Abstract
Lipids play multiple roles essential for proper mitochondrial function, from their involvement in membrane structure and fluidity, cellular energy storage, and signaling. Lipids are also major targets for reactive species, and their peroxidation byproducts themselves mediate further damage. Thousands of lipid species, from multiple classes and categories, are involved in these processes, suggesting lipid quantitative and structural analysis can help provide a better understanding of mitochondrial physiological status. Due to the diversity of lipids that contribute to and reflect mitochondrial function, analytical methods should ideally cover a wide range of lipid classes, and yield both quantitative and structural information. We developed a high resolution LC-MS method that is able to monitor the major lipid classes found in biospecimens (ie. biofluids, cells and tissues) with relative quantitation in an efficient, sensitive, and robust manner while also characterizing individual lipid side-chains, by all ion HCD fragmentation and chromatographic alignment. This method was used to profile the liver mitochondrial lipids from 192 rats undergoing a dietary macronutrient study in which changes in mitochondria function are related to changes in the major fat and glycemic index component of each diet. A total of 381 unique lipids, spanning 5 of the major LIPID MAPS defined categories, including fatty acyls, glycerophospholipids, glycerolipids, sphingolipids and prenols, were identified in mitochondria using the non-targeted LC-MS analysis in both positive and negative mode. The intention of this report is to show the breadth of this non-targeted LC-MS profiling method with regards to its ability to profile, identify and characterize the mitochondrial lipidome and the details of this will be discussed.
Keywords: mitochondria, liquid chromatography, mass spectrometry, lipidomics, dietary macronutrients
INTRODUCTION
Mitochondrial lipids are central to maintaining normal mitochondrial function at all levels, ranging from the regulation of respiratory chain activity (Barzanti et al. 1994; Lenaz 1988), to the membrane structural composition (Kiebish et al. 2009). Furthermore, many lipids, especially those containing unsaturated fatty acids, are sensitive to peroxidative damage under pathophysiological conditions, such as chronic disease and acute injury, and the byproducts of lipid peroxidation are themselves reactive and potentially toxic (Esterbauer et al. 1991; Stavrovskaya et al. 2010; Kristal et al. 2001; Burke et al. 1998; Kristal et al. 1996; Land et al. 2004; Lenaz 1988; McKenzie et al. 2004). Peroxidation has also been shown to be a major determinant of membrane fluidity in mitochondrial membranes (Chen and Yu 1994). The analytical method used to assess the composition of the mitochondrial lipidome must be qualitatively and quantitatively accurate across the inherent diversity of lipids in terms of structural class, fatty acyl (FA) side chain differences and overall lipid concentration.
Methods used to analyze mitochondrial lipids have often been targeted and non-specific, measuring the composition of one specific class at a time and ignoring the inherent diversity due to acyl side-chain composition. Analysis by thin layer chromatography (TLC), gas chromatography (GC) or liquid chromatography (LC) are effective in determining the quantitative value of different lipid classes in a sample, e.g. the total amount of phophatidylcholine (PC) present versus the amount of phosphatidylethanolamine (PE), however, the inherent diversity within the lipid class is lost. This intra-class diversity lies in the lipid side-chains, i.e., fatty acids. These fatty acid side chains can vary in the length of the acyl chain, in the degree of unsaturation (number of double bonds), and in the position of the double bonds. Each of these aspects is both structurally and functionally important, and thus is critical for understanding the lipidome and its interactions with mitochondrial physiology. Analytical methods have been developed that utilize class separation via TLC followed by hydrolysis to the free fatty acyls (FA), derivitization and subsequent separation detection by GC- mass spectrometry (MS). However, this method is insensitive, requiring large amounts of starting material; the method is also time-consuming and limiting in that the origin of the side-chains are still unknown. The application of electrospray ionization-MS methods to mitochondrial lipid analysis can be used to eliminate most of the previously mentioned drawbacks and allow for a more complete picture of the lipidome to be discerned (Bou Khalil et al. 2009).
MS-based lipidomic profiling methods vary in the degree of analytical bias toward a lipid class, set of classes or individual species, based on the goal of the study (Li et al. 2010). Any of these methods can both be used either with or without chromatographic separation prior to detection, with the latter termed shotgun lipidomics (Ejsing et al. 2006; Ejsing et al. 2009; Han and Gross 2003, 2005; Han et al. 2005; Han et al. 2006). Targeted methods, focus analysis on one or more pre-determined lipids or lipids classes, and optimize the platform, sample preparation, separation (if used) and detection, only for these specific analytes. These methods are both sensitive and informative, but they are inherently limited in that they can only measure the specific lipid(s) originally decided to be of known importance to the study (Dennis 2009). In a study where all lipid species are of potential biological importance, the most comprehensive measurement of a lipidome from a single analysis would require a non-targeted profiling approach, in which no deliberate bias is given toward any of the 8 lipid categories established and defined by LIPID MAPS (www.lipidmaps.org) (Bird et al. 2011a, 2011b; Fahy et al. 2005; Fahy et al. 2007).
Non-targeted LC-MS lipidomics profiling ideally yields comprehensive, quantitative and reproducible analytical data in an efficient and robust manner from any given biological sample. These approaches have been used to achieved differential lipid relative quantitation, identification and characterization in different biospecimen such as, mouse plasma (Hu et al. 2008), human plasma (Pietilainen et al. 2007), rat mitochondria (Bird et al. 2011a) and rat serum (Bird et al. 2011b). An advantage of discovery-based non-targeted LC-MS profiling is the ability to re-analyze experiments when neither the biological importance nor the overall lipidome complexity is completely understood prior to analysis.
In addition to quantitative profiling, lipid characterization is an important aspect of non-targeted analysis. LC-MS combined with high energy collisional dissociation (HCD) all ion fragmentation can characterize unknowns in complex mixtures by using different energies to yield both class specific and lipid specific diagnostic fragments (Bird et al. 2011a, 2011b). HCD is performed in a multipole collision cell attached to the c-trap of an Orbitrap MS. This cell provides triple quadrupole like fragments on all ions that enter the c-trap without a low molecular weight cutoff observed in conventional ion-traps. The ability to generate fragmentation data from all ions in the analysis is especially beneficial in a non-targeted study and this advantage, combined with the ability to easily re-analyze the spectra, make this approach applicable to numerous sample types and organisms where a broad-based survey of the lipidome is required.
We used an LC method with high resolution accurate mass MS detection combined with HCD fragmentation (Bird et al. 2011a) to profile the liver mitochondrial lipids from 192 rats undergoing a dietary macronutrient study in which changes in mitochondria function are related to changes in the major fat and glycemic index component of each diet. In the analytical study, a focus was placed on the characterization of the mitochondrial lipid, cardiolipin and the remaining species were only mentioned in passing. Herein we will describe the 381 unique lipids, from 5 of the major LIPID MAPS defined categories, which were identified from those original LC-MS profiling experiment in both positive and negative ionization mode. Total lipidome coverage and the possible implications each species can have on mitochondrial function will be discussed.
MATERIALS AND METHODS
Chemicals
LC-MS grade acetonitrile (ACN), methanol (MeOH), and isopropanol (IPA), as well as HPLC grade dichloromethane (DCM) and dimethyl sulfoxide (DMSO), were purchased from Fisher Scientific (Pittsburg, PA) and ammonium formate was purchased from Sigma-Aldrich (St. Louis, MO). A detailed list all lipid standards purchased as well as their abbreviations, preparation conditions and vendor sources can be found in the supplemental information of our previously published method (Bird et al. 2011a).
Preparation of Lipid Standards
Stock solutions were prepared by dissolving lipid standards in DCM:MeOH (2:1 v/v) at concentrations ranging from 1–10 mg/mL and were stored at –20°C. Working solutions were diluted in ACN/IPA/H2O (65:30:5 v/v/v) to 1 μg/ml prior to spiking studies or LC-MS analysis.
Dietary Study
Male Fisher 344 x Brown Norway F1 (FBNF1) rats (n=8), aged 7–9 weeks, were fed ad libitum one of 24 isocaloric diets that differed in fat and carbohydrate composition. This approach yielded a total of 192 animals in the study. The diets were comprised of six different fat groups, with the major constituent of each being either saturated fats (SFAs), trans fats (Trans), monounsaturated fats (MUFAs), or one of 3 groups of polyunsaturated fats (PUFAs), which vary in the ω-6/ω-3 PUFA ratios. Each type of fat was combined with one of 4 carbohydrate groups that varied based on sucrose content. The fat, carbohydrate and protein percentages in all diets were held consistent at 5, 66, and 20 (w/w), equivalent to 12, 68, and 21 (kcal %). Body weights and food composition were measured twice a week and rats were sacrificed after 8 weeks. Details of their husbandry and diets are not critical for the current report and will be presented elsewhere (manuscripts in preparation).
Liver Mitochondria Isolation
After the animals were sacrificed, each rat liver was harvested and the mitochondria isolated by the standard differential centrifugation method, using sucrose-based buffers, as described previously (Stavrovskaya et al. 2004). Mitochondrial protein concentration was determined by the Lowry method using BSA as a standard (Lowry et al. 1951). Sample aliquots containing 1 mg of protein from mitochondria were washed in 125 mM KCl, 10 mM HEPES, pH 7.4 buffer and frozen as dry pellets at −80°C before analysis. Secondary gradient purification was not performed, so some contamination with non-mitochondrial lipids is likely (see Results).
