Abstract
There is increasing evidence of the interaction of fat and bone metabolism, and the role mechanical signals may have in regulating the adaptation of these tissues. The rat hindlimb suspension model of disuse osteoporosis was used to identify genes differentially expressed relative to normal weight bearing bones, and if the relative expression of these genes is sensitive to anabolic mechanical stimuli. Ten days of hindlimb suspension suppressed Percent Labeled Surface and Bone Volume/Trabecular Volume of the proximal tibia by 46% and 69%, respectively, as compared to controls. Differential Display (DD-PCR) and Northern blot analysis identified and verified, respectively, that expression of Spot 14 (S14), an important gene in lipogenesis, was up-regulated by 4-fold in tibiae of tail suspended animals compared to long term controls. Anabolic mechanical stimulation (45Hz, 10min/day at 0.25g) didn't show statistically significant effect on S14 expression. These results indicate a potential role for lipogenic genes during bone loss caused by disuse, further supporting a link between bone and fat tissue, and considering the insensitivity of these genes to mechanical signals which promote bone formation in the skeleton, indicates the independence of resorptive and formative processes in bone.
Keywords: Osteoporosis, Disuse, Mechanical Stimulation, Spot 14
Introduction
Understanding the molecular basis of Wolff's Law, the form-follows-function paradigm of bone, is critical to understanding the etiologic basis of osteoporosis. At the tissue level, disuse is highly permissive to resorption in the skeleton [1], while new functional challenges are often met by increase in bone quantity and improvement in bone quality [2]. Certainly, identifying those genes which facilitate the resorptive process would reveal novel targets for therapeutic interventions for osteoporosis. With increasing evidence of a regulatory role of bone in the control of fat metabolism [3], and the interdependence of mechanical signals on bone and fat production [4], a specific goal of this work was to determine if genes most often associated with the regulation of adipose tissue were active in bone and influenced by changes in bone's mechanical environment.
Spot 14 (S14), predominately expressed in lipogenic tissues such as liver, fat, and the lactating mammary gland [5], was initially identified as an in vitro translated protein spot on two-dimensional gel that was rapidly induced by thyroid hormone in rat liver [6]. The S14 gene codes for a 17-kDa acidic protein that lacks any well-recognized functional motifs. Animal studies showed a close correlation between S14 expression and lipogenesis [7-10], while S14 knock out mice indicated that it is required for de novo lipogenesis in the lactating mammary gland, but not for the induction of lipogenesis in the liver by thyroid hormone [11, 12]. More recently it was reported that S14 causes an inhibition of cell proliferation and of anchorage-independent growth in human breast cancer cells [13]. Despite this evidence, the biochemical function of S14 protein remains unclear. The observation that the S14 protein is present in the nucleus of hepatics cells and that primary hepatocytes transfected with a S14 antisense oligonucleotide abolished the lipogeneic enzyme activities and lipogenesis induced by T3 and glucose suggested that S14 protein regulates lipogenesis by regulating the transcription of other lipogenic proteins [14, 15]. Alternatively, it was speculated that S14 binds to the cytoskeleton and helps to remove newly synthesized fatty acids from Fatty acid synthase (FAS) to relieve end-product inhibition [16].
Herein, we report that S14 expression was activated in tibiae of tail suspended animals, but not affected in osteogenic mechanical stimulation. Obesity protects from osteoporosis through an unknown mechanism [17-19]. Our results suggest a potential role of S14 in the protection of bone loss due to disuse, and this process is probably independent of anabolic responses caused by mechanical stimulation.
Materials and Methods
Disuse/Mechanical Stimulation
Retired female breeder Sprague-Dawley rats, 180-240 days old, were used in tail suspension and mechanical stimulation. All procedures were reviewed and approved by the Animal Care Committee of Stony Brook University, and met all guidelines for the health and welfare of the animals. For the disuse experiment, 12 rats were assigned into 2 groups: long term control (n=6) and tail suspension (n=6) according to the Morey-Holton tail suspension model [20]. The rats were individually caged with free access to food and water. The protocol ran for 10 days. For mechanical stimulation, 18 rats were assigned into 3 groups: long term control (LTC, n=6), MS1 (n=6, 10min/day 90Hz at 0.25g), MS2 (n=6, 10min/day 45Hz at 0.25g). Mechanical stimulation was performed using an oscillating platform, and the rats were individually caged with free access to food and water [21]. The protocols ran for 28 days. All animals were weighted at the beginning and end of the study. Rats were sacrificed by carbon dioxide inhalation, and tibiae were harvested. Lastly, the right tibia was used for histomorphometry, the left tibia was used for total RNA extraction.
