Summary
Clear cell sarcoma (CCS) of tendons and aponeuroses is a deadly soft-tissue malignancy resembling melanoma, with a predilection for young adults. EWS-ATF1, the fusion product of a balanced chromosomal translocation between chromosomes 22 and 12, is considered the definitional feature of the tumor. Conditional expression of the EWS-ATF1 human cDNA in the mouse generates CCS-like tumors with 100 percent penetrance. Tumors, developed through varied means of initiating expression of the fusion oncogene, model human CCS morphologically, immunohistochemically, and by genome-wide expression profiling. We also demonstrate that while fusion oncogene expression in later stages of differentiation can transform mesenchymal progenitor cells and generate tumors resembling CCS generally, expression in cells retaining stem cell markers permits the full melanoma-related phenotype.
Introduction
Clear cell sarcoma (CCS) is a soft tissue neoplasm classically arising in the extremities of young adults near tendons and aponeuroses. Despite their often small size, these tumors have high rates of recurrence and metastasis following standard local therapy, portending a poor general prognosis. CCS was first identified in 1965, then termed “Malignant Melanoma of the Soft Parts” due to its histologic appearance fitting with metastatic melanoma (Enzinger, 1965). In addition, CCS was later found to demonstrate melanocytic differentiation markers including immunohistochemical positivity for M-MITF, S100B, MelanA and HMB45 (Granter et al., 2001; Hocar et al., 2012). Until recently, the only means of differentiating CCS from the soft-tissue metastasis of a distant melanoma was its clinical history (i.e. confirmed absence of any cutaneous melanomas).
In the last decade, recognition of the characteristic t(12;22) (q13;q12) chromosomal translocation and its resultant fusion oncogene EWSR1-ATF1 (EWS-ATF1) has provided a means of defining CCS and distinguishing it from melanoma (Antonescu et al., 2002). Traditional cytogenetics, fluorescent in situ hybridization (FISH), and reverse-transcriptase polymerase chain reaction (RT-PCR) have all proven to be diagnostic tools capable of identifying this defining molecular feature of CCS (Wang et al., 2009). The type 1 fusion of EWS-ATF1, which includes exons one through eight of EWSR1 and exons four through seven of ATF1, is the most common variant of the described translocation products (Wang et al., 2009).
The EWS-ATF1 fusion protein contains the amino-terminal transcriptional activation domain of the EWSR1 protein joined to the bZIP DNA binding/dimerization domain of activating transcription factor 1 (ATF1). In the fusion protein, the EWSR1 activation domain replaces a protein kinase A phosphoacceptor site that renders endogenous ATF1 activity dependent on the presence of cyclic adenosine monophosphate (Fujimura et al., 2001). As a consequence, EWS-ATF1 is thought to be a constitutive activator of ATF1-regulated genes. A small minority of CCS cases harbor a t(2;22) (q32;q12) translocation and the alternate CREB1 fusion partner replacing ATF1, with which CREB1 shares binding of an identical consensus sequence (Wang et al., 2009).
The cell, or cells, of origin for CCS are not known. Traditionally, CCS tumors were characteristically identified near the tendons and aponeuroses of young adults, but have also recently been identified in the gastrointestinal tract (Covinsky et al., 2005; D'Amico et al., 2011; Lyle et al., 2008) and dermis (Falconieri et al., 2012; Hantschke et al., 2010). The expression of melanocyte-specific markers in CCS tumors has been variably attributed to the effects of the cell of origin or the effects of aberrant M-Mitf expression, shown to be driven by EWS-ATF1 in CCS cell lines (Davis et al., 2006). Using EWS-ATF1 as a diagnostic marker led to the identification of CCSs that do not express melanocytic markers. Further, some histologically distinct neoplasms have also been associated with this fusion oncogene, including angiomatoid fibrous histiocytoma (Somers et al., 2005) and hyalinizing clear cell carcinoma of the salivary gland (Antonescu et al., 2011). The latter tumors do not express M-Mitf or other melanocytic markers.
To investigate the role that EWS-ATF1 plays in clear cell sarcomagenesis and in tumorigenesis more broadly, we developed a mouse model that expresses the human EWS-ATF1 fusion oncogene complementary DNA (cDNA) in a conditional fashion.
Results
Generation of a targeted mouse line conditionally expressing the EWS-ATF1 oncogene
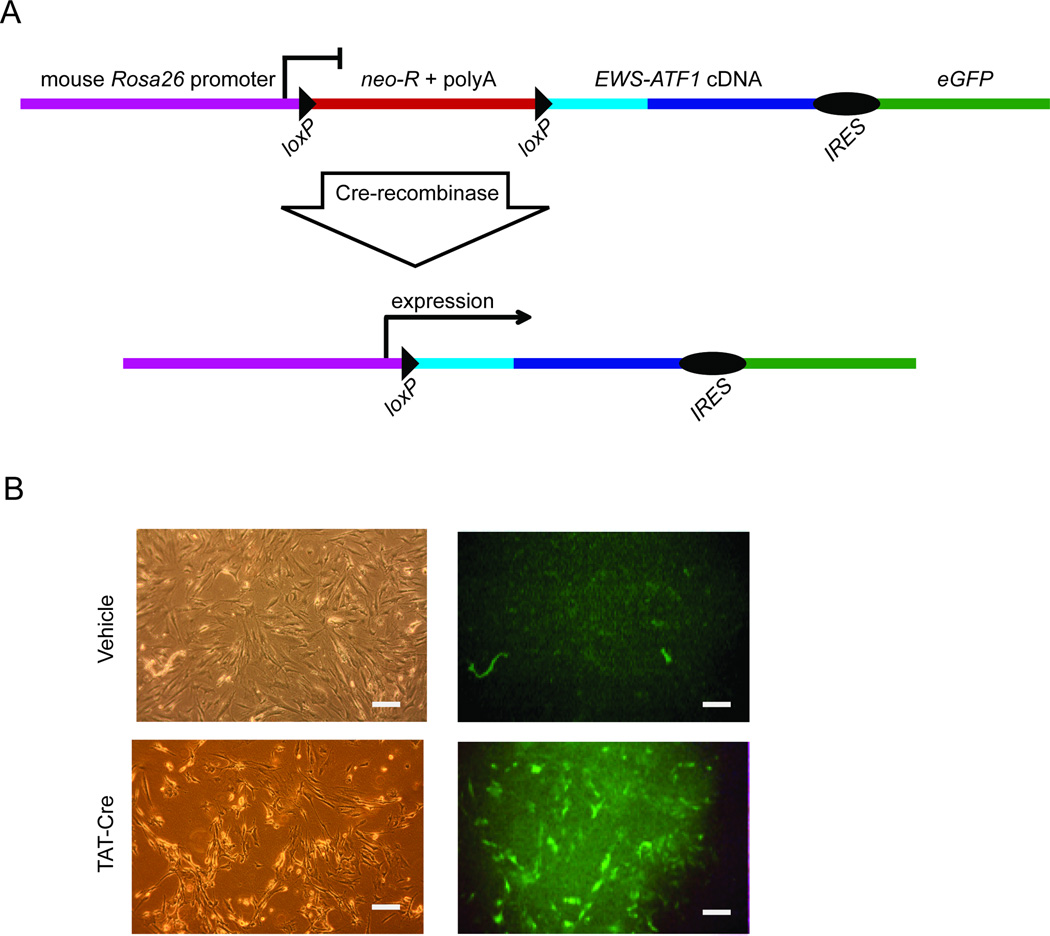
To generate the EWS-ATF1 cDNA, total RNA was isolated from human CCS tumors, reverse transcribed, and screened by PCR to identify a type 1 EWS-ATF1 fusion product. The integrity of the EWS-ATF1 cDNA was confirmed by DNA sequencing. The EWS-ATF1 cDNA was targeted to the ubiquitously expressed Rosa26 locus (Mao et al., 1999). Linked to the EWS-ATF1 cDNA via an internal ribosomal entry site (IRES) was the sequence encoding an enhanced green fluorescent protein (eGFP). To prevent transcription of the fusion gene and eGFP from the Rosa26 promoter, a neomycin resistance cassette and poly-adenylation stop signal flanked by loxP sites was inserted between the promoter and the EWS-ATF1-IRES-eGFP sequence (Figure 1A). In the absence of Cre, neither the fusion gene product nor eGFP should be expressed. Temporal, spatial, and tissue-specific control of Cre presence is possible through a variety of techniques for its genetic or protein delivery. Mouse embryonic stem cells confirmed to bear the targeted allele were injected into blastocysts to generate chimeras which were then bred to generate progeny with a germline-transmissible conditional allele of EWS-ATF1 (designated Rosa26EA1)
Figure 1. Conditional expression of the human clear cell sarcoma fusion oncogene in the mouse.
(A) Schematic showing the cDNA of the type I variant of EWS-ATF1 isolated from a human tumor and cloned into a vector designed for targeting into the mouse Rosa26 locus. neo-R: neomycin resistance cassette. polyA: polyadenylation stop sequence. IRES : internal ribosome entry site. eGFP: enhanced green fluorescent protein. Cre-mediated recombination excises the stop sequence and initiates expression of the fusion oncogene and eGFP. (B) Embryonic day 14.5 fibroblasts isolated from Rosa26EA1 heterozygous mice were exposed to TAT-Cre protein or vehicle control and images were collected 24 hr later. The left two panels show light images of fibroblasts while the right two panels show the GFP fluorescence. All Scale bars are 50 µm in length.