Mitochondrial Lipid Extraction
Immediately before extraction, each aliquot of mitochondria (containing 1 mg of protein) was dissolved in 40 μL DMSO and the membranes were disrupted by sonication. A mitochondrial pool sample was created by combining 8 μL samples from the sonicated mitochondria of each rat (n=192). The pool samples were processed for quality control (QC) and lipid identification studies.
Lipids were extracted according to the method of Bligh and Dyer (Bligh and Dyer 1959), substituting DCM for chloroform (Cequier-Sanchez et al. 2008) as described previously (Bird et al. 2011a). First, 30 μL of internal standard, which consisted of 1,1’,2,2’-tertraoleoyl cardiolipin (CL(18:1)4), 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine (PC(17:0/17:0), 1,2-dimyristoyl-sn-glycero-3-[phospho-rac-(1-glycerol)] (PG(14:0/14:0), 1-O-Hexadecyl-sn-glycero-3-phosphocholine (lysoPC(16:0)) and 1,2-dipalmitoyl-sn-glycero-3-phospho-L-serine (PS(16:0/16:0), each at the concentration of 5μg/mL, was added to each 30 μL sample, followed by 190 μL of MeOH. Next, 380 μL of DCM was added, the sample was again vortexed for 20 seconds and 120 μL of water was added to induce phase separation. The samples were then vortexed for 10 seconds and allowed to equilibrate at room temperature for 10 minutes before centrifugation at 8000 g for 10 minutes at 10 °C. A total of 370 μL of the lower lipid-rich DCM layer was then collected and the solvent evaporated to dryness under vacuum. Samples were reconstituted in 300 μL of ACN/IPA/H2O (65:30:5 v/v/v) containing PG (17:0/17:0) at a concentration of 5 μg/mL before LC-MS analysis. Ten μL of sample was injected onto the LC-MS system.
LC-MS Conditions, SIEVE Analysis and Lipid Identification
Details of the LC-MS method and SIEVE analysis have been described previously (Bird et al. 2011b, 2011a). Briefly, lipid extracts were separated on an Ascentis Express C18 2.1 x 150 mm 2.7μm column (Sigma-Aldrich, St. Louis, MO) connected to a Thermo Fisher Scientific PAL autosampler, Accela quaternary HPLC pump and an Exactive benchtop Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a heated electrospray ionization (HESI) probe. Separations ran for 30 minutes with mobile phase A and B consisting of 60:40 Water:ACN in 10mM ammonium formate and 0.1% formic acid and 90:10 IPA:ACN also with 10mM ammonium formate and 0.1% formic acid, respectively. The gradient started at 32% B for 1.5 minutes; from 1.5to 4 min increase to 45% B, from 4 to 5 min increase to 52% B, from 5 to 8 min to 58% B, from 8 to 11 min to 66% B, from 11 to 14 min to 70% B, from 14 to 18 min to 75% B, from 18 to 21 min to 97% B, during 21 to 25 min 97% B is maintained; from 25–30 min solvent B was decreased to 32% and then maintained. The column oven temperature was maintained at 45 °C and the temperature of the autosampler was set to 4 °C. The same LC conditions and buffers were used for all MS experiments with the flow rate was 260 μL/min and the scan range was between m/z 120–2000.
The spray voltage was set to 3.5 kV, whereas the heated capillary and the HESI probe were held at 250 °C and 350 °C, respectively. The sheath gas flow was set to 25 units and the auxiliary gas set to 15 units. These conditions were held constant for both positive and negative ionization mode acquisitions. The instrument was tuned by direct infusion of PG (17:0/17:0) in both positive and negative mode and external mass calibration was performed using the standard calibration mixture and protocol from ThermoFisher approximately every five days.
For full scan profiling experiments, the MS was run in high resolution mode, corresponding to a resolution of 60k and a 2 Hz scan speed. Mitochondria were profiled by injecting each sample once, in randomized order, with 17 pool samples, 17 blanks and 5 biological standard mixture (Bird et al. 2011a) (BIOSTD) samples spread throughout the analysis. This sample sequence yielded a total of 231 injections, each of which was run separately in positive and negative modes.
For lipid identification studies, HCD experiments were run on the pool, BIOSTD and blank samples only. These experiments were performed by alternating between full scan acquisitions and HCD scans, both run at 2 Hz. Three different HCD energies, 30, 60 and 100 eV, were used in separate experiments in both positive and negative mode.
All LC-MS profiling samples were analyzed using the MS label free differential analysis software package SIEVE v 1.3 (Thermo Fisher Scientific and Vast Scientific, Cambridge, MA). The framing parameters in these experiments were set at 0.01 Daltons for the m/z window and 1.00 minute for the RT window; 1000 was used at the intensity threshold. A pool from the middle of the 231 sample sequence was used as a qualitative reference and for relative quantitation, and frames built off the reference were then applied to all samples in the experiment.
For lipid identifications, the frame m/z values were used to do batch searches on the Metlin database (Smith et al. 2005), the human metabolome database (Wishart et al. 2009) (HMDB) and the LIPID MAPS database (Fahy et al. 2007) and those matches were confirmed using the molecules exact mass observed during the analysis, RT regions based on the BIOSTD elution times run throughout and HCD fragmentation. The details of these identifications are expanded in the results and discussion.
RESULTS AND DISCUSSION
The mitochondria used for this study were isolated from rat liver immediately upon sacrifice using standard differential centrifugation approaches optimized for function rather than biochemical analysis (Stavrovskaya et al. 2004). These mitochondria displayed coupled respiration with both complex I and II substrates, as well as, developed and maintained membrane potential, transported calcium, and underwent a permeability transition upon calcium overload (data not shown). While not gradient purified, analysis of equivalent preparation suggest >95% purity (Krasnikov et al. 2005). All lipids identified are thus likely primarily attributed to the mitochondrial fraction.
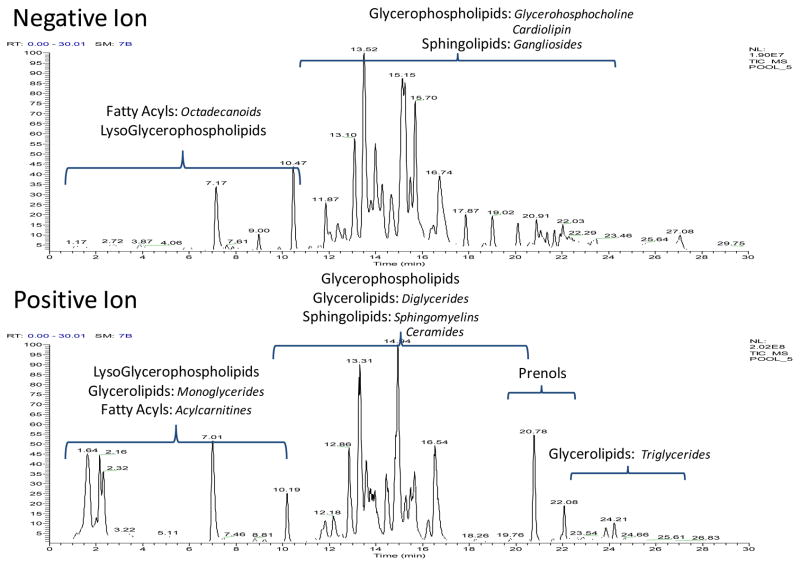
One important aspect of a successful non-targeted LC-MS lipidomic profiling method is its ability to simultaneously profile multiple classes and categories of lipids in a robust manner. The two representative mitochondrial pool total ion chromatograms (TICs) shown in Figure 1 demonstrate the breadth of which the LC-MS method employed can cover lipids from 5 LIPID MAPS categories when used in both positive and negative ionization mode. This coverage is expanded in Table 1, where the number of unique lipids from each category is defined by the class of lipids identified in our mitochondrial samples, as well as the ionization mode in which they are detected and by what adduct they are observed. Although 5 categories are seen here, this does not limit the profiling method to only these species. In other biofluids, 6 categories which include these 5 plus sterols have been observed; however, none were detected in this study. Table 1 reveals a total of 381 unique lipids identified using this non-targeted profiling technique. Details regarding the inter-category diversity, quantitative precision of the analysis, as well as, the qualitative importance of each class of lipid on proper mitochondrial function will now be described.
Figure 1. Total Lipid Coverage in Mitochondria.
Two representative mitochondria pool TICs in negative and positive mode respectively are labeled with the 5 LIPID MAPS categories observed. A few subclasses of each class is shown in italics.
Table 1. Total Mitochondrial Lipidome Coverage.
