Abstract
Mantle cell lymphoma (MCL) and chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) share many features and both arise from CD5+ B-cells, their distinction is critical as MCL is a much more aggressive neoplasm. Rarely, composite MCL and CLL/SLL have been reported. Little is known, about the nature of these cases and in particular the clonal relationship of the two lymphomas. Eleven composite MCL and CLL/SLL cases were identified. The clinical, morphologic and immunophenotypic features of the MCL and CLL/SLL were characterized. Immunoglobulin heavy chain (IGH) gene analysis was performed on microdissected MCL and CLL/SLL components to assess their clonal relationship. Ten patients had lymphadenopathy, and 7 patients had bone marrow involvement. The MCL component had the following growth patterns: in situ (n=1), mantle zone (n=3), nodular and diffuse (n=3), diffuse (n=3), and interstitial in the bone marrow (the only patient without lymphadenopathy) (n=1); 6 MCL had blastoid or pleomorphic and 5 classical cytologic features. The CLL/SLL component was internodular (n=9) or diffuse (n=2). All MCL were CD5+ and cyclin D1+ with t(11;14) translocation. All CLL/SLL were CD5+, CD23+ and negative for cyclin D1 or t(11;14). IGH gene analysis showed that the MCL and CLL/SLL components displayed different sized fragments, indicating that the MCL and CLL/SLL are likely derived from different neoplastic B-cell clones. The lack of a clonal relationship between the MCL and CLL/SLL components suggests that the MCL and CLL/SLL represent distinct disease processes and do not share a common progenitor B-cell.
Keywords: Mantle cell lymphoma, Small lymphocytic lymphoma/chronic lymphocytic leukemia, Cyclin D1
Introduction
Mantle cell lymphoma (MCL) accounts for approximately 6% of all B-cell lymphomas in Western countries [1,2]. The incidence rate is about 0.2–0.3 cases per 100,000 person-years, and the median age at diagnosis is about 60 years. Patients usually present with lymphadenopathy and commonly have involvement of the bone marrow, liver, spleen and gastrointestinal tract. Morphologically, typical cases of MCL are composed of a monotonous population of small often angulated B-cells that express CD5 and grow with a diffuse, nodular, or mantle zone pattern. MCL is usually characterized by the presence of t(11;14)(q13;q32), which juxtaposes the cyclin D1 gene with the immunoglobulin heavy chain gene (IGH) locus and results in overexpression of cyclin D1 [3]. MCL most often arises from a CD5+ B-cell without mutations in the IGH variable regions (IGHV), although a subset of MCL cases have somatically mutated IGHV [4–9].
Chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) is the most common adult blood cancer in Western countries and accounts for approximately 30% of all lymphoid neoplasms and 12% of nodal lymphomas [1,2]. The incidence rate is about 2–6 cases per 100,000 person-years and the median age at diagnosis is about 65 years [1]. Morphologically, lymph nodes usually demonstrate diffuse architectural effacement by small lymphocytes with vague, pale proliferation centers that also include paraimmunoblasts and prolymphocytes; occasionally CLL/SLL has a nodular appearance with pseudofollicles and sometimes the nodal spread is parafollicular/internodular. The predominant tumor cell type in CLL/SLL is a small CD5+, CD23+ B-lymphocyte.
Composite lymphoma is the term used to describe a single anatomic site involved by two distinct types of lymphoma. Traditionally, the two components of composite lymphoma have been thought to not be clonally related, although microdissection and molecular analysis has shown that morphologically disparate lymphomas can be clonally related or arise from a common progenitor cell at the molecular level [10–13]. A wide variety of lymphoma types have been described in composite lymphomas, however, the combination of MCL and CLL/SLL is uncommon and very few molecular studies have been performed on this rare form of composite lymphoma [14;15].
Here we report a series of 11 cases of composite lymphoma showing MCL and CLL/SLL components involving the same biopsy specimen. We describe clinical features, immunophenotype and, when possible, the results of IGH gene rearrangement analysis of each component to assess the clonal relationship of the MCL and CLL/SLL components.
Materials and Methods
Case Selection
Cases of composite MCL and CLL/SLL were identified through a multi-institutional collaboration involving five medical centers. All cases were reviewed by a group of hematopathologists (all primary center pathologists and KHY), and the diagnoses were confirmed on the basis of WHO classification criteria [2]. The current study was approved by each of the participating center Institutional Review Boards, and the overall collaborative study was approved by the Institutional Review Board at The University of Texas MD Anderson Cancer Center in Houston, Texas.
Histology and immunohistochemistry
All specimens were fixed in 10% neutral buffered formalin, embedded in paraffin, and 4 µm sections were cut and stained with hematoxylin and eosin for histological evaluation. Immunohistochemical staining was performed using formalin-fixed, paraffin-embedded tissue sections. Briefly, tissue sections were deparaffinized in xylene, rehydrated in graded alcohols, and endogenous peroxidase was blocked with 3% hydrogen peroxide. For antigen retrieval, tissue sections were immersed in citrate buffer (pH 6.0) as appropriate for each antibody used. After the sections were rinsed with phosphate-buffered saline, immunohistochemical analysis was performed for purposes of the present study using antibodies against CD3, CD10, kappa, lambda, IgA, IgG, IgM, and IgD (PS1, 56C6, 2F3.2 clones, respectively, polyclonal light and heavy chains) (Ventana, Tucson, Arizona); CD5 (4C7 clone, Novocastra, Newcastle-on-Tyne, UK); CD20 (L26 clone) and BCL6 (PG-B6p clone) (Dako, Carpinteria, California), CD23 (BU38 clone) ZAP-70 (Binding Site, Birmingham, UK), and cyclin D1 (SP4 clone, Neomarkers, Freemont, California). These stains were performed on an automated immunostainer (Benchmark XT from Ventana/Roche, Tucson, Arizona) using a streptavidin-biotin peroxidase detection system.
Flow Cytometry Studies
Lymph node and bone marrow aspirate specimens were processed according to established procedures. Briefly, mononuclear cells were isolated from the lymph node or bone marrow samples by Ficoll-Hypaque density-gradient centrifugation (Accu-Prep, Accurate Chemical, Westbury, New York). Contaminating red blood cells were removed by hypotonic lysis with 0.9% sodium chloride. Isolated cells were washed with phosphate-buffered saline (PBS) and resuspended in RPMI 1640 (Life Technologies, Grand Island, New York) supplemented with 1 mol/L of Hepes buffer, L-glutamine, penicillin, streptomycin, and 10% fetal calf serum. The separated cells were counted by using the trypan blue exclusion method, divided into aliquots, and placed in 5 mL plastic tubes at a concentration of 2 × 105 cells per tube. Cells were incubated with antibody cocktails in 200 µL of cold PBS with 0.5% bovine serum albumin for 30 minutes at 4°C. Antibodies specific for CD3, CD5, CD10, CD19, CD20, CD22, CD23, CD38, CD43, CD79a, kappa and lambda light chains, were used according to manufacturer’s recommendations. After incubation, the cells were washed with PBS, and the cell pellets were resuspended in 1% paraformaldehyde in PBS. Data were collected on a Beckman Coulter FC500 flow cytometer (Beckman Coulter, Miami, FL) using Beckman Coulter Cytomics RXP software (Applied Cytometry Systems, Dinnington, UK), and list mode files were acquired for off-line analysis.
FISH Analysis
To detect the t(11;14)(q13;q32) formalin-fixed, paraffin-embedded tissue sections were analyzed via interphase FISH analysis using the commercially available LSI IgH/CCND1 XT Dual-Color, Dual-Fusion Translocation Probe (Abbott/Vysis, Des Plaines, Illinois) as described [16].
