Abstract
CD8+ T lymphocytes recognize short peptides of ~8–10 amino acids bound to MHC class I molecules (pMHC) on the surface of antigen presenting cells. These peptides can be generated from either endogenous proteins synthesized by the biosynthetic machinery of the presenting cell or from exogenously sourced proteins. Because much of the research characterizing the MHC class I processing pathway has focused on endogenously synthesized proteins, it is not known whether differences exist in the processing pathway followed by endogenously synthesized versus exogenously sourced proteins. To highlight potential differences in the processing of endogenous versus exogenous proteins, we developed a model system to measure the efficiency of pMHC generation from nearly identical recombinant proteins expressed from vaccinia virus and Listeria monocytogenes. In these experiments, we uncovered a striking difference in the way recombinant Listeria antigens are processed and presented when compared to endogenously synthesized viral proteins. Specifically, we find that pMHC production from secreted Listeria proteins occurs at the same rate, independent of the cellular half-life of the protein from which it is derived, whereas the rate of pMHC production from endogenously synthesized viral proteins is absolutely dependent on its protein half-life. Accordingly, our data demonstrate the existence of a distinct and highly efficient MHC class I presentation pathway used for the processing of at least some exogenously synthesized proteins.
Introduction
Most, though not all, peptides presented by MHC class I molecules (pMHC) are generated by the degradation of proteins via the ubiquitin proteasome system. A general feature of this pathway is that, for a given protein, increasing the number of molecules degraded will result in a proportionally greater supply of peptides available for presentation on MHC class I molecules. Consequently, the supply of peptides available for presentation by MHC class I molecules can be increased by radically shortening the cellular half-life of a protein through the introduction of amino acid sequences that target the protein for degradation (1–3). This approach has been used in several studies and, in virtually every system tested, increasing degradation of a given protein has resulted in a concomitant increase in surface expression of pMHC class I molecules containing peptides derived from that protein (3, 4).
We have previously shown that there is an inverse correlation between protein half-life and pMHC surface levels using a recombinant vaccinia virus (rVV) expression system (4). By measuring the kinetics of surface pMHC expression, we showed that decreasing the cellular t1/2 of a substrate protein results in a concomitant increase in pMHC generation. In addition, we determined that the average efficiency of surface pMHC expression for a given peptide generated from a recombinant protein is ~1 pMHC per 3000 protein molecules degraded. An important observation made during the course of these studies was that highly similar proteins can be processed with different efficiencies. Specifically, pMHC complexes are generated more efficiently from a cellular pool of recombinant influenza nucleoprotein with a t1/2 ~ 70 min than from a nearly identical protein designed to be degraded in a near co-translational manner (t1/2 ~ 10 min).
The only other study published to date examining the efficiency of pMHC generation is that of Pamer and colleagues, which measured pMHC generation from proteins secreted by Listeria monocytogenes (Listeria) (5). Listeria is an intracellular gram positive bacterium that infects primarily professional phagocytic cells such as macrophages and dendritic cells. Upon uptake, Listeria initially reside within the host cell phagosome. Within minutes of formation of the phagosome, the Listeria begin to secrete virulence factors such as listeriolysin O (LLO) and phospholipase C that results in lysis of the phagosomal membrane and bacterial escape into the host cell cytosol. As a result, any proteins secreted by the bacterium have direct access to the MHC class I antigen processing pathway of the host cell (6). This ability of Listeria to secrete proteins directly into the cytosol of infected cells has made it a unique tool for studying class I processing of secreted bacterial proteins.
Determining processing efficiency is technologically challenging. Any values determined experimentally should be considered, at best, approximations of absolute values that might be associated with a particular model system. With that said, the studies by Pamer and colleagues showed that 1 pMHC is generated for every ~4 to 35 protein molecules degraded, an efficiency 2–3 orders of magnitude greater than that measured for recombinant proteins expressed from vaccinia virus (5, 7). It is unlikely that such large differences in processing efficiency are the result of experimental variability. What could account for the difference in processing efficiency between Listeria and vaccinia virus? One possibility is that, while the Listeria study examined presentation of peptides derived from native bacterial proteins, the vaccinia study measured presentation of a model peptide determinant generated from a recombinant protein. This would suggest that the differences in efficiency observed between the Listeria and vaccinia studies reflect differences inherent to the specific proteins and/or peptides used for these studies. To test this, we designed a novel system for expressing and secreting recombinant proteins from Listeria, which would allow us to determine the efficiency of pMHC production from a stable and rapidly degraded fusion protein in a manner analogous to our previous study using rVV.
Remarkably, we report here that the rate of pMHC generation for recombinant protein expressed by Listeria did not change, regardless of the cellular half-life of the protein. Furthermore, the efficiency of pMHC generation from recombinant Listeria proteins was at least 19-fold more efficient than that for the same recombinant proteins expressed by vaccinia. Together, our results suggest that proteins secreted from Listeria that are processed for MHC class I presentation follow, almost exclusively, a high efficiency processing pathway that is only accessed by a subset of endogenously synthesized proteins.
Materials and Methods
Cell line cultures
L929 cells, X63-Ag GM-CSF expressing cells, and the macrophage-like cell line BMA 3.1A7 (BMA3, kindly provided by Ken Rock) were grown in IMDM (Hyclone) supplemented with 10% heat-inactivated FBS (Cellgro) and 1X Glutamax (Invitrogen). All cells were maintained at 37°C in 7% CO2 atmosphere.
Bone-marrow macrophage and dendritic cell generation
Bone marrow cells were isolated from the femurs and tibiae of C57BL/6J mice (Jackson Laboratory). Bone marrow-derived macrophages (BMMΦ) and dendritic cells (BMDC) were generated by incubating bone marrow cells in growth medium consisting of IMDM with 10% heat-inactivated FBS, 1X Glutamax, 1X Penicillin and Streptomycin (Cellgro), 25 mM HEPES (Gibco, Invitrogen), and supplemented with 14% L929 cell conditioned culture medium for BMMΦ or 0.72 mM β-mercaptoethanol (2-ME, Sigma-Aldrich) and 2% X63-Ag GM-CSF cell conditioned culture medium for BMDCs. BMMΦs were used after 6 to 8 days in culture, while BMDCs were used after 8 to 9 days in culture. Mice were used in accordance with Institutional Animal Care and Use Committee guidelines. The SUNY Upstate Committee on the Humane Use of Animals approved all protocols.
Recombinant Listeria strains
Generation of ActA-deficient Listeria expressing the stable recombinant protein AU-NP-S-FLAG has been previously described (8). Rapidly degraded AU-R-NP-S-FLAG was generated as follows. DNA encoding Ub-R-NP-S-ARDG was amplified from rVV expressing Ub-R-NP-S-EGFP (1) using primers “Ub-F SOE ActA1-100” and “SIINFEKL-R with ARDG linker” (8). The resulting PCR product and the pPL2 plasmid (2) containing AU-NP-S-FLAG were digested using AflII and XmaI, ligated, and transfected into NEB5 E.coli (New England Biolabs). The resulting AU-R-NP-S-FLAG containing pPL2 plasmid was conjugated into Listeria strains as previously described (8).
The stable AU-PA-S-FLAG and rapidly degraded AU-R-PA-S-FLAG recombinant proteins were generated from rVV expressing the full-length PA protein (kindly provided by Jack Bennink and Jon Yewdell) using primers “PA Forward” 5’- CTTGTCTTAAGACTAAGAGGTGGTATGGAAGATTTTGTGCGACAATG-3’, “R-PA Forward” 5’- CTTGTCTTAAGACTAAGAGGTGGTCGAGAAGATTTTGTGCGACAATG-3’, and “PA(1–256) Reverse” 5’- TCTAGCATTTACTTCTTTGGACATTTGAGACAGCTTGCCCTC-3’. PCR sequences were extended at the 3’ end to add nucleotides encoding SIINFEKL-ARDG. Both PCR products were digested with AflII and XmaI, ligated into pPL2, and conjugated into Listeria as described previously (8). All Listeria strains used in this study express a non-secreted form of GFP or mCherry encoded by the pNF8 plasmid (kindly provided by Bruce Applegate) (8, 9).
