Abstract
As a generalization of the Dirichlet process (DP) to allow predictor dependence, we propose a local Dirichlet process (lDP). The lDP provides a prior distribution for a collection of random probability measures indexed by predictors. This is accomplished by assigning stick-breaking weights and atoms to random locations in a predictor space. The probability measure at a given predictor value is then formulated using the weights and atoms located in a neighborhood about that predictor value. This construction results in a marginal DP prior for the random measure at any specific predictor value. Dependence is induced through local sharing of random components. Theoretical properties are considered and a blocked Gibbs sampler is proposed for posterior computation in lDP mixture models. The methods are illustrated using simulated examples and an epidemiologic application.
Keywords: Dependent Dirichlet process, Blocked Gibbs sampler, Mixture model, Non-parametric Bayes, Stick-breaking representation
1 Introduction
In recent years, there has been a dramatic increase in applications of non-parametric Bayes methods, motivated largely by the availability of simple and efficient methods for posterior computation in Dirichlet process mixture (DPM) models (Lo 1984; Escobar 1994; Escobar and West 1995). The DPM models incorporate Dirichlet process (DP) priors (Ferguson 1973, 1974) for components in Bayesian hierarchical models, resulting in an extremely flexible class of models. Due to the flexibility and ease in implementation, DPM models are now routinely implemented in a wide variety of applications, ranging from machine learning (Beal et al. 2002; Blei et al. 2004) to genomics (Xing et al. 2004; Kim et al. 2006).
In many settings, it is natural to consider generalizations of the DP and DPM-based models to accommodate dependence. For example, one may be interested in studying changes in a density with predictors. Following Lo (1984), one can use a DPM for Bayes inference on a single density as follows:
| (1) |
where k(y, u) is a non-negative valued kernel defined on () such that for each and for each , with , Ω Borel subsets of Euclidean spaces and , the corresponding σ-fields, and G is a finite randomprobabilitymeasure on (Ω, ) following a DP. A natural extension for modeling of a conditional density f (y|x) for , with a Lebesgue measurable subset of , is as follows:
| (2) |
where the mixing measure Gx is now indexed by the predictor value. We are then faced with modeling a collection of random mixing measures denoted as .
Recent work on defining priors for collections of random probability measures has primarily relied on extending the stick-breaking representation of the DP (Sethuraman 1994). This literature was stimulated by the dependent Dirichlet process (DDP) framework proposed by MacEachern (1999, 2000, 2001), which replaces the atoms in the Sethuraman (1994) representation with stochastic processes. The DDP framework has been adopted to develop ANOVA-type models for random probability measures (De Iorio et al. 2004), for flexible spatial modeling (Gelfand et al. 2004), in time series applications (Caron et al. 2006), and for inferences on stochastic ordering (Dunson and Peddada 2008). The specification of the DDP used in applications incorporates dependence only through the atoms while assuming fixed weights. In other recent work, Griffin and Steel (2006) proposed an order-based DDP (πDDP) which allows varying weights, while Duan et al. (2005) developed a multivariate stick-breaking process for spatial data.
Alternatively, convex combinations of independent DPs can be used for modeling collections of dependent random measures. Müller et al. (2004) proposed this idea to allow dependence across experiments and discrete dynamic settings were considered by Pennell and Dunson (2006) and Dunson (2006). Recently, the idea has been extended to continuous covariate cases by Dunson et al. (2007) and Dunson and Park (2008).
Some desirable properties of a prior for a collection, , of predictor-dependent probability measures are: (1) increasing dependence in Gx and Gx′ with decreasing distance between x and x′; (2) simple and interpretable expressions for the expectation and variance of each Gx as well as the correlation between Gx and Gx′; (3) Gx has a marginal DP prior for all ; (4) posterior computation can proceed efficiently through a straightforward MCMC algorithm in a broad variety of applications. Although the DDP, πDDP and the prior proposed by Duan et al. (2005) achieve (1), πDDP and Duan et al. (2005) approaches are not straightforward to implement in general applications. The fixed stick-breaking weights version of the DDP tends to be easy to implement, but has the disadvantage of not allowing locally adaptive mixture weights. The kernel mixture approaches of Dunson et al. (2007) and Dunson and Park (2008) lack the marginal DP property (3). Property (3) is appealing in that there is rich theoretical literature on DPs, showing posterior consistency (Ghosal et al. 1999; Lijoi et al. 2005) and rates of convergence (Ghosal and Van der Vaart 2007).
This article proposes a simple extension of the DP, which provides an alternative to the fixed weights DDP in order to allow local adaptivity, while also achieving properties (1)–(4). The prior is constructed by first assigning stick-breaking weights and atoms to random locations in a predictor space. Each predictor-dependent random probability measure is formulated using the random weights and atoms located in a neighborhood about that predictor value. Dependence is induced by local sharing of random components. We call this prior the local Dirichlet process (lDP).
Section 2 describes stick-breaking priors (SBP) for collections of predictor-dependent random probability measures. Section 3 introduces the lDP and discusses properties. Computation is described in Sect. 4. Sections 5 and 6 include simulation studies and an epidemiologic application. Section 7 concludes with a discussion. Proofs are included in appendices.
