Abstract
Attempts to evaluate the protective effect of live attenuated SIV vaccine strains have yielded variable results depending on the route of immunization, the level of attenuation, the level of divergence between the vaccine candidate and the challenge. The protective mechanisms induced by these vaccines are still not well understood. In an effort to address whether the diversity of the CD4+ T cell repertoire in cynomolgus macaques plays a role in the immunological protection following SIVmacC8 infection, we have performed a longitudinal follow-up of the CD4 repertoire by heteroduplex tracking assay in macaques mock infected or infected with either the attenuated SIVmacC8 or its homologous SIVmacJ5 and challenged with simian-human immunodeficiency virus (SHIV89.6P). Viral load and CD4 absolute counts were determined in these animals and the presence of SHIV89.6P virus in challenged animals was evaluated by PCR and serology. In all macaques that were protected against the challenging virus, we demonstrated a reduced diversity in the CD4+ TRBV repertoire and a few dominant CD4+ T cell clones during early primary infection. In contrast, CD4 TRBV repertoire in unprotected macaques remained highly diverse. Moreover, some of the CD4 T cell clones that were expanded during primary SIV infection re-emerged after challenge suggesting their role in protection against the challenging virus. These results underline the importance of maintaining the CD4 T cell repertoire developed during acute infection and point to the restriction of the CD4 response to the vaccine as a correlate of protection.
Keywords: TRBV repertoire, CD4, primary infection, SHIV89.6P, viral challenge, protection
Introduction
Immunological responses generated against persistent viral infections are characterized by the activation and expansion of CD4 and CD8 antigen-specific T lymphocytes. Although cytotoxic T cell responses are responsible for clearance of most viral infections (Shin & wherry, 2007;Purtha, Myers et al., 2007;Kuroda, Schmitz et al., 1999), CD4 T cells play a major role in supporting antibody production, initiating and maintaining CTL activity as well as performing direct effector activity through the production of specific cytokines (Ribeiro, 2007;Davenport, Price et al., 2007). Following viral suppression through immunological control, these T cells decline in activity and number mostly through apoptosis (McHeyzer-Williams & Davis, 1995;Gupta & Gollapudi, 2007;Badovinac, Porter et al., 2002), leaving the host with a sufficient number of pathogen-specific memory T cells. The major role of memory T cells is to ensure protection upon re-exposure to pathogens through rapid clonal proliferation and functional activation. This has been demonstrated in both CD8 (Walker, Wilson et al., 1996;Levitsky, Campos-Lima et al., 1998;Maryanski, Jongeneel et al., 1996;Sourdive, Murali-Krishna et al., 1998;Blattman, Sourdive et al., 2000;Roberts & Woodland, 2004;Geginat, Lanzavecchia et al., 2003) and CD4 positive T cells (Bitmansour, Waldrop et al., 2001;seder & Ahmed, 2003;Sallusto, Geginat et al., 2004;Zaph, Uzonna et al., 2004). Several factors including the stimuli, the host or the environment can all contribute to create variations between antigen-selected repertoires. These factors can also influence whether a potent antigen-specific repertoire is selected, whether it is persistent and protective.
Attempts to generate a vaccine against human immunodeficiency virus (HIV) have led to the evaluation of attenuated strains as candidates. Macaques challenged with pathogenic simian immunodeficiency virus (SIV) several months after being infected with live attenuated SIV show better protection than monkeys immunized by any other vaccine strategy (Tenner-Racz, Stahl et al., 2004;Johnson, Lifson et al., 1999;Mori, Yasutomi et al., 2001;Koff, Johnson et al., 2006). The use of attenuated SIV viruses as vaccines has provided complete or near complete protection from challenge with wild type SIV (Koff, Johnson et al., 2006). Better protection was obtained with homologous challenge than with heterologous challenge (Johnson & Desrosiers, 1998). The most common strategy of attenuating SIV was achieved by the complete or partial deletion of nef (SIVΔnef or SIVmacC8, respectively) or the deletion of both nef and vpr (SIVΔ3). The use of these viruses has provided strong protection against pathogenic SIV challenge (Johnson & Desrosiers, 1998). Another way of attenuating SIV was achieved through the deletion of V1-V2 region of the envelope protein (Env; SIVΔV1-V2). This virus has also conferred potent protection from intravenous challenge by wild type SIVmac239 (Cole, Steckbeck et al., 2004).
The correlates and mechanisms of protection induced by attenuated viruses, however, are poorly understood. Stebbings et al. demonstrated using CD8 T cells depletion techniques, that CD8 T cells responses alone are not central to the protection against acute superinfection conferred 20 days after vaccination with attenuated SIVmacC8 (Stebbings, Berry et al., 2005). This conclusion was consistent with the findings of other investigators who were unable to identify a correlation between SIV-specific CD8 CTL responses elicited by inoculation with live attenuated SIV and protection against superinfection (Sharpe, Cope et al., 2004;Dittmer, Brooks et al., 1999).
The persistence of the CD4 helper function was shown in several model systems to be essential for the maintenance of memory CD8 responses during chronic infection as well as the generation of neutralizing antibodies to viral escape mutants and for the control of viremia (Suvas, Kumaraguru et al., 2003;Sun, Williams et al., 2004;Sun & Bevan, 2003;Matloubian, Concepcion et al., 1994;von Herrath, Yokoyama et al., 1996;Zajac, Blattman et al., 1998;Wodarz & Jansen, 2001;Ahmed, Butler et al., 1988;Kalams, Buchbinder et al., 1999). While the rapid turnover of CD4 T cells characterize HIV/SIV infection, it is not clear to what extent HIV/SIV infections can drive an expansion rather than a depletion of antigen specific CD4 T cells. Assessing T cell receptor (TCR) repertoire during HIV/SIV infection will add to our understanding of the way T cells respond to this infection and contribute in mediating protection. Chen et al. have shown using CDR3 profile and sequence analysis of CD4 T cell receptor repertoires that infection of macaques with SIV can result in prolonged periods of clonal dominance of CDR3-restricted CD4 T cell clones despite the decline of CD4 T cell count (Chen, Shen et al., 2000).
In this study we evaluated the impact of the breadth and persistence of CD4 T cell repertoire stimulated during primary infection with attenuated SIVmacC8 and pathogenic SIVmacJ5 strains on the resistance to subsequent challenge with the highly pathogenic simian-human immunodeficiency virus (SHIV89.6P) strain.
Results
Virological and immunological evolution following SIVmacJ5- or SIVmacC8-infection
In order to analyze the relationship between the breadth of the CD4 repertoire induced during primary infection (PI) by attenuated SIV strains and the susceptibility to subsequent superinfection, cynomolgus macaques were infected with either the pathogenic SIVmac strain SIVmacJ5 or the attenuated strain SIVmacC8 (Cranage, Stott et al., 1992). Twenty weeks post primary infection, animals were challenged with the pathogenic SHIV89.6P strain.
In all SIVmacJ5 infected macaques the CD4 count transiently dropped 2 weeks post infection and returned to normal values at following time points (Figure 1A). In these animals viral load peaked simultaneously to the CD4 decline (~108 copies per ml) and settled at week 6 below the limit of detection, with some occasional viral blips.
Figure 1. Disease status in SIV infected animals.