To confirm inducibility of the EWS-ATF1 fusion gene and eGFP by Cre, embryonic fibroblasts were isolated from E14.5 mouse embryos heterozygous for the Rosa26EA1 allele and exposed in culture to purified TAT-Cre protein or vehicle buffer control. TAT-Cre is an engineered Cre protein containing a short peptide sequence derived from the human immunodeficiency virus that mediates efficient endocytic uptake and nuclear localization of the protein (Joshi et al., 2002). Recombination efficiency in vitro exceeds 95 percent as reported previously (Haldar et al., 2009). Without exposure to TAT-Cre, mouse embryonic fibroblasts heterozygous for Rosa26EA1 demonstrated no green fluorescence. 24 hr after exposure to TAT-Cre (5 µM), cells began to express eGFP (Figure 1B), the percentage of fluorescing cells increased thereafter. The expression of other sarcoma fusion oncogenes from the Rosa26 locus has proven lethal for mouse embryonic fibroblasts (Haldar et al., 2007). Surprisingly, EWS-ATF1-expressing fibroblasts survived beyond the expected crisis and senescence of control fibroblasts that carried the Rosa26EA1 allele but were exposed to vehicle rather than TAT-Cre. Expression of EWS-ATF1 remained strong in the embryonic fibroblasts activated by TAT-Cre, even following long-term passage (data not shown).
Generation of tumors by in vivo exposure to TAT-Cre
Because expression of EWS-ATF1 was so well tolerated in vitro, we investigated whether the Rosa26EA1 allele might be activated in vivo first by injecting TAT-Cre into mice heterozygous for the Rosa26YFP allele. Mice receiving TAT-Cre show YFP expression within 24 hr of injection (Figure S1A). To look at this on a cellular level, mice heterozygous for Rosa26mTmG reporter allele (Muzumdar et al., 2007) were also injected with TAT-Cre. All cells in mice bearing the Rosa26mTmG allele express membrane-bound Tomato fluorescent protein prior to recombination with Cre but express a membrane-bound GFP after Cre-mediated excision of the mTomato coding sequence. A single subdermal injection of TAT-Cre resulted in dense GFP expression in the surrounding tissue (Figure S1B).
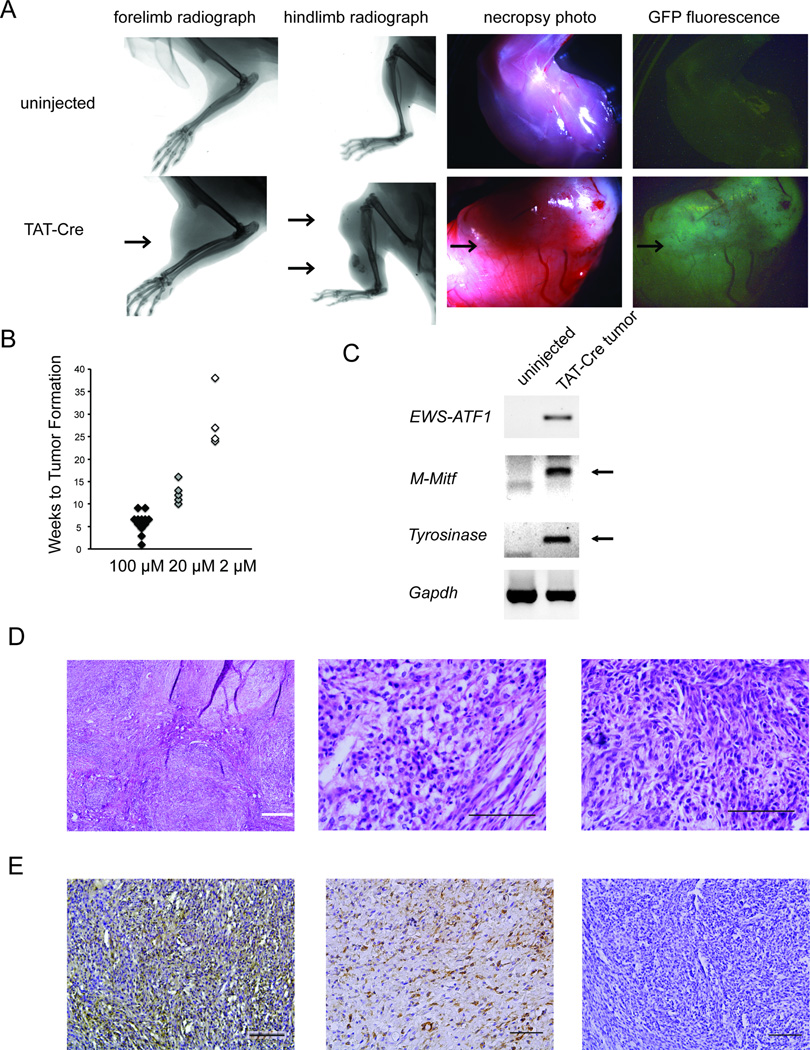
To determine the sufficiency of the EWS-AFT1 fusion gene to drive clear cell sarcomagenesis in vivo, we injected TAT-Cre protein into the anterolateral soft-tissues abutting the tibia and in the distal forelimbs of mice heterozygous for the Rosa26EA1 allele. EWS-ATF1 transcripts were detectable by RT-PCR from tissues harvested within 24 hr of the TAT-Cre injection (Figure S1C). Every injection of TAT-Cre yielded a tumor (Figure 2A), tightly localized to the injection site. Control mice, including both un-injected littermates and littermates injected with saline and followed for 15 months, never formed tumors. All mouse tumors derived from the TAT-Cre-activated Rosa26EA1 allele demonstrated eGFP fluorescence (Figure 2A).
Figure 2. TAT-Cre initiated expression ofEWS-ATF1 in vivo leads to tumorigenesis.
(A) Radiographs of limbs of Rosa26EA1 heterozygous mice without (top) or with (bottom) injection of TAT-Cre (left two columns). Necroscopy light and GFP fluorescence photo of Rosa26EA1 limb without injection (top) or of tumor extracted from the TAT-Cre injected Rosa26EA1 (bottom) (right two columns). Arrows point to individual tumors. (B) Latency to visible tumor formation for Rosa26EA1 mice injected with 100 µM, 10 µM, and 2 µM solutions of TAT-Cre. (C) RT-PCR analysis of the indicated mRNAs from uninjected control tissues and TAT-Cre induced tumors. (D) Representative histology of TAT-Cre induced tumors with H&E stain showing a multi-nodular tumor at low power (left), cells marked by round shape, clear cytoplasm and an open chromatin pattern at higher power (middle),and a tumor having a more spindled cell morphology (right). (E) Immunohistochemical stains for M-MITF (left), S100B (middle), and cytokeratin 5 (right). All black scale bars are 100 µm in length and the white bar is 500 µm.
See also Figure S1 and Table S1.
Noting an extremely brief latency to tumorigenesis, the ability to form tumors in a broad array of tissue locations, and the histological appearance of poly-clonality among the first rounds of TAT-Cre induced tumors, we investigated whether the rapid growth to large tumors might result primarily from a large initial population of induced EWS-ATF1-expressing cells. To address this question, we injected limbs of mice heterozygous for the Rosa26EA1 allele with different concentrations of TAT-Cre and found that the latency to tumor formation correlated with the concentration of TAT-Cre administered (Figure 2B). Injecting 100 □M of TAT-Cre resulted in visible tumors as quickly as three weeks post injection, with 100 percent penetrance per injection site by 6 weeks. TAT-Cre at 2 □M still produced tumors with full penetrance, but required a longer latency with visible tumors observed after a period of six to nine months. While TAT-Cre concentration impacted latency to development of a visible tumor, it did not impact the observed rate of tumor growth following visible detection of any specific tumor. Tumors appearing early from concentrated TAT-Cre or after a longer latency from diluted TAT-Cre still grew at a similarly rapid rate after detection.
The tumors that formed following TAT-Cre injection into mice heterozygous for the Rosa26EA1 allele recapitulated human CCS molecularly, with expression of the fusion oncogene and melanocytic markers M-Mitf and Tyrosinase (Figure 2C). Mouse tumors also matched human tumor histomorphology and immunohistochemical profile. In hematoxylin and eosin (H&E) stained sections, the majority of murine tumors demonstrated nuclear features typical of primitive cell types, with prominent open chromatin patterns as well as abundant clear cytoplasm (the feature for which the tumor is named) (Figure 2D). While some tumors had a pseudo-encapsulated, pushing border with surrounding tissues, others demonstrated clear infiltration into adjacent tissue planes. A minority subset of tumors (20%) demonstrated spindle cell morphology, also observed in some human CCS cases (Figure 2D). Consistent with human CCS, the tumors demonstrated immunohistochemical positivity for melanocytic markers (M-MITF and S100B) and lacked staining for cytokeratins (Figure 2E).
Unbiased expression of EWS-ATF1 also results in tumorigenesis, preferentially in mesenchyme
Because tumors rapidly invaded surrounding tissues, dissections of the limbs did not clearly indicate from which tissues they arose. In a search for tissues incompatible with transformation, TAT-Cre was injected into subcutaneous adipose, dorsal paw peritendinous, periosteal, intramuscular and mammary fat pad tissue compartments in mice between three weeks and six months of age. The latency to tumor appearance varied with age and injection site, but every injection in each of these tissue locations yielded completely penetrant tumorigenesis (Table S1). Each of these locations bears some cells of mesenchymal character, but whether these or neighboring cells actually gave rise to tumors remained unclear.