Immunoglobulin heavy chain analysis
Genomic DNA was extracted using a Genovison extraction kit (Qiagen, Valencia, California) from separately microdissected MCL and CLL/SLL components. Positively stained cells from uncovered and un-counterstained tissue slides stained for either cyclin D1 (MCL component) or CD23 (CLL/SLL component) were microdissected as described [16]. Polymerase chain reaction (PCR) based analysis for IGH gene rearrangements was performed utilizing consensus FR1, FR3 and J primers, as previously described [17]. The PCR products were examined using a high resolution fragment length analyzer (ABI 310 Genetic Analyzer, Applied Biosystems/Life Technologies, Carlsbad, California). DNA from normal polyclonal B-cell populations produced a bell-shaped curve of amplicon products (Gaussian distribution), and was used as control. Monoclonal gene rearrangements were identified as prominent, single-sized amplification products; the base pair length was recorded for each microdissected fraction. A shift of the PCR products of more than one bp between the cases was considered to indicate a clonally unrelated event.
Results
Clinical features
The clinical information is summarized in Table 1. There were 10 male and 1 female patients with a median age of 73 years (range, 55 to 80 years). Ten patients had lymphadenopathy and 1 patient presented with only bone marrow involvement. Four patients with lymphadenopathy also had bone marrow involvement. Eight patients had advanced disease at diagnosis, including 1 stage III and 7 stage IV. Three patients presented with stage II disease. Peripheral blood lymphocytosis (5.6 – 8.7 × 109/L) was present in 4 of 8 patients with a complete blood count and peripheral bloods smear available for review. Five patients died of disease, 4 patients were alive at the time of the manuscript preparation with a follow-up time between 1 and 67 months (mean 29 months), and 2 patients are lost to follow-up. The median survival was 42 months (95% CI: 27–56).
Table 1.
Clinical features of composite mantle cell lymphoma and chronic lymphocytic leukemia/small lymphocytic lymphoma
| Case | Age (year) |
Gender | Site of involvement |
Extranoda l lesion |
Stage (CTMRI- PET) |
Morphologic subtype |
B-symptom | IPI e | Peripheral blood |
Bone marrow |
Treatment regimen |
Treatment response |
Followup time |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 77 | M | Omental LN | GI, liver, spleen | IV | Blastoid | No | 3 | Abnormal | Involved | No treatment | Followup | 1 months |
| 2 | 79 | M | Right supraclavicular LN | None | III | Blastoid | No | 2 | Normal | Normal | R-CHOP | Complete remission | 35 months |
| 3 | 80 | M | Left inguinal LN | Diffuse | IV | Small lymphocytic | Yes | 3 | Abnormal | Involved | No treatment | Followup | 3 months |
| 4 | 62 | M | Left Axillary LN | Bone marrow | II | Small lymphocytic | NA | 1 | Normal | Involved | R-CHOP | Complete remission | 29 months |
| 5 | 59 | M | Cervival LN | Diffuse | IV | Blastoid | NA | NA | NA | NA | NA | NA | NA |
| 6 | 69 | M | Cervival LN | Diffuse | IV | Blastoid | NA | NA | NA | NA | NA | NA | NA |
| 7 | 70 | M | Left axillary LN | Bone marrow | IV | Small lymphocytic | Yes | 5 | Anemia | Involved | Hyper- CVAD | Complete remission | 27 months |
| 8 | 79 | M | Left inguinal LN | None | II | Blastoid | No | 1 | Normal | Involved | R-CHOP | Partial remission | 42 months |
| 9 | 55 | F | Left Axillary LN | Bone marrow | IV | Small lymphocytic | Yes | 3 | Abnormal | Involved | Hyper- CVAD | Complete remission | 59 months |
| 10 | 63 | M | Cervival LN | Bone marrow | II | Blastoid | No | 3 | Normal | Normal | R-CHOP | Complete remission | 67 months |
| 11 | 57 | M | Bone marrow | Bone marrow | IV | Small lymphocytic | Yes | 2 | Abnormal | Involved | Watch | Followup | 4 months |
LN, lymph node; NA, not available; R-CHOP, Rituxan, cyclophosphamide, doxorubicin, vincristine, and prednisone; Hyper-CVAD, cyclophosphamide, vincristine, doxorubicin, and dexamethasone; IPI, international prognostic index.
Histology, immunohistochemistry and flow cytometry analyses
Ten biopsy specimens were obtained from lymph nodes and bone marrow was biopsied in 5 patients (Tables 1–2). All biopsy specimens were reviewed by at least three hematopathologists [KHY, AT and the primary contributing pathologists], and were classified as composite lymphoma. The MCL component had blastoid or pleomorphic cytologic features in 6 patients and classical features in 5 patients (Figures 1–7, Supplemental Figure for case-7, Table 2). The pattern of the MCL component was in situ (n=1), mantlezone (n=3), nodular and diffuse (n=3), diffuse (n=3), and in the bone marrow only case, interstitial (n=1). The accompanying CLL/SLL components were parafollicular/internodular (n=9) or diffuse (n=2).
Table 2.
Morphologic and cytologic features of composite mantle cell lymphoma and chronic lymphocytic leukemia/small lymphocytic lymphoma
| Case | Morphological CLL/SLL pattern | Morphological MCL pattern | Morphological MCL subtype |
|---|---|---|---|
| 1 | Internodular | Nodular and diffuse | Blastoid |
| 2 | Internodular | Nodular and diffuse | Blastoid |
| 3 | Internodular | In Situ | Small lymphocytic |
| 4 | Internodular | Mantle zone | Small lymphocytic |
| 5 | Internodular | Nodular and diffuse | Blastoid |
| 6 | Diffuse | Focal, Patchy | Blastoid |
| 7 | Nodular | Mantle zone | Small lymphocytic |
| 8 | Internodular | Diffuse | Blastoid |
| 9 | Internodular | Diffuse | Small lymphocytic |
| 10 | Internodular | Diffuse | Blastoid |
| 11 | Nodular | Interstitial | Small lymphocytic |
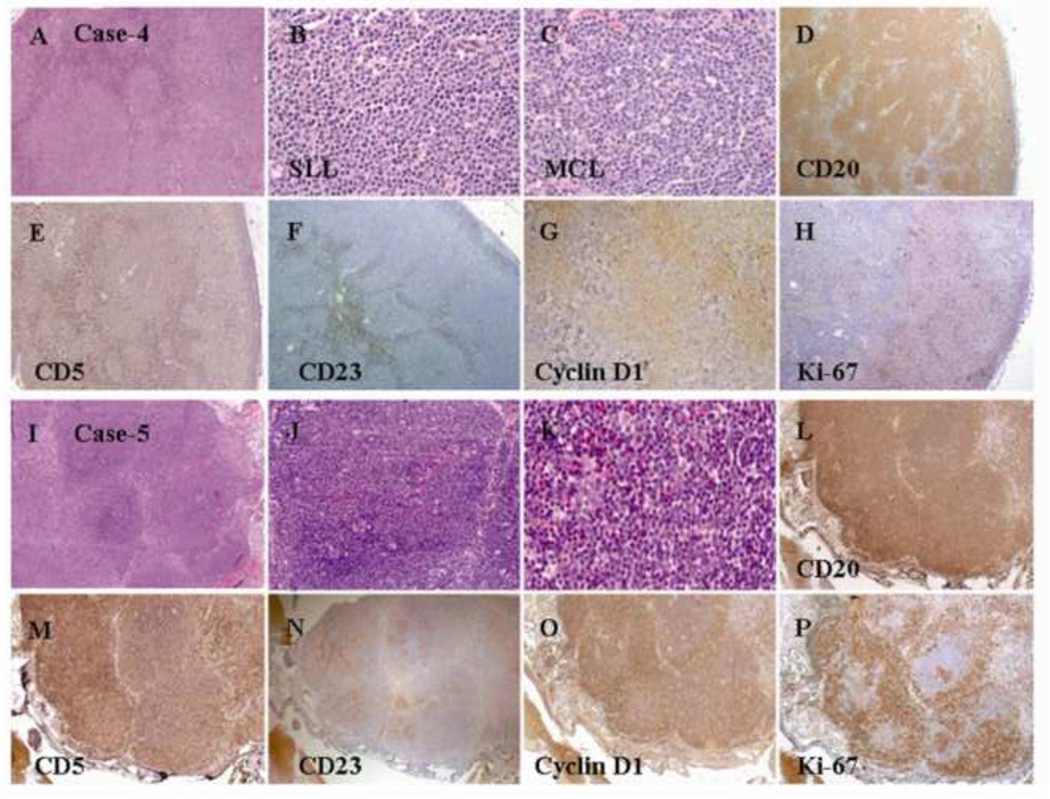
Figure 1.