2 Predictor-dependent stick-breaking priors
2.1 Stick-breaking priors
Ishwaran and James (2001) proposed a general class of SBPs for random probability measures. This class provides a useful starting point in considering extensions to allow predictor dependence.
Definition 1 A random probability measure, G, has a SBP if
| (3) |
where δθis a discrete measure concentrated at θ, are random weights with independently from with G0 a non-atomic base probability measure. For N = ∞, the condition a.s. is satisfied by Lemma 1 in Ishwaran and James (2001). For finite N, the condition is satisfied by letting VN = 1.
There are many processes that fall into this class of SBP. The DP corresponds to the special case in which N = ∞, ah = 1 and bh = α as established in Sethuraman (1994). The two-parameter Poisson-DP corresponds to the case where N = ∞, ah = 1 − a, and bh = b + ha with 0 ≤ a < 1 and b > −a (Pitman 1995, 1996). Additional special cases are listed in Ishwaran and James (2001).
2.2 Predictor-dependent stick-breaking priors
Consider an uncountable collection of predictor-dependent random probability measures, of . The predictor space is Lebesgue measurable subset Euclidian space and the random measures Gx are defined on (Ω, ) where Ω is a complete and separable metric space and is a corresponding Borel σ-algebra. Let be a probability measure on (, ) where is the space of uncountable collections of random probability measures Gx and is the corresponding Borel σ-algebra. Then, denotes that is a prior on the random collection .
We call a predictor-dependent stick-breaking prior () if can represented as:
| (4) |
where the random weights ph(x) have a stick-breaking form, ph(x) and θh(x) are predictor-dependent, and N(x) is also indexed by the predictor value x. Depending on how we form ph(x), θh(x) and N(x), different dependencies among Gx are induced. Several interesting priors, such as the DDP, πDDP and the prior proposed by Duan et al. (2005) fall into the class. In the next section, we propose a new choice of deemed the lDP.
3 Local Dirichlet process
3.1 Formulation
Formulating the lDP starts with obtaining the following three sequences of mutually independent global random components:
| (5) |
where are locations, are probability weights, and are atoms. G0 is a probability measure on (Ω, ) on which Gx will be defined and H is a probability measure on (, ) where is a Borel σ-algebra of subsets of and is a Lebesgue measurable subset of Euclidian space that may or may not correspond to the predictor space . For a given predictor space , we introduce the probability space (,, H) such that it satisfies the following regularity condition from which one can deduce :
Condition 1 For all and ψ > 0, , where is defined as a ψ-neighborhood around a point with being some distance measure.
Next, focusing on a local predictor point , we define sets of local random components for x as:
| (6) |
where is a predictor-dependent set indexing the locations belonging to the ψ-neighborhood of x, , which is defined on by ψ and d(·, ·). Hence, the sets V(x) and Θ(x) contain the random weights and atoms that are assigned to the locations Γ(x) in . Here, ψ controls the neighborhood size. For simplicity, we treat ψ as fixed throughout the paper, though one can obtain a more flexible class of priors by assuming a hyper prior for ψ.
Using the local random components in (6), we consider the following form for Gx:
| (7) |
where N(x) is the cardinality of and πl(x) is the lth ordered index in . Then, Condition 1 ensures that the following lemma holds (refer to the Proof of Lemma 1 in the Appendix).
Lemma 1 For all , N(x) = ∞ and almost surely.
By Lemma 1, it is apparent that Gx formed as in (7) is a well-defined stick-breaking random probability measure for x. It is also straightforward that we can define Gx for all by (6) and (7) using the global components in (5). Therefore, given α, G0, H, ψ) with a choice of d(·, ·), the steps from (5) to (7) defines a new choice of predictor-dependent SBP ( ) for , deemed the lDP. We use the shorthand notation ~ lDP(α, G0, H, ψ) to denote that is assigned a lDP with hyperparameters α, G0, H, ψ.
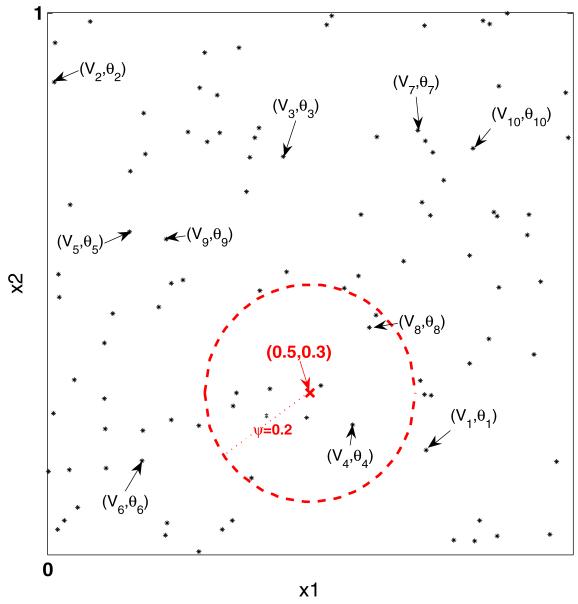
Figure 1 illustrates the lDP formulation graphically for a case where and with H =Uniform([0, 1]2) leading to and ψ = 0.2. For a simple illustration, we consider Euclidian distance for d(·, ·) for bivariate predictors. Random locations in [0, 1]2 are generated from a uniform distribution, with the first 100 locations plotted as ‘*’ in Fig. 1. The random pair of weight and atom (Vh, θh) is placed at location Γh, with the first ten pairs labeled in Fig. 1. For a predictor value x = (0.5, 0.3)′, the red dashed circle indicates the neighborhood of x, . Then, Gx at x = (0.5, 0.3)′ is constructed using the weights and atoms within the dashed circle in the order of the index to formulate the stick-breaking representation. For all other , Gx are formed following the same steps.