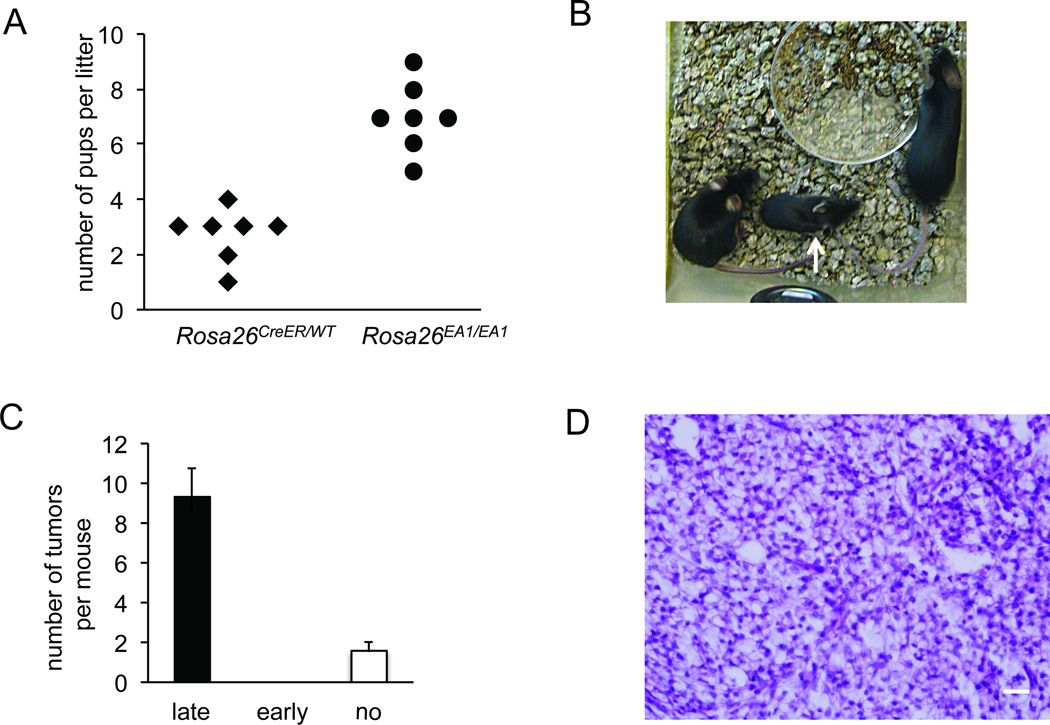
In order to broaden exposure to EWS-ATF1 fusion protein across multiple cell types and developmental periods, we next utilized Rosa26CreER to express Cre sporadically, in random cell types. While efficient CreER-mediated recombination requires the presence of tamoxifen, a very low level of CreER-mediated recombination is observed even in the absence of tamoxifen (Haldar, et al 2009). Breeding mice bearing the Rosa26CreER allele to mice bearing the Rosa26EA1 allele resulted in smaller litters than breeding Rosa26EA1 mice alone, fitting with the previously described early embryonic leakiness of this CreER and suggesting some developmental toxicity of early expression of EWS-ATF1 (Figure 3A).
Figure 3. Early expression of EWS-ATF1 in the broad Rosa26CreER-lineage renders stunted growth, but later expression drives tumorigenesis.
(A) The number of pups per litter of mice heterozygous for the Rosa26CreER allele crossed with mice homozygous for the Rosa26EA1 allele compared to that of Rosa26EA1/EA1 back-crossed controls. (B) Photograph of a 7 weeks old male Rosa26EA1/CreER mouse injected with tamoxifen at 10 days (white arrow) and two uninjected female littermates of the same genotype. (C) Chart of the number of tumors per mouse at 12 weeks age among Rosa26EA1/CreER mice injected with tamoxifen after 3 weeks (late, n = 4), before 3 weeks (early, n=3), or not at all (n = 5). Error bars denote standard deviation, t-test p < 0.05. (D) H&E stained histopathology demonstrating the classic clear cell morphology apparent in a Rosa26CreER initiated tumor after late injection of tamoxifen. Scale bar is 20 µm in length.
See also Figure S2 and Table S2.
Among the Rosa26CreER/EA1 mice that survive embryogenesis, administration of tamoxifen prior to three weeks of age results in stunted growth (Figure 3B) and death by 12 weeks of age without detectible tumor formation. Rosa26CreER/EA1 mice receiving tamoxifen after three weeks of age form more tumors than those receiving no tamoxifen, but both of these groups demonstrate complete penetrance of tumor formation by 12 weeks of age (Figure 3C). The tumors arising in Rosa26CreER/EA1 mice were histologically similar to the murine TAT-Cre induced tumors and human clear cell sarcoma (Figure 3D). Whether enhanced by later tamoxifen administration or not, tumors arose most often in the extremities, rib cage, and facial tissues of the Rosa26CreER/EA1 mice, but were also less frequently found in the dermis, liver, and bone (Table S2). As following TAT-Cre induction, the preponderance of tumors arose in mesenchymal tissue compartments. A variety of mouse cells are permissive to EWS-ATF1-driven transformation, replicating the range of tumor tissue locations observed in molecularly defined human cases of CCS, but preferentially arising in mesenchyme.
EWS-ATF1 drives an expression signature of transformation
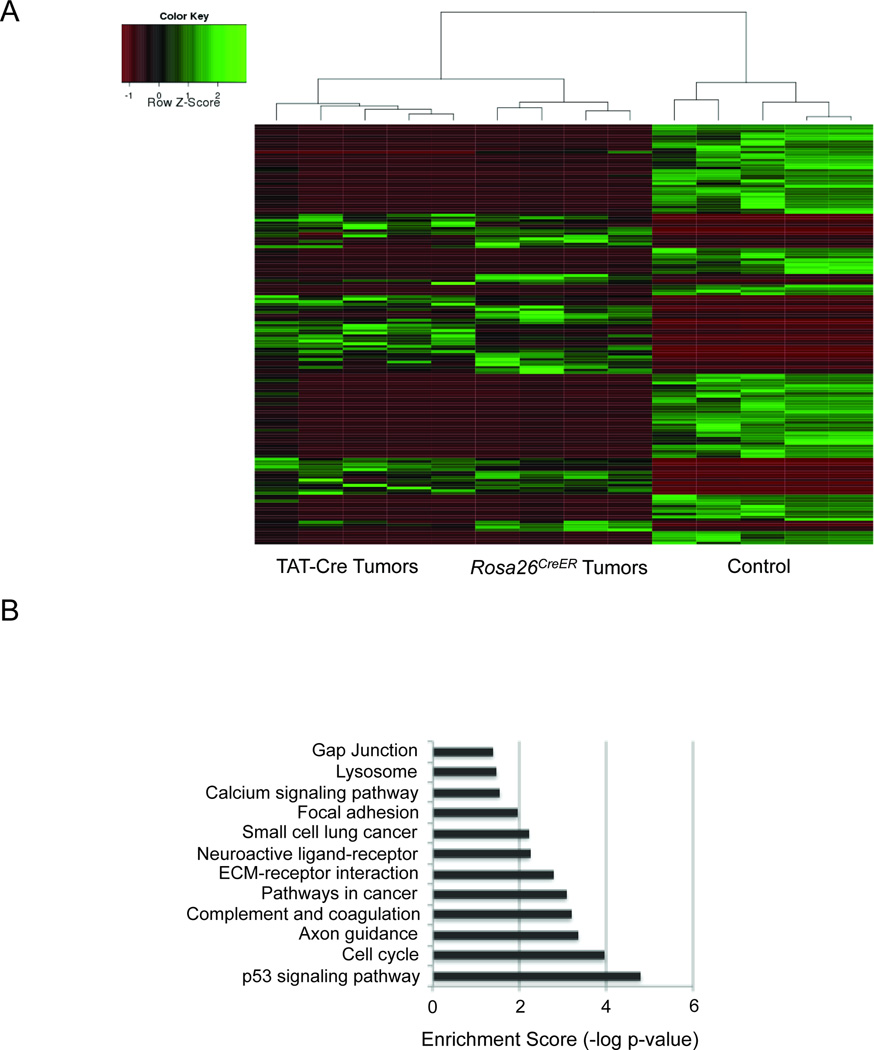
Because the brief latency to tumorigenesis in both models suggested a powerful role for EWS-ATF1 in driving transformation, we sought to define the expression signature shared by tumors from both means of inducing EWS-ATF1 expression. To this end, we harvested tumors from each cohort as well as from control mesenchymal tissue, consisting of a portion of the thoracic cage including cartilage, bone, skeletal muscle, and connective tissue, isolated total RNA from each, and performed Illumina sequencing. The samples generated an average of 24.4 ± 3.1 million reads, which aligned to 16,840 ± 778 genes at greater than or equal to 0.1 reads per kilobase per million (RPKM).
Clustering analysis demonstrated highly similar expression profiles among tumors induced by either TAT-Cre or Rosa26CreER, both distinct from control tissues. An unsupervised hierarchical clustering of the samples according to the 200 most differentially expressed genes is shown in Figure 4A (Gene lists in Tables S3, S4 and S5). DAVID analysis of the most significantly upregulated genes in the mouse tumors compared to control tissues highlighted several informative KEGG pathways, including cell cycle control, cancer, p53, and extra-cellular matrix pathways (Huang da et al., 2009a; Huang da et al., 2009b) (Figure 4B). These data provide insight into the transforming power of EWS-ATF1, with consistent alterations in gene expression profiling among different methods of activation.
Figure 4. The expression signature of EWS-ATF1-driven tumors.
(A) Heat map depicting the 200 most significantly (p-value < 0.001) differentially expressed genes between TAT-Cre or Rosa26CreER initiated tumor and control tissue, as assessed by transcriptome sequencing. (B) DAVID analysis of the most upregulated genes in tumors highlights Kegg pathways known to be involved in transformation.
Embryonic expression of EWS-ATF1 in mesenchymal tissues and predecessors causes lethality
While experiments with TAT-Cre injections and Rosa26CreER-initiated EWS-ATF1 expression suggested that a variety of cell types appear to support a program of transformation, they also suggested that the preferred clear cell sarcomagenesis tissue of origin is mesenchyme. As post-natal mesenchymal tissues derive from both neural crest and mesodermal origins, we investigated the identity of potential cells of origin by breeding mice bearing the Rosa26EA1 allele to mice bearing Cre expressed from promoters specific to these lineages. With regard to neural crest, mice bearing the Rosa26EA1 allele and Wnt1-Cre (Danielian et al., 1998) did not complete embryogenesis. Conditional EWS-ATF1 activated by Pax3-Cre (Engleka et al., 2005), Pax7-Cre (Keller et al., 2004b), Tie2-Cre (Kisanuki et al., 2001), or Prx1-Cre (Durland et al., 2008; Logan et al., 2002), all expressed in mesoderm, also resulted in embryonic lethality.