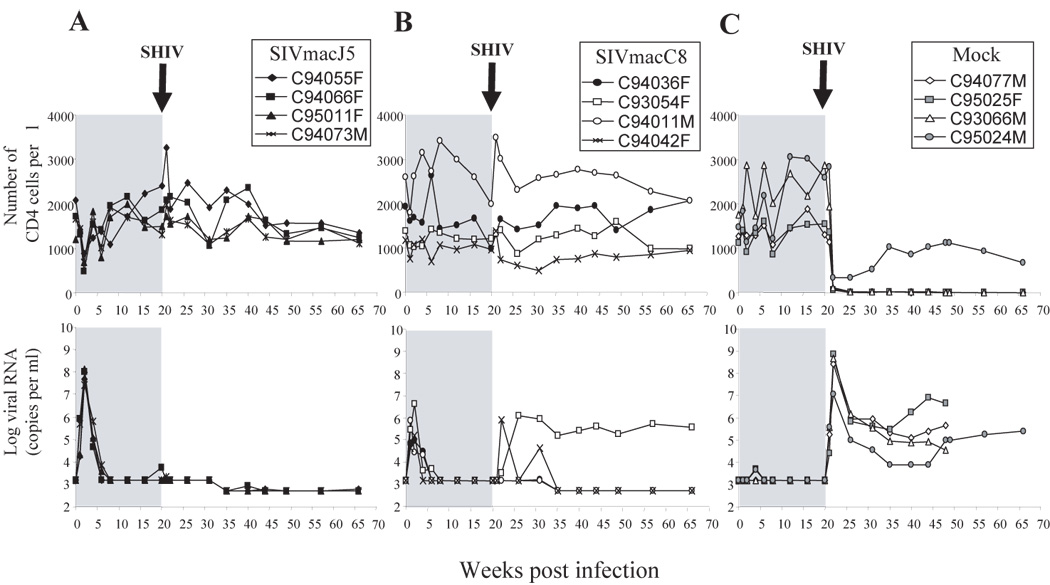
CD4 absolute count (right panels) and plasma SIV RNA detected by branched DNA (left panels) from groups A-SIVmacJ5, B-SIVmacC8, and C- Mock infected macaques. Animals infected with SIVmacJ5 or SIVmacC8 show slow progressor patterns. Following challenge at week 20 with SHIV89.6P, two SIVmacC8 infected macaques show a rebound in viral load and a progressive drop in CD4 counts. SIV viral load target probes, designed to hybridize with the pol region of the SIVmac groups of strains were used to quantify SIV viral load. Results were quantified by comparison with purified and quantified in vitro-transcribed SIV pol RNA and were plotted on a log(10) scale. The detection limit of this assay was 1500 copies of SIVmac RNA per ml until week 35 and 500 thereafter. White blood cell counts were obtained from a hematology workstation and were used to calculate the absolute CD4 counts.
In the SIVmacC8-infected group, macaques C93054F and C94042F had both lower CD4 counts at study entry compared to other macaques in that group and did not show any significant perturbations in their CD4 counts during acute infection (Figure 1B). Except for a transient peak in CD4 count at week 6 post infection, the CD4 count in macaque C94036F also remained stable during primary infection. The CD4 count in macaque C94011M demonstrated major fluctuations during the primary-infection period. After an initial drop one-week post infection followed by a transient increase until week 4-post infection, the CD4 count in that animal returned to pre-infection value by week 20-post infection. All the SIVmacC8-infected macaques showed a peak in viral load (5×105−8×106 copies per ml) at week 2 and the virus settled by week 6 below the limit of detection. Mock-infected animals had variable CD4 counts from one time point to another but remained between normal values during the follow up period (Figure 1C).
Post-infection CD4+ TRBV repertoire evolves independent of the infecting SIV strain
The diversity of the CD4 TRBV repertoire was evaluated using the heteroduplex mobility assay in two mock infected macaques, and four SIVmacC8 or SIVmacJ5 infected macaques, prior to infection (week –3 and 0) until week 8 post infection (week 1, 2, 4, 6 and 8). Seven TRBV families were analyzed (TRBV9, 20, 28, 5–1, 7, 12 and 3). As defined in Figure 2A, the CD4 TCR population for each analyzed TRBV family was classified according to the number of detectable discrete bands, TRBV families composed of 13 or more discrete bands were considered as highly diverse repertoires while the TRBV families with less than 13 bands were separated into 2 groups according to the degree of diversity (i.e. low diversity (1–6 bands) or intermediate diversity (7–12 bands)). A representative example (TRBV5-1) showing the evolution of the TCR repertoires following SIVmacJ5- and SIVmacC8-infected macaques as well as a mock-infected macaque is shown in figure 2B. The overall diversity of the analyzed TRBV repertoires varied both between animals and between samples from each animal. Figure 3 illustrates the longitudinal analysis of the CD4 repertoires in all the animals during PI. The SIVmacJ5-infected group was comprised of macaques with mixed repertoires: 3/4 animals had mainly low diversity TRBV families C94066F and C94055F and C95011F (figure 2B and 3), whereas 1/4 in that group, C94073F, had both low and high diversity TRBV families, such as TRBV20 and TRBV3 respectively. As for the SIVmacC8 group, 2/4 macaques showed a very diverse TRBV repertoire throughout the follow up period (C93054F and C94042F) and two others had mixed repertoires (C94036F and C94011M). During the same period, a persistent highly diverse CD4 TRBV repertoire characterized both mock-infected macaques tested.
Figure 2. TRBV5-1 CD4+ families in uninfected, SIVmacJ5 and SIVmacC8 infected macaques.
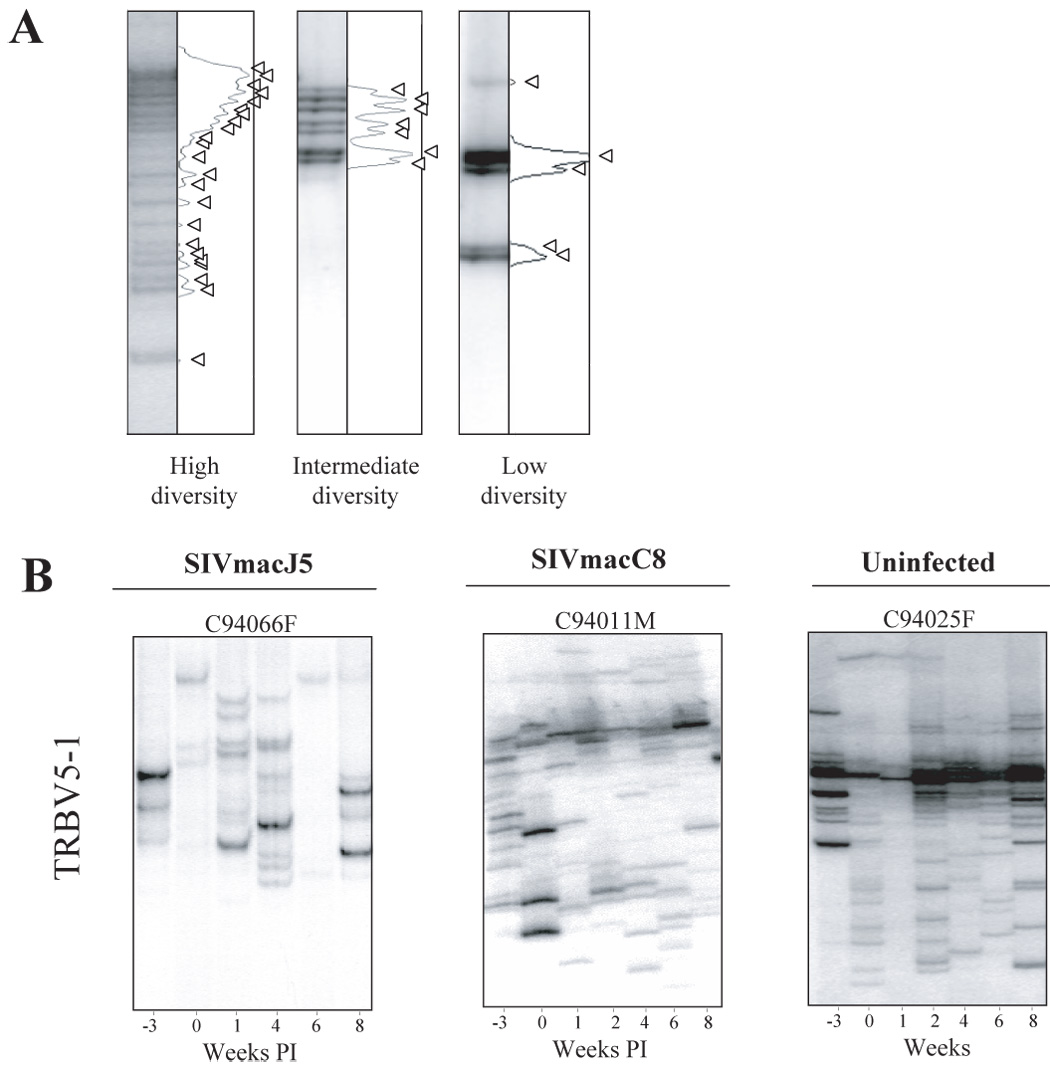
A. Computerized densitometric analysis of the HTA autoradiography. Each band on the HTA gel is transformed into a peak using the NIH image software. Each band represents a CD4 clonotype. An HTA profile is highly diverse when it is composed of more than 13 clones and with low diversity when it is composed of or less than 12 clones. Arrows indicate discrete TRBV hetero-complexes over the HTA profiles. B. Heteroduplex tracking assay using TRBV5-1 as 5’-primers and BC as 3’-primers was performed on purified CD4+ T cells collected from −3 weeks prior to SIV infection until week 8 post infection. Data from representative SIVmacJ5, SIVmacC8, and mock infected macaques is shown. SIVmacJ5 and SIVmacC8 include macaques with low and high diversity CD4 repertoires, whereas uninfected macaques have always highly diverse repertoires.