Fig. 1.
Graphical illustration for lDP formulation. Black asterisks are the first 100 random locations generated on from H = Uniform ([0,1]2). Red dashed circle indicates the neighborhood of the red crossed predictor point x = (0.5, 0.3)′ determined by Euclidian distance d(·,·) and ψ = 0.2. (Vh, θh) for h = 1, … , 10 are the first ten random pairs of weight and atom assigned to the first ten random locations Γh for h = 1, … , 10
From Fig. 1, it is apparent that the dependence between Gx and Gx′ increases as the distance between x and x′ decreases. For closer x and x′, their neighborhoods overlap more so that similar components are used for constructing Gx and Gx′, while if x and x′ are far apart, there will be at most a small area of intersection so that few to none of the random components are shared. In the non-overlapping case, Gx and are assigned independent DP priors, as is clear from Theorem 1 and the subsequent development.
Theorem 1 If , for any , Gx ~ DP(αG0).
The marginal DP property shown in Theorem 1 is appealing in allowing one to rely directly on the rich literature on properties of the DP to obtain insight into the prior for the random probability measure at any particular predictor value. However, unlike the DP, the lDP allows the probability measure to vary with predictors, while borrowing information across local regions of the predictor space. This is accomplished through incorporating shared random components. Due to the sharing and to the almost sure discreteness property of each Gx, the lDP will induce local clustering of subjects according to their predictor values. Theorem 2 illustrates this local clustering property more clearly.
Theorem 2 Suppose and , for i = 1, … , n, with xi denoting the predictor value for subject i. Then,
where is the conditional probability of Γh falling within the intersection region given .
The clustering probability κxi ,xj increases from 0 when to 1/(α + 1) when xi = xj which is the case of Pxi ,xj = 1. This implies that, for fixed α, the clustering probability under is bounded above by the clustering probability under the global DP, which takes , leading to Pr(φi = φj | α) = 1/(α + 1). Also, note that small values of the precision parameter α will induce Vh values that are close to one. This in turn causes a small number of atoms in each neighborhood to dominate, inducing few local clusters. However, when ψ is small and hence neighborhood sizes are small, there will still be many clusters across .
It is interesting to consider relationships between the lDP and other priors proposed in the literature in limiting special cases. First, note that the lDP converges to the DP as ψ → ∞, so that all the neighborhoods around each of the predictor values encompass the entire predictor space. Also, the lDP(α, G0, H, ψ) corresponds to a limiting case of the kernel stick-breaking process (KSBP) (Dunson and Park 2008), in which the kernel is defined as K(x, Γ) = 1 (d(x, Γ) < ψ) and the DP placed at each location have precision parameters → 0.
3.2 Moments and correlation
From Theorem 1 and properties of the DP, implies, for any ,
| (8) |
Next, let us consider the correlation between Gx1 and Gx2, for any x1, . First, we show the correlation conditionally on the locations Γ but marginalizing out the weights V and atoms Θ. As discussed in Sect. 3.1, if Γ is given, the lDP can be regarded as a special case of the πDDP. Hence, following Theorem 1 in Griffin and Steel (2006), for any x1, ,
| (9) |
where #S is the cardinality of the set , , , and for . In other words, #Sh is the number of indices on the locations Γ that are below h and are shared in the neighborhoods of x1 and x2, while is the number of indices that are below h and belong to the neighborhoods of either x1 or x2 but not both. For a given h, reducing #Sh by one induces adding two elements to , thus reducing the correlation, as expected. From expression (9), it is clear that the neighborhoods around x1 and x2 are increasingly overlapping and the correlation between Gx1 and Gx2 increases as x1 → x2. Expression (9) is particularly useful in being free of dependence on B.
Marginalizing the correlation in (9) over the prior for the random locations Γ is equivalent to marginalizing out the #Sh and for . In considering the correlation between Gx1 and Gx2, we can ignore the Γh for and focus on the Γh only for . Let γj be the jth ordered component of . For example, if , γ1 = 1, γ2 = 3,γ3 = 5, γ4 = 6, …. Let be an indicator for whether Γγj are shared by the neighborhoods of x1 and x2 or not. Then, the formula in (9) can be reexpressed with respect to Zγj as follows:
| (10) |
Note that it is straightforward to show that Bernoulli(Px1,x2), for j = 1, … ,∞, with the conditional probability of Γh falling within the intersection region given . Finally, marginalizing out results in the following Theorem.
Theorem 3 If , for any ,
The correlation is expressed only in terms of Px1,x2 and α. Regardless of α, the correlation is 1 if x1 = x2 which implies the neighborhoods around x1, x2 are identical and Px1,x2=1. Also, the correlation is 0 when the neighborhoods are non-overlapping with Px1,x2=0. In addition, Px1,x2 ≤ ρx1,x2 ≤ 1 and ρx1,x2 increases as α increases for fixed Px1,x2 . When α → 0, the correlation converges to Px1,x2. Meanwhile, when α → ∞, the correlation converges to .
