Figure 2. TRBV5-1 CD4+ families in uninfected, SIVmacJ5 and SIVmacC8 infected macaques.
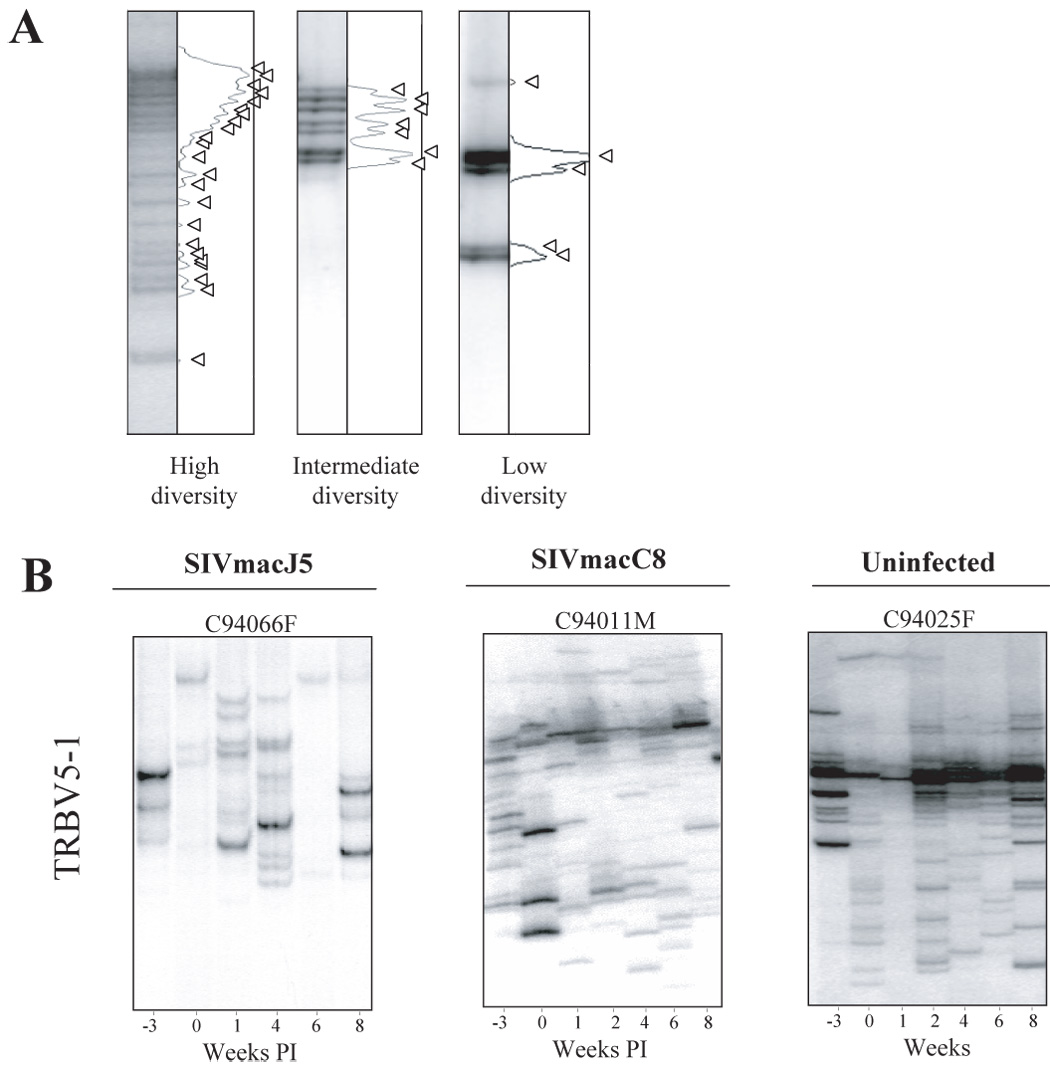
A. Computerized densitometric analysis of the HTA autoradiography. Each band on the HTA gel is transformed into a peak using the NIH image software. Each band represents a CD4 clonotype. An HTA profile is highly diverse when it is composed of more than 13 clones and with low diversity when it is composed of or less than 12 clones. Arrows indicate discrete TRBV hetero-complexes over the HTA profiles. B. Heteroduplex tracking assay using TRBV5-1 as 5’-primers and BC as 3’-primers was performed on purified CD4+ T cells collected from −3 weeks prior to SIV infection until week 8 post infection. Data from representative SIVmacJ5, SIVmacC8, and mock infected macaques is shown. SIVmacJ5 and SIVmacC8 include macaques with low and high diversity CD4 repertoires, whereas uninfected macaques have always highly diverse repertoires.