Figure 3. Summary of the longitudinal analysis of the CD4+ TRBV repertoire during primary SIV infection.
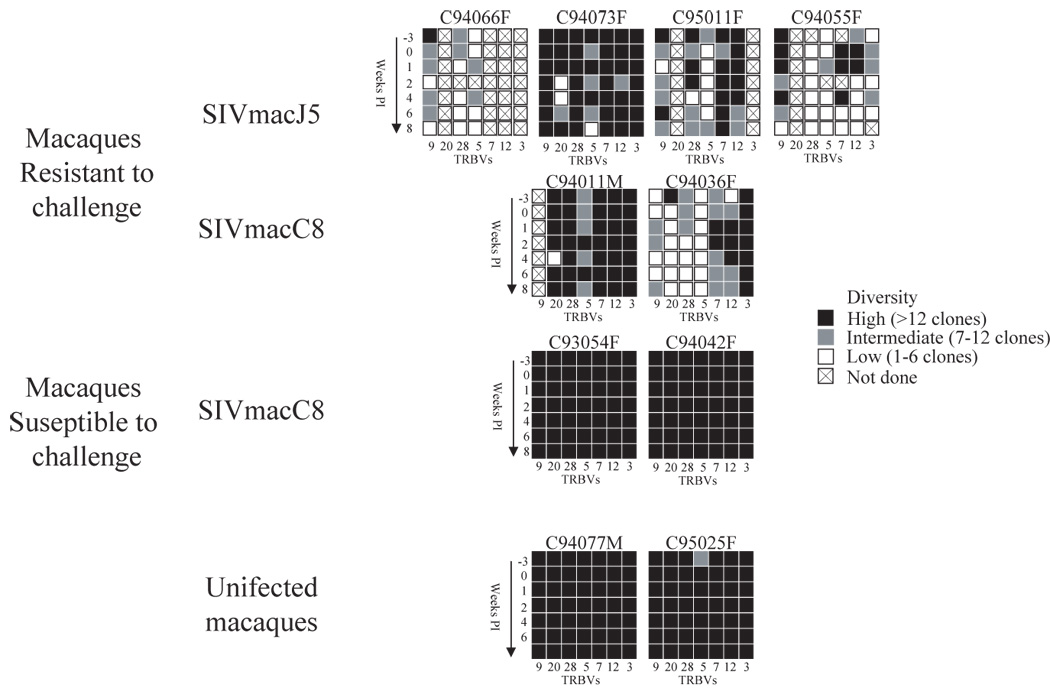
The clonality of each TRBV family at each time point and for each macaque was determined during −3 weeks prior to SIV infection until 8 weeks post infection. Heteroduplex tracking assay using a panel of TRBV as 5′-primers that include (TRBV9, 20, 28, 5–1, 7, 12, 3) and BC as 3′-primers was performed on purified CD4+ T cell clones from −3 weeks until week 8 post infection. In order to simplify the representation of these results, black boxes represent highly diverse repertoires containing 13 clones and more, grey boxes represent intermediate diversity repertoires containing between 7 and 12 clones, whereas white boxes represent low diversity repertoires with less than 6 clones. Boxes with a cross represent samples that were not done.
The development of a low diversity CD4 TRBV repertoire during PI is associated with resistance to challenge with SHIV89.6P
The animals were challenged with SHIV89.6P at week 20 post primary infection. Two macaques from the SIVmacC8-infected group (C93054F and C94042F) demonstrated evidence of superinfection (data not shown). Macaque C93054F showed a rebound in viral load at week 26 (106 copies per ml) that remained high during the follow-up period, whereas macaque C94042F had two expansions in viral load at week 24 (106 copies per ml) and at week 31 (8×104 copies per ml). None of the superinfected macaques experienced any decrease in CD4 counts as compared to their pre-challenge values, however the initial CD4 counts were lower in these animals than in the protected macaques from the same group. In the other 6 protected animals neither viral load nor CD4 counts were significantly modified following challenge (Figure 1A and B). The absence of detectable SHIV89.6P infection was confirmed by nested PCR amplification of the SHIV89.6P env gene (data not shown). The four mock-infected control animals were susceptible to SHIV89.6P infection. All four macaques within that group showed a drastic drop in CD4 counts (below 200 cells per µl) during the first week post-challenge. While CD4 counts in 3/4 mock-infected macaques remained very low, macaque C95024M showed a transient rebound in CD4 count to normal values, however by week 74 the CD4 count dropped again below 200 cells per µl (data not shown). All the control animals experienced very high viral loads (107–109 copies per ml) at week 2 post-SHIV89.6P infection. Viral set point was reached by week 24 to 35 and viral load remained between 104 and 107 copies per ml in all four animals up to the end of the study (Figure 1C).
Considering that, irrespective of the infecting virus, some macaques were protected against SHIV89.6P challenge, we investigated whether there was an association between protection against SHIV89.6P super-infection and TCR repertoire diversity in the CD4+ compartment, either prior or during primary SIV infection.
Interestingly, all the animals with low diversity CD4 TRBV repertoire following primary SIV-infection were resistant to challenge, irrespective of the SIV strain used for primary infection. Several TRBV families from protected animals had evolved overtime following SIV infection, such as TRBV9 for macaques C94055F, C94066F, C95011F as well as TRBV20 for macaques C94073F and C94011M (see figure 3). These TRBV families progressed from a highly diverse repertoire prior to infection to a very restricted repertoire composed of less than 6 clones during PI. Other TRBV families were either always restricted (such as TRBV28 for macaque C94055F) or evolved further to include less CD4 clonotypes (such as TRBV28 and TRBV5-1 for macaque C94066F). In contrast, for both the control animals and the two SIVmacC8-infected macaques susceptible to challenge (C93054F and C94042F), the TRBV repertoire was highly diverse prior to SIV infection and remained highly diverse during the 8 weeks of follow-up (Figure 3).
This longitudinal analysis of the CD4+ TRBV repertoires in animals resistant to SHIV89.6P challenge suggests that the CD4+ T cell proliferation induced during primary SIV infection in these animals was restricted to a limited number of clones.
The low diversity repertoires developed during PI in protected animals persist following challenge
In order to evaluate the possible role of CD4 T cell clones stimulated during primary infection in the resistance to the challenging virus, we investigated whether some of these clones were 1) persisting during PI and 2) reemerging following SHIV89.6P challenge.
In case of macaque C94066F, for-example, two co-migrating HTA bands were observed at week 8 post infection, and again at week 2 post challenge (Figure 4A). In order to investigate whether those co-migrating bands truly belong to the same TCR clone, the PCR products corresponding to TRBV5-1 amplifications in macaque C94066F were cloned and 25, 30 or 22 clones were sequenced for time points 4 and 8 weeks post SIVmacJ5 infection, and 2 weeks post challenge respectively. Two dominant clones (NRG and LGS) were identified at high frequency at week 8 post infection. Interestingly, among them one CD4 clonotype (NRG) was also detected at week 2 post challenge (figure 4B). To certify that the co-migrating bands observed by HTA at week 8 and week 2 post challenge belong to clone (NRG), the PCR products corresponding to TRBV5-1 at week 8 and plasmid DNA corresponding to clones NRG, LGS and as a control LGQ were run next to each other in an HTA gel. Figure 4C indicates that the upper band in the PCR sample of week 8 correspond to clone NRG, whereas the lower band corresponds to clone LGS. These results demonstrated that co-migrating bands can indeed correspond to TCRs with identical CDR3 regions. Therefore each HTA experiment showing co-migrating bands were considered as belonging to the same T cell clone only when an independent HTA experiment using series of other TRBV probes were able to confirm the co-migration.
Figure 4. Analysis of the re-emergence of CD4 clonotypes in a protected animal.