Note that Px1,x2 depends on H, ψ, and the locations x1 and x2 given a choice of d(·, ·). When for H is chosen to satisfy Condition 2, some appealing properties result.
Condition 2 For all with being p-dimensional, .
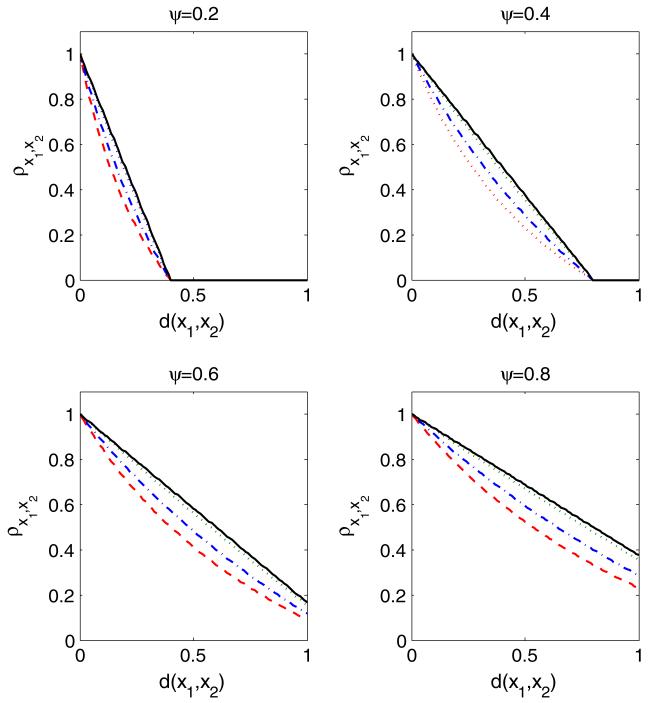
From Condition 2, one can deduce that contains all the points in within the distance of ψ from x for any . Under Condition 2, with H chosen to be a uniform probability measure on a bounded space , Px1,x2 depends only on ψ and d(x1, x2) which is the distance between x1 and x2, but not on the exact locations of x1 and x2 in . Hence, upon examination of Theorem 3, it is apparent that Condition 2 implies an isotropic correlation structure, which is an appealing default in the absence of prior knowledge of changes in the correlation structure according to the locations in . Figure 2 shows how the correlation ρx1,x2 changes as a function of d(x1, x2) in the case where and H is Uniform([−ψ, 1 + ψ]) so that and Condition 2 holds for different ψ with d(·, ·) corresponding to the Euclidian distance. The ρx1,x2 decays from 1 to 0 as d(x1, x2) increases and the decay is faster for smaller ψ. As ψ → ∞, the decay line gets closer to a horizontal line at ρx1,x2 = 1, which is the case of lDP=DP. Also, for a given ψ and d(x1, x2), the ρx1,x2 is higher as α → ∞. Although the choice of d(·, ·) being Euclidian makes the curves in Fig. 2 close to linear, the curvature can easily be changed by choosing a different distance measure d(·, ·).
Fig. 2.
Change in correlation ρx1,x2 over the change in distance d(x1, x2) for different α and ψ: α = 0.0001 (red dashed), α = 1 (blue dot-dashed), α = 10 (green dotted), α = 10, 000 (black solid)
3.3 Truncation approximation
Finite approximations to infinite SBPs form the basis for commonly used computational algorithms (Ishwaran and James 2001). In this subsection, we discuss a finite dimensional approximation to the lDP.
Since the lDP has the marginal DP property, let us recall the finite dimensional DP. Ishwaran and James (2001) defines an N-truncation of the DP (DPN) by discarding the N + 1, N + 2, … , ∞ terms and replacing pN with in the DP stick-breaking form in (3). They show that the DPN approximates the DP well in terms of the total variation (tv) norm of the marginal densities of the data obtained from the corresponding DPM models. According to their Theorem 2,
| (11) |
where || · || is tv norm, μN and μ∞ are the marginal probability measures for the data from the DPMN and DPM models, and n is the sample size. Note that the sample size has a modest effect on the bound for a reasonably large value of N and the bound decreases exponentially with N increasing, implying that even for a fairly large sample size, the DPMN approximates the DP well with moderate N.
Following a similar route, let us define an N-truncation of the lDP (lDPN) as follows:
Definition 2 For a finite N, let ΓN ={Γh, h = 1, … , N}, VN ={Vh, h =1, … , N}, and ΘN = {θh, h = 1, … , N} be the sets of global random locations, weights, and atoms, respectively. Distributional assumptions for Γh, Vh, and θh are the same as in (5) and the corresponding local sets are defined as in (6). Then, if
The Gx in Definition 3 has a similar form to obtained from the DPN except that N in G is replaced by N(x) in Gx and N in DPN is fixed while N(x) in lDPN is random. Focusing on a particular predictor value x, it is easy to show that N(x) ~ Binomial(N,Px), where N is the total number of global locations in lDPN and is the probability that a location belongs to the neighborhood around x, . Then, marginalizing out N(x) in the bound on the tv distance between the marginal densities of an observation obtained at a particular predictor value x from the lDPM and lDPMN models results in Theorem 4.