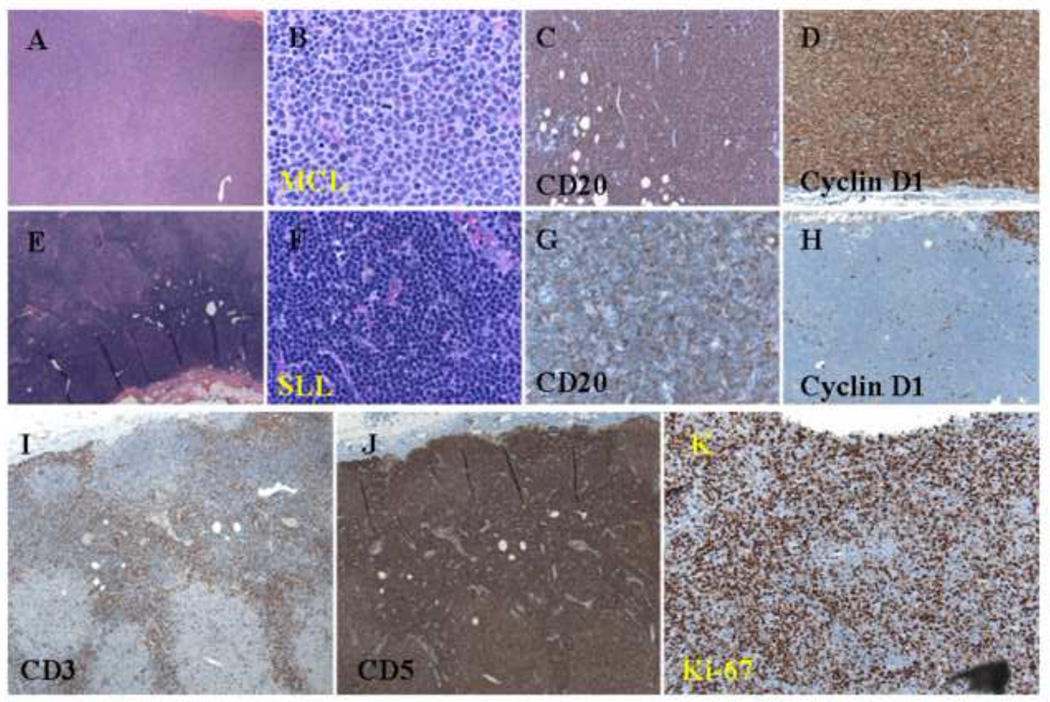
Histologic and immunophenotypic features of a composite case of MCL and CLL/SLL. (A and B) Nodular pattern of atypical small lymphoid cells with a nodular pattern (H&E staining; original magnification: 20X and 100X). (C) Positive CD20 staining of atypical small lymphoid cells in both the internodular and nodular areas (immunoperoxidase staining; original magnification: 40X). (D) Cyclin D1 staining of atypical small lymphoid cells in the nodular area (immunoperoxidase staining; original magnification: 40X). (E and F) Atypical small lymphoid cells in the internodular area (H&E staining; original magnification: 20X and 100X). (G) Weak CD20 staining of atypical small lymphoid cells in the internodular area (immunoperoxidase staining; original magnification: 40X). (H) Negative cyclin D1 staining of atypical small lymphoid cells in the internodular area (immunoperoxidase staining; original magnification: 40X). (I) CD3 staining of normal small T-cells in the internodular area (immunoperoxidase staining; original magnification: 20X). (J) CD5 staining of atypical small lymphoid cells in both the internodular and nodular areas (immunoperoxidase staining; original magnification: 20X). (K) Ki-67 staining of a relatively high proliferation index in the nodular area (immunoperoxidase staining; original magnification: 40X).
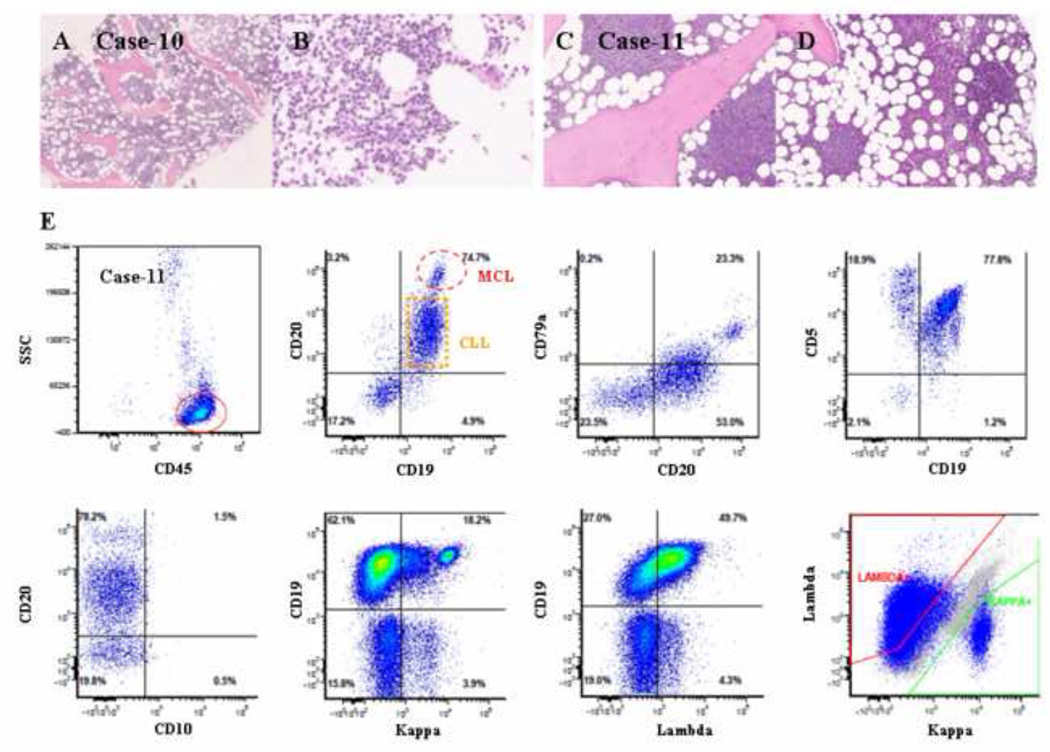
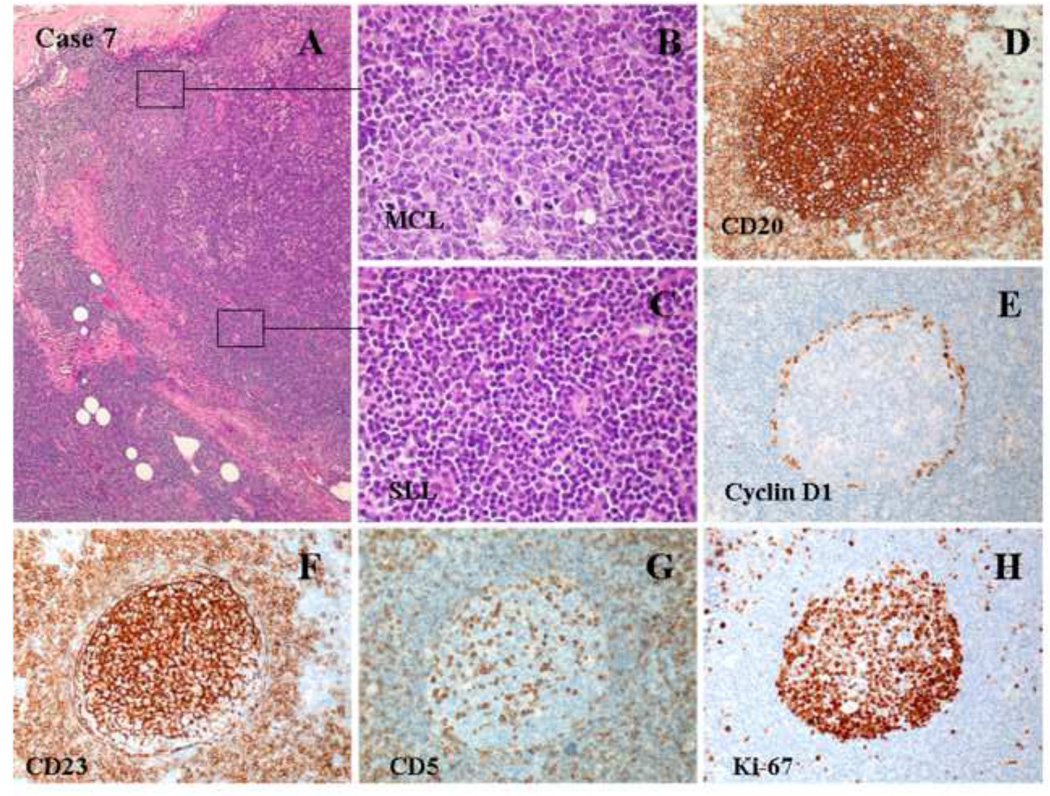
Figure 7.
Histologic and immunophenotypic features of two composite cases of MCL and CLL/SLL. (A and B) A diffuse and interstitial pattern of small atypical lymphoid infiltration (H&E staining; original magnification: 20X and 100X). (C and D) A nodular and interstitial pattern of atypical lymphoid infiltration (H&E staining; original magnification: 20X and 100X). (E) Flow cytometry revealed two clonal populations of small B-cells: a predominant population of B-cells expressed CD5, CD19, dim CD20, CD23, and dim monotypic kappa light chains, and a small population of bright CD19/CD20/CD79a-positive B-cells expressed monotypic lambda light chains. Both populations were positive for CD5 and negative for CD10.