The class and subclass breakdown of the 281 lipids identified are shown as well as the ionization mode used and main adduct that was observed.
| Lipid Class | Unique Number of Lipids** | - Ionization | + Ionization |
|---|---|---|---|
| Glycerophospholipids (GP) | |||
| Phosphocholine (PC) | 103 | [M+FormAcid] − | [M+H]+ |
| plasmalogenphosphocholine (plsPC) | 5 | [M+FormAcid] − | [M+H]+ |
| lysoPhosphocholine (LysoPC) | 19 | [M+FormAcid] − | [M+H]+ |
| Phosphoethanolamine (PE) | 62 | [M -H] − | [M+H]+ |
| plasmalogenphosphoethanolamine (plsPE) | 6 | [M -H] − | [M+H]+ |
| lysoPhosphoethanolamine (LysoPE) | 9 | [M -H] − | [M+H]+ |
| Phosphoserine (PS) | 18 | [M -H] − | |
| Phosphoinositol (PI) | 11 | [M+NH4]+ | |
| Phosphoglycerol (PG) | 31 | [M -H] − | [M+H]+ / [M+NH4]+ |
| Cardiolipin (CL) | 26 | [M -H] − | [M+NH4]+ |
| monolysocardiolipin (MLCL) | 2 | [M -H] − | [M+NH4]+ |
| Fatty Acyls (FA) | |||
| Branched Fatty acid | 1 | [M+NH4]+ | |
| Acylamide | 1 | [M -H] − | |
| Octadecanoids | 2 | [M+H]+ / [M+NH4]+ | |
| Acylcarnitines (AcCar) | 3 | [M+H]+ | |
| Sphingolipids (SL) | |||
| Sphingomyelins (SM) | 6 | [M+FormAcid] − | [M+H]+ |
| Ceramides (Cer) | 21 | [M+FormAcid] − | [M+H]+ |
| Gangliosides (Gan) | 4 | [M -H] −/[M -H20-H] − | [M+NH4]+ |
| Glycerolipids (GL) | |||
| Monoacylglycerol (MG) | 4 | [M+NH4]+ | |
| Diacylglycerol (DG) | 4 | [M+NH4]+ | |
| Triacylglycerol (TG) | 43 | [M+NH4]+ | |
| Prenol Lipids (PL) | |||
| coenzyme-Q (CoQ) | 3 | [M+H]+ / [M+NH4]+ | |
Defined as having a unique mass and retention time pair that represents a distinct species. Those species that are observed in both ionization modes are only counted once.
Prenol Lipids
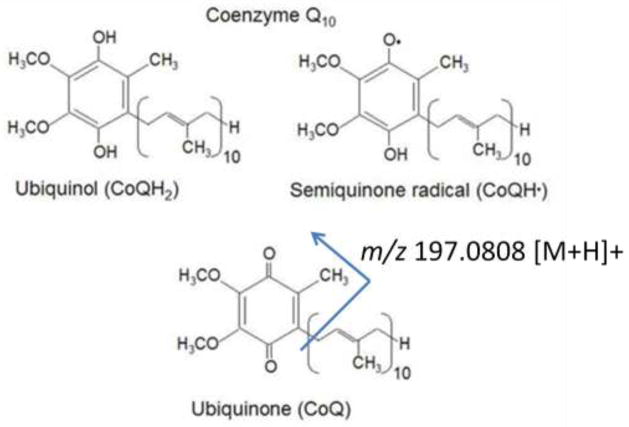
The coenzyme Q (CoQ, also known as the ubiquinones) species are a class of prenol lipids that contain a varying number of isoprene units, normally 6–10, attached to a quinoid core (Crane 2007). They exist in three redox states, as shown in figure 2 and CoQ9 and CoQ10 play a central role in electron transport during cellular respiration in rats and humans, respectively (Lenaz et al. 2007). Studies have suggested that only the CoQ9 and CoQ10 species are found in mammals, with the CoQ9 species predominantly being observed in rodents (Lenaz et al. 2007). The detection of these species is predominantly done using either LC with electrochemical detection or LC-MS in a targeted multiple reaction monitoring assay (Andreyev et al. 2010).
Figure 2. Redox states of Coenzyme Q10.
The 3 redox states of the Coenzyme Q10 quinoid core which can be found in mitochondria are shown. The full oxidized species, ubiquinone, yields a representative fragmentation of m/z 197.0808 that was used for MS characterization.
Using a non-targeted LC-MS profiling method we were able to identify and monitor CoQ10, CoQ9 and CoQ8 in our rat liver mitochondrial samples. Identifications were made based off of exact mass database searches and confirmed by matching extracted ion chromatogram (XIC) retention times to that of the CoQ10 species found in our BIOSTD. Using this assay, we are monitoring the fully oxidized ubiquinone compounds and during HCD fragmentation, these species give a diagnostic loss of the protonated head group, observed at m/z 197.0808, in positive ion mode. This fragment is shown in Figure 2 and is observed at all 3 HCD energies examined. In the mitochondrial pool samples injected over the course of the 5 day analysis, the mean intensity showed a median coefficient of variance (CV), calculated from raw peak areas, of less than 5% (n=8) for the 3 species observed. Analysis based on the mean intensity across the pools suggests that the CoQ9/CoQ10 and CoQ9/CoQ8 ratios were 11-1 and 17-1 respectively, indicating a larger pool of CoQ9 across the animals in this study. These results agree with the previous studies regarding the distribution of ubiquinones in rat and human tissues (Aberg et al. 1992; Lang and Packer 1987).
Fatty Acyls and Metabolites
Fatty acyls (FA) are long chain carboxylic acids usually derived from triacylglycerides and glycerophospholipids to be used for fuel. These species produce large quantities of ATP, in addition to forming primary metabolites used in numerous cellular processes. The detection of FAs is predominantly done using derivatization and GC-MS analysis. This method provides efficient separation between saturated and unsaturated fatty acids of different chain lengths as well as between most positional isomers (Quehenberger et al. 2011).
Despite being readily detected on this LC-MS platform, free FAs were not observed in our liver mitochondria study and merely 7 unique lipids were identified and classified to fall under the fatty acyl LIPID MAPS category. These species include a branched fatty acyl, 2 octadecanoid species, 1 acyl amide and 3 acyl carnitines. These compounds are demonstrated in table 2, as well as their retention time, mass error (given in parts per million [ppm]) and their mean intensities and CVs across the pools (n=8). The CV values were calculated from the raw peak areas established by SIEVE.
Table 2. FA Coverage in Mitochondria.
Each FA was identified by exact mass matching in the HMDB. The table outlines the RT, m/z value observed, mass error, intensity and CV (calculated from raw peak areas) for the 7 fatty acyl lipids identified in mitochondria. The error and CVs show the specificity and repeatability of the analysis platform.
| Fatty Acyls (FA) | Retention Time (min) | m/z | Error (ppm) | Intensity (n=8) | CV (n=8) |
|---|---|---|---|---|---|
| Branched Fatty acid | |||||
| methyloctadecanoic acid | 2.24 | 316.3203 | −3.70 | 8959817.06 | 3.94 |
| Acylamide | |||||
| palmitoylglycine | 4.85 | 312.2549 | 1.55 | 619.49 | 3.12 |
| Octadecanoids | |||||
| Dihydroxyoctadecanoic acid | 1.98 | 332.2790 | −1.39 | 11717.84 | 9.93 |
| Hydroxyoctadecdieneoic Acid or | 1.99 | 297.2424 | −1.79 | 6518.16 | 2.96 |
| Epoxyoctadecenoic Acid | |||||
| Acylcarnitines (AcCar) | |||||
| 9-Hexadecenoylcarnitine | 3.74 | 398.3261 | −0.67 | 22757.06 | 7.66 |
| 3-Hydroxy-9-hexadecenoylcarnitine | 3.64 | 414.3209 | −0.96 | 61001.64 | 4.06 |
| 3-Hydroxy-9Z-octadecenoylcarnitine | 3.63 | 442.3524 | −0.54 | 41318.36 | 7.99 |
Although the free FAs are not observed, the detection of some peroxidation metabolites, and acyl carnitines provide insight into the physiological state of the mitochondria regarding fatty acid metabolism and β-oxidation (Bhuiyan et al. 1992). Both octadecanoids observed, are common byproducts of ROS, consistent with mitochondrial roles as both a source and target for ROS. Additionally, acyl carnitines, which are commonly monitored in plasma due to their ability to cross the mitochondrial and cell membranes, have relatively stable concentrations and can easily reflect the intramitochondrial acyl-CoA status (Millington and Stevens 2011). The lack of additional acyl carnitine species in mitochondria was unexpected, and most likely reflects on the lower concentration of these species in the samples as opposed to an analytical bias of the platform. Based on ongoing work in our laboratory, we currently believe that this is a feature of the biological model studied. The ability to monitor these molecules provides valuable information regarding mitochondrial oxidative stress and energy production.
Sphingolipids
Sphingolipids (SL) are primarily defined by their eighteen carbon amino-alcohol backbone, and modifications to this basic structure can produce a family of lipids that play roles in mitochondrial membrane biology, cellular apoptosis and the induction of ROSs (Monette et al. 2010; Di Paola et al. 2000; Gudz et al. 1997). Different SL classes, structurally characterized by different head-groups and acyl chain lengths, can have opposing actions on cell function (Monette et al. 2010). For example, ceramides (cer) promote apoptosis whereas sphingosine-1-phosphate induces cellular survival (Hannun and Obeid 2002). Furthermore, the incorporation of different acyl chains onto the SL backbones can additionally alter the mitochondrial function of these lipid classes, making the specific characterization of each FA side-chain essential. Using our LC-MS profiling method, we were able to identify 22 unique Cer species, 6 SM, and 4 gangliosides (Gan) in rat liver mitochondria.