Histomorphometry
The proximal tibia (right) was embedded in methyl-methacrylate (Fisher Scientific, Fair Lawn, NJ) using a three-step protocol [22]. After trimming the plastic blocks, 50 μm-thick frontal sections from the central tibia were cut on a diamond wire saw (Well Wire Saws, Model 3241, Germany). Sections were mounted on an epifluorescent microscope (×10). Trabecular bone of the proximal tibial metaphysis was evaluated over an area enclosed by two lines 800 μm and 2000 μm distal of the growth plate. Twenty-four adjacent squares, each displaying 1.6 mm2, were captured by a video camera interfaced with a digitizing pad (CalComp, Anaheim, CA) and a PC. Fluorescent labels and bone surfaces were traced and morphometry software (OsteoMetrics, Atlanta, GA) was used to determine bone histomorphometric indices. Trabecular bone formation rate, with bone volume as referent (BFR•BV−1), mineralizing surface (MS•BS−1), mineral apposition rate (MAR), and bone area (BV) were determined as described previously [23]. All histomorphometric evaluations were performed without knowledge of which experimental group the bones came from.
RNA Purification
Total RNA was isolated from intact bones (which included bone marrow, articular and normal growth plate cartilage) using the ToTALLY RNA kit (Ambion) as described before [24]. The intact tibias initially were pulverized in liquid nitrogen using a pestle and mortar and then added to the denaturing solution. Each sample was homogenized with a polytron (Brinkmann Instruments, Inc., Westbury, NY, U.S.A.) and extracted once with phenol-chloroform-isoamyl alcohol using centrifugation (10,000g). The aqueous phase was then transferred to a fresh tube and a 1/10 aqueous phase volume of sodium acetate solution was added. The sample was again extracted with acid-phenol-chloroform. The aqueous phase was transferred to a fresh tube, mixed with an equal volume of isopropanol, and incubated at −20°C for at least 1 h to precipitate the RNA. Finally, the RNA was pelleted by centrifugation (10,000g), washed with 70% ethanol, air-dried, and dissolved in RNase-free water/0.1 mM EDTA. The concentration of each RNA sample was determined spectrophotometrically and the integrity of all RNA samples was monitored on agarose gels.
Differential mRNA Display (DD-PCR)
The differential mRNA display method was used as described by Liang and Pardee [25] using the RNAimage kit (GenHunter Corp.) as described before [26]. Before reverse transcription, RNA was treated with DNase I. The DNA-free total RNA was then mixed with 1 μM of each of the degenerate oligo-dT-primers, 1× reverse transcription buffer, and 20 μM dNTPs. The solution was heated for 5 minutes at 65°C and then cooled to 37°C for 10 minutes followed by the addition of 200 U of reverse transcriptase (RT). After incubation at 37°C for 1 h, the mixture was heated for 5 minutes at 95°C followed by cooling and storage at −20°C. Polymerase chain reaction (PCR) was performed in thin-walled tubes containing 0.2 vol of RT reaction, 1× PCR buffer (10 mM Tris-Cl, pH8.4, 50 mM KCl, 1.5 mM MgCl2, and 0.001% gelatin), 2 μM dNTPs,33P-dATP (0.25 μl of 1200 Ci/mmol), 1 μM of each of the degenerate oligo-dT-primers, 0.2 μM arbitrary primer, and 10 U of AmpliTaq DNA polymerase. PCR reactions were performed as follows: 94°C for 30 s, 40°C for 2 minutes, and 72°C for 30 s for 40 cycles followed by 1 cycle of extension at 72°C for 5 minutes. DNA sequencing loading buffer was added to an aliquot of each reaction and incubated at 80°C for 2 minutes. Each sample was then loaded onto a 6% denaturing DNA sequencing gel and electrophoresed at 1700 V. The gel was dried without fixation and exposed directly to Kodak Biomax film overnight at room temperature. The specific band of interest was excised from the gel, placed in 100 μl water for 10 minutes, and boiled for 15 minutes. After a 2-minute spin, the supernatant was transferred to a new tube and 10 μl of 3 M sodium acetate, 5 μl of glycogen (10 mg/ml), and 450 μl of 100% ethanol were added. After a 30-minute incubation at −80°C, the sample was centrifuged for 10 minutes at 4°C to pellet the DNA. The pellet was washed with 85% ethanol, air-dried, and dissolved in 10 μl water. Four microliters of the sample was used for reamplification using the corresponding primer set. After reamplification the cDNA fragment was checked on an agarose gel for size consistency and subcloned into the PCR-tartrate-resistant acid phosphatase (TRAP) vector (GenHunter). The cDNA fragment was subsequently sequenced.