In all cases, both the MCL and CLL/SLL components had a classical phenotype. The MCL component was CD5+, cyclin D1+, and CD23−. The CLL/SLL component was CD5+, CD23+, and cyclin D1−. IgD was expressed in the MCL component of all cases (Table 3, and Figures 1–7), as well as in the CLL/SLL component in 5 cases, although at a weaker intensity in some cases. The light chain expression was assessable in 9/11 cases and seven (cases 1, 2, 3, 5, 7, 8 and 10) showed different clonal light chain restriction in the CLL/SLL and MCL populations. ZAP70 was negative in all cases of MCL and was positive in areas densely involved by CLL/SLL in 30–100% of cells in 7 cases.
Table 3.
Immunophenotypic, molecular and genetic findings of composite mantle cell lymphoma and chronic lymphocytic leukemia/small lymphocytic lymphoma
| Case | lymphoma | CD5 | CD20 | CD23 | Cyclin D1 |
Kappa | Lambda | IgD | IgM | IgG | ZAP- 70 a |
FISH t(11;14) |
IgH FLP (bp size) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MCL | + | + | − | + | + | − | + | − | − | − | + | 127 |
| SLL | + | + | + | − | − | + | − | − | − | − | − | 83 | |
| 2 | MCL | + | + | − | + | + | − | + | + | − | − | + | 95 |
| SLL | + | + | + | − | − | + | dim + | − | − | − | − | 108 | |
| 3 | MCL | + | + | − | + | − | + | + | − | − | − | + | 103 |
| CLL | + | + | + | − | + | − | − | − | − | 40% | − | 130 | |
| 4 | MCL | + | + | − | + | ND | ND | + | − | − | − | + | 111 |
| CLL | + | + | + | − | ND | ND | dim + | − | − | 30% | − | 92 | |
| 5 | MCL | + | + | − | + | + | − | + | dim + | − | − | + | 118 b |
| CLL | + | + | + | − | − | + | − | − | − | 20% | − | 118 b | |
| 6 | MCL | + | + | − | + | ND | ND | dim + | − | − | − | + | 99 |
| SLL | + | + | + | − | ND | ND | + | + | − | 60% | − | 141 | |
| 7 | MCL | + | + | − | + | + | − | + | − | − | − | + | 112 |
| SLL | + | + | + | − | − | + | dim + | − | − | 60% | − | 98 | |
| 8 | MCL | + | + | − | + | + | − | ND | ND | ND | − | + | ND |
| CLL | + | + | + | − | − | + | ND | ND | ND | 60% | − | ND | |
| 9 | MCL | + | + | − | + | + | − | dim + | dim + | ND | − | + | ND |
| CLL | + | + | + | − | + | − | dim + | dim + | ND | − | − | ND | |
| 10 | MCL | + | + | − | + | + | − | ND | ND | ND | − | + | ND |
| CLL | + | + | + | − | − | + | ND | ND | ND | 50% | − | ND | |
| 11 | MCL | + | + | − | + | − | + | ND | ND | ND | − | + | ND |
| CLL | + | + | + | − | − | + | ND | ND | ND | 100% | − | ND |
According to Sabattini et al. 2007.