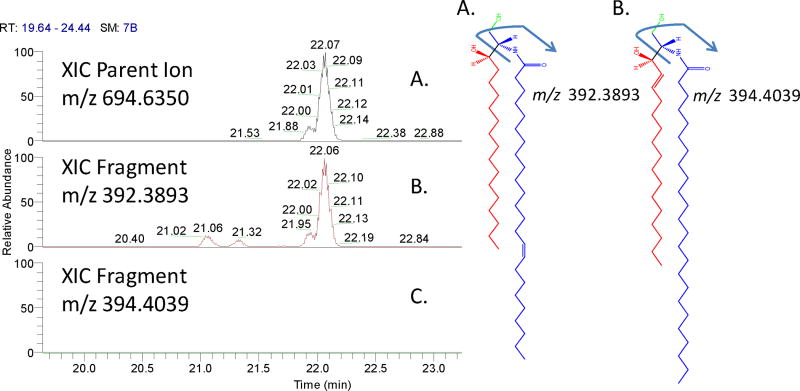
The majority of the Cers observed, 20 out of 22, were seen primarily in negative ion mode, and 8 species were observed in both positive and negative profiling experiments. Due to the increased signal in negative ion mode, the HCD characterization of the Cer lipid acyl side-chains was based on the negative ion neutral loss (NL) of the sphingoid base from the molecule. This loss provides a diagnostic fragment that confirms whether a sphingosine (d18:1) or sphinganine (d18:0) chain was present, which in turn would allow the composition of the n-acyl group to be determined. An example of this is shown in figure 3, where the XIC of the parent ion for unknown Cer with m/z 694.6365, representing the [M+Formic Acid]− adduct ion (panel A), is chromatographically aligned with the NL-H2O of the sphinganine base (panel B, and blue portion of molecule A) and not of the NL-H2O of the sphingosine base (panel C, and blue portion of molecule B). This suggests the species to be characterized as Cer (d18:0/24:1). This fragmentation scheme was applied to all 20 unique Cer identified using negative ion profiling and 17 showed this type of fragmentation, with the remaining 3 having intensities too low to discern. These data suggest that many Cer incorporated into liver mitochondria contain a sphinganine base as opposed to the sphingosine. In mitochondria, Cer are known to promote apoptosis, and recent studies have shown that Cer with the sphingosine sphingoid base are far more potent than those with sphinganine moiety (Ardail et al. 2001). In terms of the study herein, we would expect healthy animals to have a lower pool of these pro-apoptotic species and the larger incorporation of the lesser potent molecules.
Figure 3. The characterization of Mitochondrial Ceramides.
The left panel shows the XIC of the parent ion for an unknown Cer with m/z 694.6365, representing the [M+Formic Acid] − adduct ion (panel A),chromatographically aligned with the NL-H2O of the sphinganine base (panel B) and blue portion of molecule A and not of the NL-H2O of the sphingosine base (panel C) and blue portion of molecule B. This suggests the species to be characterized as Cer (d18:0/24:1) (molecule A).
In contrast, there are 8 Cer that are observed in both positive and negative ion mode, confirmed by exact mass and RT matches. When these species undergo HCD fragmentation in positive mode, the representative m/z 264.2679 established by LIPID MAPS to produce the sphingosine fragmentation is clearly observed (Sullards et al. 2007). This observation leaves the interpretation of the data on these compounds to be ambiguous. Further experiments with standards will be necessary to completely discern the fragmentations and fully characterize these species beyond exact mass matching, but, even with this limitation, our method has provided biologically relevant information regarding mitochondrial Cer distribution and quantitation. Table 3 provides an overview of the 32 SLs determined, presenting the details which were just discussed as well as the SMs and Gans which were also only identified via exact mass. This table includes each species, the ionization mode(s) in which it was observed, the m/z observed, the mass error, mean intensity and CV across the pools (n=8). The CV values were calculated from the raw peak areas established by SIEVE.
Table 3. Sphingolipids in Mitochondria.
The 31 total sphingolipids identified in mitochdonria via exact mass matches in HMDB and Metlin databases are shown with the observed variation across the pools (calculated from the raw peak areas established by SIEVE) and separated into Cer, SM, Gan subclasses.
| Sphingolipids (SL) | Ionization Mode | m/z | Error (ppm) | Intensity (n=8) | CV (n=8) |
|---|---|---|---|---|---|
| Sphingolipids (SL)Ceramides (Cer) | |||||
| Cer(d18:0/14:1) or Cer(d18:1:14:0) | − | 554.4800 | 1.77 | 218.08 | 7.51 |
| Cer(d18:0/16:0) | − | 584.5276 | 2.72 | 389.73 | 5.09 |
| Cer(d18:0/16:1) or Cer(d18:1/16:0) | −/+ | 582.5109 | 1.07 | 13011.92 | 4.17 |
| Cer(d18:0/18:1) or Cer(d18:1/18:0) | − | 610.5428 | 2.04 | 896.24 | 6.96 |
| Cer(d18:0/20:1) or Cer(d18:1/20:0) | − | 638.5739 | 1.59 | 565.03 | 8.13 |
| Cer(d18:0/22:0) or Cer(d18:1/22:1) | − | 668.6214 | 2.24 | 493.43 | 17.26 |
| Cer(d18:0/22:1) or Cer(18:1/22:0) | −/+ | 666.6050 | 1.26 | 10829.24 | 5.10 |
| Cer(d18:0/22:2) or Cer(d18:1/22:1) | − | 664.5903 | 2.76 | 1571.52 | 7.34 |
| Cer(d18:0/23:0) | −/+ | 682.6370 | 2.23 | 406.10 | 16.99 |
| Cer(d18:0/23:1) or Cer(d18:1/23:0) | − | 680.6205 | 0.86 | 14505.47 | 6.29 |
| Cer(d18:0/24:0) | −/+ | 696.6528 | 2.40 | 1782.43 | 7.34 |
| Cer(d18:0/24:1) or Cer(d18:1/24:0) | −/+ | 694.6365 | 1.49 | 77735.14 | 6.12 |
| Cer(d18:0/24:2) or Cer(d18:1/24:1) | − | 692.6205 | 0.84 | 31587.01 | 5.28 |
| Cer(d18:0/25:1) or Cer(d18:1/25:0) | −/+ | 708.6527 | 2.27 | 12455.20 | 4.22 |
| Cer(d18:0/26:0) | − | 724.6841 | 2.18 | 478.34 | 32.65 |
| Cer(d18:0/26:1) or Cer(d18:1/26:0) | −/+ | 722.6682 | 2.09 | 2093.79 | 6.65 |
| Cer(d18:0/26:2) or Cer(d18:1/26:1) | −/+ | 720.6525 | 1.90 | 2185.09 | 6.47 |
| GlcCer(d18:0/16:1) or GlcCer(d18:1/16:0) | − | 700.5724 | 0.23 | 5487.51 | 7.04 |
| GlcCer(d18:0/24:1) or GlcCer(d18:1/24:0) | −/+ | 856.6898 | 1.66 | 9681.37 | 5.97 |
| GlcCer(d18:0/24:2) or GlcCer(d18:1/24:1) | − | 854.6752 | 2.96 | 1372.72 | 4.98 |
| GlcCer(d18:0/25:1) or GlcCer(d18:1/25:0) | − | 870.7060 | 2.29 | 497.52 | 6.24 |
| Sphingomyelin (SM) | |||||
| SM(d18:0/24:2) or SM(d18:1/24:1) | − | 857.6774 | 2.48 | 5909.13 | 5.24 |
| SM(d18:0/18:1) or SM(d18:1/18:0) | − | 775.5988 | 2.13 | 528.09 | 6.70 |
| SM(d18:0/22:1) or SM(d18:1/22:0) | − | 831.6624 | 3.26 | 3535.84 | 4.52 |
| SM(d18:0/22:2) or SM(d18:1/22:1) | + | 787.6673 | −1.88 | 33719.26 | 5.97 |
| SM(d18:0/23:1) or SM(d18:1/23:0) | − | 845.6770 | 2.01 | 3103.26 | 5.95 |
| SM(d18:0/24:1) or SM(d18:1/24:0) | −/+ | 859.6932 | 2.65 | 14886.09 | 6.70 |
| Gangliosides (Gan) | |||||
| Gan(d18:0/18:2) or Gan(d18:1/18:1) | − | 1399.8468 | 3.41 | 5175.93 | 7.46 |
| Gan(d18:0/24:1) or Gan(d18:1/24:0) | − | 1511.9214 | 0.45 | 1001.41 | 4.46 |
| Gan(d18:0/26:0) | − | 1541.9683 | 0.36 | 998.71 | 7.50 |
| Gan(d18:0/26:1) or Gan(d18:1/26:0) | + | 1539.9506 | 0.87 | 1610.44 | 8.12 |
Glycerolipids
The glycerolipids (GL) class is mainly comprised of 3 subclasses, the monoacylglycerols (MGs), diacylglycerols (DG) and triacylglycerols (TGs); these 3 groups differ by the number of acyl chains connected to the primary backbone. The acyl group composition of these species, especially the TGs, can be very diverse and the chemical properties associated with each side-chain (i.e. chain length and degree of saturation) greatly affect the biological properties of the molecules. In the cell, TGs are stored energy and they are metabolized to free fatty acids used during β-oxidation in mitochondria.