Theorem 4 Define a model (2) with as local Dirichlet process mixture (lDPM) model). lDPMN corresponds to (2) with . Suppose an observation is obtained from lDPMN and lDPM models at x. Then,
where μN (x) and μ∞(x) are the marginal probability measures for the observation. Notice that the bound decreases exponentially with N increasing, suggesting that we can obtain a good approximation to the lDP using a moderate N, as long as α is small and the neighborhood size is not too small. In particular, we require a large N for a given level of accuracy as ψ → 0, since Px decreases as the size of decreases.
4 Posterior computation
We develop an MCMC algorithm based on the blocked Gibbs sampler (Ishwaran and James 2001) for an lDPMN model. For simplicity in exposition, we describe a Gibbs sampling algorithm for a particular hierarchical model, though the approach can be easily adapted for computation in a broad variety of other settings. We let
| (12) |
where f (yi|xi, βi, τ) = N(yi; , βi, τ−1) βi = (βi1, … ,βip)′. For simplicity, we consider a univariate predictor case where p = 2 and with d(·, ·) Euclidian distance but the generalization to multiple predictors or to using different distance metric is straightforward. G0 is assumed to be Np(μβ, Σβ), H is assumed to be Uniform(aΓ, bΓ) and additional conjugate priors are assigned for τ, α, μβ and Σβ.
Let Ki be an indicator variable denoting that Ki = h implies ith subject is assigned to the hth mixture component. Then, the hierarchical structure of the model (12) with respect to the random variables is recast as follows.
| (13) |
where , K = {Ki, i = 1 …, n}, V = {Vh, h = 1, …, N}, and Γ = {Γh, h = 1, …, N}. The full conditionals for each of the random components are based on the following joint distribution.
| (14) |
Then, the Gibbs sampler proceeds by sampling from the following conditional posterior distributions:
- Conditional for Ki, i = 1, … , n
- Conditional for Vh, h = 1, … , N
- Conditional for Γh, h = 1, … , N
- Conditional for
where yih is nh × 1 response vector and is nh × p design matrix for the subjects with Ki = h and nh is the number of those subjects. - Conditional for μβ
- Conditional for
- Conditional for τ
- Conditional for α
Note that this Gibbs sampling algorithm consists only of simple steps for sampling from standard distributions and is no more complex than blocked Gibbs samplers for DPMs. In addition, we have observed good computational performance, in terms of mixing and convergence rates, in simulated and real data applications.
5 Simulation examples
We obtained data from two simulated examples, where n = 500 and a univariate predictor xi was simulated from Uniform(0,1). Case 1 was a null case where yi was generated from a normal regression model N(yi; −1 + 2xi, 0.01). Case 2 was a mixture of two normal linear regression models, with the mixture weights depending on the predictor, with the error variance differing, and with a nonlinear mean function for the second component:
| (15) |
We applied the lDPMN model in (12) to the simulated data with N = 50. Based on the results, N = 50 seems to be chosen to be large enough since the higher clusters having higher indices are not used in any of the subjects or are used in only a small proportion of them. Also, repeating the analysis for twice N, we obtained very similar results, suggesting that the results are robust to the choice of N, as long as N is not chosen to be small.
For the hyperparameters, we let ν1 = ν2 = 0.01, η1 = η2 = 2, ν0 = p, Σ0 = Ip, μ0 = 0, Σμ = n(X′X)−1, aΓ = −0.05, and bΓ = 1.05. The neighborhood size ψ = 0.05 was chosen such that the average number of subjects belonging to the neighborhoods around each predictor value in the sample is ≈n/10. We analyzed the simulated data using the proposed Gibbs sampling algorithm run for 10,000 iterations with a 5,000 iteration burn-in. The convergence and mixing of the MCMC algorithm were good (trace plots not shown). Also, results tended to be robust to repeating analysis with reasonable alternative hyperparameter values.
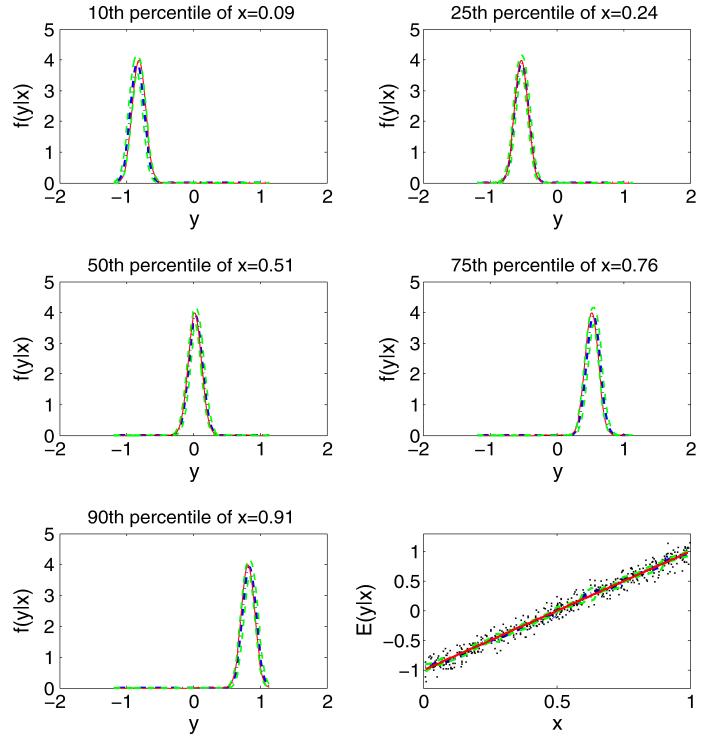
For Case 1, as shown in Fig. 3, the predictive mean regression curve (blue dashed, right bottom panel), the true linear regression function (red solid), and the pointwise 95% credible intervals (green dashed) were almost the same. Figure 3 also shows the predictive densities (blue dashed) at the 10th, 25th, 50th, 75th, and 90th sample percentiles of xi, with these densities almost indistinguishable from the true densities (red solid).