Since the polymerase chain reaction was designed to detect the t(11;14) translocation as well and this translocation could not be detected in the cyclin D1+ microdissected fraction, a “contamination” could not be excluded; thus, a clonal relationship between MCL and CLL/SLL should not be considered proven. ND, not done; FLP/ IgH, Immunoglobulin heavy chain variable region fragment length polymorphism analysis
IGH Analysis by PCR
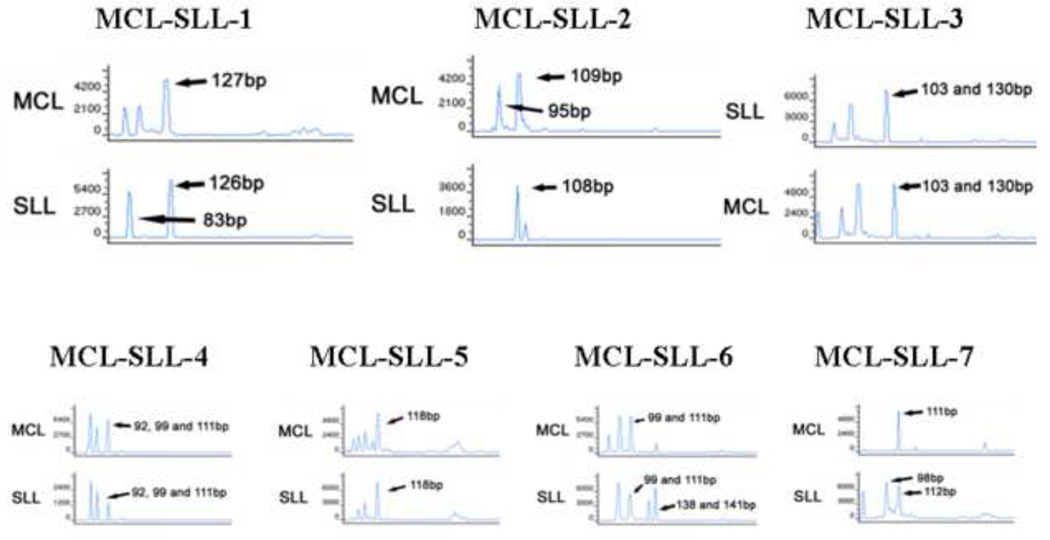
The IGH results obtained from microdissected cyclin D1+ MCL cells or CD23+ CLL/SLL cells (Figure 8) revealed that out of the 7 cases with sufficient material for analysis 6 had a monoclonal peak in the respective microdissected components. In all 6 cases, the MCL and CLL/SLL cells displayed differently sized amplicons, indicating that the MCL and CLL/SLL components were derived from different B-cell clones (Table 3). Signals indicating at least two amplicons of different sizes (one the size of the other microdissected component, and another of a different size) in one of the microdissected components were identifiable in 5 cases (5 in CLL/SLL components and 3 in MCL components) indicated that, despite microdissection, isolated tumor cells of either neoplastic components had “contaminated” the specimens. Nevertheless, in all cases except 1, the relative peak heights as well as the amplicon sizes and the concurrent detection of the t(11;14) with the chosen multiplex PCR design (detectable in all 5 in all MCL components) allowed for reliable result interpretation. In case 5, the fragment length of 118 bp was the same in both components, but since the PCR was designed to detect the t(11;14) as well, and this failed in the cyclin D1+ microdissected fraction due to technical reason, poor quality of extracted DNA or due to “contamination” with CLL/SLL cells, a clonal relationship could not be regarded proven in this case. However, FISH confirmed the presence of t(11;14) translocation in the MCL of case 5, but not in the CLL/SLL component.
Figure 8.
IGH gene rearrangement analysis of six composite cases of MCL and CLL/SLL revealed that the two lymphoid populations were clonally unrelated in five cases. Similar but also distinct clonal peaks are seen in the separate PCR runs of both components in cases 1, 2, 6 and 7. Since biclonal (biallelic) rearrangements in one, especially in the SLL, component of these composite cases are very unprobable, this indicates that, despite microdissection, isolated tumor cells particularly of the MCL component had “contaminated” the specimens. In case 6 the SLL component presents with peaks at 138 and 141 bp, which is rather attributable to a typical PCR anomaly - 3bp increment of the same IGH gene PCR product because of in frame code - than to a biallelic rearrangement. In case 4 there are 3 peaks, with a considerable decrease of the relative peak size of the 111bp component in the CD23+ microdissected cells, indicating that this peak more probably belongs to the MCL component. For case 5 see explanations in the text. For clarity only larges peaks’ sizes are indicated.
Repeated IGH results obtained from microdissected cyclin D1+ MCL cells or CD23+ CLL/SLL cells from cases 1, 2 and 7 achieved similar results. However, purified DNA peak products from PCR agarose gel failed in IGH sequencing analysis.
Discussion
We present the largest series of composite lymphomas consisting of MCL and CLL/SLL. In our literature review, we found only four previous case reports in the literature that presented similar observations [14,15,18,19]. Importantly, none of the MCL components in these four cases were shown to be clonally related to the CLL/SLL component. These findings support our results, since the IGH rearrangements amplified were, yet limited by technical difficulties of probable “contamination” of the microdissection specimens by each other component of the composite lymphoma, of different size in the MCL and CLL/SLL components in 5/6 analyzed cases. These observations are further supported by the different light chain restriction of both components in 3/5 informative cases. These cases can be therefore considered as bona fide examples of composite lymphoma consisting clonally unrelated MCL and CLL/SLL. This result is perhaps surprising, since MCL and CLL/SLL share morphological and immunophenotypic characteristics and are thought to arise from CD5+ B-cells that can undergo somatic mutation of the IGHV genes [4–9]. Nevertheless, the unanimous results in a total of 9 cases analyzed (our 5 and those reported in the literature), makes a strong case for a lack of a clonal relationship in cases of composite MCL and CLL/SLL.
Our actual observation of lacking clonal relationship between MCL and CLL/SLL is clearly different to what we recently observed in MCL with a plasma cell component [16], showing evidently a clonal relationship in a subset of the cases, which we supposed might be due to a retained MCL propensity to “mature” towards neoplastic plasma cells in such rare occasions rather than representing composite neoplasms sensu strictu. Along with the retained ability to undergo somatic IGHV hypermutations, the observed plasmacytic “maturation” of MCL points towards some plasticity of the latter. Nevertheless, our present data support mutually exclusive MCL or CLL/SLL lymphomagenic pathways of naïve CD5+ B-cells; MCL being tightly linked to cell cycle dysregulation through cyclin D1 overexpression and accumulation of additional genomic aberrations to inactivate DNA damage response pathways, whereas CLL/SLL being more linked to apoptosis arrest and NF-κB activation [20,21].
The question remains whether the simultaneous presence of MCL and CLL/SLL is simply co-incidence or can be attributed to a common biological denominator. Importantly, patients with a history of CLL/SLL have a two-fold increased risk of developing other malignancies such as skin, prostate, breast and colon cancer, as well as a 10-fold increased risk of developing other lymphomas, including Hodgkin lymphoma [22]. This risk in CLL/SLL patients could be explained by environmental and/or genetic factors [23–28], increasing the individual susceptibility for lymphomagenesis.
From a diagnostic point of view, the results of this study show that it is essential in “small B-cell lymphomas” to perform immunohistochemical studies for CD5, CD23 and cyclin D1 in order to detect morphologically unexpected co-incidental MCL (including in situ MCL) in a CLL/SLL case or vice versa. The detection of a second, clonally unrelated lymphoma may be helpful for therapeutic decisions or prognostic stratification.
In summary, we have reported 11 cases of composite MCL and CLL/SLL with assessment of the clonal relationship of the two components. Patients with composite MCL and CLL/SLL usually present with lymphadenopathy, high stage disease, and have a poor prognosis. In at least 7 cases we report, as well as in 4 cases reported by others, the MCL and CLL/SLL components were not clonally related as manifested by different immunoglobulin light chain expression or other immunophenotypic features, variable IGH rearrangements amplified, t(11;14) translocation, and unique morphologic features.
