Abstract
Yapsin 1 is an aspartic protease from Saccharomyces cerevisiae and belongs to a class of aspartic proteases that demonstrate specificity for basic amino acids. It is capable of processing prohormone substrates at specific basic residue cleavage sites, similar to that of the prohormone convertases, to generate bioactive peptide hormones. An antibody raised against yapsin 1 was previously shown to immuno-stain endocrine cells of rat pituitary and brain as well as lysates from bovine pituitary secretory granules demonstrating the existence of yapsin 1-like aspartic proteases in mammalian endocrine tissues, potentially involved in peptide hormone production. Here we show the specific staining of yapsin 1 immunoreactivity in the alpha cells of human pancreatic islets. No staining was observed in the beta or delta cells indicating a specificity of the staining for glucagon producing and not insulin or somatostatin producing cells. Purified yapsin 1 was also shown to process proglucagon into authentic glucagon in vitro demonstrating that the prototypical enzyme of this sub-class of enzymes can correctly process proglucagon to glucagon. These findings suggest the existence of a yapsin 1-like enzyme exclusively in the alpha cells of the islets of Langerhans in humans and may play a role in the production of glucagon in that tissue.
Keywords: Aspartic protease, proglucagon processing, human Islets, yapsin antibody
Introduction
Aspartic proteases represent a class of proteases that utilize two catalytic triads composed of Asp-Thr-Gly as the functional component of their active site for catalysis. X-ray crystallography and specificity studies revealed that their active sites are generally positioned to accept hydrophobic amino acids; hence, the cleavage specificity of aspartic proteases was thought to be predominantly for bulky hydrophobic residues within proteins (Tang 1963), although there are examples of fungal aspartic proteases that can cleave other residues. Coupled with the presence of chymosin (Foltmann 1992), and pepsins (Ryle and Porter 1959) in the stomach to aid digestion and of cathepsin D in the lysosome to aid protein degradation (Erickson and Blobel 1979), it was generally believed that aspartic proteases fell within a group classified as “non-specific” degradation enzymes. This is not the case however for all aspartic proteases, as evident from the specific processing roles that aspartic proteases play; for example memapsin 2 (BACE) (Lin, et al. 2000; Vassar, et al. 1999) and γ-secretase (Wolfe, et al. 1999) in the pathogenesis of Alzheimer’s disease, or of the role of renin in the generation of angiotensin I (Fukamizu and Murakami 1995) and its function in the regulation of blood pressure. The yapsins (Cawley and Loh 2011) may also represent an exception to this classification.
The yapsin family of aspartic proteases includes yapsin 1–3 and yapsin 6 and 7 from Saccharomyces cerevisiae while other members have been identified in Candida albicans (SAP8 and 9, (Monod, et al. 1998)), Saccharomyces pombe (Sp Yps1, (Ladds and Davey 2000)), Candida glabrata (Cg YPS1, (Dujon, et al. 2004)) and Aspergillus oryzae (opsB, (Kunihiro, et al. 2002)). Yapsin 1 is involved in the maintenance of cell wall integrity through incorporation and/or retention of glucan (Krysan, et al. 2005), processing of mucins that activate the MAP kinase signaling pathway (Vadaie, et al. 2008) and cleavage of GPI-anchored proteins which are shed into the growth medium (Gagnon-Arsenault, et al. 2008). Unique to the yapsins are their ability to cleave proteins at basic amino acids as opposed to the generally preferred hydrophobic amino acids of other aspartic proteases. Yapsin 1 has been shown to be able to cleave several prohormones including pro-opiomelanocortin (POMC), proinsulin and adrenocorticotropin (ACTH) (Cawley, et al. 1996a) with catalytic efficiencies increasing with increasing basic residues around the scissile bond (Olsen, et al. 1998). In yeast, yapsin 1 was cloned based on its ability to correctly process pro-α-mating factor at its basic residue cleavage sites (Egel-Mitani, et al. 1990). Since this process is usually performed by Kex2, a subtilisin-like serine protease, it was speculated that yapsins may be backup enzymes for the serine proteases involved in prohormone processing.