Listeria and rVV infections
Listeria infection of BMA3 cells was as previously described (8). Briefly, BMA3 cells were infected with log-phase Listeria at a multiplicity of infection (MOI) of 20 for 1 h. Cells were washed, resuspended in IMDM containing 5 µg̃mL gentamicin (Cellgro) to kill any remaining extracellular bacteria (4) and incubated at 37°C for the duration of the experiment. This protocol was used for Listeria infection of BMMΦs and BMDCs except that cells were lifted using 2 mM EDTA (Gibco, Invitrogen) in PBS, and BMMΦs were infected at MOI of 10.
Infection of BMA3 cells with rVV was as previously described (8). Briefly, BMA3 cells were resuspended in HBSS (Cellgro) containing 0.1% BSA (Fisher Scientific) and rVV at MOI of 10 for 15 min with constant agitation, then resuspended in IMDM and incubated at 37°C for the duration of the experiment.
Extracellular (IMDM) secretion rates
To determine secretion rates of recombinant proteins, 1 × 109 CFU of log-phase Listeria were washed in ice-cold IMDM, resuspended in 5 ml IMDM at 37°C and grown on an orbital shaker at 37°C at 225 rpm for 2 h. Aliquots were taken at 0.5, 1, and 2 h post-inoculation and Listeria were removed by centrifugation. Culture supernatants were stored at −80°C for use in Western blot analysis.
Intracellular secretion rate
Adherent BMA3 cells were lifted by treatment with trypsin (Cellgro), resuspended in IMDM and treated with 5 µM epoxomicin (Enzo life Sciences) or DMSO vehicle (ATCC) for 30 min at 37°C. BMA3 cells were then washed in IMDM to remove excess drug and infected with Listeria. Three hours post-infection, aliquots were lysed at 4 × 106 cell equivalents per ml using RIPA buffer [150 mM NaCl (Fisher Scientific), 50 mM Tris pH 8.0 (Invitrogen), 1 mM EDTA (Gibco, Invitrogen), 0.1% SDS (Gibco, Invitrogen), 0.5% deoxycholate (Sigma-Aldrich), 1% Igepal® CO-630 (Sigma-Aldrich)]. Lysates were cleared of insoluble material by centrifugation and stored at −80°C for use in Western blot analysis.
Western blot analysis
The following antibodies were used for western blot analysis: M2 Anti-FLAG (Sigma-Aldrich), Anti-β-Actin clone AC-15 (Sigma-Aldrich), culture supernatant from the anti-Influenza NP hybridoma H19-S24-C16 (kindly provided by Laurence Eisenlohr) (5), and HRP conjugated goat anti-mouse IgG (Biolegend). Samples for Western blot analysis were mixed with LDS sample buffer (Invitrogen) and 2-ME, then heated to 95°C for 5 min. Samples were resolved on NuPAGE® Novex 4–12% Bis-Tris gels (Invitrogen) using NuPAGE® MOPS buffer (Invitrogen) and transferred to PVDF membrane (Pierce) using NuPAGE® transfer buffer (Invitrogen). Membranes were blocked using StartingBlock blocking buffer (Pierce) and probed with the appropriate antibodies.
The AU-NP-S-FLAG Western blot standard was prepared by expressing AU-NP-S-FLAG from the NF-L1177 strain of Listeria (kindly provided by Nancy Freitag), which has constitutive expression of pRFA-regulated genes, such as those encoding LLO, ActA, and AU-NP-S-FLAG (10). Aliquots of G145S-NP (AU-NP-S-FLAG expressed from NF-L1177) for use as a protein standard were prepared from supernatants of overnight cultures and stored at −80°C. Absolute concentrations of G145S-NP were determined by comparing with a known NP standard prepared from purified influenza virus A/Puerto Rico/8/34 (Charles River).
Flow cytometry
BMA3 and BMDC cells were incubated in 2.4G2 hybridoma supernatant to block CD16/CD32 Fcγ receptors (7) and stained with Alexa Flour 647 (Invitrogen) conjugated 25-D1.16 mAb (8). BMDCs were additionally stained with PE anti-CD11b and PE-Cy7 anti-CD11c (both from Biolegend) and gated on CD11b+ CD11c+ cells for all analysis. BMMΦ cells were prepared similarly to BMA3 cells with the exception that 20% mouse serum (Sigma-Aldrich) was added during the blocking step. Fluorescence was determined using an LSR II Flow cytometer (BD Biosciences). Mean fluorescent intensity was correlated to the number of surface Kb-SIINFEKL molecules using Dako FluoroSpheres as described previously (8). Data were analyzed using FlowJo software (Treestar).
Results
N-end rule degrons secreted from Listeria are rapidly degraded by the host cell
Listeria secretes a protein called ActA, which polymerizes cellular actin, allowing the bacterium to escape the host cell and infect neighboring cells (11). We chose to use an ActA-deficient form of Listeria for our in vitro kinetic studies because it is unable to polymerize host cell actin. This prevents Listeria from spreading from cell to cell during the course of the assay and, potentially, introducing greater variability in determining the kinetics of surface pMHC expression. By using this ActA deficient form of Listeria, we were able to restrict the timing of the infection to the initial period of incubation with the attenuated Listeria.
To evaluate the role of protein half-life in pMHC production for recombinant proteins expressed from Listeria, we generated two attenuated ActA-deficient Listeria constructs expressing recombinant proteins with vastly different cellular stabilities (Fig. 1A). The first construct, termed AU-NP-S-FLAG (8), is a fusion protein consisting of the first 100 amino acids of the Listeria ActA protein, followed by a ubiquitin moiety (Ub), full-length influenza nucleoprotein (NP), the model MHC Class I Kb binding peptide SIINFEKL (S), and a 3×-FLAG tag. ActA1–100 contains a bacterial secretion signal that enables efficient entry of the fusion protein into the host cell but, importantly, lacks the actin binding domain (12). Inclusion of the Ub moiety allows for rapid and efficient liberation of NP-S-FLAG by resident ubiquitin hydrolases in the host cell cytosol (13). Immediately following Ub is influenza NP, a model protein with a half-life measured in days (4). The inclusion of the SIINFEKL peptide allows for the precise quantification of surface pMHC production using the 25-D1.16 mAb, which specifically detects SIINFEKL peptide bound to the murine MHC class I Kb molecule (14). Lastly, the 3×-FLAG tag facilitates detection of the fusion protein by Western blot analysis. Moreover, including a motif at the carboxy-terminus of the SIINFEKL moiety ensures that liberation of SIINFEKL is dependent on the activity of host cell proteasomes.