We have previously demonstrated that the LC-MS profiling and HCD fragmentation methods employed can be used to profile a greater number of TGs than targeted methods in rat serum, and that these methods provide individual acyl side-chain compositions in a sensitive and robust manner (Bird et al. 2011b). The HCD fragmentations were chromatographically aligned with the parent ion, as shown for Cer in Figure 3, to structurally assign the acyl side-chains to each molecule where the signal was great enough to determine the fragments. This method, applied to the mitochondria samples, yielded 45 unique TG species, 41 of which are fully characterized. The subjectivity of the chromatographic alignment can cause problems when assessing a large pool of species which elute over a short period of time, such as with the TGs and this is the reason all TGs are not characterized in this study. The identified mitochondrial TGs are shown in Table 4, including each individual lipid mass, error, average intensity and CV from the pool (n=8). The MG and DG molecules present in mitochondria where only identified using exact mass, and are shown in Table 5 providing the same statistics as for the TGs, which highlight the accuracy and reproducibility of the method.
Table 5. MGs and DGs found in Mitochondria.
The MG and DG glycerolipids identified in mitochondria by exact mass matching and their statistics across the pools, calculated from raw peak areas are highlighted.
| Glycerolipids (GLs) | Retention Time (min) | m/z [M+NH4]+ | Error (ppm) | Intensity (n=8) | CV (n=8) |
|---|---|---|---|---|---|
| Monoacylglycerols (MGs) | |||||
| MG(0:0/18:3) | 3.73 | 370.2947 | −1.05 | 30076.64 | 3.63 |
| MG(0:0/20:1) | 3.31 | 402.3573 | −0.99 | 7149.69 | 4.37 |
| MG(0:0/22:1) | 5.10 | 430.3884 | −1.61 | 14928.54 | 3.46 |
| MG(plasmalogen18:0) | 2.34 | 360.3466 | −1.61 | 126268.17 | 3.93 |
| Diacylglycerols (DGs) | |||||
| DG(36:1) | 12.49 | 640.5872 | 0.59 | 8454.21 | 4.09 |
| DG(38:1) | 13.99 | 668.6191 | 0.53 | 7049.42 | 5.74 |
| DG(38:5) | 17.21 | 660.5557 | −0.42 | 5559.73 | 5.26 |
| DG(40:10) | 15.52 | 696.6491 | −1.22 | 6229.87 | 4.47 |
Glycerolphospholipids
Glycerolphospholipids (PL) were once regarded as merely structurally important species, providing only membrane integrity, which greatly underestimates the importance these species play in mainlining proper mitochondrial function. PLs are now known to participate in important mitochondrial processes that extend to cell signaling, bioenergetics, protein transport and apoptosis (Osman et al. 2011). Additionally, observations regarding changes to mitochondrial PL levels have been linked to human disease states (Chicco and Sparagna 2007) indicating that these species are tightly regulated and that changes to this composition, either in concentration or structural make-up, are not well tolerated.
Generally, changes to PLs are assessed by aggregate amount, meaning the total of each subclass, such as phosphocholine (PC) or phosphoethanolamine (PE), found in the sample. This method is limiting in that it ignores the inherent diversity of each aggregate pool and assumes each lipid in those subclasses is regulated in the same way. Additionally, as was displayed in the TG analysis, the FA side-chain composition can be quite varied and changes to those side-chains may be related to biological function. Monitoring lipids with that level of specificity can provide more information regarding mitochondrial PL regulation and proper mitochondrial function.
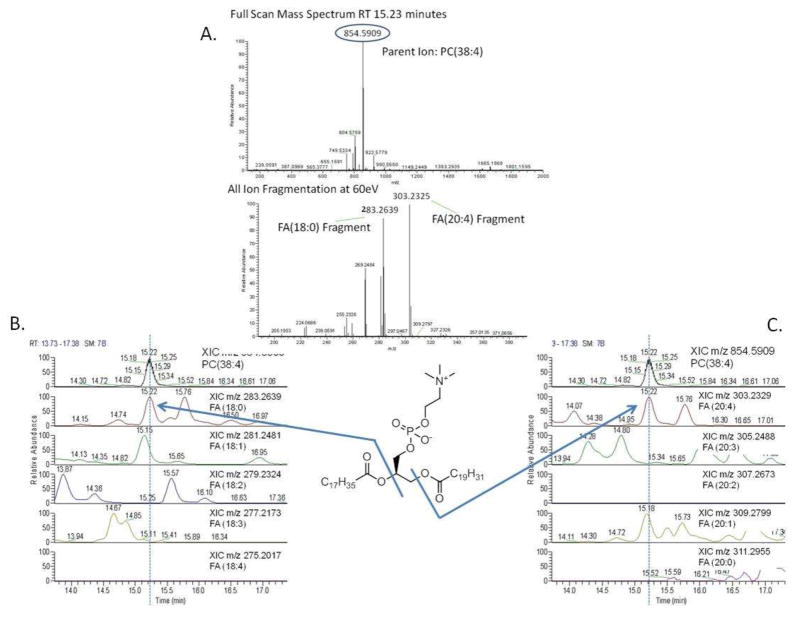
All PL species were identified and characterized using exact mass profiling and HCD fragmentation to cleave the individual fatty acyl side-chains from each molecule and chromatographically align those pieces with the parent for characterization. An example of this is shown in Figure 4, where an unknown PC with a m/z 854.5909 for the [M+Formic Acid]-species was observed at 15.23 minutes. Exact mass calculation identifies this mass as a PC with a total of 38 carbons and 4 double bonds, suggesting at least 5 combinations of fatty acyl side-chains which could comprise this molecule. HCD fragmentation at this time, provides diagnostic fragments of m/z 283.2639 and m/z 303.2325, representing the [M-H]-ions for fatty acid 18:0 and 20:4 respectively. Chromatographic alignment of these masses, as well as the other possibilities which could comprise this PC are also shown in Figure 4, where only the fragments for 18:0 and 20:4 were aligned with the parent. This characterization method was applied to all PLs identified in mitochondria, and just likes with the TGs only a percentage of the total PLs were characterized. This is often due to chromatographic overlap of high and low abundance PLs, which causes the fragment alignment for the low abundant peaks to be indistinguishable from the high abundant species. At times when the FA side-chains are unable to be characterized via HCD fragmentation, the lipids are distinguished by their total acyl carbon and double bonds as calculated from exact mass matches.
Figure 4.
Characterization of PC(18:0/20:4) Panel A shows an unknown PC with a m/z 854.5909 for the [M+Formic Acid]-parent ion and HCD fragmentation, providing diagnostic fragments of m/z 283.2639 and m/z 303.2325, representing the [M-H]-ions for fatty acid 18:0 and 20:4 respectively. Chromatographic alignment of these masses, as well as the other possibilities that could comprise this PC are also shown in panels B and C, where only the fragments for 18:0 and 20:4 were aligned with the parent.
Using the LC-MS profiling method described, 289 unique species from 11 PL subclasses are observed. This number represents approximately 75% of the total lipids that have been presently identified in this mitochondrial study. The composition of the total PL pool is presented in Tables 6a, 6b and 6c where all lipids from each subclass are highlighted. Due to the large number of species determined, the information regarding the ionization mode(s) in which they were observed, their exact mass, mass error, mean intensity and CV (n=8) are presented in subsequent charts in the supplemental information. The cardiolipin (CL) and monolysocardiolipin (MLCL) species are not outlined in this table, as they have been characterized and discussed in detail elsewhere.(Bird et al. 2011a) Consistent with previous reports (Andreyev et al. 2010; Osman et al. 2011; Vance 2008), the majority of the PLs were PCs, 43%, followed by PE at 26%, PG at 11%, CL at 10%, and PS and PI at 6% and 4% respectively.
Table 6a. PC subclasses found in mitochondria.
Species that shown with their FA side-chains were identified using HCD fragmentation and chromatographic alignment in negative ion mode; the species that only show the total number of carbon:double bonds in the molecule, were identified via exact mass matching to one of the databases (HMDB, Metlin, LIPID MAPS) searched in positive ion mode.