Mammalian aspartic proteases with similar properties to the yapsins have been characterized from bovine pituitary intermediate lobe secretory granules (Loh, et al. 1985) and bovine adrenal chromaffin granules (Azaryan, et al. 1995). Also, an anglerfish aspartic protease capable of processing prosomatostatin at a basic residue cleavage site has been described (Mackin, et al. 1991). Indeed, an antibody against yapsin 1 has been used to immunologically identify mammalian yapsin 1-like proteins in bovine and mouse endocrine and neuroendocrine tissue (Cawley, et al. 1996b). Here we extend those findings and show yapsin 1-like immunoreactivity exclusively in human pancreatic islet α-cells and that purified yapsin 1 can generate glucagon by processing proglucagon. These results suggest the presence of a yapsin 1-like endoprotease in human pancreatic α-cells with an ability to produce glucagon.
Material and Methods
Immunohistochemistry
Tissue specimens from six adult human pancreata were obtained from surgical samples removed at operation for pancreatic adenocarcinoma. The study was approved by the local ethics committee at Lisbon University Hospital. The patients had no evidence of any endocrine disease. The specimens were taken from macro- and microscopically normal glandular regions from the body-tail region at least 3 cm distant from the neoplasm. The mean size of the specimens was 1 × 2 cm. The specimens were routinely fixed in 10 % buffered neutral formalin for 18 to 20 h at room temperature (RT) and embedded in paraffin. Sections, 5 µm thick, were cut and attached to positively charged (Superfrost+; Menzel, Braunschweig, Germany) glass slides. Hematoxylin-eosin was used as a routine staining.
Single staining
Single immunofluorescence staining and the indirect two-step peroxidase labeled dextran-polymer technique (EnVision™, DakoCytomation, Glostrup, Denmark; (Sabattini, et al. 1998)) with diaminobenzidine as chromogen, using a Dako Autostainer (DakoCytomation), were performed to reveal the distribution pattern of the different endocrine cell types and yapsin 1-immunoreactivity (IR) in the pancreas, as well as to perform the control staining specified below. The sections were pre-treated in a microwave oven (Philips Whirlpool Nordic AB, Stockholm, Sweden) for 2×5 min at 750 W, using a citrate buffer, pH 6.0, as a retrieval solution.
Double staining
Co-localization studies were performed using immunofluorescence methods with the yapsin 1 antibody and antibodies to various islet hormones, without microwave pretreatment. The yapsin 1 immunofluorescence staining was enhanced by the catalyzed reporter deposition (CARD) method with biotinyl tyramide. For details of these methods, see Portela-Gomes et al. (2000) (Portela-Gomes, et al. 2000). The secondary antibodies were pre-incubated overnight at 4 °C with non-immune serum, both from the animal species recognized by the other secondary antibody and from the species producing that antibody, at a dilution of 1:10.
The control staining included omission of the primary antibody and replacement of the first layer of antibody by pre-immune or non-immune serum diluted 1:10 or by the diluent alone. To confirm the specificity of the yapsin 1 antibody, absorption tests were carried out by overnight incubation at 4 °C of 5 µl of primary antibody with 45 µl of culture media containing yapsin 1, before application to the sections. The culture media for the absorption control was obtained from a yeast over-expression system inducible by galactose, as described previously (Azaryan, et al. 1993; Cawley, et al. 1995). The other primary antibodies were preincubated with the relevant antigen (10 nM per ml diluted antibody solution, respectively) before application to the sections. The hormone antigens used were obtained from Sigma Chemical Co. (St. Louis, MO).