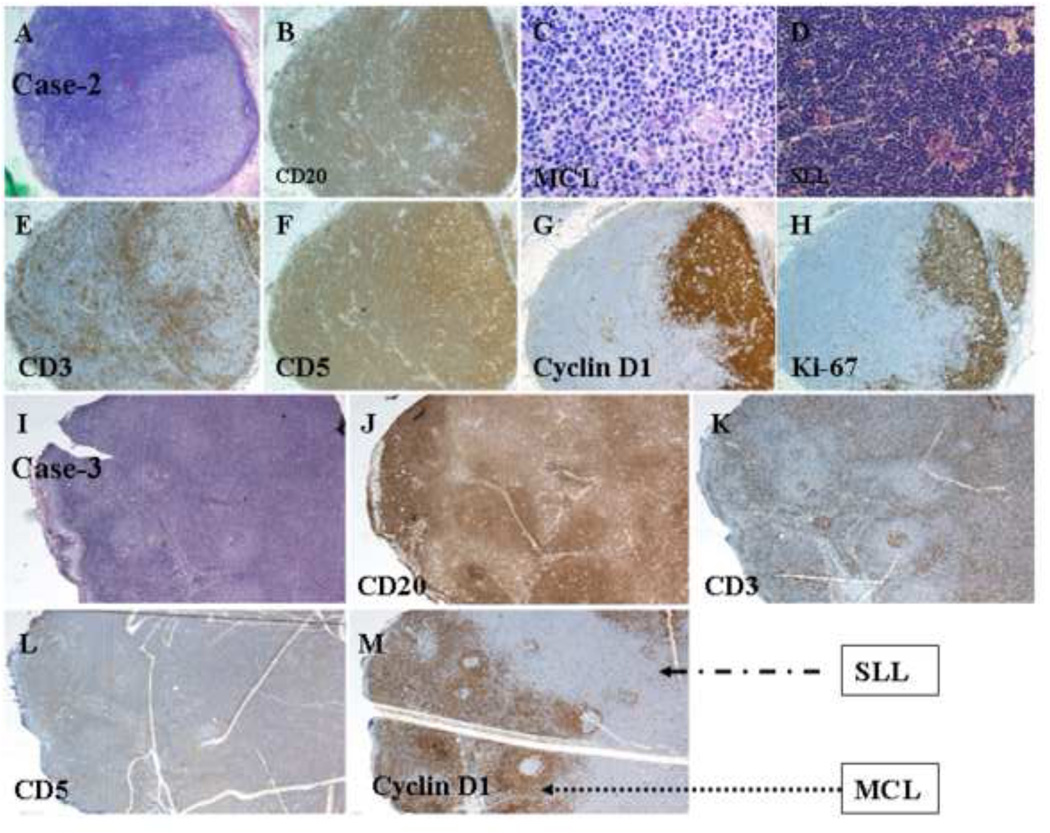
Figure 2.
Histologic and immunophenotypic features of a composite case of MCL and CLL/SLL. (A) Two irregular patterns of atypical small lymphoid infiltration in an effaced lymph node; one is nodular, and the other is diffuse (H&E staining; original magnification: 20X). (B) CD20 staining of atypical small lymphoid cells in both nodular and diffuse areas (immunoperoxidase staining; original magnification: 20X). (C) Atypical MCL infiltration (H&E staining; original magnification: 100X). (D) Atypical SLL infiltration (H&E staining; original magnification: 40X). (E) CD3 staining of normal small T-cells in the internodular area (immunoperoxidase staining; original magnification: 20X). (F) CD5 staining of atypical small lymphoid cells in both the internodular and nodular areas (immunoperoxidase staining; original magnification: 20X). (G) Cyclin D1 staining of atypical small lymphoid cells in the MCL area (immunoperoxidase staining; original magnification: 20X). (H) Ki-67 staining of a high proliferation index in the MCL area (immunoperoxidase staining; original magnification: 20X). (I) Nodular pattern of atypical lymphoid cells with expanded mantle zone and internodular areas (H&E staining; original magnification: 20X). (J) Differential CD20 staining of atypical lymphoid cells in the nodular and internodular areas (immunoperoxidase staining; original magnification: 20X). (K) CD3 staining of small T-cells in the internodular areas (immunoperoxidase staining; original magnification: 20X). (L) Weak CD5 staining of atypical lymphoid cells in the nodular and internodular areas (immunoperoxidase staining; original magnification: 20X). (M) Cyclin D1 staining of atypical lymphoid cells in the expanded mantle zone and internodular areas with a focal negative area of SLL (immunoperoxidase staining; original magnification: 20X).
Figure 3.
Histologic and immunophenotypic features of a composite case of MCL and CLL/SLL. (A) Nodular pattern of atypical small lymphoid infiltration in an effaced lymph node (H&E staining; original magnification: 20X). (B) Atypical SLL infiltration (H&E staining; original magnification: 100X). (C) Atypical MCL infiltration (H&E staining; original magnification: 100X). (D) CD20 staining of atypical small lymphoid cells in both the internodular and nodular areas (immunoperoxidase staining; original magnification: 20X). (E) CD5 staining of atypical lymphoid cells in the nodular and internodular areas (immunoperoxidase staining; original magnification: 20X). (F) CD23 staining of atypical lymphoid cells in the internodular SLL areas (immunoperoxidase staining; original magnification: 20X). (G) Cyclin D1 staining of atypical lymphoid cells in the mantle zone and nodular areas with a focal negative area of SLL (immunoperoxidase staining; original magnification: 20X). (H) Ki-67 staining of a high proliferation index in the MCL area (immunoperoxidase staining; original magnification: 20X). (I) Nodular pattern of atypical small lymphoid infiltration in an effaced lymph node (H&E staining; original magnification: 20X). (J) Atypical lymphoid infiltration (H&E staining; original magnification: 40X). (K) Atypical MCL infiltration (H&E staining; original magnification: 100X). (L) CD20 staining of atypical small lymphoid cells in both the internodular and nodular areas (immunoperoxidase staining; original magnification: 20X). (M) CD5 staining of atypical lymphoid cells in the nodular and internodular areas (immunoperoxidase staining; original magnification: 20X). (N) CD23 staining of atypical lymphoid cells within the nodules of SLL areas (immunoperoxidase staining; original magnification: 20X). (O) Cyclin D1 staining of atypical lymphoid cells in the mantle zone and nodular areas with a focal negative area of SLL (immunoperoxidase staining; original magnification: 20X). (P) Ki-67 staining of a high proliferation index in the mantle zone area (immunoperoxidase staining; original magnification: 20X).
Figure 4.
Histologic and immunophenotypic features of a composite case of MCL and CLL/SLL. (A) Two irregular patterns of atypical small lymphoid infiltration in an effaced lymph node; one is nodular with intermediate-sized cells, and the other is diffuse with smallsized cells (H&E staining; original magnification: 20X). (B) Atypical SLL infiltration (H&E staining; original magnification: 100X). (C) Atypical MCL infiltration (H&E staining; original magnification: 100X). (D) CD20 staining of atypical small lymphoid cells in both nodular and diffuse areas (immunoperoxidase staining; original magnification: 20X). (E) CD5 staining of atypical small lymphoid cells in both the internodular and nodular areas (immunoperoxidase staining; original magnification: 20X). (F) CD23 staining of SLL cells in the nodular area (immunoperoxidase staining; original magnification: 20X). (G) Cyclin D1 staining of atypical small lymphoid cells in the MCL area (immunoperoxidase staining; original magnification: 20X). (H) Ki-67 staining of a high proliferation index in the SLL area (immunoperoxidase staining; original magnification: 20X).
Figure 5.
Histologic and immunophenotypic features of a composite case of MCL and CLL/SLL. (A) A nodular pattern of atypical small lymphoid infiltration in an effaced lymph node (H&E staining; original magnification: 20X). (B) Atypical mantle-zone growth pattern (H&E staining; original magnification: 100X). (C) Atypical small lymphoid infiltration in the internodular area (H&E staining; original magnification: 100X). (D) CD20 staining of atypical lymphoid nodules (immunoperoxidase staining; original magnification: 40X). (E) Cyclin D1 staining of atypical small lymphoid cells in the mantle zone (immunoperoxidase staining; original magnification: 40X). (F) CD23 staining of SLL cells in the nodular area (immunoperoxidase staining; original magnification: 20X). (G) CD5 staining of atypical small lymphoid cells in the internodular areas and a few small T-cells within the follicles (immunoperoxidase staining; original magnification: 40X). (H) Ki-67 staining of a high proliferation index in the reactive follicles (immunoperoxidase staining; original magnification: 40X).