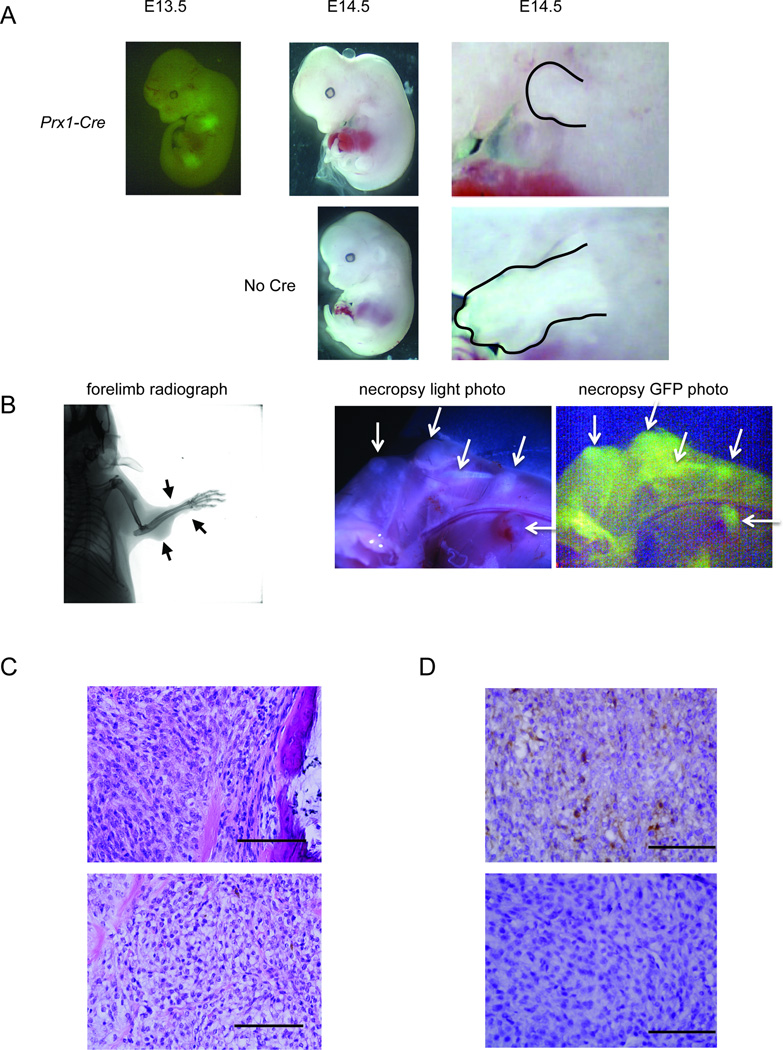
At E13.5, the Prx1-Cre lineage demonstrated GFP fluorescence in Prx1-Cre;Rosa26EA1 mouse embryos, indicating expression of eGFP from the IRES on the EWS-ATF1 transcript (Figure 5A). As late as E14.5 Prx1-Cre;Rosa26EA1 mouse embryos remained viable, but demonstrated severe limb deformities (Figure 5A). It suggests that EWS-ATF1 was toxic to these cells.
Figure 5. EWS-ATF1 drives apoptosis in the embryonic Prx1-lineage and tumorigenesis in the post-weaning Prx1-lineage.
(A) GFP fluorescence image of Prx1-Cre;Rosa26EA1 embryos at embryonic day 13.5 (far left). Prx1-Cre;Rosa26EA1 at E14.5 (top middle), compared to littermate controls lacking Prx1-Cre (bottom middle), the limbs of mutant and control are enlarged to show difference in size (outlined in right panels) (B) Radiograph of a Prx1-CreERT2;Rosa26EA1 mouse 8 weeks after a single tamoxifen injection at 4 weeks of age (left), gross necropsy photos with halogen light (middle) or GFP fluorescence (right) of Prx1-reERT2;Rosa26EA1 limb 8 weeks post tamoxifen injection (arrows point to tumors). (C) H&E stained histolopathology of Prx1-reERT2;Rosa26EA1 tumor immediately adjacent to bone (upper) and tumor exhibiting classic clear cell sarcoma morphology (lower). (D) Immunohistochemistry for M-MITF on Prx1-CreERT2;Rosa26EA1 derived tumors. Scale bars are 100 µm in length.
Pax7-Cre; Rosa26EA1 did not produce live progeny but embyros were retrieved as late as E18.5. These embryos showed severe craniofacial deformation with no visible eGFP which suggests that the lineage cannot survive expression of EWS-ATF1 (Figure S2A).
Mice bearing the Rosa26EA1 allele and Myf5-Cre, which activated the fusion gene expression in myoblasts (Haldar et al., 2007), did not form tumors but demonstrated another phenotype in which EWS-ATF1 was also apparently toxic to cells. These mice survived to birth and lived to approximately three months of age, but were very small in size and severely myopathic, with eGFP expression detectable in the remaining muscle fibers (Figure S2B and C).
These data demonstrate that even though the expression of EWS-ATF1 is tolerated better than the expression of other sarcoma-related fusion oncogenes in mouse embryonic fibroblasts in vitro and many tissues in vivo when induced after weaning, it is of significant toxicity in specific tissue settings during development.
EWS-ATF1 expression in post-natal mesenchymal progenitors generates two CCS tumor types
Noting that early tamoxifen administration in Rosa26CreER/EA1 mice did not result in tumorigenesis but later administration did (Figure 3), we hypothesized that the toxicity prompted in the mesenchymal tissues embryonically might be avoided at later stages of development. In order to test this hypothesis, we bred mice bearing the Rosa26EA1 allele to mice bearing either Pax7CreERT2 (Murphy et al., 2011) or Prx1-CreERT2 (Hasson et al., 2007), then administered tamoxifen after three weeks of age. By 12 months post-tamoxifen, myopathy consistently developed in the Pax7CreERT2;Rosa26EA1 mice but no tumors were observed (Figures S2D, S2E). The absence of tumor formation argues against muscle satellite cells being a potential cell of origin for EWS-ATF1 induced tumors. In the embryo, Prx1-Cre is widely expressed in the mesenchyme of the developing mammalian limb and head (Durland et al., 2008), whereas Prx1-CreERT2 expression is reported to be restricted post-natally to a smaller progenitor cell population in the same anatomic regions, still capable of osteochondrogenic differentiation (Kawanami et al., 2009).
Prx1-CreERT2;Rosa26EA1 mice developed tumors (Figure 5B) by eight weeks post-tamoxifen injection with 100% penetrance. Most of these tumors developed in the extremities and head, consistent with the expected anatomic distribution of Prx1 post-natal expression. The tumors showed GFP fluorescence marking expression of the EWS-ATF1-IRES-eGFP transcript (Figure 5B). The Prx1-CreERT2 induced tumors appeared to arise from the periosteal/perichondrial layer as well as from within the musculature as judged by gross microscopy and histology (Figure 5B and C). The tumors induced in the Prx1 lineage demonstrated either the clear cell morphology with lightly stained eosinophilic cytoplasm or spindle cell morphology, both closely resembling tumors induced by TAT-Cre.
Notably, some tumors induced by Prx1-CreERT2 expressed M-MITF but others did not (Figure 5D). CCS in humans often but not always expresses M-MITF (Granter et al., 2001). Thus it appears that induction of EWS-ATF1 expression with Prx1-CreERT2 generated both subtypes of CCS.
EWS-ATF1 expression in post-natal mesenchymal stem cells generates CCSs with a more consistent phenotype
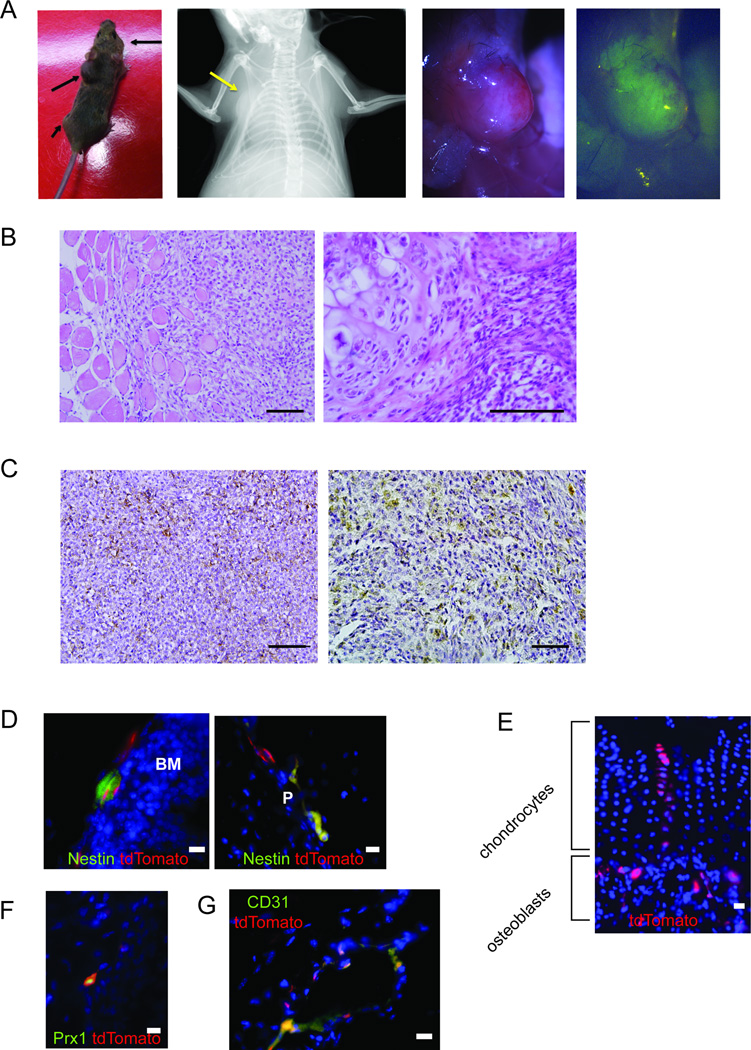
In order to interrogate possible cells of origin in the undifferentiated progenitor cell population that precedes Prx1 expression post-natally, we bred mice bearing the Rosa26EA1 allele to mice bearing Bmi1CreERT2 (Sangiorgi and Capecchi, 2008). Bmi1 is a general marker for stem cells and has been demonstrated to label intestinal, neural, epidermal and hematopoietic stem cells (Claudinot et al., 2005; Leung et al., 2004; Park et al., 2003; Sangiorgi and Capecchi, 2008). A single peritoneal injection of tamoxifen into Bmi1CreERT2;Rosa26EA1 mice after 3 weeks of age resulted in fully-penetrant tumorigenesis. Every mouse developed tumors in the deeper mesenchymal tissues of the limb and trunk (Figure 6A). Some tumors formed completely within and surrounded by muscle (Figure 6B), similar to a subset of the Prx1-lineage tumors. A larger portion formed adjacent to bone, arising from the periosteum/perichondrium, also similar to several of the Prx1-lineage tumors (Figure 6B). The Bmi1-lineage tumors consistently matched the clear cell morphology and immunohistochemical profile of human CCS tumors, uniformly demonstrating M-MITF and S100B (Figure 6B and C).