The yapsin 1 antibody (MW283) (Cawley et al. 1995) was used at 1:1600 for the dextran-polymer technique and 1:80 for immunofluorescence. The other primary antibodies were as follows: mouse monoclonal antibodies against human somatostatin (Novo Nordisk S/A, Bagsvaerd, Denmark; clone Som-018; 1:50), guinea pig antibodies against human insulin (P. Westermark, Dept. of Genetics and Pathology, Uppsala, Sweden; code #Ma37; 1:200); and chicken antibodies against human glucagon (1:800) and pancreatic polypeptide (1:100) (A. Larsson, Dept. of Medical Sciences, Clinical Chemistry; Uppsala, Sweden). The labeled secondary antisera were: biotinylated swine anti-rabbit IgG (DAKO; Glostrup, Denmark), Alexa Fluor 594-labeled streptavidin, Alexa Fluor 488-conjugated goat anti-mouse, anti-guinea pig, and anti-chicken IgG (Molecular Probes, Invitrogen Life Technologies Corporation, Carlsbad, CA) and biotinyl tyramide (TSA Biotin System; PerkinElmer, Waltham, MA). For co-localization studies, the sections were examined in a Zeiss META confocal microscope (Zeiss GmbH, Jena, Germany).
DNA constructs and transfection
The cDNA of full-length guinea pig proglucagon in the mammalian expression vector pcDNA 3.1(+) was a gift from Dr. S. Dhanvantari (Lawson Health Research Institute, Canada). The transfection of the plasmid was accomplished with program T-24 using the Nucleofector 1 device according to the manufacturer (Lonza Inc. Allendale, NJ).
Cell culture and proglucagon expression
Neuro2a (N2a) cells were transfected with proglucagon or empty vector plasmid and maintained in DMEM containing 10 % fetal bovine serum for 24 hours. The cells were then washed and incubated with serum free DMEM (SFM) for 4 hours. The conditioned media were collected and concentrated with Amicon Ultracel-3K centrifuge filter units following the protocol of the manufacturer (Millipore, Billerica, MA) and aliquots frozen at −20 °C until used.
In vitro processing
Conditioned media containing proglucagon (~1.3 µM) was incubated with and without purified yapsin 1 (3.7–6.2 µM), obtained in 1998 (Cawley, et al. 1998), in 50 mM sodium acetate, pH 5.5 supplemented with fresh 1X complete protease inhibitor cocktail (G-Biosciences, St. Louis, MO) and 50 µM AEBSF. The samples were incubated for 18 h at 37°C after which they were stopped by freezing on dry ice until analysis.
Analysis of proglucagon processing
Aliquots of the reactions were analyzed by Western blot using rabbit monoclonal antibody against glucagon (Abcam, Inc, Cambridge, MA,) and MW283, both at a dilution of 1:5000. Glucagon-IR was also determined in the samples using the glucagon Enzyme Immunoassay (EIA) (Phoenix Pharmaceuticals, Inc, Burlingame, CA) to determine levels of recovery.
Glucagon Analysis by HPLC and EIA
Forty µl of the samples were separated by HPLC on a 4.6 × 250 mm 5 µm reverse phase Jupiter C18 column (Phenomenex, Torrance, CA). Buffer A was 0.1 % TFA and Buffer B was 100 % acetonitrile/0.1 % TFA and the gradient was 20 to 60 % B in 30 min. Fractions #16–26 were collected, lyophilized, and reconstituted in EIA buffer and assayed by EIA for glucagon-like IR. Under these conditions, glucagon standard (Phoenix Pharmaceuticals, Inc.) eluted in fraction 19.