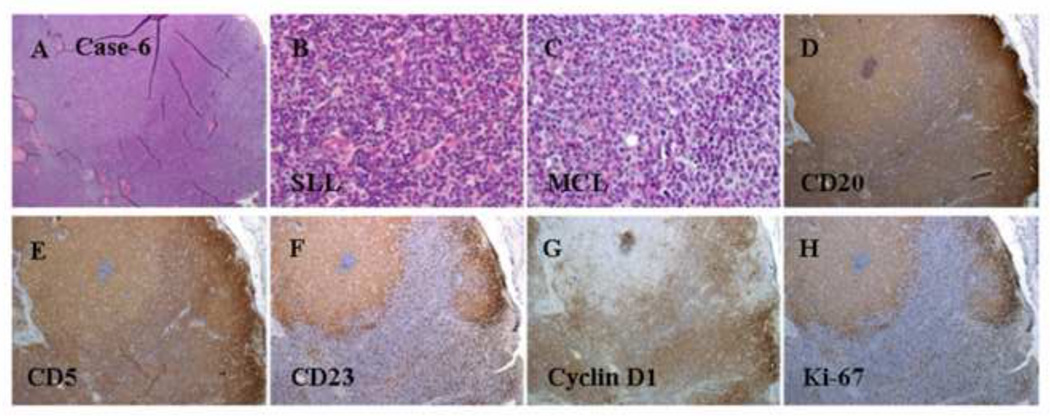
Figure 6.
Histologic and immunophenotypic features of a composite case of MCL and CLL/SLL. (A-C) A diffuse pattern of atypical lymphoid infiltration (H&E staining; original magnification: 20X and 100X). (D) CD20 staining of atypical lymphoid cells (immunoperoxidase staining; original magnification: 20X). (E) CD3 staining of normal small T-cells (immunoperoxidase staining; original magnification: 20X). (F) CD5 staining of atypical small lymphoid cells (immunoperoxidase staining; original magnification: 20X). (G) Cyclin D1 staining of atypical small lymphoid cells (immunoperoxidase staining; original magnification: 20X). (H) Ki-67 staining of a low proliferation index in the MCL area (immunoperoxidase staining; original magnification: 20X). (I) Bone marrow aspirate smear showing a few scattered atypical lymphoid cells with a clumped chromatin pattern. Flow cytometry revealed that these atypical lymphoid cells were positive for CD5, dim CD20, CD23, and monotypic kappa light chains, whereas cyclin D1 staining was negative based on the immunostaining of the bone marrow biopsy (data not shown).
Acknowledgement
SH is an honorable pathology scholar supported by University of Basel Hospital and The University of Texas MD Anderson Cancer Center. KHY is supported by The University of Texas MD Anderson Cancer Center Institutional R & D Fund, MD Anderson Institutional Research Program Award, MD Anderson SPORE Development Research Program Award, Forward Lymphoma Fund and Gundersen Lutheran Medical Foundation Award. This study is also partially supported by the Stiftung zur Krebsbekämpfung (#269) and NCI/NIH (R01CA138688 and 1RC1CA146299).
Technical and publication editing supports from Markeda L. Wade from the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center are greatly appreciated.
Footnotes
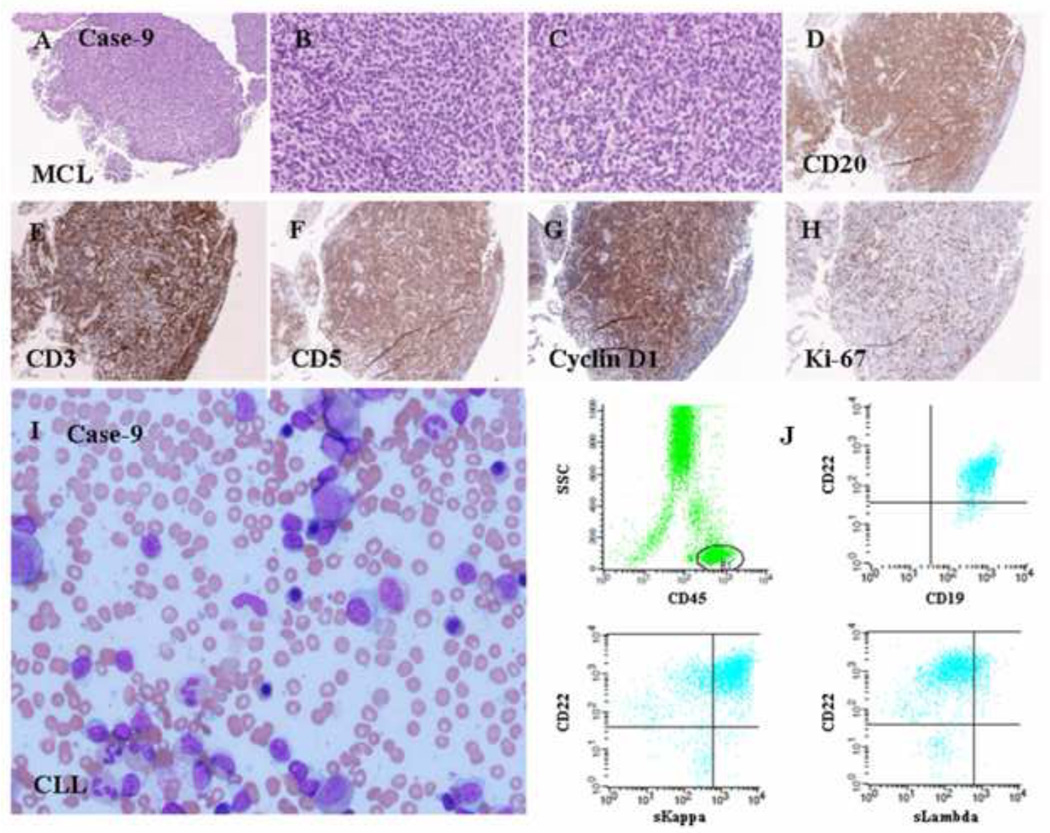
Supplemental Figure (Case 7): Imunophenotypic features of a composite case of MCL and CLL/SLL. (A) MCL component with expression of CD5, CD19, bright CD20, CD22, CD38, and bright monotypic kappa light chains. (B) CLL/SLL component with expression of CD5, CD19, dim CD20, CD22, CD23, CD38, and dim monotypic lambda light chains.