Northern Blot Analysis
Northern analysis was performed as previously described [27]. Total RNA (20 μg) from multiple samples was prepared, fractionated on a 1% formaldehyde/agarose gel, transferred to a nylon membrane (Nytran), and UV cross-linked according to standard procedures. cDNA probes were random labeled with 32P-dCTP and hybridized to the membrane at 65°C overnight in a solution containing 15% formamide, 200 mM NaPO4 (pH 7.2), 1 mM EDTA, 7% SDS, and 1% BSA. Following hybridization, the blot was washed in a solution of 2× SSC/1%SDS at 50°C for 30 min, 0.2× SSC/1%SDS at 50°C for 30 min, and 0.2× SSC/0.1%SDS at 65°C for 30 min. Finally, the blot was exposed to Kodak Biomax film at −80 °C. The amount of bound probe was quantitated by scanning the x-ray film and measuring the integrated optical density (IOD) of each band using Image-Pro Plus software (Media Cybernetics). The values of bound probe were normalized to the corresponding levels of 18S plus 28S rRNA (on membrane), and plotted as the ratio of probe to 18S plus 28S rRNA in arbitrary units.
Statistics
Two-tailed t-tests were used to compare histomorphometric indices between Control, Mechanical Stimulation, and Disuse groups. Northern analysis ratios were also analyzed by two-tailed t-test. Changes in body mass between day 0 and sacrifice day were evaluated via paired t-tests within groups. All data were represented as Mean ± SD.
Results
Histomorphometry
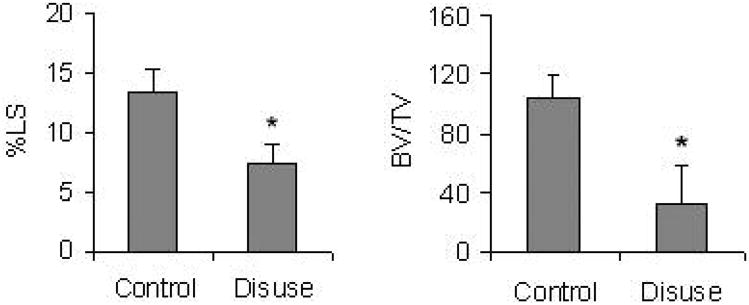
During the course of the study, no significant weight changes were observed in any of the groups. Two of the mechanical stimuli (90Hz and 45 Hz, 28d) showed a significant increase in Percent Labeled Surface (LS) (+89%; +135% respectively, p<0.05) and Bone Volume/Trabecular Volume (BV/TV) (+107%; +97% respectively, p<0.05) over long term control (Table 1). In contrast to the anabolic nature of the mechanical stimuli, 10d of tail suspension caused both LS and BV/TV to drop precipitously, as compared to control (−46%; −69% respectively, p<0.05) (Fig. 1).
Table 1.
The osteogenic potential of the mechanical signal was dependent on specific parameters of intensity, duration and frequency. As compared to long term control, a 10 minute exposure was strongly anabolic if induced at 90Hz or 45Hz. Data were represented as Mean ± SD, * indicates p < 0.05.
| LTC | 90Hz | 45Hz | |
|---|---|---|---|
| %LS | 11.7 ± 1.6 * | 22.2 ± 2.5 * | 27.6 ± 3.4 * |
| BV/TV | 90 ± 18 * | 187 ± 32 * | 178 ± 27 * |
%LS, Percent Labeled Surface; BV/TV, Bone Volume/Trabecular Volume; LTC, long term control.
Fig.1.