Figure 6. The Bmi1 lineage, including mesenchymal stem cells, enables more consistent development of clear cell sarcomas with melanocytic features.
(A) Living image of Bmi1CreERT2;Rosa26EA1 mouse with black arrows indicating tumor formation (far left), radiograph (left), necropsy photo (right) and GFP fluorescence (far right) image of tumor formed in the chest wall (yellow arrow showing location of the tumor). (B) H&E stained histopathology of Bmi1CreERT2-initiated tumors, including both intra-muscular (left) and periosteal/perichondrial (right) tumors. (C) Immunohistochemistry of Bmi1CreERT2-initiated tumors stained for M-MITF (left) and S100B (right). (D) Immunofluorescence for Nestin (green) and Bmi1CreERT2-induced expression of tdTomato (red) in Bmi1CreERT2; Rosa26tdTomato mice 30 days after injection of tamoxifen shown in the bone marrow BM (left) and periosteum P (right). (E) Bmi1-lineage tracing from Bmi1CreERT2;Rosa26tdTomato mouse at 60 days post tamoxifen demonstrates tdTomato fluorescence (red) in osteoblasts and chondrocytes. (F) Immunofluorescence against Prx1 (green) and Bmi1-lineage defined by tdTomato (red) in Bmi1CreERT2;Rosa26tdTomato mice 30 days post tamoxifen injection. (G) Immunofluorescence against CD31 (green) and tdTomato Bmi1-lineage cells (red) at 60 days post-tamoxifen All Scale bars are 100 µm in length.
See also Figure S3.
Prior reports have suggested a role for Bmi1 in mesenchymal stem cells based on depletion of the pool of osteochondroprogenitors in mice bearing homozygous disruption of Bmi1 (Zhang et al., 2010). To follow up these studies and more fully characterize the Bmi1 lineage in mesenchyme, mice bearing Bmi1CreERT2 were crossed to a robust reporter mouse expressing tdTomato conditionally from the Rosa26 locus. Bmi1CreER;Rosa26tdTomato mice were injected with a single dose of tamoxifen at six weeks of age and their limb tissues were harvested 1, 2, and 9 months post-injection for analysis.
We analyzed the early Bmi1 lineage focused on the mesenchymal progenitor population within the bone marrow and endosteum. Nestin has recently been shown to be an accurate marker of the multipotent mesenchymal stem cell subpopulation in this location, whereas no specific markers are agreed upon in the periosteum (Mendez-Ferrer et al., 2010). Co-labeling experiments on tissue sections revealed the co-localization of the early Bmi1 lineage marker to the Nestin-expressing cells both at the endosteal surface in the bone marrow and in periosteum (Figure 6D), indicating that Bmi1 is indeed expressed in this mesenchymal stem cell population. Lineage tracing experiments from tissues harvested 60 days and 9 months following tamoxifen administration revealed that Bmi1 expressing cells contribute to both osteoblast and chondrocyte lineages (Figure 6E), further affirming Bmi1 as a marker of very early, stem-like cells in mesenchyme. The previously described Prx1-CreERT2-defined lineage marks a similar, but smaller population of these differentiated mesenchymal cells (Kawanami et al., 2009). Using an antibody against Prx1 to locate cells actively expressing the marker, we found a portion but not all of the cells in the Bmi1CreER;Rosa26tdTomato lineage expressed Prx1, suggesting that differentiating cells from the Bmi1 lineage express Prx1 (Figure 6F). Lineage tracing with Prx1-CreERT2 mice and immunofluorescence against Nestin confirmed that a subset of Prx1-lineage cells still express Nestin as well (Figure S3A).
While mesenchymal stem cell and progenitor populations within and adjacent to the bone explain the location of some tumors arising in both the Bmi1 and Prx1 lineages, other tumors in both groups arose within the muscle compartments. Since it has previously been demonstrated that Bmi1 is expressed in Pax7-positive satellite cells within muscle (Robson et al., 2011), it is possible that the intramuscular Bmi1-lineage tumors could arise from this cell population. However, we already discussed that Pax7CreERT2; Rosa26EA1 do not form tumors, arguing against the possibility that satellite cells provide a potential cell of origin for CCS (Figure S2D and E).
Further analysis of the Bmi1-lineage within the muscle compartments identified not only satellite cells, but also CD31-expressing endothelial cells (Figure 6G). Lineage tracing with Prx1-CreERT2 mice identified CD31+ endothelial cells within its lineage as well (Figure S3B). A perivascular population of cells that has expressed both Bmi1 and Prx1, perhaps at different stages of differentiation, may serve as the originating cells of the CCS tumors that appear within the muscle compartments in these two groups.
Murine CCS tumors fit the human CCS expression profile; variations appear to reflect different cells of origin
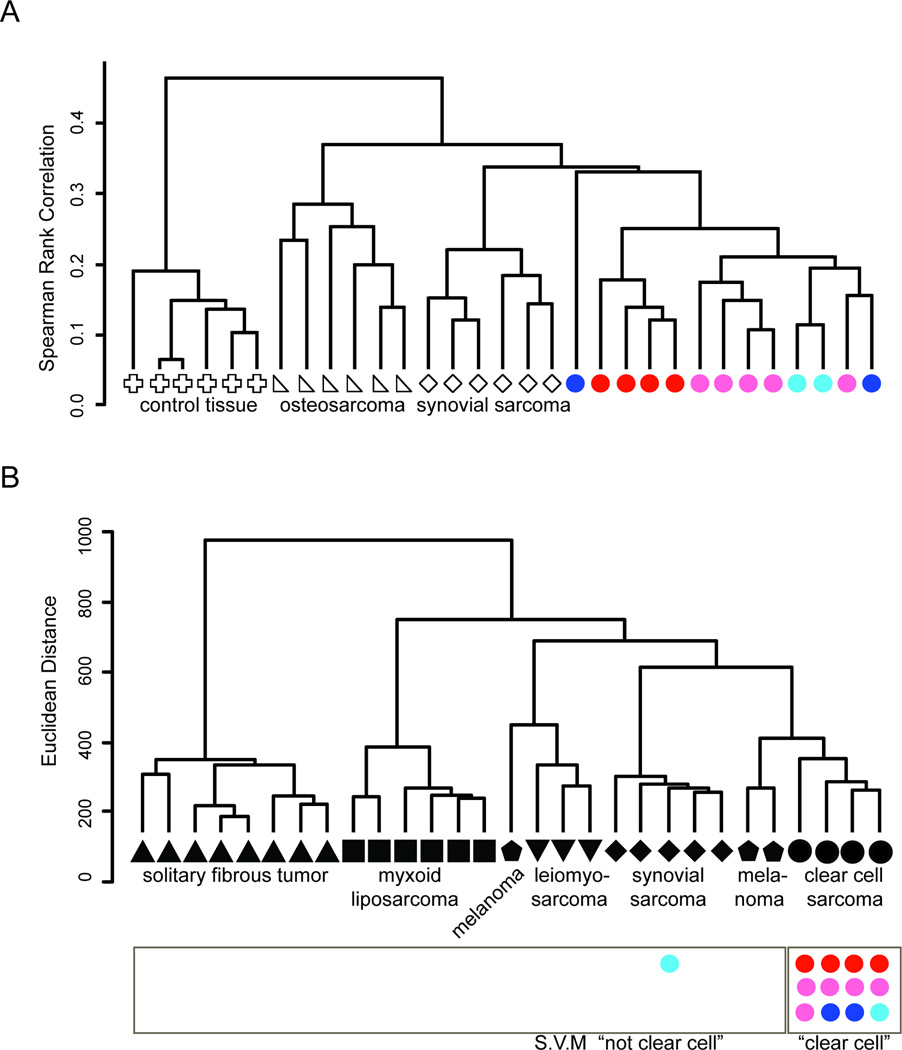
In order to determine the impact of these varied potential cells of origin on the resultant tumors, we performed RNA sequencing on a panel of mouse CCS tumors, induced by TAT-Cre, Rosa26CreER, Bmi1CreERT2, and Prx1-CreERT2. We simultaneously sequenced RNA from mouse synovial sarcomas and osteosarcomas, as well as control mesenchymal tissue. Using Spearman correlation as the distance metric and an average linkage, CCS from all induction methods clustered closely and separately from control tissue and the other two sarcoma types (Figure 7A).
Figure 7. Tumors derived from EWS-ATF1 expression in different cells of origin cluster together according to expression profile and fit human clear cell sarcoma profiles.