| Glycerophospholipids (PLs) | ||||
|---|---|---|---|---|
| Phosphocholine (PC) | ||||
| PC(14:1/22:5) | PC(18:1/18:1) | PC(20:5/16:0) | PC(34:4) or PE(37:4) | PC(42:9) |
| PC(14:1/22:5) | PC(18:1/18:1) | PC(22:5/15:0) | PC(35:1) or PE(38:1) | PC(44:11) |
| PC(15:0/15:0) | PC(18:2/16:0) | PC(22:5/15:0) | PC(35:1) or PE(38:1) | PC(44:12) |
| PC(15:0/16:1) | PC(18:2/16:1) | PC(22:5/16:0) | PC(35:2) or PE(38:2) | |
| PC(15:0/18:1) | PC(18:2/18:0) | PC(22:5/16:0) | PC(35:3) or PE(38:3) | |
| PC(15:0/18:2) | PC(18:2/20:4) | PC(22:6/15:0) | PC(36:3) | |
| PC(15:0/20:2) | PC(18:3/14:0) | PC(22:6/16:0) | PC(36:3) | |
| PC(15:0/20:3) | PC(18:3/16:0) | PC(22:6/16:1) | PC(36:4) | |
| PC(15:0/20:3) | PC(18:3/18:4) | PC(22:6/20:4) | PC(38:0) | |
| PC(15:0/20:4) | PC(18:3/22:4) | PC(32:1) or PE(35:1) | PC(38:0) | |
| PC(15:0/20:4) | PC(18;1/16:0) | PC(34:0) | PC(38:8) | |
| PC(15:0/22:1) | PC(20:1/18:0) | PC(34:3) | PC(38:8) | |
| PC(16:0/16:0) | PC(20:2/18:0) | PC(38:7) | PC(38:9) | |
| PC(16:0/20:5) | PC(20:2/18:0) | PC(38:7) | PC(40:10) | |
| PC(16:0/20:5) | PC(20:3/18:0) | PC(40:4) | PC(40:10) | |
| PC(16:1/14:0) | PC(20:3/18:0) | PC(40:6) | PC(40:3) | |
| PC(17:0/20:4) | PC(20:3/18:0) | PC(40:6) | PC(40:8) | |
| PC(18:0/14:1) | PC(20:4/14:0) | PC(40:7) | PC(42:10) | |
| PC(18:0/18:0) | PC(20:4/14:1) | PC (32:1) or PE (35:1) | PC(42:11) | |
| PC(18:0/22:4) | PC(20:4/16:0) | PC (32:1) or PE (35:1) | PC(42:7) | |
| PC(18:0/22:5) | PC(20:4/16:1) | PC (32:2) or PE (35:2) | PC(42:8) | |
| PC(18:0/22:5) | PC(20:4/18:0) | PC (32:4) or PE (35:4) | PC(42:8) | |
| PC(18:0/22:6) | PC(20:4/18:0) | PC (32:4) or PE (35:4) | PC(42:8) | |
| PC(18:1/14:1) | PC(20:4/20:4) | PC(34:2) or PE(37:2) | PC(42:9) | |
| PC(18:1/18:0) | PC(20:4/20:5) | PC(34:4) or PE(37:4) | PC(42:9) | |
| plasmalogenphosphocholine (plsPC) | lysoPhosphocholine (LysoPC) | |||
| PC plasmalogen(38:4) | LysoPAF(C16) | lysoPC(20:0) | lysoPC(18:3) | |
| PC(plasmalogen(18:3/18:0) | LysoPC (16:0) | lysoPC(20:3) | LysoPC(22:0) | |
| PC(plasmalogen36:5) | LysoPC (16:1) | LysoPC(20:4) | lysoPC(4:0) | |
| PCplasmalogen(34:0) | lysoPC (18:0) | lysoPC(20:5) | lysoPC(0 12:0) | |
| PCplasmalogen(38:3) | lysoPC (18:1) | lysoPC(22:5) | lysoPC(0 –12:0/o –1:0) | |
| lysoPC (18:2) | lysoPC(22:6) | lysoPC(0 –12:0/o -1:0) | ||
| lysoPC(14:0) | lysoPAF(C18:0) | |||
Table 6b. PE subclasses found in mitochondria.
Species that shown with their FA side-chains were identified using HCD fragmentation and chromatographic alignment in negative ion mode; the species that only show the total number of carbon:double bonds in the molecule, were identified via exact mass matching to one of the databases (HMDB, Metlin, LIPID MAPS) searched also in negative ion mode.
| Glycerophospholipids (PLs) | ||||
|---|---|---|---|---|
| Phosphoethanolamine (PE) | plasmalogenphosphoethanolamine (plsPE) | |||
| PE(14:0/20:3) | PE(22:5/16:0) | PE(40:1) | PE(20:4/plasmalogen18:0) | |
| PE(15:0/15:0) | PE(22:5/18:0) | PE(40:2) | PE(20:4/plasmalogen18:1) | |
| PE(16:0/16:1) | PE(22:5/18:2) | PE(40:2) | PE(22:4/plasmalogen16:0) | |
| PE(16:0/18:0) | PE(22:6/15:0) | PE(40:5) | PEplasmalogen(38:6) | |
| PE(16:0/20:4) | PE(22:6/16:0) | PE(40:8) | PEplasmalogen(36:4) | |
| PE(18:0/22:4) or PE(20:4/20:0) | PE(22:6/18:0) | PE(42:4) | ||
| PE(18:0/22:4) or PE(20:4/20:0) | PE(22:6/18:2) | PE(42:4) | ||
| PE(18:1/16:0) | PE(22:6/20:0) | PE(42:5) | lysoPhosphoethanolamine (LysoPE) | |
| PE(18:1/18:0) | PE(22:6/22:6) | PE(44:5) | LysoPE(16:0) | LysoPE(22:0) |
| PE(18:2/16:0) | PE(32:0) | PE(44:6) | LysoPE(18:0) | LysoPE(22:5) |
| PE(18:2/18:0) | PE(32:2) | LysoPE(18:1) | LysoPE(22:6) | |
| PE(18:3/18:0) | PE(33:1) | LysoPE(18:2) | LysoPE(20:4) | |
| PE(18:3/18:2) | PE(34:4) | lysoPE(20:4) | ||
| PE(20:3/18:0) | PE(34:4) | |||
| PE(20:3/18:0) | PE(36:0) | |||
| PE(20:3/18:4) | PE(36:1) | |||
| PE(20:4/15:0) | PE(36:3) | |||
| PE(20:4/16:1) | PE(40:3) | |||
| PE(20:4/18:0) | PE(40:3) | |||
| PE(20:4/22:1) | PE(40:6) | |||
| PE(20:4/22:6) or (20:5:22:5) | PE (34:1) or PC (31:1) | |||
| PE(20:5/18:0) | PE (34:3) | |||
| PE(20:5/18:2) | PE(36:2) or PC(33:2) | |||
| PE(22:4/15:0) | PE(36:2) or PC(33:2) | |||
| PE(22:4/16:0) | PE(38:8) | |||
Table 6c. PG, PS, PI and CL subclass species found in mitochondria.
Species that shown with their FA side-chains were identified using HCD fragmentation and chromatographic alignment in negative ion mode; the species that only show the total number of carbon:double bonds in the molecule, were identified via exact mass matching to one of the databases (HMDB, Metlin, LIPID MAPS) searched. PS species were only observed in negative mode, while all PI species were exact mass matched using positive ion mode.
| Glycerophospholipids (PLs) | |||
|---|---|---|---|
| Phosphoglycerol (PG) | Phosphoserine (PS) | Phosphoinositol (PI) | |
| PG(16:0/16:0) | PG(32:2) or PA(36:2) | PS(16:0/16:0) | PI(36:2) |
| PG(16:0/16:1) | PG (34:0) | PS(16:0/18:3) | PI(36:4) |
| PG(16:0/20:4) | PG (34:2) | PS(16:0/20:3) | PI(38:3) |
| PG(16:1/16:1) | PG (36:0) | PS(16:0/22:6) | PI(38:3) |
| PG(16:1/18:2) | PGP(34:2) | PS(16:0/22:6) | PI(38:4) |
| PG(18:0/18:1) | PGP(36:0) | PS(16:1/22:6) | PI(38:4) |
| PG(18:0/18:2) | PGP(36:1) | PS(18:0/22:6) | PI(38:5) |
| PG(18:0/18:3) | PGP(36:1) | PS(18:1/22:6) | PI(38:5) |
| PG(18:1/16:0) | PGP(36:2) | PS(18:2/18:2) | PI(40:4) |
| PG(18:1/18:1) | PGP(36:3) | PS(18:2/22:6) | PI(40:5) |
| PG(18:1/20:3) | PGP(38:4) | PS(18:2/22:6) or PS(20:3/20:4) | PIP(38:2) |
| PG(18:1/22:5) | PGP(40:6) | PS(20:0/18:0) | |
| PG(18:2/18:2) | PGP(40:6) | PS(20:3/18:1) | |
| PG(18:2/20:3) | PS(20:3/18:2) | ||
| PG(18:2/22:6) | PS(20:3/18:2) | ||
| PG(22:5/16:1) | PS(22:5/18:0) | ||
| PG(22:6/16:1) | PS(22:5/28:0) | ||
| PG(22:6/18:1) | PS(22:6/18:3) | ||
| Cardiolipin (CL) | monolysocardiolipin (MLCL) | ||
| CL (14:0)(16:1)(16:1)(18:2) | CL (18:2)(18:2)(18:2)(22:6) | CL (18:1)(18:1)(18:2)(18:2) | MLCL(18:2)3 |
| CL (18:2)(18:3)(20:4)(22:6) | CL (18:1)(18:1)(18:1)(18:1) | CL(18:2)(18:2)(18:2)(16:1) | MLCL(18:2)2(18:1) |
| CL (18:1)(18:2)(20:3)(22:6) | CL (18:1)(18:1)(18:2)(22:6) | CL (18:2)(18:2)(18:2)(20:3) | |
| CL (18:1)(18:1)(18:1)(18:2) | CL (14:0)(16:0)(18:2)(20:4) | CL(18:2)(18:2)(18:1)(16:1) | |
| CL (16:0)(18:1)(18:1)(18:2) | CL (18:2)18:2)(20:3)(20:4) | CL (18:2)(18:2)(18:2)(18:2) | |
| CL (18:2)(18:2)(20:3)(22:6) | CL (16:1)(18:2)(18:2)(22:6) | CL (18:2)(18:2)(18:2)(18:1) | |
| CL (14:0)(16:1)(18:1)(18:2) | CL(16:1)(16:1)(18:1)(18:2) | ||
| CL (18:2)(18:2)(20:4)(22:6) | CL (18:2)(18:1)(18:2)(20:3) | ||
| CL (14:0)(16:1)(18:2)(18:2) | CL (16:1)(18:1)(18:1)(18:2) | ||
| CL (16:0)(16:1)(18:1)(18:2) | CL (18:2)(18:2)(18:2)(18:3) | ||
CONCLUSION
The intention of this report is to show the extent to which our non-targeted LC-MS profiling method, with HCD, could profile, identify and characterize the mitochondrial lipidome. We present a full in depth analysis of the rat liver mitochondrial lipidome using both positive and negative ionization profiling. This analysis yielded a total of 381 unique lipid species across 5 LIPID MAPS categories with 236 characterized by FA side-chain using HCD fragmentation and chromatographic alignment. For a non-targeted analysis, this coverage is diverse and specific, providing lipid details that can be used to contribute to understanding of mitochondrial membrane function, oxidative stress, apoptosis, bioenergetics and cellular respiration. The data presented highlighted the use of all ion fragmentation and chromatographic alignment for characterization, showing times where the method falls short as in the case of the Cer and when it excels in providing information key to any biological analysis, as seen in the PL analysis. The key to this type of non-targeted study is to provide reproducible, robust and sensitive analytical results which can address a variety of biological questions in a single analysis. Additionally, all data acquired can be queried again as future questions arise. The sensitivity, specificity and robustness of this method suggests its application to other systems can provide discovery information necessary to answer biological questions unable to be studied by previous methodologies.