Fig. 3.
Results for simulation Case 1: true conditional densities of y|x (red solid), predictive conditional densities (blue dot-dashed), and 95% pointwise credible intervals (green dashed). The lower right panel shows the data (black dots), along with true (red solid) and estimated mean (blue dashed) regression curves superimposed with 95% credible line (green dashed)
For Case 2, Fig. 4 shows an x – y plot (right bottom panel) of the data along with the estimated predictive mean curve (blue dashed), which closely follows the true mean curve (red solid). Figure 4 also shows the estimated predictive densities (blue dashed) correspond approximately to the true densities (red solid) in most cases and the 95% credible intervals (green dashed) closely cover the true densities in all cases.
Fig. 4.
Results for simulation Case 2: true conditional densities of y|x (red solid), predictive conditional densities (blue dot-dashed), and 95% pointwise credible intervals (green dashed). The lower right panel shows the data (black dots), along with true (red solid) and estimated mean (blue dashed) regression curves superimposed with 95% credible line (green dashed)
Repeating the analysis for Case 2, but with and G ~ DP(αG0), we obtained poor results estimates diverged substantially from true densities, posterior mean curve failed to capture true nonlinear function), suggesting that a DPM model is inadequate.
6 Epidemiological application
6.1 Background and motivation
In diabetic studies, interest often focuses on the relationship between 2-h serum insulin levels (indicator for insulin sensitivity/resistence) and 2-h plasma glucose levels (indicator for diabetic risk) that are measured in the oral glucose tolerance test (OGTT). Although most studies examine the mean change of the 2-h insulin versus 2-h glucose, it would be more interesting to assess the whole distributional change of the 2-h insulin level across the range of the 2-h glucose levels.
We obtained data from a study which followed a sample of Pima Indians from a population near Phoenix, Arizona since 1965. This study was conducted by the National Institute of Diabetes and Digestive and Kidney Disease, with the Pima Indians chosen because of their high risk of diabetes. Using these data, our goal is conducting inferences on changes in the 2-h serum insulin distribution with changes in 2-h glucose level without making restrictive assumptions, such as normality or a constant residual variation. Certainly, it is biologically plausible that the insulin distribution is non-normal and should change as the glucose level changes not only in mean but also in other features such as skewness, residual variation, and modality.
6.2 Analysis and results
For woman i (i = 1, … , 393), let yi correspond to the 2-h serum insulin level measured in μU/ml (micro Units per milliliter) and let xi denote the 2-h plasma glucose level measured in mg/dl (milligrams per deciliter). We applied the lDPMN model described in (12), after scaling y and x by dividing by 100. Hyperparameters were set to be the same as in the simulation study except that ψ = 0.08 such that n/10 subjects belong to each neighborhood on average and aΓ = min(xi) − ψ, and bΓ = max(xi) + ψ) such that the edge effects are avoided in the inference. We analyzed the data using the proposed Gibbs sampling algorithm run for 10,000 iterations with a 5,000 iteration burn-in. The convergence and mixing of the MCMC algorithm were good (Trace plots not shown) and results were robust with reasonable alternative hyperparameter values.
Figure 5 shows the predictive distributions for the insulin level at various empirical percentiles of the glucose level. As the glucose level increases, there is a slightly nonlinear change in the mean insulin level (right bottom panel) and a dramatic increase in the heaviness of the right tail of the insulin distribution. Also, some multi-modality in the insulin distribution appears as the glucose level falls into the pre-diabetes range (140–200mg/dl) and closer to the cut point (200mg/dl) for the diagnosis of diabetes. This shift in the shape of the insulin distribution biologically implies that the women with pre-diabetes are expected to have different insulin sensitivities, which may further induce different diabetic risks even for the same glucose level. This may be due to unadjusted covariates or unmeasured risk factors. Such distributional changes in response induced by predictors (e.g. risk factor, exposure, treatment, and, etc.) is pervasive in epidemiologic studies, but is not at all well characterized by standard regression models that do not allow the whole distribution to flexibly change with predictors.
Fig. 5.
Results for Pima Indian Example: predictive conditional densities (blue dot-dashed), and 95% pointwise credible intervals (green dashed). The lower right panel shows the data (black dots), along with estimated mean (blue dashed) regression curves superimposed with 95% credible line (green dashed)
7 Discussion
This article proposed a new SBP for the collection of predictor-dependent random probability measures. The prior, called the lDP, is a useful alternative to recently developed prior models that induce predictor-dependence among distributions. Its marginal DP structure should be useful in considering theoretical properties, such as posterior consistency and rates of convergence. A related formulation was independently developed by Griffin and Steel (2008) although the lDP is appealing in its simplicity for construction and computation. In particular, the construction is intuitive and leads to simple expressions for the dependence in random measures at different locations, while also leading to straightforward posterior computation relying on truncation with a fair amount of accuracy.