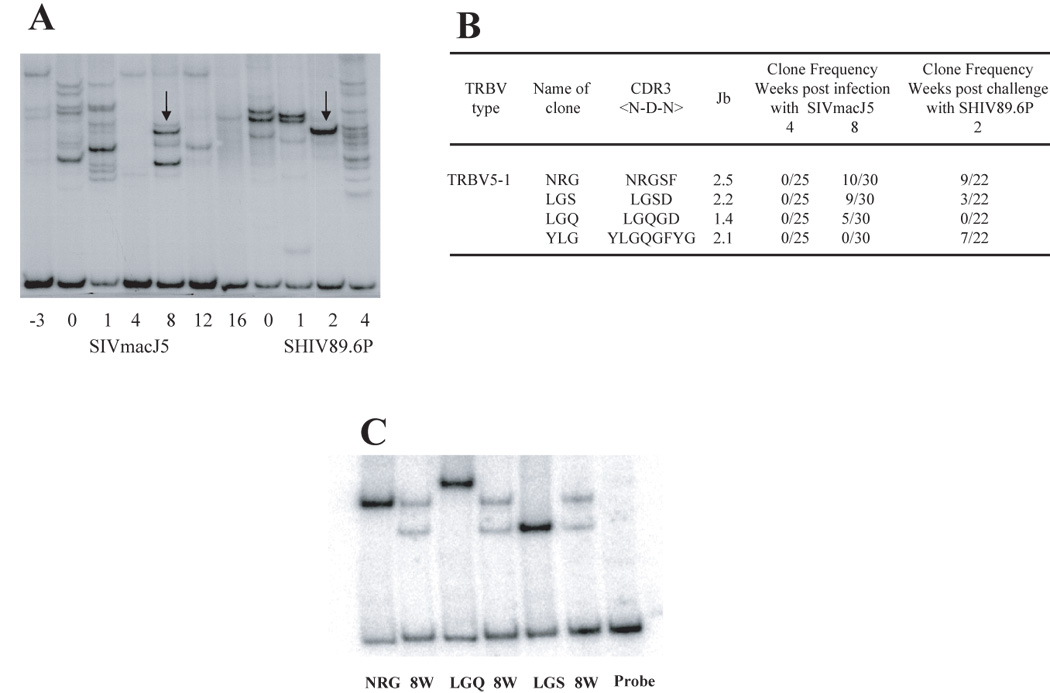
A- TRBV5-1+ CD4 + clonotypes in macaque C94066F was analyzed by HTA from week −3 to week 4 post challenge. Arrows represent co-migrating bands at week 8 and week 2 post challenge. B- The PCR products from time points 4, 8 and 22 weeks (or 2 weeks post challenge) as shown by the arrows in figure A, were cloned and sequenced to identify their CDR3 regions as described in materials and methods. At week 8 two dominant CDR3 regions were identified, NRGSF with 10/25 clones and LGSD with 9/25 clones. At week 2 post challenge those two dominant TCR clones were detected. C- Co-migration of the DNA plasmids encoding the two dominant clones detected at week 8, NRG and LGS together with the PCR product from week 8. These results demonstrate that clones co-migrating at the same level are identical.
We first analyzed the persistence of expanded CD4+ T cell clones in the SIVmacJ5-, SIVmacC8-infected and control animals during the first 12 weeks of follow-up. Interestingly, almost all the protected animals, whether they were infected by SIVmacJ5 or SIVmacC8 viruses, had at least two TRBV families with persistent CD4 clonotypes during PI (Table 1). In contrast, unprotected and mock-infected animals showed little (C94042F in TRBV28) or no persistence of CD4 clonotypes during PI. Although some TRBV families in unprotected or uninfected macaques were composed of a few dominant clones at some time points post-infection, these dominant clones did not persist (i.e. TRBV5-1 for macaque C93054F) suggesting that in these animals, the CD4+ T cells clonally expanded during primary infection were rapidly lost during the following weeks (Mattapallil, Douek et al., 2005;Li, Duan et al., 2005;Picker & Watkins, 2005).
Table 1.
Clonotype persistence during PI
| Macaque | Virus | Time points showing persistent clones during PI (weeks)1 | Status | ||||||
|---|---|---|---|---|---|---|---|---|---|
| TRBV9 | TRBV20 | TRBV28 | TRBV5-1 | TRBV7 | TRBV12 | TRBV3 | |||
| C94055F | SIVmacJ5 | 1,42 | 4,6 | Resistant to SHIV89.6P challenge 3 | |||||
| C94066F | |||||||||
| C94073F | 4,6 | 1,4 | |||||||
| C95011F | 4,6 | 1,4; 6,8,12 | |||||||
| C94036F | SIVmacC8 | 6,8 | 4,6 | 4,6 | 6,8 | ||||
| C94011M | 4,8 | 1,2,4 | |||||||
| C94042F | SIVmacC8 | 4,6 | Susceptible to SHIV89.6P challenge4 | ||||||
| C93054F | |||||||||
| C94077M | uninfected | ||||||||
| C95025F | |||||||||
Two TRBV probes with different CDR3 and BJ regions were used to confirm co-migration of bands
Weeks after infection
Macaques in which no SHIV89.6P viral RNA can be detected by PCR, serology.
Macaques in which SHIV89.6P viral RNA can be detected by PCR and serology
We then evaluated whether some of the expanded clones identified during primary infection could participate in the resistance to super-infection by SHIV89.6P. We longitudinally analyzed the CD4+ TRBV repertoire during the first four weeks following challenge and compared the clonally expanded clones to those stimulated during primary infection (figure 5). Following challenge, the CD4+ TRBV repertoires were either of high or low diversity in the different macaques.
Figure 5. Evolution of the CD4+ TRBV repertoire in animals protected and unprotected from SHIV89.6P superinfection.
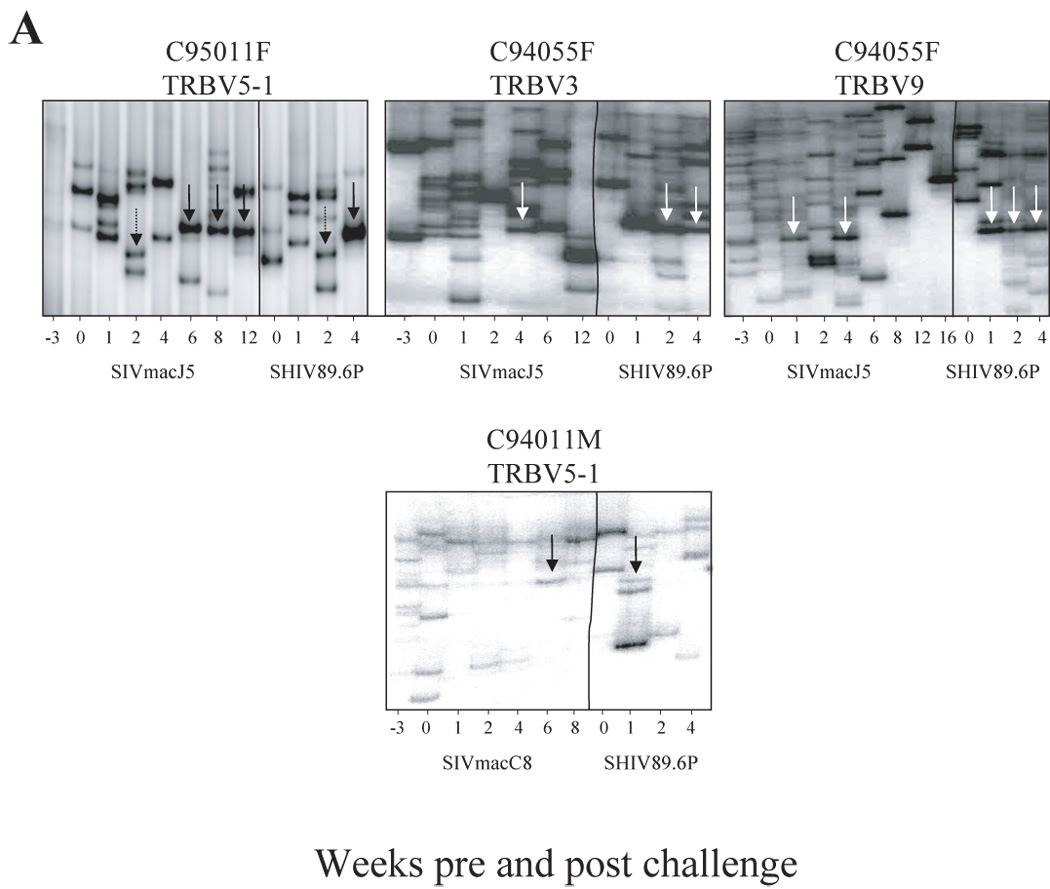
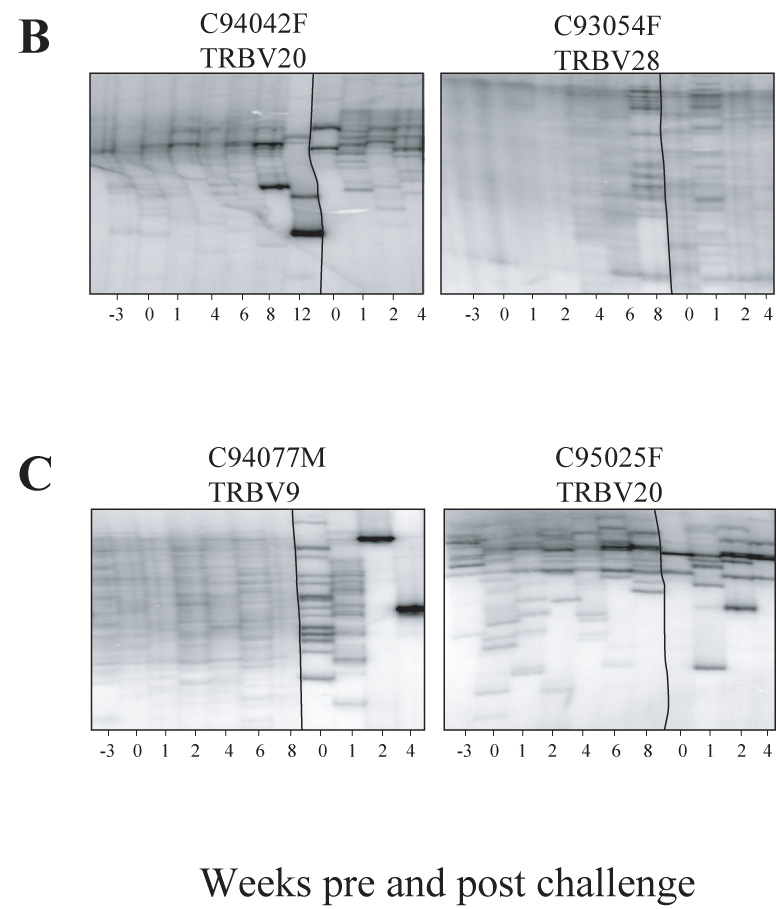
Longitudinal analysis of the CD4 TCR repertoire prior to, following SIV and SHIV89.6P challenge at week 20. Vertical lines delineates between pre-challenge and post-challenge time points. A- Animals resistant to SHIV89.6P super-infection (refer also to figure 4A for animal C94066F, TRBV5-1). B- Animals susceptible to SHIV89.6P super-infection. C-Mock infected animals infected with SHIV89.6P at week 20. Whereas animals susceptible to SHIV89.6P super-infected have a highly diverse repertoire pre and post challenge, macaques resistant to super-infection have restricted repertoire post challenge, and some are composed of dominant CD4 clonotypes that have been generated pre-challenge during acute SIV infection.