Supplementary Material
Table 4. TGs in Mitochondria.
The 43 TGs identified in mitochondria and their statistics across the pools, calculated from raw peak area values are highlighted. 4 TGs were identified via exact mass matching, shown by giving the total number of carbon and double bonds in the molecule, e.g., TG 51:1, which the remaining 39 were characterized via HCD fragmentation and chromatographic alignment as explained in the text.
| Glycerolipids (GLs) | Retention Time (min) | m/z [M+NH4]+ | Error (ppm) | Intensity (n=8) | CV (n=8) |
|---|---|---|---|---|---|
| Triacylglycerols (TG) | |||||
| TG(42:0) | 23.14 | 740.6751 | −1.50 | 8842.53 | 2.37 |
| TG(44:1) | 23.18 | 766.6910 | 0.53 | 14431.06 | 1.75 |
| TG(44:0) | 23.54 | 768.7069 | −0.84 | 21076.17 | 2.43 |
| TG(16:1/14:0/16:1) | 23.24 | 792.7066 | −1.25 | 18642.42 | 1.10 |
| TG(16:0/14:0/16:1) | 23.58 | 794.7220 | −1.46 | 46902.02 | 2.32 |
| TG(16:0/14:0/16:0) | 23.94 | 796.7380 | −0.96 | 54969.44 | 2.51 |
| TG(16:1/14:0/18:2) | 23.30 | 818.7223 | −1.12 | 12904.38 | 1.72 |
| TG(16:0/16:1/16:1) | 23.55 | 820.7373 | −1.87 | 66223.40 | 2.99 |
| TG(16:0/16:0/16:1) | 23.94 | 822.7530 | −1.82 | 129812.51 | 2.73 |
| TG(16:0/16:0/16:0) | 24.34 | 824.7683 | −2.16 | 82666.03 | 3.12 |
| TG(15:0/18:1/16:1) | 23.79 | 834.7535 | −1.24 | 36149.02 | 2.26 |
| TG(15:0/18:1/16:0) | 24.12 | 836.7687 | −1.71 | 53382.01 | 2.71 |
| TG(16:1/16:2/18:1) | 23.45 | 844.7381 | −0.88 | 7706.02 | 1.94 |
| TG(16:0/16:1/18:2) | 23.58 | 846.7528 | −2.01 | 44360.38 | 3.33 |
| TG(16:1/16:0/18:1) | 23.90 | 848.7679 | −2.57 | 221864.86 | 3.40 |
| TG(16:0/16:0/18:1) | 24.32 | 850.7839 | −2.23 | 223290.46 | 3.55 |
| TG(16:0/16:0/18:0) | 24.76 | 852.8001 | −1.59 | 28738.42 | 1.90 |
| TG(51:3) | 23.79 | 860.7691 | −1.19 | 13782.24 | 3.08 |
| TG(15:0/18:1/18:1) | 24.12 | 862.7845 | −1.54 | 35190.99 | 2.08 |
| TG(18:1/14:0/20:4) | 23.44 | 870.7535 | −1.18 | 15613.91 | 2.59 |
| TG(18:1/16:0/18:3) | 23.63 | 872.7685 | −1.88 | 48442.48 | 2.81 |
| TG(18:1/16:0/18:2) | 23.94 | 874.7832 | −2.99 | 163329.11 | 3.04 |
| TG(18:1/16:0/18:1) | 24.30 | 876.7988 | −3.07 | 420036.40 | 4.32 |
| TG(16:0/16:0/20:1) | 24.73 | 878.8151 | −2.28 | 68296.23 | 1.60 |
| TG(18:2/18:2/18:3) | 22.98 | 894.7538 | −0.81 | 9858.46 | 2.29 |
| TG(16:0/16:0/22:6) | 23.31 | 896.7683 | −2.00 | 41949.09 | 2.03 |
| TG(18:1/18:1/18:3) | 23.82 | 898.7835 | −2.58 | 92349.53 | 3.15 |
| TG(18:2/16:0/20:2) | 23.94 | 900.7990 | −2.81 | 89876.78 | 3.14 |
| TG(18:1/16:0/20:2) | 24.30 | 902.8140 | −3.46 | 163085.51 | 3.92 |
| TG(16:0/18:1/20:1) | 24.29 | 904.8300 | −3.12 | 72646.91 | 3.27 |
| TG(16:0/18:1/20:1) | 24.80 | 904.8282 | −5.14 | 62395.92 | 6.30 |
| TG(16:0/18:0/20:1) | 25.18 | 906.8469 | −1.70 | 24588.81 | 2.80 |
| TG(18:2/16:0/22:6 | 23.39 | 920.7688 | −1.46 | 4161.86 | 2.98 |
| TG(18:1/16:0/22:6) | 23.65 | 922.7837 | −2.31 | 7296.70 | 4.16 |
| TG(18:1/16:0/22:5) | 23.82 | 924.7993 | −2.34 | 17143.46 | 3.47 |
| TG(18:1/18:1/20:3) | 23.82 | 926.8153 | −1.97 | 15708.78 | 3.09 |
| TG(18:1/18:1/20:2) | 24.30 | 928.8311 | −1.88 | 16358.28 | 3.94 |
| TG(16:1/20:1/20:1) | 24.27 | 930.8467 | −1.82 | 14833.24 | 3.57 |
| TG(18:1/18:1/20:1) | 24.77 | 930.8470 | −1.55 | 16618.48 | 4.55 |
| TG(16:0/20:1/20:1) | 25.15 | 932.8628 | −1.35 | 14641.88 | 2.03 |
| TG(18:0/18:0/20:1) | 25.73 | 934.8788 | −1.08 | 12148.54 | 1.71 |
| TG(18:1/18:1/22:6) | 23.75 | 948.7991 | −2.49 | 4051.18 | 3.82 |
| TG(18:0/20:1/20:1) | 25.62 | 960.8943 | −0.99 | 8638.94 | 2.35 |
Acknowledgments
The studies reported were funded by U01-ES16048 (BSK, PI), a part of the NIH Genes and Environment Initiative (GEI). The authors also thank ThermoFisher for the loan of an Exactive Benchtop Orbitrap for demonstration testing and financial support for scientific meeting attendance.