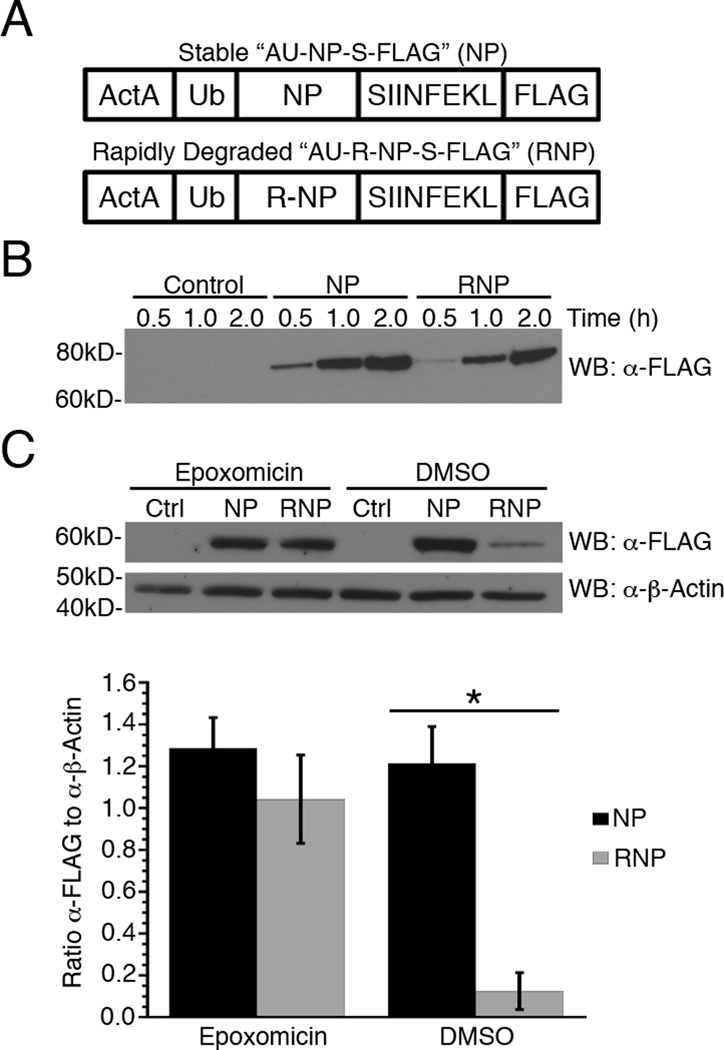
Figure 1. Generation of stable and rapidly degraded forms of a recombinant protein expressed by Listeria.
(A) Schematic representation of recombinant proteins expressed from Listeria. The NP construct is comprised of the first 100 amino acids of ActA, followed by full-length ubiquitin (Ub) and influenza nucleoprotein (NP), the minimal Kb-binding peptide SIINFEKL (S) and a 3×FLAG® tag (FLAG). The RNP construct is identical to NP except the amino-terminal methionine residue of NP has been replaced with arginine. (B) NP and RNP synthesis and secretion from recombinant Listeria in eukaryotic cell culture medium were determined by quantitative Western blot analysis using a monoclonal anti-FLAG antibody. The control is eukaryotic cell culture medium inoculated with Listeria that does not express a FLAG-tagged protein. (C) Secretion of NP and RNP into Listeria infected BMA3 cells was determined in the presence or absence of 5 μM epoxomicin by quantitative Western blot analysis as in (B) above. FLAG concentrations were normalized to β-actin for three independent experiments and data are shown as mean ± SEM, *,p<0.05.
Except for the addition of an arginine (R) at the amino-terminus of the NP moiety, AU-R-NP-S-FLAG is identical to AU-NP-S-FLAG. This single-amino acid addition is used to generate a version of the NP-S-FLAG protein with a cellular half-life of ~10 min compared to days for the original NP-S-FLAG construct. Arginine residues at the amino-terminus of eukaryotic proteins act as N-end rule degrons, targeting the proteins for rapid degradation in a proteasome-dependent manner (1). Upon entry of the recombinant protein into the host cell, eukaryotic ubiquitin hydrolases remove the amino-terminal ActA-Ub moiety, exposing the N-terminal arginine of the recombinant R-NP-S-FLAG protein.
Since both proteins are expressed using the pPL2 integration vector, only one copy of each gene is expressed per bacterium (15). Importantly, this helps reduce, if not eliminate, the variability in the amount of protein secreted from different recombinant strains of Listeria. We established that both the AU-NP-S-FLAG and AU-R-NP-S-FLAG proteins were synthesized and secreted by Listeria at the same rate by inoculating eukaryotic cell culture medium with Listeria expressing either construct and then measuring protein levels in the growth medium at several time points post-inoculation. Eukaryotic growth medium (IMDM) is used for these in vitro secretion assays as it initiates translation of proteins under the control of virulence promoters, including the ActA promoter used for the constructs in this study (12, 16). As shown in Fig. 1B, AU-NP-S-FLAG and AU-R-NP-S-FLAG are secreted at nearly identical rates in a cell-free system.
To determine whether AU-R-NP-S-FLAG is secreted in infected host cells at the same rate as AU-NP-S-FLAG, we treated the macrophage-like cell line BMA3.1A7 (BMA3) with 5 µM of the irreversible proteasome inhibitor epoxomicin (8) to prevent degradation of the R-NP-S-FLAG protein, then infected the BMA3 cells for 3 h with Listeria expressing either AU-NP-S-FLAG or AU-R-NP-S-FLAG. Infected cells were lysed and protein levels determined by Western blot analysis (Fig. 1C), confirming AU-NP-S-FLAG and AU-R-NP-S-FLAG are secreted in Listeria infected BMA3 cells at comparable rates. Importantly, in cells with functional proteasomes, R-NP-S-FLAG exhibits a significantly higher rate (p<0.05) of degradation than NP-S-FLAG, demonstrating it behaves as an N-end rule substrate comparable to those generated from host cell ribosomes.
Generation of pMHC from recombinant Listeria-derived NP is independent of the cellular half-life of the protein
We previously showed that proteins synthesized by host cell ribosomes, such as those encoded by recombinant vaccinia virus (rVV), generate pMHC at a rate proportional to the amount of protein degraded (4). We confirmed this correlation between protein degradation and pMHC generation for endogenously synthesized proteins in BMA3 cells by infecting with rVV expressing stable and rapidly degraded forms of NP, comparable to those expressed from our recombinant Listeria (Fig. 2B). It is important to note here that surface pMHC values were measured only in live, infected cells. In both the Listeria and rVV systems used for these experiments, we begin to see significant levels of cell death by 5–6 h post-infection. Therefore, to minimize the potential impact of cell death on our pMHC measurements, we limited all kinetic analyses to the first 5 h of infection.