(A) Unsupervised hierarchical clustering of mouse samples by sequencing-defined transcriptional profiling, osteosarcomas (triangle), synovial sarcomas (diamond), EWS-ATF1-driven tumors by TAT-Cre (red circles), Rosa26CreER (pink circles), Bmi1CreERT2 (dark blue circles), and Prx1-CreERT2 (light blue circles) and control samples from chest-wall mesenchymal tissues (cross). (B) Unsupervised hierarchical clustering of human tumors profiled by microarray and used to train the supervised machine-learning module, solitary fibrous tumor (triangle), myxoid liposarcoma (square), melanoma (pentagon), leiomyosarcoma (triangle), synovial sarcoma (diamond) and clear cell sarcoma (circle). The mouse EWS-ATF1-driven tumors are placed under the category of human tumors they most identified with on the supervised machine-learning module, TAT-Cre (red circles), Rosa26CreER (pink circles), Bmi1CreERT2 (dark blue circles), and Prx1-CreERT2 (light blue circles).
To test the extent of molecular similarity between human CCS and the tumors induced by expression of EWS-ATF1 in mice across the range of tissues of origin, we compared mRNA sequencing expression analysis of mouse CCS tumors to microarray-derived expression data from a panel of related human tumors. A support vector machine (SVM), or supervised machine learning, technique was created to predict the identity of a tumor using a training platform built by human HEEBO microarray data from samples of CCS, melanoma (MEL), solitary fibrous tumor (SFT), leiomyosarcoma (LMS), myxoid liposarcoma (MLS) and synovial sarcoma (SS), all of which except CCS were previously reported (Nielsen et al., 2002). We first filtered the human data by mapping the HEEBO probes to the mm9 mouse reference genome, retaining only the 12,246 probes whose alignments demonstrated at least 80 percent homology across the probe length. The human expression data across this subset of probes was used to train the linear SVM model by creating CCS and non-CCS classes (n = 4 and 25, respectively). The normalized RPKM mouse data were input into the SVM as read depths per probe and classified into one of the two classes. Twelve of 13 murine CCS tumors were predicted to be human CCS, rather than one of the other tumor types (Figure 7B). As a control, 0 of 5 RNA sequencing data sets from wildtype mouse mesenchymal tissue controls were identified as human CCS when input into the same SVM. These data confirm that mouse tumors derived from conditional expression of EWS-ATF1 resemble the expression profile of human CCS more closely than the other soft tissue tumors or melanoma.
Notably, the one mouse CCS that did not cluster closely to the human CCS SVM prediction was a Prx1-CreERT2-initiated tumor that did not express M-Mitf. In contrast, another Prx1-CreERT2-initiated tumor that expressed M-Mitf did cluster with human clear cell sarcoma.
Further comparative analysis of the RNA sequencing expression profiles of the two Prx1-lineage and the two Bmi1-lineage tumors, both sets from the same anatomic locations, identified a number of consistently differentially expressed genes. Three of the top ten genes more highly expressed in Prx1-CreERT2;Rosa26EA1 were Crlf1, which is present in osteoblasts and chondrocytes, along with Saa1 and Saa2, which are upregulated in differentiated compared to undifferentiated mesenchymal stem cells (Clancy et al., 2003; Kovacevic et al., 2008).
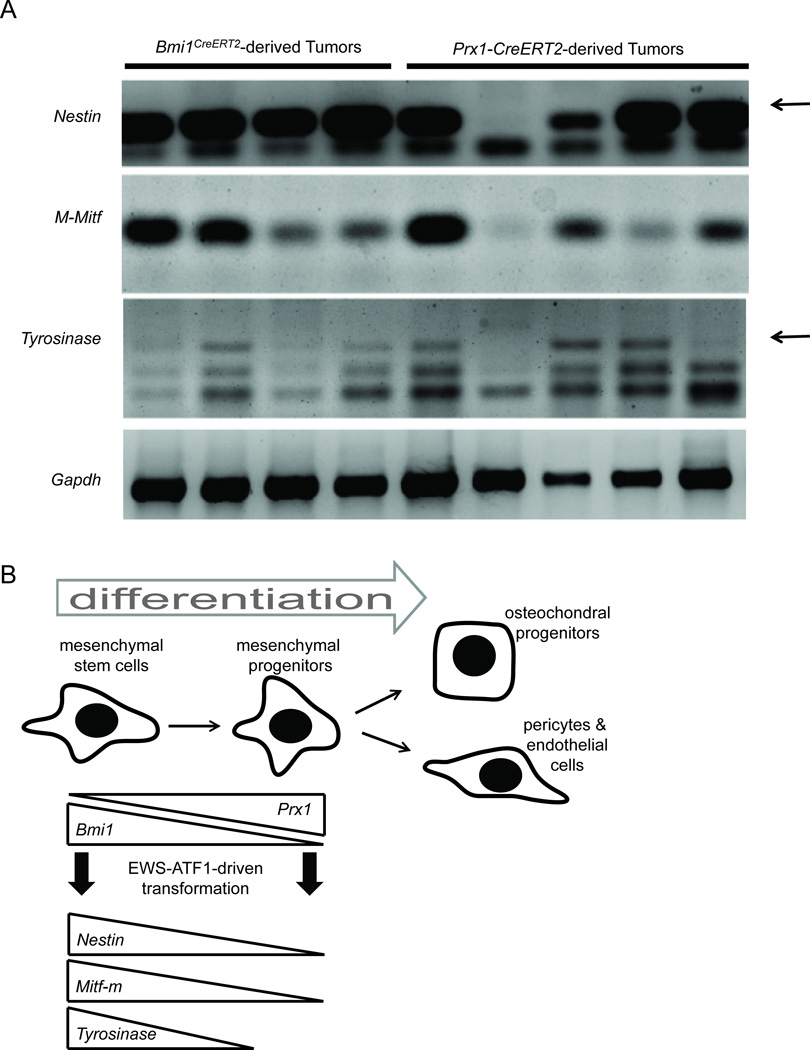
To better evaluate the difference between the Bmi1CreERT2;Rosa26EA1 and Prx1-CreERT2;Rosa26EA1 tumors, we performed RT-PCR on a panel of additional samples. This revealed that some Prx1-lineage tumors indeed expressed M-Mitf and Tyrosinase, but not all (Figure 8A). All Bmi1-lineage tumors expressed these melanocytic markers. Interestingly, the Prx1-lineage tumor that did not express Nestin also did not express M-Mitf or Tyrosinase. An additional Prx1-lineage tumor failed to express Tyrosinase only. This may indicate that between the stem-like state of Bmi1 expression and the progenitor state of Prx1 expression Nestin, M-Mitf, and Tyrosinase are epigentically silenced (Figure 8B).
Figure 8. Tumors arising from EWS-ATF1 expression initiated in the Prx1- or Bmi1-lineage differ by expression profile.
(A) RT-PCR analysis of the indicated transcripts on total RNA isolated from Bmi1- and Prx1-lineage tumors (arrow points to the correct band) (B) Working model of the impact of early differentiation within multipotent mesenchymal progenitors on the possibility of transdifferentiation to express melanocytic markers upon clear cell sarcomagenesis driven by EWS-ATF1.
Discussion
A few mouse genetic models of translocation-associated sarcomas have been described over the last decade, including models of alveolar rhabdomyosarcoma expressing PAX3-FKHR (Keller et al., 2004a; Keller and Capecchi, 2005; Keller et al., 2004b), myxoid liposarcoma expressing FUS-CHOP (Perez-Losada et al., 2000) and synovial sarcoma expressing SS18-SSX2 (Haldar et al., 2007; Haldar et al., 2009; Haldar et al., 2008). The last of these used means of transcriptional control of the fusion oncogene similar to those used in the current study, with expression in Cre/loxP conditional fashion from the Rosa26 locus. The model of synovial sarcoma also demonstrated fully penetrant tumorigenesis when initiated by Rosa26CreER or Myf5Cre (Haldar et al., 2009). Comparison of the two models highlights the brief latency to tumorigenesis following expression of EWS-ATF1 initiated by Rosa26CreER. The synovial sarcoma model develops tumors after a year, rather than within 3 months. Further, expression of SS18-SSX2 in most cells, in vitro or in vivo, was toxic. The very rapid onset of visible tumors in the mouse CCS model relative to synovial sarcoma suggest that fewer additional hits may be required to foster progression of the tumor. This may represent a fundamental difference between the biology of the two fusion oncogenes, as all other aspects of the models are the same.
It has not escaped our notice that the extreme oncogenicity of EWS-ATF1 in mice contrasts with the rare incidence of formally diagnosed CCS in humans. A potential explanations for this discrepancy is the possibility that EWS-ATF1 involvement in human tumorigenesis remains underappreciated. t(12;22) (q13;q12) chromosomal translocation and resultant fusion oncogenes are identified only when pursued in clinical tumors. The widening range of tumors recently found to bear EWS-ATF1, such as angiomatoid fibrous histiocytoma and clear cell carcinoma of the salivary gland (Antonescu et al., 2011; Chen et al., 2011; Mangham et al., 2010; Ren et al., 2009), suggests that more may yet be identified in the near future. Second, the observed toxicity to embryonic development in the mouse by the early expression of the EWS-ATF1 fusion gene may greatly limit the acceptable timeframe for the formation of the required t(12;22) chromosomal translocation in humans. Finally, not all balanced chromosomal translocations arise with equal frequency and the formation of the t(12;22) translocation responsible for CCS may be the rate limiting step for this malignancy.