In protected macaques (Figure 5A), many CD4+ T cell clones identified during primary SIV infection persisted following challenge. In some cases, such as TRBV5-1 for macaque C95011F, the repertoire was constantly restricted throughout the study. In other cases, the CD4 repertoires contracted overtime and, by week 8, up to 6 CD4 clonotypes were present (for-example: TRBV9 for macaque C94055F, TRBV3 for macaque C94055F). Following challenge at week 1 through 4, some clones that were present during PI dominated the repertoire (Figure 5A, arrows). Occasionally the repertoire returned to high diversity following challenge such as TRBV5-1 at week 4 post challenge in macaque C94066F (Figure 4A).
Interestingly, the unprotected as well as the mock-infected animals demonstrated CD4+ T cell clone expansions following challenge (Figure 5B). However, none of these expanded clones were detected before challenge. At week 12 and contrary to the preceding samples, a low diversity CD4+ repertoire was identified for TRBV20 in macaque C94042F. However, these clones did not correspond to earlier CD4 clonotypes, nor did they persist at later time points, or re-expand after challenge. Following infection of control animals with SHIV89.6P, CD4 TRBV repertoires shifted to low diversity. However, at week 2, the CD4 counts in control animals dropped below 200 cells per µl, therefore the contraction of the CD4 repertoires at that time point, which includes very few clones, could represent the limited CD4 clones still remaining in these animals (Figure 5C).
Discussion
In this study we analyzed the breadth of the CD4 TRBV repertoire during primary SIV-infection in cynomolgus macaques and evaluated its impact on the resistance to subsequent challenge. We determined the dominance of CD4 T cell clones initiated during primary infection with attenuated SIVmacC8 and pathogenic SIVmacJ5, and their persistence following subsequent challenge with the highly pathogenic SHIV89.6P strain. Our results indicated that the development of CD4 clonotypes early following SIV infection and their maintenance over time is associated with protection against subsequent challenge with virulent SHIV89.6P.
Cynomolgus macaques infected with SIVmacJ5 or SIVmacC8 developed different types of CD4 T cell repertoires irrespective of the infecting strain (Figure 2 and 3). While some animals demonstrated a very restricted diversity in all the studied TRBV families within a few weeks post-infection, others showed a low diversity repertoire in only some TRBV families or even remained completely diverse. Such a heterogeneous response was clearly neither due to the infecting strain, nor was it a consequence of the intensity of the acute phase.
An interesting observation was the association between resistance to superinfection by SHI89.6P and the contraction of some CD4 TRBV subsets following peak viremia to include a handful of clones by week 8 post infection. Furthermore, upon challenge with SHIV89.6P, the same clonotypes that were present during primary SIV infection became dominant as early as one week post challenge in some TRBV families. These results suggest that immuno-surveillance against SIV infection rests with few CD4 clonotypes recognizing a limited array of SIV determinants. This further supports the idea that these CD4 clonotypes were generated against SIV epitopes and were re-activated after encountering SHIV89.6P, which shares several homologies with SIVmac (Reimann, Li et al., 1996;Karlsson, Halloran et al., 1997). The Sequential association of these clonal cell populations with infection and the dominance of these cell populations following challenge with SHIV89.6P suggest that these cells are SIV specific. Several studies have shown that memory CD4 (Bitmansour, Waldrop et al., 2001) or CD8 (Levitsky, Campos-Lima et al., 1998;Maryanski, Jongeneel et al., 1996;Sourdive, Murali-Krishna et al., 1998;Blattman, Sourdive et al., 2000) responses largely reflect the repertoire selected by the peak of the primary response. Fasso et al. demonstrated that a TCR repertoire selection process occurs early in the immune response in vivo, resulting in a relatively homogenous population of CD4+ T cells (Fasso, Anandasabapathy et al., 2000). Moreover, In their pigeon cytochrome c (PCC) immunization model, McHeyzer-Williams et al. demonstrated that the secondary CD4+ T cell response to PCC was more homogeneous than the primary response in terms of CDR3 loop length and amino acid composition (McHeyzer-Williams & Davis, 1995;Heyzer-Williams, Altman et al., 1996a;Heyzer-Williams, Panus et al., 1999;Heyzer-Williams, Altman et al., 1996b). Through a maturation process, it is possible that the second immune response has the ability at this stage to prevent the establishment of a second round of viral infection upon challenge, a property that primary T cell responses do not have.
However, not all animals had a contraction of the TRBV repertoire upon infection, in fact those animals already demonstrated a low diversity repertoire at W-3 and/or W0 (TRBV5 in C94066F, TRBV28 and TRBV5 in C94055F, TRBV5 in C94036F). In these cases it is possible that the state of activation at the time of infection influenced the subsequent repertoire diversity.
The most intriguing finding was the association between highly diverse CD4 T cell repertoire during PI and susceptibility to superinfection with SHIV89.6P. All the animals that developed a restricted repertoire remained resistant to challenge with SHIV89.6P, while those that had a highly diverse repertoire were susceptible (Figure 3). The high diversity of the CD4 T cell repertoire in unprotected animals following challenge, suggests that the SIV-specific CD4 T cells stimulated during PI were not able to survive or to clonally expand. This could be due to the specific destruction of activated CD4 T cells during early primary SIV-infection as demonstrated in humans (Mattapallil, Douek et al., 2005). Our results suggest that the ability to develop clonally expanded CD4 T cells during PI, a combination of both activation/proliferation of the CD4 T cell clones and death/anergy/inhibition of proliferation due to viral infection, is a major component of subsequent disease development or resistance to super-infection.