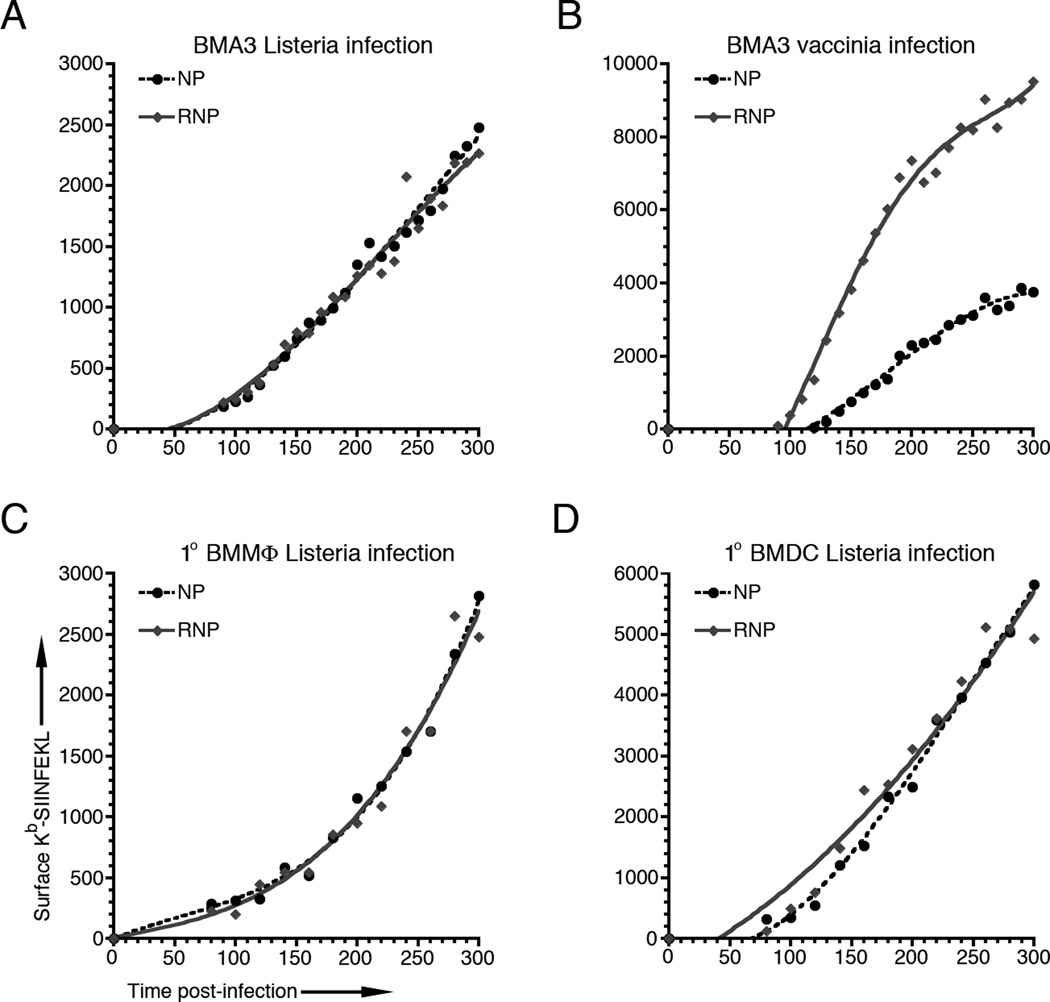
Figure 2. Presentation of Kb-SIINFEKL from Listeria-derived recombinant proteins is independent of protein half-life.
(A) BMA3 cells were infected with Listeria expressing AU-NP-S-FLAG or AU-R-NP-S-FLAG and surface Kb-SIINFEKL levels were measured at 10 min intervals from 90 to 300 min post-infection. (B) BMA3 cells were infected with rVV expressing NP-S-eGFP or Ub-R-NP-S-eGFP and surface Kb-SIINFEKL levels were measured as in (A), above. (C and D) Bone marrow-derived macrophages (BMMΦ) and dendritic cells (BMDC) were infected with Listeria expressing AU-NP-S-FLAG or AU-R-NP-S-FLAG and surface Kb-SIINFEKL levels were measured at 20 min intervals from 80 to 300 min post-infection. All graphs are representative of 3 independent experiments.
To determine whether protein stability plays a role in pMHC generation from secreted Listeria proteins, we infected BMA3 cells with Listeria expressing the stable AU-NP-S-FLAG or rapidly degraded AU-R-NP-S-FLAG forms of recombinant protein. Strikingly, both recombinant proteins generated surface Kb-SIINFEKL with similar kinetics, irrespective of the cellular half-life of the protein from which they were generated (Fig. 2A). To ensure that this apparent lack of correlation between protein half-life and pMHC generation was not limited to the BMA3 cell line, we infected primary bone marrow-derived macrophages (BMMΦ) (Fig. 2C) and bone marrow-derived dendritic cells (BMDC) (Fig. 2D) from C57BL/6 mice with either the AU-NP-S-FLAG or AU-R-NP-S-FLAG expressing Listeria constructs. Again, we found the kinetics of surface Kb-SIINFEKL was completely independent of the cellular half-life of the protein from which they were derived.
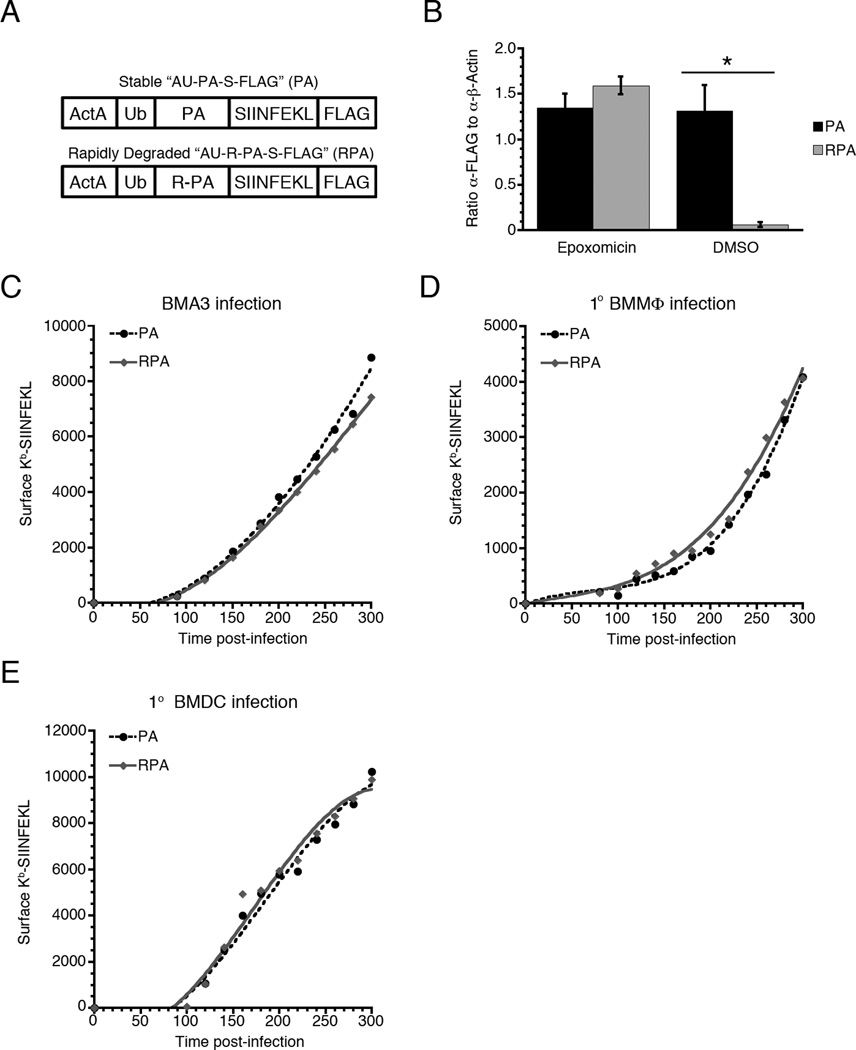
It is possible that the lack of correlation between the amount of protein degraded and surface pMHC generated is specific for the recombinant protein construct used for these assays. To address this, we replaced the influenza NP portion of the recombinant protein with the amino-terminal domain of the influenza acid polymerase protein (PA1–256), generating stable AU-PA-S-FLAG and rapidly degraded AU-R-PA-S-FLAG versions of the protein (Fig. 3A). We limited this construct to the first domain of PA, as we have found that increasing the size of a secreted Listeria protein can significantly reduce protein expression levels (8). As with the NP constructs, we confirmed that the two recombinant PA proteins are secreted at the same rate and that the amino-terminal arginine acted as a destabilizing N-end rule degron (Fig. 3B). We then measured the kinetics of surface Kb-SIINFEKL production in BMA3 cells (Fig. 3C), BMMΦs (Fig. 3D), and BMDCs (Fig. 3E). In all cases, the rate of surface Kb-SIINFEKL production from the stable and rapidly degraded recombinant proteins was equal and, importantly, independent of the cellular stability of the secreted recombinant protein.
Figure 3. The lack of correlation between Kb-SIINFEKL surface expression and protein half-life is not limited to chimeric nucleoprotein constructs.