Dynamic histomorphometry was used to quantify the bone tissue response to ten days of tail suspension. Both labeled surface (%LS) and bone volume/trabecular volume (BV/TV) were reduced significantly by disuse (p < 0.05). Data were represented as Mean ± SD. 140×70mm (72 × 72 DPI)
Transcriptional sensitivity of Spot 14 to disuse and mechanical signals
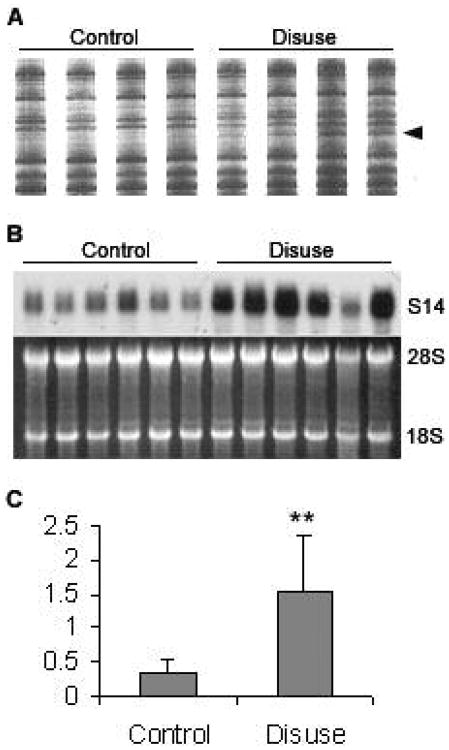
In order to identify differentially expressed genes, we employed DD-PCR of mRNA isolated from the tibiae of the tail suspended animals and compared it to RNA isolated from control animals. Using specific combinations of primers, we identified a 383-bp cDNA fragments with increased expression in disuse samples (Fig 2A). The cDNA were isolated from the gel, purified, reamplified using the corresponding primer pair, subcloned, and sequenced. BLAST search found the sequence 99% homologous to the Rat Spot 14 cDNA (GenBank K01934).
Fig.2.
Up-regulation of S14 gene in disuse samples. (A) Differential Display result. PCR products from four control (left) and four disuse (right) tibiae RNA's was loaded side by side on differential display gel, the bands marked by arrow was cut from the gel and was identified as S14 cDNA. (B) Northern blot of S14 and agarose gel image. The left six lanes are from six different long term control animals, and the right six lanes are from six animals subject to disuse. (C) Quantification of Northern blots. Using optical density methods and normalized to 18S plus 28S rRNA, S14 expression was up-regulated by about 4-fold (p < 0.01) when following ten days of disuse. Data were represented as Mean ± SD. 84×142mm (72 × 72 DPI)
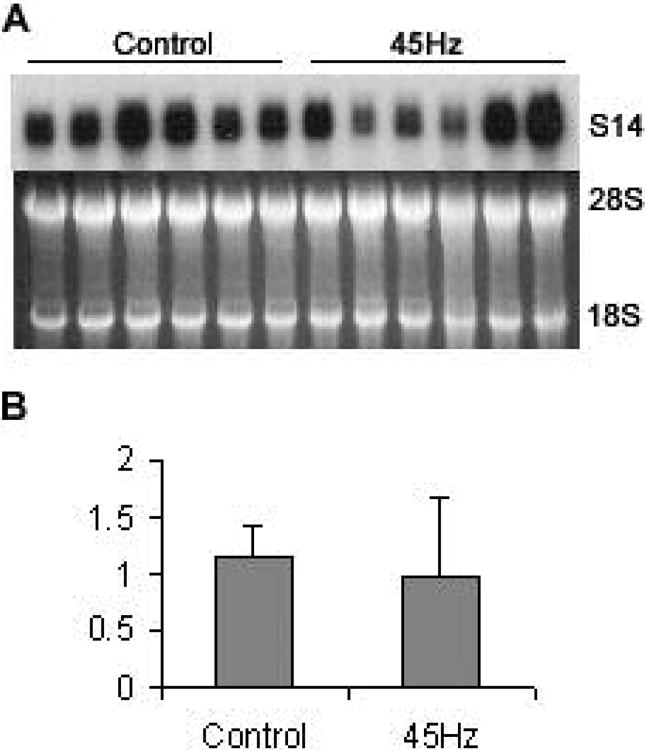
To confirm the results obtained from the differential mRNA display, we used the subcloned cDNAs as a probe for Northern analysis. The hybridization signals from the Northern blot were determined by integrated optical density measurements and normalized to 18S plus 28S rRNA levels (Fig. 2B, 2C). Result show that S14 mRNA transcript was up-regulated by about 4-fold (p<0.01) in the disuse samples (lanes 7-12, 1.54 ± 0.80) as compared to controls (lanes 1-6, 0.37 ± 0.17). We also measured S14 expression under 45Hz mechanical stimulus using Northern hybridization, this osteogenic stimulus didn't show statistically significant effect on S14 expression (Fig. 3, 1.15 ± 0.28 in long term control, 0.97 ± 0.71 in 45Hz mechanical stimulation).
Fig.3.