References
- 1.Mitterlechner T, Fiegl M, Muhlbock H, Oberaigner W, Dirnhofer S, Tzankov A. Epidemiology of non-Hodgkin lymphomas in Tyrol/Austria from 1991 to 2000. J Clin Pathol. 2006;59:48–55. doi: 10.1136/jcp.2005.026815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabattini E, Bacci F, Sagramoso C, Pileri SA. WHO classification of tumours of haematopoietic and lymphoid tissues in 2008: an overview. Pathologica. 2010;102:83–87. [PubMed] [Google Scholar]
- 3.Williams ME, Westermann CD, Swerdlow SH. Genotypic characterization of centrocytic lymphoma: frequent rearrangement of the chromosome 11 bcl-1 locus. Blood. 1990;76:1387–1391. [PubMed] [Google Scholar]
- 4.Babbage G, Garand R, Robillard N, Zojer N, Stevenson FK, Sahota SS. Mantle cell lymphoma with t(11;14) and unmutated or mutated VH genes expresses AID and undergoes isotype switch events. Blood. 2004;103:2795–2798. doi: 10.1182/blood-2003-05-1632. [DOI] [PubMed] [Google Scholar]
- 5.Orchard J, Garand R, Davis Z, et al. A subset of t(11;14) lymphoma with mantle cell features displays mutated IgVH genes and includes patients with good prognosis, nonnodal disease. Blood. 2003;101:4975–4981. doi: 10.1182/blood-2002-06-1864. [DOI] [PubMed] [Google Scholar]
- 6.Walsh SH, Thorselius M, Johnson A, et al. Mutated VH genes and preferential VH3-21 use define new subsets of mantle cell lymphoma. Blood. 2003;101:4047–54. doi: 10.1182/blood-2002-11-3479. [DOI] [PubMed] [Google Scholar]
- 7.Thorselius M, Walsh S, Eriksson I, et al. Somatic hypermutation and V(H) gene usage in mantle cell lymphoma. Eur J Haematol. 2002;68:217–224. doi: 10.1034/j.1600-0609.2002.01662.x. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura N, Kuze T, Hashimoto Y, et al. Analysis of the immunoglobulin heavy chain gene variable region of 101 cases with peripheral B cell neoplasms and B cell chronic lymphocytic leukemia in the japanese population. Pathol Int. 1999;49:595–600. doi: 10.1046/j.1440-1827.1999.00911.x. [DOI] [PubMed] [Google Scholar]
- 9.Cogliatti SB, Bertoni F, Zimmermann DR, et al. IgV H mutations in blastoid mantle cell lymphoma characterize a subgroup with a tendency to more favourable clinical outcome. J Pathol. 2005;206:320–327. doi: 10.1002/path.1781. [DOI] [PubMed] [Google Scholar]
- 10.Caleo A, Sanchez-Aguilera A, Rodriguez S, et al. Composite Hodgkin lymphoma and mantle cell lymphoma: two clonally unrelated tumors. Am J Surg Pathol. 2003;27:1577–1580. doi: 10.1097/00000478-200312000-00012. [DOI] [PubMed] [Google Scholar]
- 11.Ho AK, Teman CJ, Smith GP, Nightingale DR, Miles RR. Composite mantle cell and diffuse large B-cell lymphoma: report of two cases. Int J Surg Pathol. 2011;19:643–648. doi: 10.1177/1066896911405656. [DOI] [PubMed] [Google Scholar]
- 12.Zamo A, Zanotti R, Lestani M, et al. Molecular characterization of composite mantle cell and follicular lymphoma. Virchows Arch. 2006;448:639–643. doi: 10.1007/s00428-006-0158-9. [DOI] [PubMed] [Google Scholar]
- 13.Traverse-Glehen A, Pittaluga S, Gaulard P, et al. Mediastinal gray zone lymphoma: the missing link between classic Hodgkin's lymphoma and mediastinal large B-cell lymphoma. Am J Surg Pathol. 2005;29:1411–1421. doi: 10.1097/01.pas.0000180856.74572.73. [DOI] [PubMed] [Google Scholar]
- 14.Lima M, Pinto L, Dos Anjos TM, et al. Guess what: Chronic 13q14.3+/CD5-/CD23+ lymphocytic leukemia in blood and t(11;14)(q13;q32)+/CD5+/ Cytometry B Clin Cytom. 2003;51:41–44. doi: 10.1002/cyto.b.10005. [DOI] [PubMed] [Google Scholar]
- 15.Fend F, Quintanilla-Martinez L, Kumar S, et al. Composite low grade B-cell lymphomas with two immunophenotypically distinct cell populations are true biclonal lymphomas. A molecular analysis using laser capture microdissection. Am J Pathol. 1999;154:1857–1866. doi: 10.1016/S0002-9440(10)65443-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Visco C, Hoeller S, Malik JT, et al. Molecular characteristics of mantle cell lymphoma presenting with clonal plasma cell component. Am J Surg Pathol. 2011;35:177–189. doi: 10.1097/PAS.0b013e3182049a9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier VS, Rufle A, Gudat F. Simultaneous evaluation of T- and B-cell clonality, t(11;14) and t(14;18), in a single reaction by a four-color multiplex polymerase chain reaction assay and automated high-resolution fragment analysis: a method for the rapid molecular diagnosis of lymphoproliferative disorders applicable to fresh frozen and formalin-fixed, paraffin-embedded tissues, blood, and bone marrow aspirates. Am J Pathol. 2001;159:2031–2043. doi: 10.1016/S0002-9440(10)63055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Addada J, Anoop P, Swansbury JG, Wotherspoon A, Thomas JM, Matutes E. Synchronous mantle cell lymphoma, chronic lymphocytic leukaemia and melanoma in a single lymph node. Acta Haematol. 2010;123:194–196. doi: 10.1159/000297525. [DOI] [PubMed] [Google Scholar]
- 19.Kourelis TV, Kahl BS, Benn P, Delach JA, Bilgrami SF. Treatment of synchronous mantle cell lymphoma and small lymphocytic lymphoma with bendamustine and rituximab. Acta Haematol. 2011;126:40–43. doi: 10.1159/000324193. [DOI] [PubMed] [Google Scholar]
- 20.Pekarsky Y, Zanesi N, Croce CM. Molecular basis of CLL. Semin Cancer Biol. 2010;20:370–376. doi: 10.1016/j.semcancer.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jares P, Campo E. Advances in the understanding of mantle cell lymphoma. Br J Haematol. 2008;142:149–165. doi: 10.1111/j.1365-2141.2008.07124.x. [DOI] [PubMed] [Google Scholar]
- 22.Tsimberidou AM, Wen S, McLaughlin P, et al. Other malignancies in chronic lymphocytic leukemia/small lymphocytic lymphoma. J Clin Oncol. 2009;27:904–910. doi: 10.1200/JCO.2008.17.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capalbo S, Callea V, Musolino C, et al. Familial B-cell chronic lymphocytic leukemia in a population of patients from Southern Italy. Int J Hematol. 2004;79:354–357. doi: 10.1532/ijh97.e0304. [DOI] [PubMed] [Google Scholar]
- 24.Yuille MR, Matutes E, Marossy A, Hilditch B, Catovsky D, Houlston RS. Familial chronic lymphocytic leukaemia: a survey and review of published studies. Br J Haematol. 2000;109:794–799. doi: 10.1046/j.1365-2141.2000.02111.x. [DOI] [PubMed] [Google Scholar]
- 25.Goldin LR, Pfeiffer RM, Li X, Hemminki K. Familial risk of lymphoproliferative tumors in families of patients with chronic lymphocytic leukemia: results from the Swedish Family-Cancer Database. Blood. 2004;104:1850–1854. doi: 10.1182/blood-2004-01-0341. [DOI] [PubMed] [Google Scholar]
- 26.Caporaso N, Marti GE, Goldin L. Perspectives on familial chronic lymphocytic leukemia: genes and the environment. Semin Hematol. 2004;41:201–206. doi: 10.1053/j.seminhematol.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Goldin LR, Landgren O, Marti GE, Caporaso NE. Familial Aspects of Chronic Lymphocytic Leukemia, Monoclonal B-Cell Lymphocytosis (MBL), and Related Lymphomas. European J Clin Med Oncol. 2010;2:119–126. [PMC free article] [PubMed] [Google Scholar]
- 28.Goldin LR, Slager SL. Familial CLL: genes and environment. Hematology Am Soc Hematol Educ Program. 2007:339–345. doi: 10.1182/asheducation-2007.1.339. [DOI] [PubMed] [Google Scholar]