(A) The NP moiety of AU-NP-S-FLAG and AU-R-NP-S-FLAG was replaced with the amino-terminal portion of influenza acid polymerase (PA) to generate AU-PA-S-FLAG or AU-R-PA-S-FLAG. (B) BMA3 cells were infected with Listeria expressing AU-PA-S-FLAG or AU-R-PA-S-FLAG in the presence or absence of 5 µM expoxomicin. At 3 h post-infection, cells were lysed and immunoblotted for FLAG and β-actin. FLAG staining was normalized to β-actin for three independent experiments. Data are shown as mean ± SEM, *,p<0.05. (C) BMA3 cells, (D) BMMΦ, and (E) BMDC were infected with Listeria expressing AU-PA-S-FLAG or AU-R-PA-S-FLAG and surface Kb-SIINFEKL levels were measured at 20 min intervals from 80 to 300 min post-infection. Graphs are representative of 3 independent experiments.
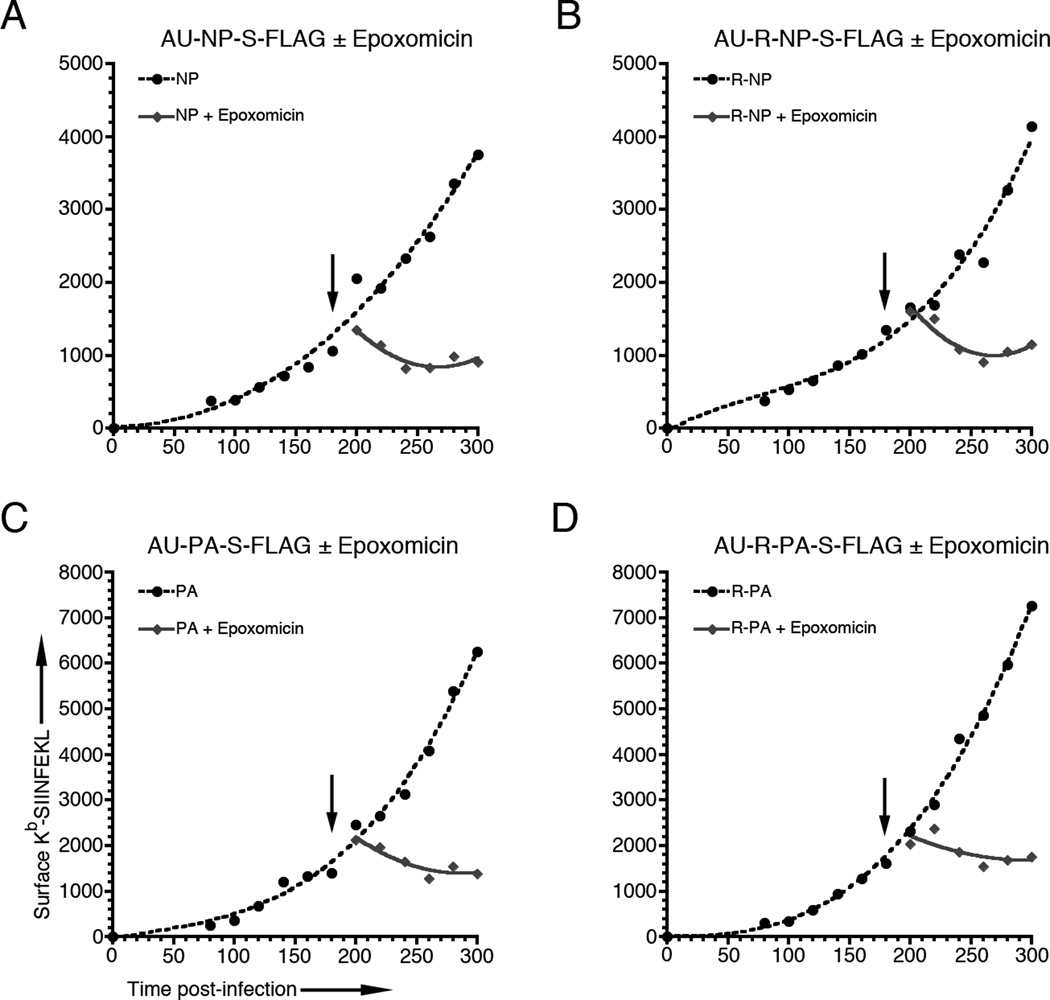
Although we demonstrated that degradation of the N-end rule R-NP and R-PA degrons expressed from Listeria was dependent on proteasomal activity in infected cells (Figs. 1C & 3B), we wanted to determine if pMHC generation from Listeria-derived proteins followed the classical, proteasome dependent MHC class I antigen processing pathway. We therefore infected BMA3 cells with AU-NP-S-FLAG (Fig 4A), AU-R-NP-S-FLAG (Fig 4B), AU-PA-S-FLAG (Fig 4C), or AU-R-PA-S-FLAG (Fig 4D). At 180 min post-infection, each sample was split and treated either with 5 μM epoxomicin or DMSO vehicle and surface Kb-SIINFEKL levels were measured for an additional 2 h. For each construct, surface Kb-SIINFEKL expression was completely abrogated by the addition of the proteasome inhibitor, but unaffected by the DMSO vehicle. Importantly, the 20 to 40 min delay between treatment with epoxomicin and cessation of pMHC presentation matches the time delay seen in our previous findings for rVV-derived recombinant proteins (4). In addition, we infected BMMΦs from TAP−/− mice with our recombinant Listeria constructs. The complete absence of surface Kb-SIINFEKL expression in these cells confirmed that antigen presentation was TAP dependent (data not shown). We also treated infected BMA3 cells with brefeldin A, which completely abrogated surface Kb-SIINFEKL expression, to demonstrate that presentation of Kb-SIINFEKL was dependent on Golgi transport (data not shown).
Figure 4. Presentation of Kb-SIINFEKL from Listeria-derived recombinant proteins is dependent on proteasome function.
BMA3 cells were infected with Listeria expressing (A) AU-NP-S-FLAG, (B) AU-R-NP-S-FLAG, (C) AU-PA-S-FLAG or (D) AU-R-PA-S-FLAG and surface Kb-SIINFEKL levels were measured at 20 min intervals from 80 to 300 min post-infection. Cells were split at 180 min post-infection and treated either with 5 µM epoxomicin or DMSO vehicle (arrows) and surface Kb-SIINFEKL levels were measured for an additional 2 h. Graphs are representative of 3 independent experiments.