No change in S14 expression during mechanical stimulation. (A) Northern blot of S14 and agarose gel image. The left six lanes are from six different long term control animals, and the right six lanes are from six animals subject to 45Hz mechanical simulation. (B) Quantification of Northern blots. Using optical density methods and normalized to 18S plus 28S rRNA, S14 expression showed no statistically significant change. Data were represented as Mean ± SD. 84×100mm (72 × 72 DPI)
Discussion
The discovery of genes that are involved in mechanically stimulated bone formation as well as disuse induced bone loss will be critical for discovering the underlying mechanisms of Wolff's Law. In vitro and in vivo studies have been conducted to screen or evaluate genes responsive to mechanical loading and disuse/microgravity unloading [28, 29]. In this study we report the S14 gene, which plays an important role in lipogenesis, is up-regulated in the hindlimb disuse model, but has no response to anabolic mechanical stimulation.
It is well known that a decrease in bone volume associated with osteoporosis and age-related osteopenia is accompanied by increase in marrow adipose tissue [30, 31]. Indeed, an increase in marrow adipocytes is observed in conditions that lead to bone loss, such as ovariectomy [32], immobilization [33], or treatment with glucocorticoids [34]. Our discovery of increased S14 gene expression in disuse rats is consistent with these observations. It is also known that obesity protects mammals from osteoporosis [17-19]. Considering the important role S14 plays in lipogenesis, it is conceivable that it can play a significant role in this protective process. Recently it was reported that human spot 14 protein interacts with thyroid hormone receptor (TR) and suppresses the malic enzyme promoter activity enhanced by liganded TR [35]. Considering the negative effects of thyroid hormone on bone remodeling [36, 37], it is reasonable to presume that S14 helps prevent bone resorption by reducing the negative effect of TR on the skeleton.
The mechanism that could account for the apparent reciprocal relationship between decreased bone density and increased fat formation is beginning to be understood. The bone marrow stroma is a complex system composed of mesenchymal cells (MSCs) which can replicate as undifferentiated cells, as well as differentiate into different lineages of mensenchymal tissues, including bone, cartilage, fat, muscle, and marrow stroma [38]. Increasing evidence of transdifferentiation of these cells suggests a large degree of plasticity between osteoblasts and adipocytes [29, 38-41]. Although our result did not show significant reduction of S14 expression following anabolic mechanical stimulation, we could not eliminate the possibility that S14 may also play a role in the plasticity between osteoblastogenesis and adipogenesis. To support this speculation, we looked into the expression of S14 gene in fracture callus at different ages and time points using publicly available data (NCBI GEO GSE594) [42]. S14 expression at early stages of fracture (3d, 1w, 2w) was reduced by at least 4-fold compared to intact (0d) and later stage fractures (4w, 6w).
It is unknown how disuse elevates the expression of S14. PPARγ (peroxisome proliferators-activated receptor γ) is considered as a master regulator of the adipocyte differentiation program and induces LXR (liver X receptor) [43, 44], which then stimulates SREBP-1 and S14 expression sequentially [45-47]. It was previously reported that dietary PUFA (polyunsaturated fatty acids) suppresses fatty acid synthase transcription by decreasing nuclear SREBP-1 content and altering the binding of NF-Y to promoters of PUFA-response genes [48]. Further, in vitro experiments demonstrated that exposure of hMSC to 7d of modeled microgravity increases PPARγ expression [29]. It is possible that during disuse (an in vivo microgravity simulation), the PPARγ pathway can be activated and thus induce S14 expression. Thus, further investigations are needed to clearly define the molecular events involved in S14 expression and its involvement in the skeleton's adaptation to the absence of mechanical loading.
Acknowledgments
This study was kindly funded by NASA NAG 53950.