However, SIVmacC8 infected macaques susceptible to SHIV89.6P superinfection did not experience the same rate of disease progression as did control animals following SHIV89.6P infection. This suggests that in unprotected SIVmacC8-infected animals, other factors may be involved in preventing a rapid disease progression, which is usually caused by virulent strains such as SHIV89.6P. Following SHIV89.6P infection, the pool of potentially infectable CD4 T cells is less important in SIVmacC8-infected animals than they are in mock infected controls, which would cause a higher SHIV89.6P replication potential in the latter and as a consequence, a poor disease outcome. Moreover, infection with SIVmacC8 may have induced CD8 specific responses that might play a role in decreasing suceptibility to superinfection compared to naïve animals. Although unprotected animals do not succumb to disease as rapidly as mock infected controls, one of the two unprotected macaques (C93054F) died by week 105-post challenge and the other C94042F had two peaks in viremia following challenge and showed a poor prognosis at later time points. Interestingly, while the 12 bp deletion was maintained in macaque C94042F, the nef gene in macaque C93054F experienced a recombination between the nef of SHIV89.6P and SIVmacC8 (Rud et al. unpublished observations). This recombination was due to superinfection and not a self repair of SIVmacC8 as observed by Whatmore et al. (Whatmore, Cook et al., 1995) and resulted in a repair of the 12 bp deletion which most likely led to the rapid CD4 T-cell loss.
The association between highly diverse CD4 T cell repertoires and susceptibility to super-infection is contrasted to what was described in the CD8 compartment by Price et al. (Price, West et al., 2004). While a polyclonal SIV-specific CD8 T cell repertoire characterized by highly conserved CDR3 motifs was associated with mutation in the viral epitope, a diverse CDR3 usage in the clonotypic population of SIV-specific CD8 response was not associated with viral escape mutants. It is important to point out that the major difference between CD4 and CD8 T cell responses is the fine balance between activation of functional SIV-specific CD4 T cell clones and the expansion of putative SIV targets, which is only observed in the CD4 compartment of SIV infected animals. Therefore, the association between highly diverse CD4 repertoires and superinfection by SHIV89.6P, could be explained by the fact that these animals have a larger pool of naïve CD4 T cells due to the lack of SIV-specific CD4 responses that can be a target for SHIV89.6P. These reservoirs can then facilitate even further SIV replication and ultimately accelerating disease progression (Pierson, McArthur et al., 2000). Ideally, the narrowing of the CD4+ T cell repertoire following SIV infection through the presence of few dominant SIV-specific CD4 T cell clones would minimize viral output and re-infection by having fewer targets available. Altogether, the association between resistance to superinfection with SHIV89.6P and the restriction and maturation of CD4 TRBV repertoires in cynomolgus macaques following SIV infection, underlines the importance of narrowing the CD4 TRBV repertoire early in infection: First, to build the anti-viral immune response against SIV, and second, to limit the pool of CD4 infectable targets.
Materials and Methods
Viruses
Viral isolates include: SIVmacJ5 and SIVmacC8 originating from SIVmac32H(pJ5) and SIVmac32H(pC8) molecular clones respectively and SHIV89.6P, a pathogenic chimeric SHIV (Reimann, Li et al., 1996). The molecular clones, SIVmac32H(pJ5) and SIVmac32H(pC8), were isolated from cells infected with the 11/88 pool of SIVmac32H, an isolate derived by in vivo passage of SIVmac251 in rhesus macaque 32H (Cranage, Stott et al., 1992). The attenuated SIVmacC8 virus clone differs from the wild-type SIVmacJ5 clone by a 12-bp deletion and two non synonymous nucleotide changes, resulting in conservative amino acid changes in the nef open reading frame (Rud, Cranage et al., 1994). In contrast to SIVmacJ5, which induces an AIDS-like pathology in infected macaques, SIVmacC8 shows an attenuated phenotype in monkeys ((Cranage, Stott et al., 1992) and G. Hall, personal communication). SHIV89.6P infection results in a rapid decline in CD4+ T cell counts in rhesus and cynomolgus macaques within a couple of months of infection (Reimann, Li et al., 1996). The SIVmacJ5- and SIVmacC8-infected animals along with a group of four naïve macaques (mock infected) were challenged with SHIV89.6P (a gift from Dr Keith Reimann) at week 20.
Animals
Animals used in this study were part of a large European concerted action on AIDS vaccine (Vogel, Fournier et al., 1998) and Rud, unpublished data). Three groups of four juvenile (approx. 3 year old) cynomolgus macaques (Macaca fascicularis) (these cynomolgus macaques are from a breeding colony derived from the Philippines in 1983 and not the commercially available cynomolgus macaques from Mauritius) were infected via the intravenous route. All animals used in this study were colony bred within the Non-human Primate Breeding Colony of Health Canada under Canadian Council of Animal Care approved conditions. All animals were serologically negative for Herpes B virus, STLV-1, SRV-1, 2, 5 and SIV (The Virus Reference Laboratory, Inc., San Antonio, TX). The percentage of CD4-positive T-lymphocytes was determined using a FACScan® (Becton Dickinson, CA, USA) flow cytometer and CELLQuest software. Whole blood collected in EDTA was analyzed for lymphocyte subsets by incubation with anti-human CD2 (Fluorescein Isothiocyanate (FITC)-labeled, Becton Dickinson), antihuman CD4 (Phycoerythrin (PE)-labeled, Becton Dickinson). White blood cell counts were obtained from a hematology workstation (Coulter Counter S-PLUS IV, FL, USA) and were used to calculate the lymphocyte subset absolute counts.
Plasma viral Load
Plasma SIV RNA concentrations were determined using a branched DNA signal amplification method developed by Dr. Peter Dailey (Chiron Corporation, Emeryville, CA) for SIV using the same approach as Quantiplex HIV RNA branched DNA method (Pachl, Todd et al., 1995). For quantitating SIV viral load, target probes designed to hybridize with the pol region of the SIVmac strains were used. In each sample, SIV RNA concentration was quantified by comparison with purified and in vitro-transcribed SIV pol RNA. The lower limit of detection of this assay was 15000 copies of SIVmac RNA per ml plasma in all the animals, at the initiation of the study but was further improved to detect down to 500 copies at 35 weeks.
CD4 T cell purification
Macaque PBMCs (10×106 cells) collected at described time points were used to isolate CD4 T cells by staining with microbeads-conjugated CD4 antibodies (Miltenyi Biotech) and subjecting them to MACS MS columns and MACS magnetic cell sorting apparatus (Miltenyi Biotech) as recommended by the manufacturer. Purified CD4+ T cells were then washed in PBS and resuspended into Trizol® (Life technologies, Burlington, Canada) for RNA extraction. CD4 T cell purity (>90%) was verified for each sample by staining with PE- conjugated anti-CD4 and PERCP- conjugated anti-CD8 antibodies (Becton Dickinson, CA, USA) and analyzed on a FACScan® using the Cell Quest® Software.
Heteroduplex tracking assay
Heteroduplex tracking assays (HTA) were performed as described (Shen, Doukhan et al., 1998) for all SIV infected and two uninfected macaques in this study at the bleeding time points (−3, 0, 1, 2, 4, 6, 8, 12, 16, 20, 21, 22 and 24 weeks post infection with SIVmacJ5 or SIVmacC8). Briefly, total RNA was purified from CD4 T cells as mentioned above using Trizol® and was used to synthesize complementary DNA (cDNA) using oligo dT (12–18) primers. Nested polymerase chain reaction (nPCR) was performed on cDNA samples from sequential time points using TRBVout and Cβout as outer primers and TRBVin and Cβin as inner primers for TRBV9, 20, 28, 5–1, 7, 12, 3 (Chen, Kou et al., 1993). These TRBV families were chosen because they were found to be the most dominant TRBV families in cynomolgus macaques after analysis of the whole TCR repertoire in four healthy cynomolgus macaques using the same nPCR strategy (data not shown). The uniformity of yield and amplification efficacy of each primer pair was assessed by agarose gel electrophoresis. In parallel, 32−P labeled probes were produced by PCR amplification of two TRBV-Cβ inserts previously cloned in Bluescript (Stratagene Cloning Systems, La Jolla, Ca), using 32−P-labeled Cβin primer and unlabeled TRBVin primers. Radiolabeled DNA heteroduplexes were obtained by mixing 4.5 µl from second round PCR with 0.5 µl of probe in annealing buffer (0.16 M NaCl, 16mM Tris [pH 8.0], 3.3 mM EDTA). DNA was denatured at 95° C for 2 min, after which they were reannealed at 55°C for 5 min and rapidly cooled to room temperature in a thermocycler. Heteroduplexes were resolved on a 20-cm high, 5% polyacrylamide gel (29:1 acrylamide:bisacrylamide) at 200V for 3h. HTA gels were vacuum-dried and a PhosphorImager (Amersham Pharmacia Biotech, NJ, USA) plate was used for signal detection. The clonality of each TRBV family in each sample was analyzed using the NIH image software as shown in figure 2A. A TRBV repertoire composed of 13 clones or more was considered as highly diverse, whereas 12 clones or less was considered as not diverse (restricted). Moreover, within restricted repertoires, we distinguished between populations composed of less than 7 clones (low diversity) and those containing 7 to 12 clones (intermediate diversity).