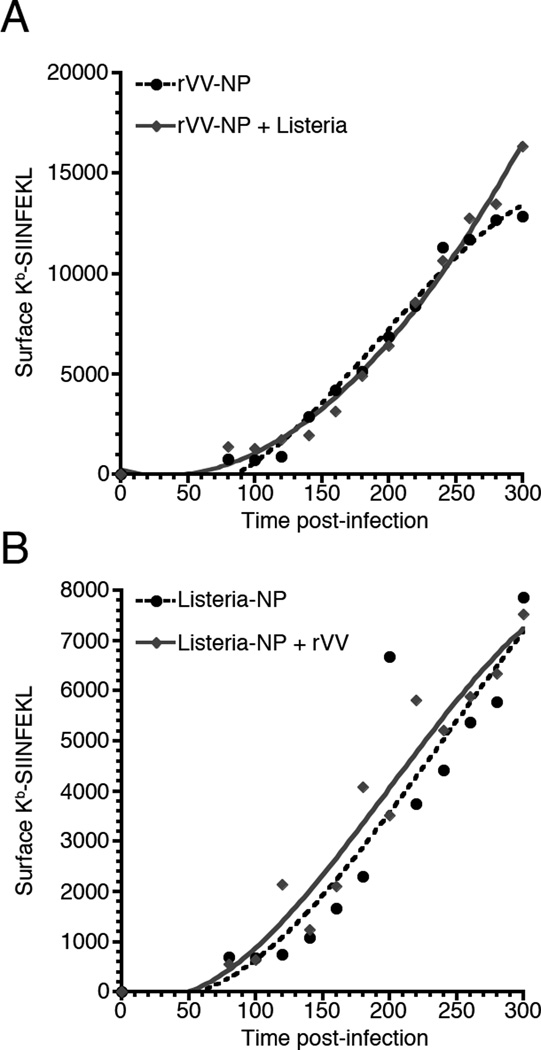
It is possible that Listeria infection causes global changes in the antigen processing pathway that affects pMHC generation independent of the origin of the protein degraded. To address this possibility, we co-infected BMA3 cells with Listeria and rVV, with only one of the vectors acting as a source of peptide, and determined the kinetics of Kb-SIINFEKL surface expression. In the first case, we infected BMA3 cells with rVV expressing NP-S-eGFP as the source of the SIINFEKL peptide and Listeria expressing a non-secreted form of the red fluorescent mCherry protein (Fig. 5A). In the reciprocal experiment, BMA3 cells were infected with Listeria expressing a non-secreted form of mCherry and AU-NP-S-FLAG as the source of SIINFEKL and rVV expressing only the yellow fluorescent Venus protein (Fig. 5B). Expressing red and green or yellow fluorescent proteins from Listeria and rVV, respectively, allowed us to evaluate Kb-SIINFEKL surface levels only on those cells co-infected with both vectors. In both cases, the kinetics of presentation were determined by the vector from which the recombinant protein was expressed, and, importantly, was independent of co-infection with the complimentary vector.
Figure 5. Co-infection with Listeria and rVV does not alter Kb-SIINFEKL presentation kinetics from either vector.
(A) Surface Kb-SIINFEKL levels were determined for BMA3 cells infected with rVV expressing NP-S-eGFP alone or BMA3 cells co-infected with Listeria not expressing the SIINFEKL peptide. (B) Surface Kb-SIINFEKL levels were determined for BMA3 cells infected with Listeria expressing AU-NP-S-FLAG alone or BMA3 cells co-infected with rVV not expressing SIINFEKL peptide. In both infections, Listeria expressed the red fluorescent mCherry protein and rVV expressed either eGFP (A) or Venus (B) to limit Kb-SIINFEKL analysis to BMA3 cells infected with both vectors. Each graph is representative of 3 independent experiments.
The efficiency of pMHC generation from recombinant proteins secreted from Listeria is higher than for comparable endogenously synthesized proteins
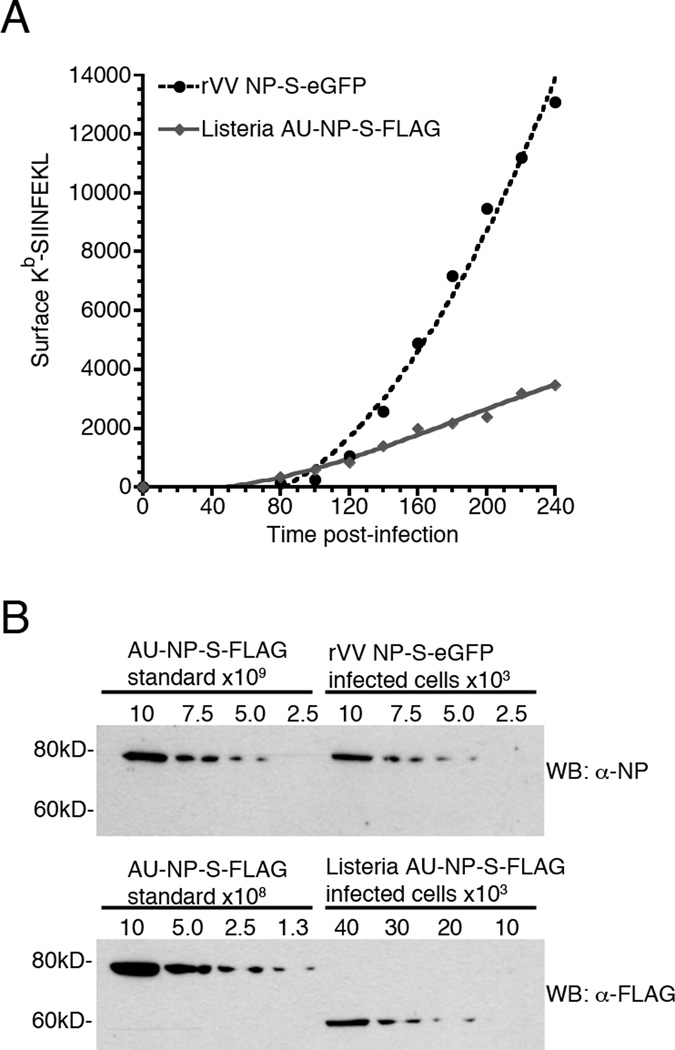
In light of the highly disparate efficiencies measured for pMHC generation from rVV and native Listeria proteins (4, 5, 7, 17), we sought to determine the efficiency of pMHC generation from recombinant NP expressed from Listeria. However, to calculate the efficiency of pMHC generation using the method employed for our rVV studies requires there be a proportional relationship between the amount of protein degraded and the amount of surface pMHC generated. Because pMHC generation from the secreted Listeria-derived proteins in these studies proved to be independent of the amount of protein degraded, we were unable to use this method to determine the efficiency of Kb-SIINFEKL production from Listeria. Instead, we chose to establish what the minimal efficiency of surface Kb-SIINFEKL production from Listeria would be if every recombinant protein secreted into the host cell was degraded and available as a source of MHC class I binding peptide. To accomplish this, we compared recombinant protein levels and surface Kb-SIINFEKL production from both rVV and Listeria by infecting BMA3 cells with either Listeria expressing AU-NP-S-FLAG or rVV expressing NP-S-eGFP. At 1 h post-infection, infected cells were either treated with epoxomicin to inhibit proteasome mediated degradation or DMSO in the control sample. The kinetics of Kb-SIINFEKL production for DMSO treated Listeria and rVV infected cells is shown in Fig. 6A. Consistent with our earlier experiments [Fig. 4 and (4)], epoxomicin treatment completely abrogated Kb-SIINFEKL production in both Listeria and rVV infected BMA3 cells (data not shown). To estimate the relative efficiency of surface Kb-SIINFEKL production from rVV and Listeria expressed proteins, we restricted our measurements to time points from 180 to 240 min post-infection, when pMHC generation was linear for both vectors. This approach is consistent with the method we used to determine processing efficiency in earlier experiments using proteins expressed from rVV (4). We found that there was little variation in the ratio of surface Kb-SIINFEKL generated from rVV and Listeria expressed proteins for each of two individual experiments (Table I). Using these measurements of surface pMHC complexes, we estimated that Kb-SIINFEKL generated from rVV derived NP-S-eGFP was ~2.8-fold greater than surface Kb-SIINFEKL levels generated from Listeria-derived AU-NP-S-FLAG. We then used lysates of the epoxomicin-treated cells to measure the levels of total protein synthesized in rVV and Listeria infected cells at 3 h post-infection (Fig. 6B). We have previously shown that processing and presentation of peptides derived from recombinant proteins expressed from both rVV and Listeria requires ~40–50 min (4, 8). Therefore, the ratio of protein levels measured at 3 h post-infection corresponds to the ratio of pMHC levels measured at 180–240 min post-infection. We compared each lysate to an NP protein standard to determine that the concentration of NP-S-eGFP in rVV-infected cells was ~54-fold higher than the concentration of AU-NP-S-FLAG in Listeria-infected cells (Table II).