References
- Aberg F, Appelkvist EL, Dallner G, Ernster L. Distribution and redox state of ubiquinones in rat and human tissues. Arch Biochem Biophys. 1992;295(2):230–234. doi: 10.1016/0003-9861(92)90511-t. [DOI] [PubMed] [Google Scholar]
- Andreyev AY, Fahy E, Guan Z, Kelly S, Li X, McDonald JG, et al. Subcellular organelle lipidomics in TLR-4-activated macrophages. J Lipid Res. 2010;51(9):2785–2797. doi: 10.1194/jlr.M008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardail D, Popa I, Alcantara K, Pons A, Zanetta JP, Louisot P, et al. Occurrence of ceramides and neutral glycolipids with unusual long-chain base composition in purified rat liver mitochondria. FEBS Lett. 2001;488(3):160–164. doi: 10.1016/s0014-5793(00)02332-2. [DOI] [PubMed] [Google Scholar]
- Barzanti V, Battino M, Baracca A, Cavazzoni M, Cocchi M, Noble R, et al. The effect of dietary lipid changes on the fatty acid composition and function of liver, heart and brain mitochondria in the rat at different ages. Br J Nutr. 1994;71(2):193–202. doi: 10.1079/bjn19940126. [DOI] [PubMed] [Google Scholar]
- Bhuiyan AK, Jackson S, Turnbull DM, Aynsley-Green A, Leonard JV, Bartlett K. The measurement of carnitine and acyl-carnitines: application to the investigation of patients with suspected inherited disorders of mitochondrial fatty acid oxidation. Clin Chim Acta. 1992;207(3):185–204. doi: 10.1016/0009-8981(92)90118-a. [DOI] [PubMed] [Google Scholar]
- Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Lipidomics profiling by high-resolution LC-MS and high-energy collisional dissociation fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins. Anal Chem. 2011a;83(3):940–949. doi: 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS. Serum Lipidomics Profiling Using LC-MS and High-Energy Collisional Dissociation Fragmentation: Focus on Triglyceride Detection and Characterization. Anal Chem. 2011b;83(17):6648–6657. doi: 10.1021/ac201195d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bou Khalil M, Hou W, Zhou H, Elisma F, Swayne LA, Blanchard AP, et al. Lipidomics era: accomplishments and challenges. Mass Spectrom Rev. 2009;29(6):877–929. doi: 10.1002/mas.20294. [DOI] [PubMed] [Google Scholar]
- Burke WJ, Kristal BS, Yu BP, Li SW, Lin TS. Norepinephrine transmitter metabolite generates free radicals and activates mitochondrial permeability transition: a mechanism for DOPEGAL-induced apoptosis. Brain Res. 1998;787(2):328–332. doi: 10.1016/s0006-8993(97)01488-1. [DOI] [PubMed] [Google Scholar]
- Cequier-Sanchez E, Rodriguez C, Ravelo AG, Zarate R. Dichloromethane as a solvent for lipid extraction and assessment of lipid classes and fatty acids from samples of different natures. J Agric Food Chem. 2008;56(12):4297–4303. doi: 10.1021/jf073471e. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Yu BP. Alterations in mitochondrial membrane fluidity by lipid peroxidation products. Free Radic Biol Med. 1994;17(5):411–418. doi: 10.1016/0891-5849(94)90167-8. [DOI] [PubMed] [Google Scholar]
- Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. Am J Physiol Cell Physiol. 2007;292(1):C33–44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- Crane FL. Discovery of ubiquinone (coenzyme Q) and an overview of function. Mitochondrion. 2007;7(Suppl):S2–7. doi: 10.1016/j.mito.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Dennis EA. Lipidomics joins the omics evolution. Proc Natl Acad Sci U S A. 2009;106(7):2089–2090. doi: 10.1073/pnas.0812636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paola M, Cocco T, Lorusso M. Ceramide interaction with the respiratory chain of heart mitochondria. Biochemistry. 2000;39(22):6660–6668. doi: 10.1021/bi9924415. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Moehring T, Bahr U, Duchoslav E, Karas M, Simons K, et al. Collision-induced dissociation pathways of yeast sphingolipids and their molecular profiling in total lipid extracts: a study by quadrupole TOF and linear ion trap-orbitrap mass spectrometry. J Mass Spectrom. 2006;41(3):372–389. doi: 10.1002/jms.997. [DOI] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106(7):2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11(1):81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr, Murphy RC, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46(5):839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35(Web Server issue):W606–612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudz TI, Tserng KY, Hoppel CL. Direct inhibition of mitochondrial respiratory chain complex III by cell-permeable ceramide. J Biol Chem. 1997;272(39):24154–24158. doi: 10.1074/jbc.272.39.24154. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J Lipid Res. 2003;44(6):1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- Han X, Gross RW. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom Rev. 2005;24(3):367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- Han X, Yang J, Cheng H, Yang K, Abendschein DR, Gross RW. Shotgun lipidomics identifies cardiolipin depletion in diabetic myocardium linking altered substrate utilization with mitochondrial dysfunction. Biochemistry. 2005;44(50):16684–16694. doi: 10.1021/bi051908a. [DOI] [PubMed] [Google Scholar]
- Han X, Yang K, Yang J, Cheng H, Gross RW. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. J Lipid Res. 2006;47(4):864–879. doi: 10.1194/jlr.D500044-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. The Ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J Biol Chem. 2002;277(29):25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- Hu C, van Dommelen J, van der Heijden R, Spijksma G, Reijmers TH, Wang M, et al. RPLC-ion-trap-FTMS method for lipid profiling of plasma: method validation and application to p53 mutant mouse model. J Proteome Res. 2008;7(11):4982–4991. doi: 10.1021/pr800373m. [DOI] [PubMed] [Google Scholar]
- Kiebish MA, Han X, Seyfried TN. Examination of the brain mitochondrial lipidome using shotgun lipidomics. Methods Mol Biol. 2009;579:3–18. doi: 10.1007/978-1-60761-322-0_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasnikov BF, Kim SY, McConoughey SJ, Ryu H, Xu H, Stavrovskaya I, et al. Transglutaminase activity is present in highly purified nonsynaptosomal mouse brain and liver mitochondria. Biochemistry. 2005;44(21):7830–7843. doi: 10.1021/bi0500877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristal BS, Conway AD, Brown AM, Jain JC, Ulluci PA, Li SW, et al. Selective dopaminergic vulnerability: 3,4-dihydroxyphenylacetaldehyde targets mitochondria. Free Radic Biol Med. 2001;30(8):924–931. doi: 10.1016/s0891-5849(01)00484-1. [DOI] [PubMed] [Google Scholar]
- Kristal BS, Park BK, Yu BP. 4-Hydroxyhexenal is a potent inducer of the mitochondrial permeability transition. J Biol Chem. 1996;271(11):6033–6038. doi: 10.1074/jbc.271.11.6033. [DOI] [PubMed] [Google Scholar]
- Land JM, Morgan-Hughes JA, Hargreaves I, Heales SJ. Mitochondrial disease: a historical, biochemical, and London perspective. Neurochem Res. 2004;29(3):483–491. doi: 10.1023/b:nere.0000014819.53972.b0. [DOI] [PubMed] [Google Scholar]
- Lang JK, Packer L. Quantitative determination of vitamin E and oxidized and reduced coenzyme Q by high-performance liquid chromatography with in-line ultraviolet and electrochemical detection. J Chromatogr. 1987;385:109–117. doi: 10.1016/s0021-9673(01)94626-6. [DOI] [PubMed] [Google Scholar]
- Lenaz G. Role of mobility of redox components in the inner mitochondrial membrane. J Membr Biol. 1988;104(3):193–209. doi: 10.1007/BF01872322. [DOI] [PubMed] [Google Scholar]
- Lenaz G, Fato R, Formiggini G, Genova ML. The role of Coenzyme Q in mitochondrial electron transport. Mitochondrion. 2007;7(Suppl):S8–33. doi: 10.1016/j.mito.2007.03.009. [DOI] [PubMed] [Google Scholar]
- Li M, Zhou Z, Nie H, Bai Y, Liu H. Recent advances of chromatography and mass spectrometry in lipidomics. Anal Bioanal Chem. 2010;399(1):243–249. doi: 10.1007/s00216-010-4327-y. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193(1):265–275. [PubMed] [Google Scholar]
- McKenzie M, Liolitsa D, Hanna MG. Mitochondrial disease: mutations and mechanisms. Neurochem Res. 2004;29(3):589–600. doi: 10.1023/b:nere.0000014829.42364.dd. [DOI] [PubMed] [Google Scholar]
- Millington DS, Stevens RD. Acylcarnitines: analysis in plasma and whole blood using tandem mass spectrometry. Methods Mol Biol. 2011;708:55–72. doi: 10.1007/978-1-61737-985-7_3. [DOI] [PubMed] [Google Scholar]
- Monette JS, Gomez LA, Moreau RF, Bemer BA, Taylor AW, Hagen TM. Characteristics of the rat cardiac sphingolipid pool in two mitochondrial subpopulations. Biochem Biophys Res Commun. 2010;398(2):272–277. doi: 10.1016/j.bbrc.2010.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. J Cell Biol. 2011;192(1):7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietilainen KH, Sysi-Aho M, Rissanen A, Seppanen-Laakso T, Yki-Jarvinen H, Kaprio J, et al. Acquired obesity is associated with changes in the serum lipidomic profile independent of genetic effects - a monozygotic twin study. PLoS ONE. 2007;2(2):e218. doi: 10.1371/journal.pone.0000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quehenberger O, Armando AM, Dennis EA. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbalip.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27(6):747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Stavrovskaya IG, Baranov SV, Guo X, Davies SS, Roberts LJ, 2nd, Kristal BS. Reactive gamma-ketoaldehydes formed via the isoprostane pathway disrupt mitochondrial respiration and calcium homeostasis. Free Radic Biol Med. 2010;49(4):567–579. doi: 10.1016/j.freeradbiomed.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrovskaya IG, Narayanan MV, Zhang W, Krasnikov BF, Heemskerk J, Young SS, et al. Clinically approved heterocyclics act on a mitochondrial target and reduce stroke-induced pathology. J Exp Med. 2004;200(2):211–222. doi: 10.1084/jem.20032053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullards MC, Allegood JC, Kelly S, Wang E, Haynes CA, Park H, et al. Structure-specific, quantitative methods for analysis of sphingolipids by liquid chromatography-tandem mass spectrometry: "inside-out" sphingolipidomics. Methods Enzymol. 2007;432:83–115. doi: 10.1016/S0076-6879(07)32004-1. [DOI] [PubMed] [Google Scholar]
- Vance JE. Phosphatidylserine and phosphatidylethanolamine in mammalian cells: two metabolically related aminophospholipids. J Lipid Res. 2008;49(7):1377–1387. doi: 10.1194/jlr.R700020-JLR200. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37(Database issue):D603–610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.