Different cells of origin impacted features of the tumors resulting from EWS-ATF1 expression. Tumors from both the Bmi1- and the Prx1- post-natal lineages developed in nearly identical anatomic locations (in the muscle and from the periosteal surface of bones) and fit the histologic appearance of human CCS, but the Prx1-lineage tumors did not consistently express the melanocytic markers M-Mitf, and Tyrosinase. The M-Mitf-expressing Prx1-lineage tumors fit the general expression signature of human CCS. Those not expressing M-Mitf did not. This observation may explain the fact that not all human CCSs clearly express melanoma markers. All four human tumors used for expression profiling were typical, M-Mitf-expressing, melanomalike CCSs. Perhaps the Prx1-induced mouse tumor that did not express M-Mitf might have fit better with a broader group of human CCSs.
Expression of Nestin has previously been reported in human CCS cell lines (Dimas et al., 2008). All Bmi1-lineage tumors expressed Nestin and the melanocytic markers while variable Nestin expression in Prx1-lineage tumors predicted expression of the melanocytic markers. Nestin has previously been proven to be a marker of stemness in mesenchyme (Mendez-Ferrer et al., 2010), and overlapped with the Bmi1-lineage and partly with the Prx1-lineage. This evidence along with the co-labeling experiments showing Prx1 expression within the Bmi1 lineage suggests that the differentiation steps between Bmi1-expressing cells and Prx1-expressing cells include a gradual loss of stemness. The epigenetic state may render the melanocytic markers unavailable for upregulation by ATF constitutive activation alone (Figure 8B).
Many have argued in the past about the origin of the melanocytic features of human CCS, variably attributing them to either cell of origin or transformation by EWS-ATF1, which also upregulates M-Mitf. Although we have not ruled out the possibility that melanocytes or their precursors might also offer sufficient cells of origin, the tumors we have induced in mesenchyme actually bolster both prior arguments. CCS transformation can enable transdifferentiation that includes melanocytic markers, but only from certain cells of origin. Perhaps a cell's reprogrammability, even during aggressive transformation from expression of a powerful fusion oncogene, remains at least partly checked by its antecedent differentiation state.
This mouse model will enable additional dissection of the ATF pathway's impact on oncogenesis in vivo. Further, the model is ideally suited to pre-clinical testing of targeted therapies for this and other CCS pathways. Administration of TAT-Cre produces visible tumors within a few weeks and avoids any pleiotropic developmental effects from toxicity of the fusion oncogene expressed across an entire tissue. These tumors are well-localized, offering a reasonable model for the study of metastasis. Unlike delivery of Cre with a virus, a single molecule of TAT-Cre cannot readily diffuse to a great distance or travel through the blood stream to a distant site and induce recombination, as at least two molecules of the TAT-Cre protein must enter any given cell to catalyze the recombination event (Joshi et al., 2002).
Due to the low frequency of sarcomas, the most critically scarce resource for developing more effective therapeutic strategies for these malignancies is patients themselves. It is not easy to assemble sufficient numbers of patients for carrying out subtype-specific sarcoma clinical trials. Therapeutic strategies must be optimized and prioritized in the pre-clinical arena before advancing to clinical trial. Authenticated mouse models could serve this purpose. Genomic sequencing and profiling has vastly increased the ability to test the authenticity of a mouse model's recapitulation of human malignancy. Once established as an excellent facsimile to the human cancer, a mouse model can be used not only to interrogate cancer mechanisms and identify pertinent therapeutic targets, but also as a platform for assessment of drug efficacy. Successful candidate therapeutic strategies could be then moved to human trials with greater assurance of success.
In summary, we have shown that the oncogenic fusion protein that characterizes CCS is sufficient to initiate CCS-like tumors in mice that recapitulate human CCS in terms of cell morphology, immunohistochemistry and genome-wide expression. This fusion gene generates two distinguishable tumor types in mice that also reflect similar variance in morphology and tissue distribution seen in humans (i.e. principally falling in two classes, those that resemble melanomas because of the expression of M-Mitf and its target genes, and those that do not). The apparent difference in cells of origin of these two potential subclasses described herein with mice may provide an explanation of why human CCS commonly, but inconsistently, displays the classical melanocytic features.
Experimental procedures
Targeted mouse line production
Human EWS-ATF1 cDNA was obtained by RT-PCR of total RNA from 8 histologically and immunohistochemically confirmed clear cell sarcoma tumors . The total RNA was obtained as deidentified patient sample through an approved University of Utah Institutional Review Board Protocol. PCR was used to identify tumors that contained the type 1 EWS-ATF1 fusion gene. After screening, the cDNA sample was used to amplify the entire fusion oncogene. The cDNA clone was subcloned into the Rosa26UA plasmid using AscI and Fse1 Sites. This plasmid contained the Loxp-pgk-Neo-tPA-Loxp-AscI-FheI-IRES-EGFP within the Rosa26 homology arms and the final targeting construct contained Loxp-pgk-Neo-tPA-Loxp-EWS-ATF1-IRES-EGFP. Genotyping protocol and further details on gene targeting used can be found in Supplemental Experimental Procedures.
Animals, radiograph, tissue preparation, and immunohistochemistry
All mouse experiments were performed in accordance with humane practices, national and international regulations, and with the approval of the University of Utah Institutional Animal Care and Use committee. Radiographs were obtained post-asphyxiation using a Carestream 4000 Pro-Fx instrument (Carestream Molecular Imaging, Woodbridge, Connecticut).
Mouse tumors were extracted after asphyxiation and were fixed overnight in 4% paraformaldehyde prior to embedding in paraffin. Immunostaining and tissue preparation along with antibodies used are described in the Supplemental Experimental Procedures.
Transcriptome analysis
A portion of each tumor was snap frozen for delayed total RNA extraction using the Qiagen RNeasy Mini kit (Qiagen, Inc., Valencia, California). RT-PCR was performed with random hexamer primers to generate cDNAs, followed by PCR for specific transcripts (Supplemental Experimental Procedures).
For RNA sequencing, total RNA was prepared using an Illumina TruSeq RNA sample prep kit (Illumina, Inc., San Diego, California) and quality checked with an Agilent Bioanalyzer RNA 6000 chip (Agilent Technologies, Santa Clara, California). mRNAs were captured by oligodT magnetic beads and fragmented. Library quality was then checked by Nanodrop analysis (Thermo Scientific, Wilmington, Delaware), qPCR quantitation using Illumina primers, and another bioanalyzer run. Sequencing was performed on an Illumina HiSeq 2000 machine (Illumina, Inc., San Diego, California) using a 50 cycle single end read. PhiX control library reads were added to each lane for quality assurance. Reads were then aligned with the mm9 mouse genome. Basic clusterings were performed using GeneSifter software (Geospiza, Inc. Seattle, Washington). Method used to classify tumor type using RNA-seq data can be found in the Supplemental Experimental Procedures.
Accession Numbers
The RNA-seq data have been deposited in NCBI’s Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through the GEO series accession number GSE41293.
Human microarray data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through the GEO series accession number GSE43045.
Supplemental Data
The Supplemental Data include Supplemental Experimental Procedures, three supplemental figures, and five supplemental tables.
Supplementary Material
Highlights.
Clear cell sarcoma fusion gene EWS-ATF1 expression produces tumors in 100% of mice.
EWS-ATF1-driven mouse tumors model human tumors histology and gene expression profile.
A range of mouse cells tolerates EWS-ATF1 expression and subsequently yields tumors.
The differentiation state of the transforming cells impacts tumor characteristics.
Significance.
CCS, a rare soft tissue malignancy typically arising in the extremities of young adults, closely resembles melanoma. EWS-ATF1, a fusion oncogene resulting from a t(12;22) chromosomal translocation, defines the neoplasm. Conditional expression of the human EWS-ATF1 cDNA in the mouse demonstrates its profound transformational impact. While a number of cell types can be rendered tumorigenic via expression of the fusion oncogene, the undifferentiated cells appear to enable the full melanoma-related phenotype. This model deciphers part of the enigma of the melanoma expression profile of CCS and explains the range of human tumors derived from this powerful translocation product. The mouse model should provide a robust platform for interrogating molecular mechanisms and developing more effective therapies for CCS.