Figure 6. Surface Kb-SIINFEKL expression is more efficient for peptides derived from recombinant proteins expressed by Listeria than by rVV.
BMA3 cells were infected with either Listeria expressing AU-NP-S-FLAG or rVV expressing NP-S-eGFP and treated with either 5 μM Epoxomicin or DMSO vehicle. (A) Surface Kb-SIINFEKL levels were measured at 20 min intervals from 80 to 240 min post-infection in cells treated with DMSO vehicle. (B) Recombinant protein levels were determined in epoxomicin treated cells 3 h post-infection by quantitative Western blot analysis using an AU-NP-S-FLAG of known concentration as a standard. The graph and Western blots are representative of two independent experiments.
Table I.
Surface Kb-SIINFEKL complexes used for calculating the efficiency of Kb-SIINFEKL generation from recombinant Listeria proteins
| Experiment 1 | Experiment 2 | ||||||
|---|---|---|---|---|---|---|---|
| Minutes post- infection |
rVV NP-S- eGFP |
Listeria NP- S-FLAG |
Ratio rVV/Lm |
rVV NP-S- eGFP |
Listeria NP- S-FLAG |
Ratio rVV/Lm |
|
| 180 | 7189 | 2171 | 3.3 | 13087 | 6458 | 2.0 | |
| 200 | 9451 | 2392 | 4.0 | 16349 | 7781 | 2.1 | |
| 220 | 11193 | 3198 | 3.5 | 19978 | 9524 | 2.1 | |
| 240 | 13091 | 3484 | 3.8 | 20213 | 11096 | 1.8 | |
| Average: | 3.6 | 2.0 | |||||
Table II.
Molecules of rVV-derived NP-S-eGFP or Listeria-derived AU-NP-S-FLAG protein per cell at 3 h post-infection
| rVV NP-S-eGFP | Listeria NP-S-FLAG | Ratio rVV/Lm | |
|---|---|---|---|
| Experiment 1 | 8.49 × 105 | 2.44 × 104 | 34.8 |
| Experiment 2 | 9.12 × 105 | 1.25 × 104 | 73.0 |
| Average: | 54 |
The efficiency of Kb-SIINFEKL production from recombinant Listeria derived protein was calculated by measuring the total recombinant protein, either synthesized in rVV infected cells or secreted by Listeria into the cytosol of infected cells, that is necessary to generate equivalent levels of surface Kb-SIINFEKL complexes. We then divided this fold-difference in protein level (54-fold higher from rVV than from Listeria) by the fold-difference in surface Kb-SIINFEKL (2.8-fold higher from rVV than from Listeria) for rVV and Listeria infected cells. We determined that at least 19 recombinant NP molecules must be synthesized in rVV-infected cells for every 1 recombinant NP molecule secreted by Listeria to generate equivalent levels of surface Kb-SIINFEKL expression in infected cells. Because the efficiency of Kb-SIINFEKL production from rVV-derived NP-S-eGFP is known (~1 surface pMHC per 3000 protein molecules degraded) (4), we can use this difference in efficiency to arrive at a minimal processing efficiency for secreted recombinant Listeria proteins. By dividing the efficiency for rVV-infected cells (~1 in 3000) by 19 we can calculate that the efficiency of Kb-SIINFEKL generation in Listeria infected cells would be 1 surface complex for every 160 AU-NP-S-FLAG molecules entering the host cell. It is important to note here that our calculations reflect the minimum efficiency possible in our experimental system, since it is based on the degradation of every AU-NP-S-FLAG molecule secreted by the Listeria into the host cell cytosol. However, we know that only a fraction of the total AU-NP-S-FLAG synthesized is degraded. Therefore, the efficiency of pMHC generation must be significantly greater than 1 pMHC per 160 protein molecules degraded, and may well be within the range calculated by the Pamer group for native Listeria proteins (1 pMHC per 3.5–35 proteins degraded) (5, 7).
Discussion
To date, there has been only one report of MHC class I antigen presentation occurring independently of the degradation rate of the protein from which the peptides presented are derived. In this study from Shastri and colleagues, cells were transfected with DNA encoding fusion proteins containing an amino-terminal ubiquitin moiety followed by recombinant proteins expressed either as stable proteins or N-end rule degrons (18). The one feature this study has in common with our own experimental approach is the expression of protein constructs containing Ub-protein motifs. While it is conceivable that expression of these recombinant proteins as Ub-protein chimeras may target the protein to a high efficiency processing pathway, the use of these motifs in rVV expression systems suggests otherwise. When Ub-R-NP-S-eGFP is expressed from rVV, it increases surface pMHC ~3-fold relative to presentation from NP-S-eGFP, which corresponds directly with the difference in the amount of protein degraded for these two constructs (4). If the Ub-protein motif were responsible for targeting protein to the high efficiency pathway, then pMHC generation from Ub-R-NP-S-eGFP should be significantly greater than the ~3-fold difference from NP-S-eGFP. As such, these data strongly suggest that it is expression from Listeria that is the determining factor in the targeting of recombinant protein to the high efficiency processing pathway.
It is possible that the observed difference in processing efficiency for proteins expressed from rVV and Listeria is the result of the significantly higher levels of recombinant protein synthesized in rVV vs. Listeria infected cells. However, a study by Wherry et al., in which vaccinia virus was engineered to express different levels of a recombinant NP-SIINFEKL protein construct, suggests this is not the case (19). In this study, the level of NP-SIINFEKL expression between the lowest and highest expressing rVV differed by 200-fold, a range that is 4-fold greater than the difference in protein expression levels between the rVV and Listeria constructs used for the efficiency studies we report here. Wherry, et al. demonstrated a direct correlation between the level of protein synthesized and surface Kb-SIINFEKL expression across the entire 200-fold range of protein expression tested. It is important to note here that the levels of Kb-SIINFEKL expression tested in the vaccinia study by Wherry et al. were comparable to those in our study using a similar NP-SIINFEKL construct and extended to levels several fold lower than those measured in our Listeria system when vaccinia with lowered protein expression were evaluated.
Approximately 10-fold more AU-R-NP-S-FLAG and AU-R-PA-S-FLAG is degraded when expressed from Listeria than from the corresponding stable protein constructs. Because the rapidly degraded proteins generate similar levels of surface Kb-SIINFEKL as the stable forms, it is possible that pMHC is generated from the rapidly degraded proteins with at least a 10-fold lower efficiency than from the stable proteins. While it is possible that processing occurs with different efficiencies for proteins with different cellular half-lives, we do not believe this is likely considering the nearly identical presentation kinetics we observe for stable and rapidly degraded forms of two distinct proteins, one of which localizes to the cytosol (PA) and the other to the nucleus (NP). Instead, we hypothesize that only a certain proportion of Listeria-derived proteins are able to access a high efficiency pMHC-processing pathway, and that access to this pathway is independent of the cellular half-life of the protein. Importantly, high-efficiency processing occurs almost immediately upon secretion of protein substrate by the Listeria into the host cell cytosol. Any Listeria-derived proteins that do not enter this high efficiency pathway enter the cellular pool of protein and are subsequently processed at an efficiency low enough that it will not contribute significantly to surface pMHC levels. A recently published study by Grauling-Halama et al. supports our findings. In this study, the authors use regression analysis of previously published data from Pamer and colleagues to demonstrate a tight linkage between Listeria protein synthesis and pMHC generation. Although they do not directly measure production of pMHC complexes in Listeria infected cells, their analysis of the Pamer groups data is completely consistent with our observations (20).