References
- 1.Lang T, LeBlanc A, Evans H, Lu Y, Genant H, Yu A. Cortical and trabecular bone mineral loss from the spine and hip in long-duration spaceflight. J Bone Miner Res. 2004;19:1006–1012. doi: 10.1359/JBMR.040307. [DOI] [PubMed] [Google Scholar]
- 2.Mackay DL, Tesar PJ, Liang LN, Haynesworth SE. Characterizing medullary and human mesenchymal stem cell-derived adipocytes. J Cell Physiol. 2006;207:722–728. doi: 10.1002/jcp.20617. [DOI] [PubMed] [Google Scholar]
- 3.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, Zhang Z, Kim JK, Mauvais-Jarvis F, Ducy P, Karsenty G. Endocrine regulation of energy metabolism by the skeleton. Cell. 2007;130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubin CT, Capilla E, Luu YK, Busa B, Crawford H, Nolan DJ, Mittal V, Rosen CJ, Pessin JE, Judex S. Adipogenesis is inhibited by brief, daily exposure to high-frequency, extremely low-magnitude mechanical signals. Proc Natl Acad Sci U S A. 2007;104:17879–17884. doi: 10.1073/pnas.0708467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jump DB, Oppenheimer JH. High basal expression and 3,5,3′-triiodothyronine regulation of messenger ribonucleic acid S14 in lipogenic tissues. Endocrinology. 1985;117:2259–2266. doi: 10.1210/endo-117-6-2259. [DOI] [PubMed] [Google Scholar]
- 6.Seelig S, Liaw C, Towle HC, Oppenheimer JH. Thyroid hormone attenuates and augments hepatic gene expression at a pretranslational level. Proc Natl Acad Sci U S A. 1981;78:4733–4737. doi: 10.1073/pnas.78.8.4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freake HC, Oppenheimer JH. Stimulation of S14 mRNA and lipogenesis in brown fat by hypothyroidism, cold exposure, and cafeteria feeding: evidence supporting a general role for S14 in lipogenesis and lipogenesis in the maintenance of thermogenesis. Proc Natl Acad Sci U S A. 1987;84:3070–3074. doi: 10.1073/pnas.84.9.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinlaw WB, Perez-Castillo AM, Fish LH, Mariash CN, Schwartz HL, Oppenheimer JH. Interaction of dietary carbohydrate and glucagon in regulation of rat hepatic messenger ribonucleic acid S14 expression: role of circadian factors and 3′,5′-cyclic adenosine monophosphate. Mol Endocrinol. 1987;1:609–613. doi: 10.1210/mend-1-9-609. [DOI] [PubMed] [Google Scholar]
- 9.Perez-Castillo A, Schwartz HL, Oppenheimer JH. Rat hepatic mRNA-S14 and lipogenic enzymes during weaning: role of S14 in lipogenesis. Am J Physiol. 1987;253:E536–542. doi: 10.1152/ajpendo.1987.253.5.E536. [DOI] [PubMed] [Google Scholar]
- 10.Sudo Y, Goto Y, Mariash CN. Location of a glucose-dependent response region in the rat S14 promoter. Endocrinology. 1993;133:1221–1229. doi: 10.1210/endo.133.3.8365364. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Q, Anderson GW, Mucha GT, Parks EJ, Metkowski JK, Mariash CN. The Spot 14 protein is required for de novo lipid synthesis in the lactating mammary gland. Endocrinology. 2005;146:3343–3350. doi: 10.1210/en.2005-0204. [DOI] [PubMed] [Google Scholar]
- 12.Zhu Q, Mariash A, Margosian MR, Gopinath S, Fareed MT, Anderson GW, Mariash CN. Spot 14 gene deletion increases hepatic de novo lipogenesis. Endocrinology. 2001;142:4363–4370. doi: 10.1210/endo.142.10.8431. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Rodriguez J, Kaninda-Tshilumbu JP, Santos A, Perez-Castillo A. The spot 14 protein inhibits growth and induces differentiation and cell death of human MCF-7 breast cancer cells. Biochem J. 2005;390:57–65. doi: 10.1042/BJ20042080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinlaw WB, Church JL, Harmon J, Mariash CN. Direct evidence for a role of the “spot 14” protein in the regulation of lipid synthesis. J Biol Chem. 1995;270:16615–16618. doi: 10.1074/jbc.270.28.16615. [DOI] [PubMed] [Google Scholar]
- 15.Kinlaw WB, Tron P, Friedmann AS. Nuclear localization and hepatic zonation of rat “spot 14” protein: immunohistochemical investigation employing anti-fusion protein antibodies. Endocrinology. 1992;131:3120–3122. doi: 10.1210/endo.131.6.1446647. [DOI] [PubMed] [Google Scholar]