Results
Immunohistochemistry
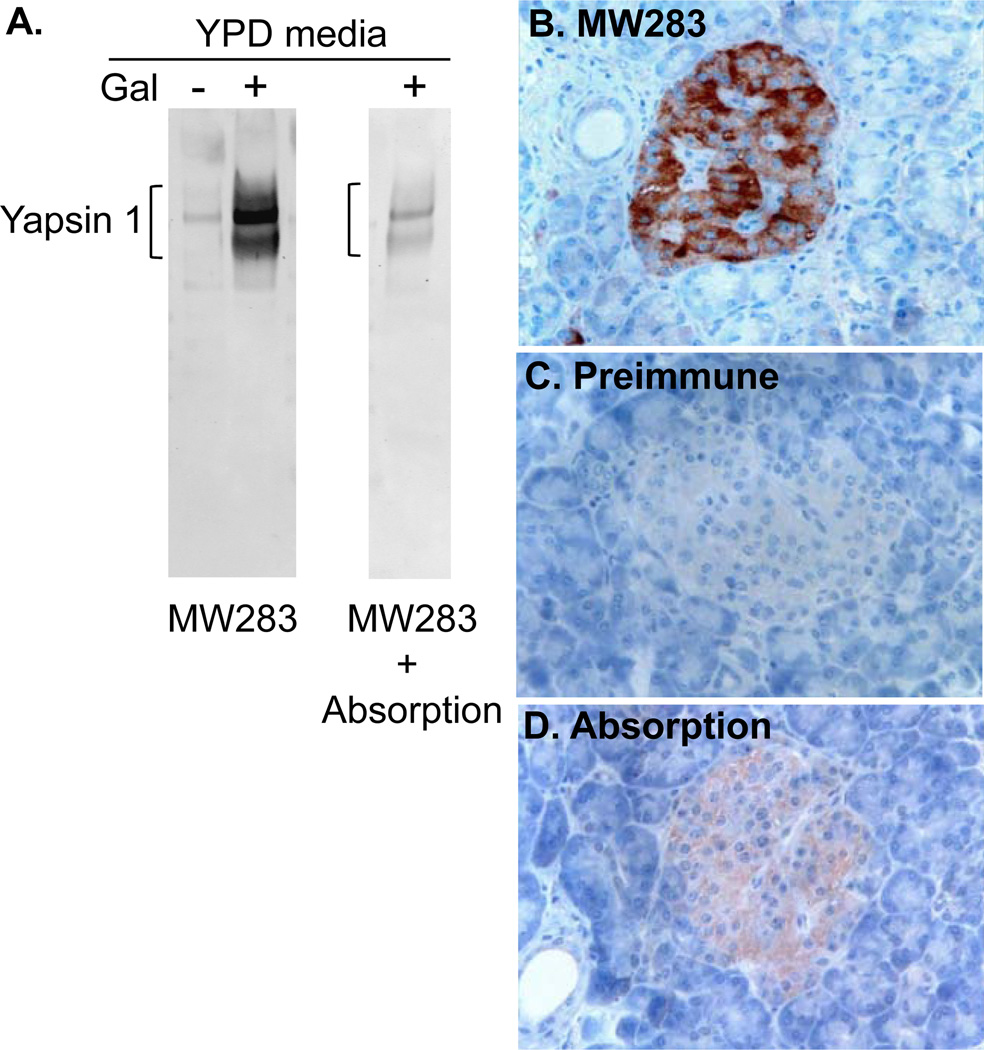
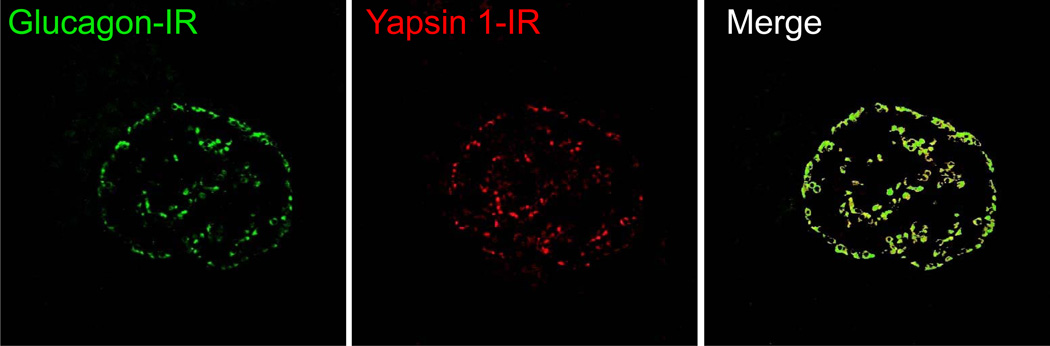
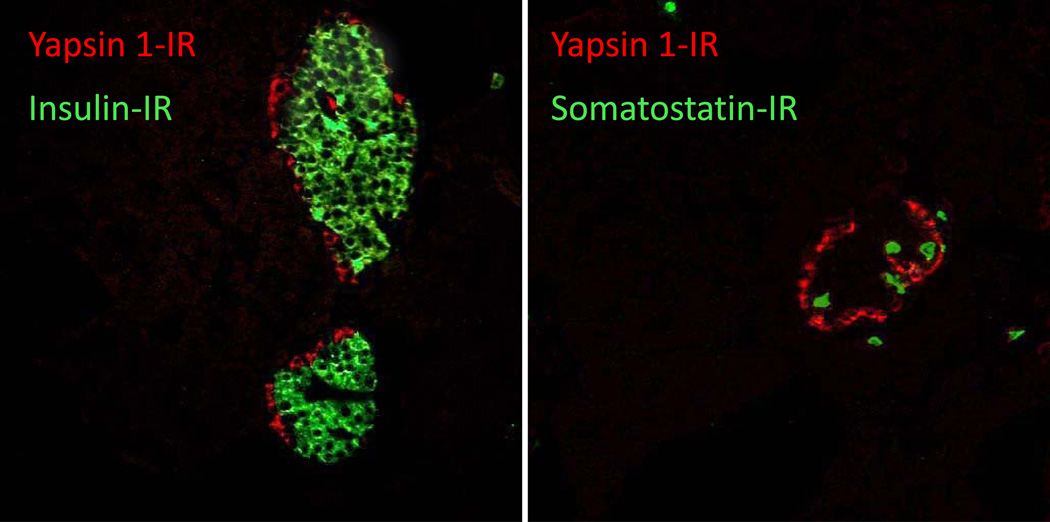
Distinct yapsin 1-immunoreactive cells were seen in the human pancreatic islets. The immunoreactivity was strong and diffusely distributed throughout the cytoplasm, involving cell processes when present. The cells belonging to the exocrine parenchyma did not show any immunoreactivity (Fig. 1B–D). Yapsin 1-IR was found in virtually all glucagon-immunoreactive cells and vice-versa (Fig. 2). The intracytoplasmic distributions of the yapsin 1-IR and glucagon immunoreactivities were in agreement, but the yapsin 1-IR often appeared weaker than that of glucagon. Yapsin 1-IR was not present in any of the insulin-, pancreatic polypeptide-(not shown) or somatostatin-immunoreactive cells (Fig. 3). In double immunofluorescence staining, our control tests showed that omission of one of the primary antibodies gave a staining pattern corresponding to that of the remaining primary antibody. No immunoreactivity was observed in islets stained with the yapsin 1 preimmune serum. The yapsin 1 antibody, preincubated with galactose-induced medium containing yapsin 1 gave rise to only weak immunoreactive cells, demonstrating the specific reduction of the yapsin 1-IR signal by the pre-absorbed antiserum (Fig. 1A).
Figure 1.