While it is possible that this high efficiency pathway is confined to proteins expressed from Listeria, results from our original rVV efficiency study (4), as well as more recent data from Dolan et al., suggests otherwise (3, 4). In the rVV study, we observed higher presentation efficiency for peptides derived from a cellular form of the NP-S-eGFP protein containing a destabilizing KEKE motif. This suggested to us that presentation efficiency may be higher for proteins that enter the cellular pool when compared to those processed in a near co-translational manner, such as endogenously synthesized N-end rule degrons or proteins processed as defective ribosomal products (DRiPs) (21, 22). The study by Dolan et al. used biochemical techniques to distinguish between pMHC generated from proteins that have entered the cellular pool and pMHC generated from newly synthesized DRiPs. In this study, >50% of all pMHC were generated from a very small fraction of nascent, endogenously synthesized DRiPs. The authors conclude that “retired” proteins are an inefficient source of peptides for pMHC presentation and that pMHC generated from newly synthesized DRiPs are presented with at least a 6.5-fold greater efficiency than “retirees”.
The results we present in this study are consistent with those reported by Dolan et al., with one important difference. While we observed the same high efficiency processing of a small fraction of total protein synthesized, the source of these proteins cannot, by definition, be DRiPs. Listeria proteins are synthesized by bacterial ribosomes and cannot access the antigen processing machinery of the host cell until secreted by the bacterium. Therefore, entry of Listeria-derived proteins to a high efficiency processing pathway cannot be the result of a direct association between the biosynthetic machinery and the antigen processing pathway of the host cell, as has been suggested for DRiPs (3, 23, 24). Rather, our results demonstrate that the high efficiency processing pathway is accessible to both endogenously synthesized and exogenously derived proteins. Importantly, this suggests that the high efficiency processing pathway is accessible to multiple sources of antigenic protein, which may include proteins entering the cross-presentation pathway of professional antigen presenting cells. The specific factors that target a protein for processing by the high efficiency pathway remain to be elucidated. However, demonstrating that virtually all pMHC generation from Listeria-derived proteins follows this pathway should greatly facilitate our ability to define differences in the MHC class I processing pathway that result in high- and low-efficiency presentation of specific antigenic proteins.
Acknowledgments
We thank Dr. Sandy Hayes for critical reading of the manuscript and Chiharu Kumagai for technical assistance.
This work was supported in part by American Heart Association Scientist Development Grant AHA0830010N and National Institutes of Health Grant 1R21AI090516 to MFP.
References
- 1.Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- 2.Decatur AL, Portnoy DA. A PEST-like sequence in listeriolysin O essential for Listeria monocytogenes pathogenicity. Science. 2000;290:992–995. doi: 10.1126/science.290.5493.992. [DOI] [PubMed] [Google Scholar]
- 3.Dolan BP, Li L, Veltri CA, Ireland CM, Bennink JR, Yewdell JW. Distinct Pathways Generate Peptides from Defective Ribosomal Products for CD8+ T Cell Immunosurveillance. J. Immunol. 2011;186:2065–2072. doi: 10.4049/jimmunol.1003096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Princiotta MF, Finzi D, Qian S-B, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- 5.Villanueva MS, Fischer P, Feen K, Pamer EG. Efficiency of MHC class I antigen processing: a quantitative analysis. Immunity. 1994;1:479–489. doi: 10.1016/1074-7613(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 6.Pamer EG, Sijts AJ, Villanueva MS, Busch DH, Vijh S. MHC class I antigen processing of Listeria monocytogenes proteins: implications for dominant and subdominant CTL responses. Immunol. Rev. 1997;158:129–136. doi: 10.1111/j.1600-065x.1997.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 7.Villanueva MS, Sijts AJ, Pamer EG. Listeriolysin is processed efficiently into an MHC class I-associated epitope in Listeria monocytogenes-infected cells. J. Immunol. 1995;155:5227–5233. [PubMed] [Google Scholar]
- 8.Wolf BJ, Princiotta MF. Viral and bacterial minigene products are presented by MHC class I molecules with similar efficiencies. Mol. Immunol. 2011;48:463–471. doi: 10.1016/j.molimm.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Fortineau N, Trieu-Cuot P, Gaillot O, Pellegrini E, Berche P, Gaillard JL. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res. Microbiol. 2000;151:353–360. doi: 10.1016/s0923-2508(00)00158-3. [DOI] [PubMed] [Google Scholar]
- 10.Miner MD, Port GC, Freitag NE. Functional impact of mutational activation on the Listeria monocytogenes central virulence regulator PrfA. Microbiology. 2008;154:3579–3589. doi: 10.1099/mic.0.2008/021063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevens JM, Galyov EE, Stevens MP. Actin-dependent movement of bacterial pathogens. Nat Rev Micro. 2006;4:91–101. doi: 10.1038/nrmicro1320. [DOI] [PubMed] [Google Scholar]
- 12.Moors MA, Levitt B, Youngman P, Portnoy DA. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect Immun. 1999;67:131–139. doi: 10.1128/iai.67.1.131-139.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fruci D, Lauvau G, Saveanu L, Amicosante M, Butler RH, Polack A, Ginhoux F, Lemonnier F, Firat H, van Endert PM. Quantifying recruitment of cytosolic peptides for HLA class I presentation: impact of TAP transport. J. Immunol. 2003;170:2977–2984. doi: 10.4049/jimmunol.170.6.2977. [DOI] [PubMed] [Google Scholar]
- 14.Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- 15.Lauer P, Chow MYN, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohne J, Sokolovic Z, Goebel W. Transcriptional regulation of prfA and PrfA-regulated virulence genes in Listeria monocytogenes. Mol. Microbiol. 1994;11:1141–1150. doi: 10.1111/j.1365-2958.1994.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 17.Sijts AJ, Neisig A, Neefjes J, Pamer EG. Two Listeria monocytogenes CTL epitopes are processed from the same antigen with different efficiencies. J. Immunol. 1996;156:683–692. [PubMed] [Google Scholar]
- 18.Goth S, Nguyen V, Shastri N. Generation of naturally processed peptide/MHC class I complexes is independent of the stability of endogenously synthesized precursors. J. Immunol. 1996;157:1894–1904. [PubMed] [Google Scholar]
- 19.Wherry EJ, Puorro KA, Porgador A, Eisenlohr LC. The induction of virus-specific CTL as a function of increasing epitope expression: responses rise steadily until excessively high levels of epitope are attained. J. Immunol. 1999;163:3735–3745. [PubMed] [Google Scholar]
- 20.Grauling-Halama S, Schenk S, Bubert A, Geginat G. Linkage of Bacterial Protein Synthesis and Presentation of MHC Class I-Restricted Listeria monocytogenes-Derived Antigenic Peptides. PLoS ONE. 2012;7:e33335. doi: 10.1371/journal.pone.0033335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yewdell JW. DRiPs solidify: progress in understanding endogenous MHC class I antigen processing. Trends Immunol. 2011;32:548–558. doi: 10.1016/j.it.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yewdell JW, Anton LC, Bennink JR. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J. Immunol. 1996;157:1823–1826. [PubMed] [Google Scholar]
- 23.Lev A, Princiotta MF, Zanker D, Takeda K, Gibbs JS, Kumagai C, Waffarn E, Dolan BP, Burgevin A, van Endert P, Chen W, Bennink JR, Yewdell JW. Compartmentalized MHC class I antigen processing enhances immunosurveillance by circumventing the law of mass action. Proc. Natl. Acad. Sci. U.S.A. 2010;107:6964–6969. doi: 10.1073/pnas.0910997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yewdell JW. The seven dirty little secrets of major histocompatibility complex class I antigen processing. Immunol. Rev. 2005;207:8–18. doi: 10.1111/j.0105-2896.2005.00309.x. [DOI] [PubMed] [Google Scholar]