Molecular cloning and sequencing of the CDR3 regions
Amplification products were cloned into Bluescript vector and transformed into E.coli. (DH5α). Plasmids containing the cloned TCR inserts were sequenced using T7 DNA polymerase (Amersham Pharmacia Biotech, NJ, USA) or automated sequencing (Sheldon Biotechnology Center, McGill University, Montreal).
Acknowledgements
This work was supported by grants awarded to R.-P.S. from the US National Institutes of Health, the Canadian Institutes of Health Research, the Canadian Network for Vaccine and Immunotherapeutics, Genome Québec, Genome Canada and the Réseau SIDA FRSQ and to R.H from the Canadian Institutes of Health Research. R.-P.S. is the Canada Research Chair in Human Immunology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Ahmed R, Butler LD, Bhatti L. T4+ T helper cell function in vivo: differential requirement for induction of antiviral cytotoxic T-cell and antibody responses. J.Virol. 1988;62:2102–2106. doi: 10.1128/jvi.62.6.2102-2106.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badovinac VP, Porter BB, Harty JT. Programmed contraction of CD8(+) T cells after infection. Nat.Immunol. 2002;3:619–626. doi: 10.1038/ni804. [DOI] [PubMed] [Google Scholar]
- Bitmansour AD, Waldrop SL, Pitcher CJ, Khatamzas E, Kern F, Maino VC, Picker LJ. Clonotypic structure of the human CD4+ memory T cell response to cytomegalovirus. J.Immunol. 2001;167:1151–1163. doi: 10.4049/jimmunol.167.3.1151. [DOI] [PubMed] [Google Scholar]
- Blattman JN, Sourdive DJ, Murali-Krishna K, Ahmed R, Altman JD. Evolution of the T cell repertoire during primary, memory, and recall responses to viral infection. J.Immunol. 2000;165:6081–6090. doi: 10.4049/jimmunol.165.11.6081. [DOI] [PubMed] [Google Scholar]
- Chen ZW, Kou ZC, Shen L, Reimann KA, Letvin NL. Conserved T-cell receptor repertoire in simian immunodeficiency virus-infected rhesus monkeys. J.Immunol. 1993;151:2177–2187. [PubMed] [Google Scholar]
- Chen ZW, Shen Y, Kou Z, Ibegbu C, Zhou D, Shen L, Morrison P, Bogle C, McClure HM, Nahmias AJ, Sehgal PK, Letvin NL. Prolonged dominance of clonally restricted CD4(+) T cells in macaques infected with simian immunodeficiency viruses. J.Virol. 2000;74:7442–7450. doi: 10.1128/jvi.74.16.7442-7450.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole KS, Steckbeck JD, Rowles JL, Desrosiers RC, Montelaro RC. Removal of N-linked glycosylation sites in the V1 region of simian immunodeficiency virus gp120 results in redirection of B-cell responses to V3. J.Virol. 2004;78:1525–1539. doi: 10.1128/JVI.78.3.1525-1539.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranage M, Stott J, Mills K, Ashworth T, Taffs F, Farrar G, Chan L, Dennis M, Putkonen P, Biberfeld G. Vaccine studies with the 32H reisolate of SIVmac251: an overview. AIDS Res.Hum.Retroviruses. 1992;8:1479–1481. doi: 10.1089/aid.1992.8.1479. [DOI] [PubMed] [Google Scholar]
- Davenport MP, Price DA, McMichael AJ. The T cell repertoire in infection and vaccination: implications for control of persistent viruses. Curr.Opin.Immunol. 2007;19:294–300. doi: 10.1016/j.coi.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Dittmer U, Brooks DM, Hasenkrug KJ. Requirement for multiple lymphocyte subsets in protection by a live attenuated vaccine against retroviral infection. Nat.Med. 1999;5:189–193. doi: 10.1038/5550. [DOI] [PubMed] [Google Scholar]
- Fasso M, Anandasabapathy N, Crawford F, Kappler J, Fathman CG, Ridgway WM. T cell receptor (TCR)-mediated repertoire selection and loss of TCR vbeta diversity during the initiation of a CD4(+) T cell response in vivo. J.Exp.Med. 2000;192:1719–1730. doi: 10.1084/jem.192.12.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geginat J, Lanzavecchia A, Sallusto F. Proliferation and differentiation potential of human CD8+ memory T-cell subsets in response to antigen or homeostatic cytokines. Blood. 2003;101:4260–4266. doi: 10.1182/blood-2002-11-3577. [DOI] [PubMed] [Google Scholar]
- Gupta S, Gollapudi S. Effector memory CD8+ T cells are resistant to apoptosis. Ann.N.Y.Acad.Sci. 2007;1109:145–150. doi: 10.1196/annals.1398.017. [DOI] [PubMed] [Google Scholar]
- Heyzer-Williams LJ, Panus JF, Mikszta JA, Heyzer-Williams MG. Evolution of antigen-specific T cell receptors in vivo: preimmune and antigen-driven selection of preferred complementarity-determining region 3 (CDR3) motifs. J.Exp.Med. 1999;189:1823–1838. doi: 10.1084/jem.189.11.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyzer-Williams MG, Altman JD, Davis MM. Enumeration and characterization of memory cells in the TH compartment. Immunol.Rev. 1996a;150:5–21. doi: 10.1111/j.1600-065x.1996.tb00693.x. [DOI] [PubMed] [Google Scholar]
- Heyzer-Williams MG, Altman JD, Davis MM. Tracking antigen-specific helper T cell responses. Curr.Opin.Immunol. 1996b;8:278–284. doi: 10.1016/s0952-7915(96)80068-9. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Desrosiers RC. Protective immunity induced by live attenuated simian immunodeficiency virus. Curr.Opin.Immunol. 1998;10:436–443. doi: 10.1016/s0952-7915(98)80118-0. [DOI] [PubMed] [Google Scholar]
- Johnson RP, Lifson JD, Czajak SC, Cole KS, Manson KH, Glickman R, Yang J, Montefiori DC, Montelaro R, Wyand MS, Desrosiers RC. Highly attenuated vaccine strains of simian immunodeficiency virus protect against vaginal challenge: inverse relationship of degree of protection with level of attenuation. J.Virol. 1999;73:4952–4961. doi: 10.1128/jvi.73.6.4952-4961.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalams SA, Buchbinder SP, Rosenberg ES, Billingsley JM, Colbert DS, Jones NG, Shea AK, Trocha AK, Walker BD. Association between virus-specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J.Virol. 1999;73:6715–6720. doi: 10.1128/jvi.73.8.6715-6720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson GB, Halloran M, Li J, Park IW, Gomila R, Reimann KA, Axthelm MK, Iliff SA, Letvin NL, Sodroski J. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J.Virol. 1997;71:4218–4225. doi: 10.1128/jvi.71.6.4218-4225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff WC, Johnson PR, Watkins DI, Burton DR, Lifson JD, Hasenkrug KJ, McDermott AB, Schultz A, Zamb TJ, Boyle R, Desrosiers RC. HIV vaccine design: insights from live attenuated SIV vaccines. Nat.Immunol. 2006;7:19–23. doi: 10.1038/ni1296. [DOI] [PubMed] [Google Scholar]
- Kuroda MJ, Schmitz JE, Charini WA, Nickerson CE, Lifton MA, Lord CI, Forman MA, Letvin NL. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J.Immunol. 1999;162:5127–5133. [PubMed] [Google Scholar]
- Levitsky V, Campos-Lima PO, Frisan T, Masucci MG. The clonal composition of a peptide-specific oligoclonal CTL repertoire selected in response to persistent EBV infection is stable over time. J.Immunol. 1998;161:594–601. [PubMed] [Google Scholar]
- Li Q, Duan L, Estes JD, Ma ZM, Rourke T, Wang Y, Reilly C, Carlis J, Miller CJ, Haase AT. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature. 2005;434:1148–1152. doi: 10.1038/nature03513. [DOI] [PubMed] [Google Scholar]
- Maryanski JL, Jongeneel CV, Bucher P, Casanova JL, Walker PR. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: a high magnitude CD8 response is comprised of very few clones. Immunity. 1996;4:47–55. doi: 10.1016/s1074-7613(00)80297-6. [DOI] [PubMed] [Google Scholar]