Acknowledgements
The authors gratefully acknowledge Matt Hockin (University of Utah) for producing the TAT-Cre, Allie Grossmann (University of Utah) for assistance with pathology analysis, Jay S. Wunder and Irene L. Andrulis (University of Toronto) for providing the human CCS samples for isolation of the cDNA, Brian Dalley and Brett Milash (University of Utah) for assistance with RNA sequencing and analysis. K.M.S. receives graduate student support from the Howard Hughes Med2Grad program. K.B.J. receives career development support from the National Cancer Institute (NIH) K08CA138764K08. This work was partly supported by P30CA042014 from the National Cancer Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonescu CR, Katabi N, Zhang L, Sung YS, Seethala RR, Jordan RC, Perez-Ordonez B, Have C, Asa SL, Leong IT, et al. EWSR1-ATF1 fusion is a novel and consistent finding in hyalinizing clear-cell carcinoma of salivary gland. Genes, chromosomes & cancer. 2011;50:559–570. doi: 10.1002/gcc.20881. [DOI] [PubMed] [Google Scholar]
- Antonescu CR, Tschernyavsky SJ, Woodruff JM, Jungbluth AA, Brennan MF, Ladanyi M. Molecular diagnosis of clear cell sarcoma: detection of EWS-ATF1 and M-MITF transcripts and histopathological and ultrastructural analysis of 12 cases. The Journal of molecular diagnostics : JMD. 2002;4:44–52. doi: 10.1016/S1525-1578(10)60679-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Folpe AL, Colby TV, Sittampalam K, Patey M, Chen MG, Chan JK. Angiomatoid fibrous histiocytoma: unusual sites and unusual morphology. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24:1560–1570. doi: 10.1038/modpathol.2011.126. [DOI] [PubMed] [Google Scholar]
- Clancy BM, Johnson JD, Lambert AJ, Rezvankhah S, Wong A, Resmini C, Feldman JL, Leppanen S, Pittman DD. A gene expression profile for endochondral bone formation: oligonucleotide microarrays establish novel connections between known genes and BMP-2-induced bone formation in mouse quadriceps. Bone. 2003;33:46–63. doi: 10.1016/s8756-3282(03)00116-9. [DOI] [PubMed] [Google Scholar]
- Claudinot S, Nicolas M, Oshima H, Rochat A, Barrandon Y. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14677–14682. doi: 10.1073/pnas.0507250102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covinsky M, Gong S, Rajaram V, Perry A, Pfeifer J. EWS-ATF1 fusion transcripts in gastrointestinal tumors previously diagnosed as malignant melanoma. Human pathology. 2005;36:74–81. doi: 10.1016/j.humpath.2004.10.015. [DOI] [PubMed] [Google Scholar]
- D'Amico FE, Ruffolo C, Romeo S, Massani M, Dei Tos AP, Bassi N, Bassi N. Clear Cell Sarcoma of the Ileum: Report of a Case and Review of the Literature. International journal of surgical pathology. 2011 doi: 10.1177/1066896911428073. [DOI] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Davis IJ, Kim JJ, Ozsolak F, Widlund HR, Rozenblatt-Rosen O, Granter SR, Du J, Fletcher JA, Denny CT, Lessnick SL, et al. Oncogenic MITF dysregulation in clear cell sarcoma: defining the MiT family of human cancers. Cancer cell. 2006;9:473–484. doi: 10.1016/j.ccr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Dimas K, Tsimplouli C, Anagnostopoulos AK, Mahaira L, Iliopoulou E, Perez S, Vougas K, Tsangaris GT. The proteome profile of two cell lines and their xenografts isolated from a patient with clear cell sarcoma (soft tissue melanoma) Cancer genomics & proteomics. 2008;5:175–237. [PubMed] [Google Scholar]
- Durland JL, Sferlazzo M, Logan M, Burke AC. Visualizing the lateral somitic frontier in the Prx1Cre transgenic mouse. Journal of anatomy. 2008;212:590–602. doi: 10.1111/j.1469-7580.2008.00879.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleka KA, Gitler AD, Zhang M, Zhou DD, High FA, Epstein JA. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Enzinger FM. CLEAR-CELL SARCOMA OF TENDONS AND APONEUROSES. AN ANALYSIS OF 21 CASES. Cancer. 1965;18:1163–1174. doi: 10.1002/1097-0142(196509)18:9<1163::aid-cncr2820180916>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Falconieri G, Bacchi CE, Luzar B. Cutaneous Clear Cell Sarcoma: Report of Three Cases of a Potentially Underestimated Mimicker of Spindle Cell Melanoma. The American Journal of dermatopathology. 2012 doi: 10.1097/DAD.0b013e3182473190. [DOI] [PubMed] [Google Scholar]
- Fujimura Y, Siddique H, Lee L, Rao VN, Reddy ES. EWS-ATF-1 chimeric protein in soft tissue clear cell sarcoma associates with CREB-binding protein and interferes with p53-mediated trans-activation function. Oncogene. 2001;20:6653–6659. doi: 10.1038/sj.onc.1204684. [DOI] [PubMed] [Google Scholar]
- Granter SR, Weilbaecher KN, Quigley C, Fletcher CD, Fisher DE. Clear cell sarcoma shows immunoreactivity for microphthalmia transcription factor: further evidence for melanocytic differentiation. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2001;14:6–9. doi: 10.1038/modpathol.3880249. [DOI] [PubMed] [Google Scholar]
- Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- Haldar M, Hedberg ML, Hockin MF, Capecchi MR. A CreER-based random induction strategy for modeling translocation-associated sarcomas in mice. Cancer Res. 2009;69:3657–3664. doi: 10.1158/0008-5472.CAN-08-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar M, Randall RL, Capecchi MR. Synovial sarcoma: from genetics to genetic-based animal modeling. Clinical orthopaedics and related research. 2008;466:2156–2167. doi: 10.1007/s11999-008-0340-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantschke M, Mentzel T, Rutten A, Palmedo G, Calonje E, Lazar AJ, Kutzner H. Cutaneous clear cell sarcoma: a clinicopathologic, immunohistochemical, and molecular analysis of 12 cases emphasizing its distinction from dermal melanoma. The American journal of surgical pathology. 2010;34:216–222. doi: 10.1097/PAS.0b013e3181c7d8b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson P, Del Buono J, Logan MP. Tbx5 is dispensable for forelimb outgrowth. Development. 2007;134:85–92. doi: 10.1242/dev.02622. [DOI] [PubMed] [Google Scholar]
- Hocar O, Le Cesne A, Berissi S, Terrier P, Bonvalot S, Vanel D, Auperin A, Le Pechoux C, Bui B, Coindre JM, Robert C. Clear cell sarcoma (malignant melanoma) of soft parts: a clinicopathologic study of 52 cases. Dermatology research and practice. 2012;2012 doi: 10.1155/2012/984096. 984096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic acids research. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Joshi SK, Hashimoto K, Koni PA. Induced DNA recombination by Cre recombinase protein transduction. Genesis. 2002;33:48–54. doi: 10.1002/gene.10089. [DOI] [PubMed] [Google Scholar]
- Kawanami A, Matsushita T, Chan YY, Murakami S. Mice expressing GFP and CreER in osteochondro progenitor cells in the periosteum. Biochemical and biophysical research communications. 2009;386:477–482. doi: 10.1016/j.bbrc.2009.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Arenkiel BR, Coffin CM, El-Bardeesy N, DePinho RA, Capecchi MR. Alveolar rhabdomyosarcomas in conditional Pax3:Fkhr mice: cooperativity of Ink4a/ARF and Trp53 loss of function. Genes & development. 2004a;18:2614–2626. doi: 10.1101/gad.1244004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C, Capecchi MR. New genetic tactics to model alveolar rhabdomyosarcoma in the mouse. Cancer research. 2005;65:7530–7532. doi: 10.1158/0008-5472.CAN-05-0477. [DOI] [PubMed] [Google Scholar]
- Keller C, Hansen MS, Coffin CM, Capecchi MR. Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: implications for alveolar rhabdomyosarcoma cell of origin. Genes & development. 2004b;18:2608–2613. doi: 10.1101/gad.1243904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- Kovacevic A, Hammer A, Stadelmeyer E, Windischhofer W, Sundl M, Ray A, Schweighofer N, Friedl G, Windhager R, Sattler W, Malle E. Expression of serum amyloid A transcripts in human bone tissues, differentiated osteoblast-like stem cells and human osteosarcoma cell lines. Journal of cellular biochemistry. 2008;103:994–1004. doi: 10.1002/jcb.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C, Lingbeek M, Shakhova O, Liu J, Tanger E, Saremaslani P, Van Lohuizen M, Marino S. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428:337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Lyle PL, Amato CM, Fitzpatrick JE, Robinson WA. Gastrointestinal melanoma or clear cell sarcoma? Molecular evaluation of 7 cases previously diagnosed as malignant melanoma. The American journal of surgical pathology. 2008;32:858–866. doi: 10.1097/PAS.0b013e31815b8288. [DOI] [PubMed] [Google Scholar]
- Mangham DC, Williams A, Lalam RK, Brundler MA, Leahy MG, Cool WP. Angiomatoid fibrous histiocytoma of bone: a calcifying sclerosing variant mimicking osteosarcoma. The American journal of surgical pathology. 2010;34:279–285. doi: 10.1097/PAS.0b013e3181cb4017. [DOI] [PubMed] [Google Scholar]
- Mao X, Fujiwara Y, Orkin SH. Improved reporter strain for monitoring Cre recombinase-mediated DNA excisions in mice. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5037–5042. doi: 10.1073/pnas.96.9.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma'ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development (Cambridge, England) 2011;138:3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O'Connell JX, Zhu S, Fero M, Sherlock G, Pollack JR, et al. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J, Pintado B, Gutierrez-Adan A, Flores T, Banares-Gonzalez B, del Campo JC, Martin-Martin JF, Battaner E, Sanchez-Garcia I. The chimeric FUS/TLS-CHOP fusion protein specifically induces liposarcomas in transgenic mice. Oncogene. 2000;19:2413–2422. doi: 10.1038/sj.onc.1203572. [DOI] [PubMed] [Google Scholar]
- Ren L, Guo SP, Zhou XG, Chan JK. Angiomatoid fibrous histiocytoma: first report of primary pulmonary origin. The American journal of surgical pathology. 2009;33:1570–1574. doi: 10.1097/PAS.0b013e3181b20e64. [DOI] [PubMed] [Google Scholar]
- Robson LG, Di Foggia V, Radunovic A, Bird K, Zhang X, Marino S. Bmi1 is expressed in postnatal myogenic satellite cells, controls their maintenance and plays an essential role in repeated muscle regeneration. PloS one. 2011;6:e27116. doi: 10.1371/journal.pone.0027116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nature genetics. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers GR, Viero S, Nathan PC, Teshima I, Pereira C, Zielenska M. Association of the t(12;22)(q13;q12) EWS/ATF1 rearrangement with polyphenotypic round cell sarcoma of bone: a case report. The American journal of surgical pathology. 2005;29:1673–1679. doi: 10.1097/01.pas.0000176434.19197.c1. [DOI] [PubMed] [Google Scholar]
- Wang WL, Mayordomo E, Zhang W, Hernandez VS, Tuvin D, Garcia L, Lev DC, Lazar AJ, Lopez-Terrada D. Detection and characterization of EWSR1/ATF1 and EWSR1/CREB1 chimeric transcripts in clear cell sarcoma (melanoma of soft parts) Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2009;22:1201–1209. doi: 10.1038/modpathol.2009.85. [DOI] [PubMed] [Google Scholar]
- Zhang HW, Ding J, Jin JL, Guo J, Liu JN, Karaplis A, Goltzman D, Miao D. Defects in mesenchymal stem cell self-renewal and cell fate determination lead to an osteopenic phenotype in Bmi-1 null mice. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2010;25:640–652. doi: 10.1359/jbmr.090812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.