Although we have focused on a conditional density estimation application, there are many interesting applications of the lDP to be considered in future work. First, the DP is widely used to induce a prior on a random partition or clustering structure (Quintana 2006; Kim et al. 2006). In such settings, the DP has the potential disadvantage of requiring an exchangeability assumption, which may be violated when predictors are available that can inform about the clustering. The lDP provides a straightforward mechanism for local, predictor-dependent clustering, which can be used as an alternative to product partition models (Quintana and Iglesias 2003) and model-based clustering approaches (Fraley and Raftery 2002). It is of interest to explore the theoretical properties of the induced prior on the random partition. In this respect, it is likely that the hyperparameter ψ plays a key role. Hence, as a more robust data-driven approach one may consider fully Bayes or empirical Bayes methods for allowing uncertainty in ψ.
Acknowledgments
This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Appendix
Proof of Lemma 1 An infinite number of locations Γ = {Γh, h = 1, …, ∞} are generated from H on . Any ψ-neighborhood of x defined as with ψ > 0 is a subsect of . The regularity Condition 1 for H ensures that there is a positive probability for a location Γh to be generated in any . Therefore, there are also an infinite number of locations in for all and ψ > 0, which implies N(x) = ∞. Then, almost surely by Lemma 1 in Ishwaran and James (2001).
Proof of Theorem 1 Assume that . Then, from the definition of the lDP in (5)–(7), we can reexpress (7) as , where is the lth element of V(x) and is the lth element of Θ(x). Note that it follows from the proof of Lemma 1 that N(x) = ∞. Since the random weights and atoms are generated by iid sampling from Beta(1, α) and G0, respectively, independently from the location, we have independently from , for l = 1, … , ∞. Hence, it follows directly from Sethuraman’s (1994) representation of the DP, that .
Proof of Theorem 2 Given Γ and V,
For the definition of and , refer to the Eq. (9) in Sect. 3.2. Marginalizing out V over the Beta distribution,
In order to marginalize out and , we introduce as described in the formulations from (9) to (10) in Sect. 3.2. Then,
(After marginalizing out the as in the Proof of Theorem 3, we obtain:
Proof of Theorem 3 From (10),
where Zγj are iid draws from Bernoulli(Px1, x2). Taking expectation of with respect to Bernoulli((Px1, x2)),
where Yj ~ Binomial(j − 1, Px1,x2). Using the Binomial Theorem, the expectation on the right is marginalized out with respect to Binomial(j – 1, Px1,x2), which results in
Since , the infinite sum on the right converges. Then,
Proof of Theorem 4 Due to the marginal DP property and using the inequality on the left in (11) with n = 1, we get , where μN, μ∞, N in (11) are replaced by μN(x), μ∞(x), N(x), respectively, and n is substituted by 1. Here, N(x) is random differently from N in (11). Conditioned on N(x) but marginalizing out ph, we get . Note that N(x) ~ Binomial(N, Px) as discussed in Sect. 3.3. Then, using the Binomial Theorem, we obtain .
Contributor Information
Yeonseung Chung, Department of Biostatistics, Harvard School of Public Health, 655 Huntington Ave. Bldg 2, Room 435A, Boston, MA 02115, USA.
David B. Dunson, Department of Statistical Science, Duke University, 219A Old Chemistry Bldg, Box 90251, Durham, NC 27708, USA, dunson@stat.duke.edu
References
- Beal M, Ghahramani Z, Rasmussen C. Neural information processing systems. Vol. 14. MIT Press; Cambridge: 2002. The infinite hidden Markov model. [Google Scholar]
- Blei D, Griffiths T, Jordan M, Tenenbaum J. Neural information processing systems. Vol. 16. MIT Press; Cambridge: 2004. Hierarchical topic models and the nested Chinese restaurant process. [Google Scholar]
- Caron F, Davy M, Doucet A, Duflos E, Vanheeghe P. Bayesian inference for dynamic models with Dirichet process mixtures; International conference on information fusion; Italia. 2006.Jul 10-13, [Google Scholar]
- De Iorio M, Müller P, Rosner GL, MacEachern SN. An Anova model for dependent random measures. Journal of the American Statistical Association. 2004;99:205–215. [Google Scholar]
- Dowse KG, Zimmet PZ, Alberti GMM, Bringham L, Carlin JB, Tuomlehto J, Knight LT, Gareeboo H. Serum insulin distributions and reproducibility of the relationship between 2- hour insulin and plasma glucose levels in Asian Indian, Creole, and Chinese Mauritians. Metabolism. 1993;42:1232–1241. doi: 10.1016/0026-0495(93)90119-9. [DOI] [PubMed] [Google Scholar]
- Duan JA, Guidani M, Gelfand AE. ISDS Discussion Paper, 05-23. Duke University; Durham: 2005. Generalized spatial Dirichlet process models. [Google Scholar]
- Dunson DB. Bayesian dynamic modeling of latent trait distributions. Biostatistics. 2006;7:551–568. doi: 10.1093/biostatistics/kxj025. [DOI] [PubMed] [Google Scholar]