- Matloubian M, Concepcion RJ, Ahmed R. CD4+ T cells are required to sustain CD8+ cytotoxic T-cell responses during chronic viral infection. J.Virol. 1994;68:8056–8063. doi: 10.1128/jvi.68.12.8056-8063.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattapallil JJ, Douek DC, Hill B, Nishimura Y, Martin M, Roederer M. Massive infection and loss of memory CD4+ T cells in multiple tissues during acute SIV infection. Nature. 2005;434:1093–1097. doi: 10.1038/nature03501. [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams MG, Davis MM. Antigen-specific development of primary and memory T cells in vivo. Science. 1995;268:106–111. doi: 10.1126/science.7535476. [DOI] [PubMed] [Google Scholar]
- Mori K, Yasutomi Y, Ohgimoto S, Nakasone T, Takamura S, Shioda T, Nagai Y. Quintuple deglycosylation mutant of simian immunodeficiency virus SIVmac239 in rhesus macaques: robust primary replication, tightly contained chronic infection, and elicitation of potent immunity against the parental wild-type strain. J.Virol. 2001;75:4023–4028. doi: 10.1128/JVI.75.9.4023-4028.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachl C, Todd JA, Kern DG, Sheridan PJ, Fong SJ, Stempien M, Hoo B, Besemer D, Yeghiazarian T, Irvine B. Rapid and precise quantification of HIV-1 RNA in plasma using a branched DNA signal amplification assay. J.Acquir.Immune.Defic.Syndr.Hum.Retrovirol. 1995;8:446–454. doi: 10.1097/00042560-199504120-00003. [DOI] [PubMed] [Google Scholar]
- Picker LJ, Watkins DI. HIV pathogenesis: the first cut is the deepest. Nat.Immunol. 2005;6:430–432. doi: 10.1038/ni0505-430. [DOI] [PubMed] [Google Scholar]
- Pierson T, McArthur J, Siliciano RF. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu.Rev.Immunol. 2000;18:665–708. doi: 10.1146/annurev.immunol.18.1.665. [DOI] [PubMed] [Google Scholar]
- Price DA, West SM, Betts MR, Ruff LE, Brenchley JM, Ambrozak DR, Edghill-Smith Y, Kuroda MJ, Bogdan D, Kunstman K, Letvin NL, Franchini G, Wolinsky SM, Koup RA, Douek DC. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21:793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- Purtha WE, Myers N, Mitaksov V, Sitati E, Connolly J, Fremont DH, Hansen TH, Diamond MS. Antigen-specific cytotoxic T lymphocytes protect against lethal West Nile virus encephalitis. Eur.J.Immunol. 2007;37:1845–1854. doi: 10.1002/eji.200737192. [DOI] [PubMed] [Google Scholar]
- Reimann KA, Li JT, Veazey R, Halloran M, Park IW, Karlsson GB, Sodroski J, Letvin NL. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J.Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro RM. Dynamics of CD4+ T cells in HIV-1 infection. Immunol.Cell Biol. 2007;85:287–294. doi: 10.1038/sj.icb.7100056. [DOI] [PubMed] [Google Scholar]
- Roberts AD, Woodland DL. Cutting edge: effector memory CD8+ T cells play a prominent role in recall responses to secondary viral infection in the lung. J.Immunol. 2004;172:6533–6537. doi: 10.4049/jimmunol.172.11.6533. [DOI] [PubMed] [Google Scholar]
- Rud EW, Cranage M, Yon J, Quirk J, Ogilvie L, Cook N, Webster S, Dennis M, Clarke BE. Molecular and biological characterization of simian immunodeficiency virus macaque strain 32H proviral clones containing nef size variants. J.Gen.Virol. 1994;75(Pt 3):529–543. doi: 10.1099/0022-1317-75-3-529. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu.Rev.Immunol. 2004;22:745–763. doi: 10.1146/annurev.immunol.22.012703.104702. [DOI] [PubMed] [Google Scholar]
- seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat.Immunol. 2003;4:835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- Sharpe SA, Cope A, Dowall S, Berry N, Ham C, Heeney JL, Hopkins D, Easterbrook L, Dennis M, Almond N, Cranage M. Macaques infected long-term with attenuated simian immunodeficiency virus (SIVmac) remain resistant to wild-type challenge, despite declining cytotoxic T lymphocyte responses to an immunodominant epitope. J.Gen.Virol. 2004;85:2591–2602. doi: 10.1099/vir.0.80050-0. [DOI] [PubMed] [Google Scholar]
- Shen DF, Doukhan L, Kalams S, Delwart E. High-resolution analysis of T-cell receptor beta-chain repertoires using DNA heteroduplex tracking: generally stable, clonal CD8+ expansions in all healthy young adults. J.Immunol.Methods. 1998;215:113–121. doi: 10.1016/s0022-1759(98)00066-0. [DOI] [PubMed] [Google Scholar]
- Shin H, wherry EJ. CD8 T cell dysfunction during chronic viral infection. Curr.Opin.Immunol. 2007;19:408–415. doi: 10.1016/j.coi.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Sourdive DJ, Murali-Krishna K, Altman JD, Zajac AJ, Whitmire JK, Pannetier C, Kourilsky P, Evavold B, Sette A, Ahmed R. Conserved T cell receptor repertoire in primary and memory CD8 T cell responses to an acute viral infection. J.Exp.Med. 1998;188:71–82. doi: 10.1084/jem.188.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbings R, Berry N, Waldmann H, Bird P, Hale G, Stott J, North D, Hull R, Hall J, Lines J, Brown S, D'Arcy N, Davis L, Elsley W, Edwards C, Ferguson D, Allen J, Almond N. CD8+ lymphocytes do not mediate protection against acute superinfection 20 days after vaccination with a live attenuated simian immunodeficiency virus. J.Virol. 2005;79:12264–12272. doi: 10.1128/JVI.79.19.12264-12272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–342. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JC, Williams MA, Bevan MJ. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat.Immunol. 2004;5:927–933. doi: 10.1038/ni1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvas S, Kumaraguru U, Pack CD, Lee S, Rouse BT. CD4+ CD25+ T cells regulate virus-specific primary and memory CD8+ T cell responses. J.Exp.Med. 2003;198:889–901. doi: 10.1084/jem.20030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenner-Racz K, Stahl HC, Uberla K, Stoiber H, Ignatius R, Heeney J, Steinman RM, Racz P. Early protection against pathogenic virus infection at a mucosal challenge site after vaccination with attenuated simian immunodeficiency virus. Proc.Natl.Acad.Sci.U.S.A. 2004;101:3017–3022. doi: 10.1073/pnas.0308677101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel TU, Fournier J, Sherring A, Ko D, Parenteau M, Bogdanovic D, Mihowich J, Rud EW. Presence of circulating CTL induced by infection with wild-type or attenuated SIV and their correlation with protection from pathogenic SHIV challenge. J.Med.Primatol. 1998;27:65–72. doi: 10.1111/j.1600-0684.1998.tb00228.x. [DOI] [PubMed] [Google Scholar]
- von Herrath MG, Yokoyama M, Dockter J, Oldstone MB, Whitton JL. CD4-deficient mice have reduced levels of memory cytotoxic T lymphocytes after immunization and show diminished resistance to subsequent virus challenge. J.Virol. 1996;70:1072–1079. doi: 10.1128/jvi.70.2.1072-1079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker PR, Wilson A, Bucher P, Maryanski JL. Memory TCR repertoires analyzed long-term reflect those selected during the primary response. Int.Immunol. 1996;8:1131–1138. doi: 10.1093/intimm/8.7.1131. [DOI] [PubMed] [Google Scholar]
- Whatmore AM, Cook N, Hall GA, Sharpe S, Rud EW, Cranage MP. Repair and evolution of nef in vivo modulates simian immunodeficiency virus virulence. J.Virol. 1995;69:5117–5123. doi: 10.1128/jvi.69.8.5117-5123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz D, Jansen VA. The role of T cell help for anti-viral CTL responses. J.Theor.Biol. 2001;211:419–432. doi: 10.1006/jtbi.2001.2358. [DOI] [PubMed] [Google Scholar]
- Zajac AJ, Blattman JN, Murali-Krishna K, Sourdive DJ, Suresh M, Altman JD, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J.Exp.Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaph C, Uzonna J, Beverley SM, Scott P. Central memory T cells mediate long-term immunity to Leishmania major in the absence of persistent parasites. Nat.Med. 2004;10:1104–1110. doi: 10.1038/nm1108. [DOI] [PubMed] [Google Scholar]