- 16.LaFave LT, Augustin LB, Mariash CN. S14: insights from knockout mice. Endocrinology. 2006;147:4044–4047. doi: 10.1210/en.2006-0473. [DOI] [PubMed] [Google Scholar]
- 17.Felson DT, Zhang Y, Hannan MT, Anderson JJ. Effects of weight and body mass index on bone mineral density in men and women: the Framingham study. J Bone Miner Res. 1993;8:567–573. doi: 10.1002/jbmr.5650080507. [DOI] [PubMed] [Google Scholar]
- 18.Ravn P, Cizza G, Bjarnason NH, Thompson D, Daley M, Wasnich RD, McClung M, Hosking D, Yates AJ, Christiansen C. Low body mass index is an important risk factor for low bone mass and increased bone loss in early postmenopausal women. Early Postmenopausal Intervention Cohort (EPIC) study group. J Bone Miner Res. 1999;14:1622–1627. doi: 10.1359/jbmr.1999.14.9.1622. [DOI] [PubMed] [Google Scholar]
- 19.Tremollieres FA, Pouilles JM, Ribot C. Vertebral postmenopausal bone loss is reduced in overweight women: a longitudinal study in 155 early postmenopausal women. J Clin Endocrinol Metab. 1993;77:683–686. doi: 10.1210/jcem.77.3.8370689. [DOI] [PubMed] [Google Scholar]
- 20.Morey-Holton ER, Globus RK. Hindlimb unloading rodent model: technical aspects. J Appl Physiol. 2002;92:1367–1377. doi: 10.1152/japplphysiol.00969.2001. [DOI] [PubMed] [Google Scholar]
- 21.Rubin C, Xu G, Judex S. The anabolic activity of bone tissue, suppressed by disuse, is normalized by brief exposure to extremely low-magnitude mechanical stimuli. Faseb J. 2001;15:2225–2229. doi: 10.1096/fj.01-0166com. [DOI] [PubMed] [Google Scholar]
- 22.Erben RG. Embedding of bone samples in methylmethacrylate: an improved method suitable for bone histomorphometry, histochemistry, and immunohistochemistry. J Histochem Cytochem. 1997;45:307–313. doi: 10.1177/002215549704500215. [DOI] [PubMed] [Google Scholar]
- 23.Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 1987;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 24.Hadjiargyrou M, Halsey MF, Ahrens W, Rightmire EP, McLeod KJ, Rubin CT. Cloning of a novel cDNA expressed during the early stages of fracture healing. Biochem Biophys Res Commun. 1998;249:879–884. doi: 10.1006/bbrc.1998.9167. [DOI] [PubMed] [Google Scholar]
- 25.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 26.Hadjiargyrou M, Ahrens W, Rubin CT. Temporal expression of the chondrogenic and angiogenic growth factor CYR61 during fracture repair. J Bone Miner Res. 2000;15:1014–1023. doi: 10.1359/jbmr.2000.15.6.1014. [DOI] [PubMed] [Google Scholar]
- 27.Zhi J, Sommerfeldt DW, Rubin CT, Hadjiargyrou M. Differential expression of neuroleukin in osseous tissues and its involvement in mineralization during osteoblast differentiation. J Bone Miner Res. 2001;16:1994–2004. doi: 10.1359/jbmr.2001.16.11.1994. [DOI] [PubMed] [Google Scholar]
- 28.Judex S, Zhong N, Squire ME, Ye K, Donahue LR, Hadjiargyrou M, Rubin CT. Mechanical modulation of molecular signals which regulate anabolic and catabolic activity in bone tissue. J Cell Biochem. 2005;94:982–994. doi: 10.1002/jcb.20363. [DOI] [PubMed] [Google Scholar]
- 29.Zayzafoon M, Gathings WE, McDonald JM. Modeled microgravity inhibits osteogenic differentiation of human mesenchymal stem cells and increases adipogenesis. Endocrinology. 2004;145:2421–2432. doi: 10.1210/en.2003-1156. [DOI] [PubMed] [Google Scholar]
- 30.Burkhardt R, Kettner G, Bohm W, Schmidmeier M, Schlag R, Frisch B, Mallmann B, Eisenmenger W, Gilg T. Changes in trabecular bone, hematopoiesis and bone marrow vessels in aplastic anemia, primary osteoporosis, and old age: a comparative histomorphometric study. Bone. 1987;8:157–164. doi: 10.1016/8756-3282(87)90015-9. [DOI] [PubMed] [Google Scholar]
- 31.Meunier P, Aaron J, Edouard C, Vignon G. Osteoporosis and the replacement of cell populations of the marrow by adipose tissue. A quantitative study of 84 iliac bone biopsies. Clin Orthop Relat Res. 1971;80:147–154. doi: 10.1097/00003086-197110000-00021. [DOI] [PubMed] [Google Scholar]
- 32.Wronski TJ, Walsh CC, Ignaszewski LA. Histologic evidence for osteopenia and increased bone turnover in ovariectomized rats. Bone. 1986;7:119–123. doi: 10.1016/8756-3282(86)90683-6. [DOI] [PubMed] [Google Scholar]