- Dunson DB, Park J-H. Kernel stick-breaking process. Biometrika. 2008;95:307–323. doi: 10.1093/biomet/asn012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunson DB, Peddada SD. Bayesian nonparametric inference on stochastic ordering. Biometrika. 2008;95:859–874. doi: 10.1093/biomet/asn043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunson DB, Pillai N, Park J-H. Bayesian density regression. Journal of the Royal Statistical Society, Series B. 2007;69:163–183. [Google Scholar]
- Escobar MD. Estimating normal means with a Dirichlet process prior. Journal of the American Statistical Association. 1994;89:268–277. [Google Scholar]
- Escobar MD, West M. Bayesian density estimation and inference using mixtures. Journal of the American Statistical Association. 1995;90:577–588. [Google Scholar]
- Ferguson TS. A Bayesian analysis of some nonparametric problems. The Annals of Statistics. 1973;1:209–230. [Google Scholar]
- Ferguson TS. Prior distributions on spaces of probability measures. The Annals of Statistics. 1974;2:615–629. [Google Scholar]
- Fraley C, Raftery AE. Model-based clustering, discriminant analysis, and density estimation. Journal of the American Statistical Association. 2002;97:611–631. [Google Scholar]
- Gelfand AE, Kottas A, MacEachern SN. Bayesian nonparametric spatial modeling with Dirichlet process mixing. Journal of the American Statistical Association. 2004;100:1021–1035. [Google Scholar]
- Ghosal S, Van der Vaart AW. Posterior convergence rates of Dirichlet mixtures at smooth densities. The Annals of Statistics. 2007;35(2):697–723. [Google Scholar]
- Ghosal S, Ghosh JK, Ramamoorthi RV. Posterior consistency of Dirichlet mixtures in density estimation. The Annals of Statistics. 1999;27:143–158. [Google Scholar]
- Griffin JE, Steel MFJ. Order-based dependent Dirichlet processes. Journal of the American Statistical Association. 2006;101:179–194. [Google Scholar]
- Griffin JE, Steel MFJ. Bayesian nonparametric modeling with the Dirichlet process regression smoother. Technical Report. University of Warwick; 2008. [Google Scholar]
- Ishwaran H, James LF. Gibbs sampling methods for stick-breaking priors. Journal of the American Statistical Association. 2001;96:161–173. [Google Scholar]
- Kim S, Tadesse MG, Vannucci M. Variable selection in clustering via Dirichlet process mixture models. Biometrika. 2006;94:877–893. [Google Scholar]
- Lijoi A, Prünster I, Walker SG. On consistency of non-parametric normal mixtures for Bayesian density estimation. Journal of the American Statistical Association. 2005;100:1292–1296. [Google Scholar]
- Lo AY. On a class of Bayesian nonparametric estimates: I. Density estimates. The Annals of Statistics. 1984;12:351–357. [Google Scholar]
- MacEachern SN. Dependent nonparametric processes; ASA proceedings of the section on bayesian statistical science; Alexandria. 1999; American Statistical Association; [Google Scholar]
- MacEachern SN. Dependent Dirichlet processes. Department of Statistics, The Ohio State University; 2000. Unpublished manuscript. [Google Scholar]
- MacEachern SN. Decision theoretic aspects of dependent nonparametric processes. In: George E, editor. Bayesian methods with applications to science, policy and official statistics. ISBA; Creta: 2001. pp. 551–560. [Google Scholar]
- Müller P, Quintana F, Rosner G. A method for combining inference across related nonparamet- ricBayesian models. Journal of the Royal Statistical Society B. 2004;66:735–749. [Google Scholar]
- Pennell ML, Dunson DB. Bayesian semiparametric dynamic frailty models for multiple event time data. Biometrics. 2006;62:1044–1052. doi: 10.1111/j.1541-0420.2006.00571.x. [DOI] [PubMed] [Google Scholar]
- Pitman J. Exchangeable and partially exchangeable random partitions. Probability Theory and Related Fields. 1995;102:145–158. [Google Scholar]
- Pitman J. Some developments of the Blackwell-MacQueen urn scheme. In: Ferguson TS, Shapley LS, MacQueen JB, editors. Statistics, probability and game theory. Vol. 30. Institute of Mathematical Statistics; Hayward: 1996. pp. 245–267. IMS Lecture Notes-Monograph Series. [Google Scholar]
- Quintana FA. A predictive view of Bayesian clustering. Journal of Statistical Planning and Inference. 2006;136:2407–2429. [Google Scholar]
- Quintana FA, Iglesias PL. Bayesian Clustering and product partition models. Journal of the Royal Statistical Society B. 2003;65:557–574. [Google Scholar]
- Sethuraman J. A constructive definition of the Dirichlet process prior. Statistica Sinica. 1994;2:639–650. [Google Scholar]
- Smith JW, Everhart JE, Dickson WC, Knowler WC, Johannes RS. Using the ADAP learning algorithm to forecast the onset of diabetes mellitus; Proceedings of the symposium on computer applications in medical care; 1988.pp. 261–265. [Google Scholar]
- Xing EP, Sharan R, Jordan M. Bayesian haplotype inference via the Dirichlet process; Proceedings of the international conference on machine learning (ICML); 2004. [DOI] [PubMed] [Google Scholar]