role of NK cells during the induction of anticancer responses by dendritic cells. Mol Immunol. 2005;42:535–9. doi: 10.1016/j.molimm.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 42.Vieira PL, de Jong EC, Wierenga EA, Kapsenberg ML, Kalinski P. Development of Th1-inducing capacity in myeloid dendritic cells requires environmental instruction. J Immunol. 2000;164:4507–12. doi: 10.4049/jimmunol.164.9.4507. [DOI] [PubMed] [Google Scholar]
- 43.Jacobs B, Ullrich E. The Interaction of NK Cells and Dendritic Cells in the Tumor Environment: How to Enforce NK Cell & DC Action Under Immunosuppressive Conditions? Curr Med Chem. 2012;19:1771–9. doi: 10.2174/092986712800099857. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen-Pham TN, Yang DH, Nguyen TA, Lim MS, Hong CY, Kim MH, et al. Optimal culture conditions for the generation of natural killer cell-induced dendritic cells for cancer immunotherapy. Cell Mol Immunol. 2012;9:45–53. doi: 10.1038/cmi.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wehner R, Dietze K, Bachmann M, Schmitz M. The bidirectional crosstalk between human dendritic cells and natural killer cells. J Innate Immun. 2011;3:258–63. doi: 10.1159/000323923. [DOI] [PubMed] [Google Scholar]
- 46.Walzer T, Dalod M, Robbins SH, Zitvogel L, Vivier E. Natural-killer cells and dendritic cells: “l’union fait la force”. Blood. 2005;106:2252–8. doi: 10.1182/blood-2005-03-1154. [DOI] [PubMed] [Google Scholar]
- 47.Mailliard RB, Alber SM, Shen H, Watkins SC, Kirkwood JM, Herberman RB, et al. IL-18-induced CD83+CCR7+ NK helper cells. J Exp Med. 2005;202:941–53. doi: 10.1084/jem.20050128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mailliard RB, Son YI, Redlinger R, Coates PT, Giermasz A, Morel PA, et al. Dendritic cells mediate NK cell help for Th1 and CTL responses: two-signal requirement for the induction of NK cell helper function. J Immunol. 2003;171:2366–73. doi: 10.4049/jimmunol.171.5.2366. [DOI] [PubMed] [Google Scholar]
- 49.Vanneman M, Dranoff G. Combining immunotherapy and targeted therapies in cancer treatment. Nat Rev Cancer. 2012;12:237–51. doi: 10.1038/nrc3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ferris RL, Whiteside TL, Ferrone S. Immune escape associated with functional defects in antigen-processing machinery in head and neck cancer. Clin Cancer Res. 2006;12:3890–5. doi: 10.1158/1078-0432.CCR-05-2750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.