- 33.Minaire P, Neunier P, Edouard C, Bernard J, Courpron P, Bourret J. Quantitative histological data on disuse osteoporosis: comparison with biological data. Calcif Tissue Res. 1974;17:57–73. doi: 10.1007/BF02547214. [DOI] [PubMed] [Google Scholar]
- 34.Wang GJ, Sweet DE, Reger SI, Thompson RC. Fat-cell changes as a mechanism of avascular necrosis of the femoral head in cortisone-treated rabbits. J Bone Joint Surg Am. 1977;59:729–735. [PubMed] [Google Scholar]
- 35.Chou WY, Cheng YS, Ho CL, Liu ST, Liu PY, Kuo CC, Chang HP, Chen YH, Chang GG, Huang SM. Human spot 14 protein interacts physically and functionally with the thyroid receptor. Biochem Biophys Res Commun. 2007;357:133–138. doi: 10.1016/j.bbrc.2007.03.103. [DOI] [PubMed] [Google Scholar]
- 36.Bassett JH, O'Shea PJ, Sriskantharajah S, Rabier B, Boyde A, Howell PG, Weiss RE, Roux JP, Malaval L, Clement-Lacroix P, Samarut J, Chassande O, Williams GR. Thyroid hormone excess rather than thyrotropin deficiency induces osteoporosis in hyperthyroidism. Mol Endocrinol. 2007;21:1095–1107. doi: 10.1210/me.2007-0033. [DOI] [PubMed] [Google Scholar]
- 37.Murphy E, Williams GR. The thyroid and the skeleton. Clin Endocrinol (Oxf) 2004;61:285–298. doi: 10.1111/j.1365-2265.2004.02053.x. [DOI] [PubMed] [Google Scholar]
- 38.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 39.Beresford JN, Bennett JH, Devlin C, Leboy PS, Owen ME. Evidence for an inverse relationship between the differentiation of adipocytic and osteogenic cells in rat marrow stromal cell cultures. J Cell Sci. 1992;102(Pt 2):341–351. doi: 10.1242/jcs.102.2.341. [DOI] [PubMed] [Google Scholar]
- 40.Nuttall ME, Patton AJ, Olivera DL, Nadeau DP, Gowen M. Human trabecular bone cells are able to express both osteoblastic and adipocytic phenotype: implications for osteopenic disorders. J Bone Miner Res. 1998;13:371–382. doi: 10.1359/jbmr.1998.13.3.371. [DOI] [PubMed] [Google Scholar]
- 41.Dorheim MA, Sullivan M, Dandapani V, Wu X, Hudson J, Segarini PR, Rosen DM, Aulthouse AL, Gimble JM. Osteoblastic gene expression during adipogenesis in hematopoietic supporting murine bone marrow stromal cells. J Cell Physiol. 1993;154:317–328. doi: 10.1002/jcp.1041540215. [DOI] [PubMed] [Google Scholar]
- 42.Meyer MH, Etienne W, Meyer RA., Jr Altered mRNA expression of genes related to nerve cell activity in the fracture callus of older rats: A randomized, controlled, microarray study. BMC Musculoskelet Disord. 2004;5:24. doi: 10.1186/1471-2474-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chawla A, Boisvert WA, Lee CH, Laffitte BA, Barak Y, Joseph SB, Liao D, Nagy L, Edwards PA, Curtiss LK, Evans RM, Tontonoz P. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol Cell. 2001;7:161–171. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- 44.Laffitte BA, Joseph SB, Walczak R, Pei L, Wilpitz DC, Collins JL, Tontonoz P. Autoregulation of the human liver X receptor alpha promoter. Mol Cell Biol. 2001;21:7558–7568. doi: 10.1128/MCB.21.22.7558-7568.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foretz M, Pacot C, Dugail I, Lemarchand P, Guichard C, Le Liepvre X, Berthelier-Lubrano C, Spiegelman B, Kim JB, Ferre P, Foufelle F. ADD1/SREBP-1c is required in the activation of hepatic lipogenic gene expression by glucose. Mol Cell Biol. 1999;19:3760–3768. doi: 10.1128/mcb.19.5.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hummasti S, Laffitte BA, Watson MA, Galardi C, Chao LC, Ramamurthy L, Moore JT, Tontonoz P. Liver X receptors are regulators of adipocyte gene expression but not differentiation: identification of apoD as a direct target. J Lipid Res. 2004;45:616–625. doi: 10.1194/jlr.M300312-JLR200. [DOI] [PubMed] [Google Scholar]
- 48.Teran-Garcia M, Adamson AW, Yu G, Rufo C, Suchankova G, Dreesen TD, Tekle M, Clarke SD, Gettys TW. Polyunsaturated fatty acid suppression of fatty acid synthase (FASN): evidence for dietary modulation of NF-Y binding to the Fasn promoter by SREBP-1c. Biochem J. 2007;402:591–600. doi: 10.1042/BJ20061722. [DOI] [PMC free article] [PubMed] [Google Scholar]