A. Western blot of uninduced (−) and galactose-induced (+) culture media for yapsin 1 stained with the yapsin 1 antibody MW283 alone or absorbed (+ Absorption) with galactose-induced culture medium. Note the reduction in yapsin 1 signal by > 80 % in the absorbed antibody sample. B–D. Normal human pancreatic islets immunostained with the indirect dextran-polymer technique (EnVision) with diaminobenzidine as chromogen using (B) MW283, (C) MW283 preimmune serum, (D) MW283 preincubated with galactose-induced culture medium containing yapsin 1. The slides were counterstained with Mayer’s hematoxylin. Note that yapsin 1-IR was seen in cells of the Islets using the immune serum that were not present when stained with pre-immune serum or where the immune serum was preabsorbed with yapsin 1 protein.
Figure 2.
Normal human pancreatic islet double immunostained for yapsin 1 (Alexa Fluor 594, red) and glucagon (Alexa Fluor 488, green). All glucagon α-cells show yellow to yellow-green color indicating co-localization of glucagon and yapsin 1-IR in human islets.
Figure 3.
Normal human pancreatic islet double immunostained for yapsin 1 (Alexa Fluor 594, red) and insulin (Alexa Fluor 488, green) (left panel) or somatostatin (Alexa Fluor 488, green) (right panel). Insulin (β) or somatostatin (δ) positive cells (green) do not co-stain with yapsin 1-IR (red) in human islets.
Production and processing of proglucagon
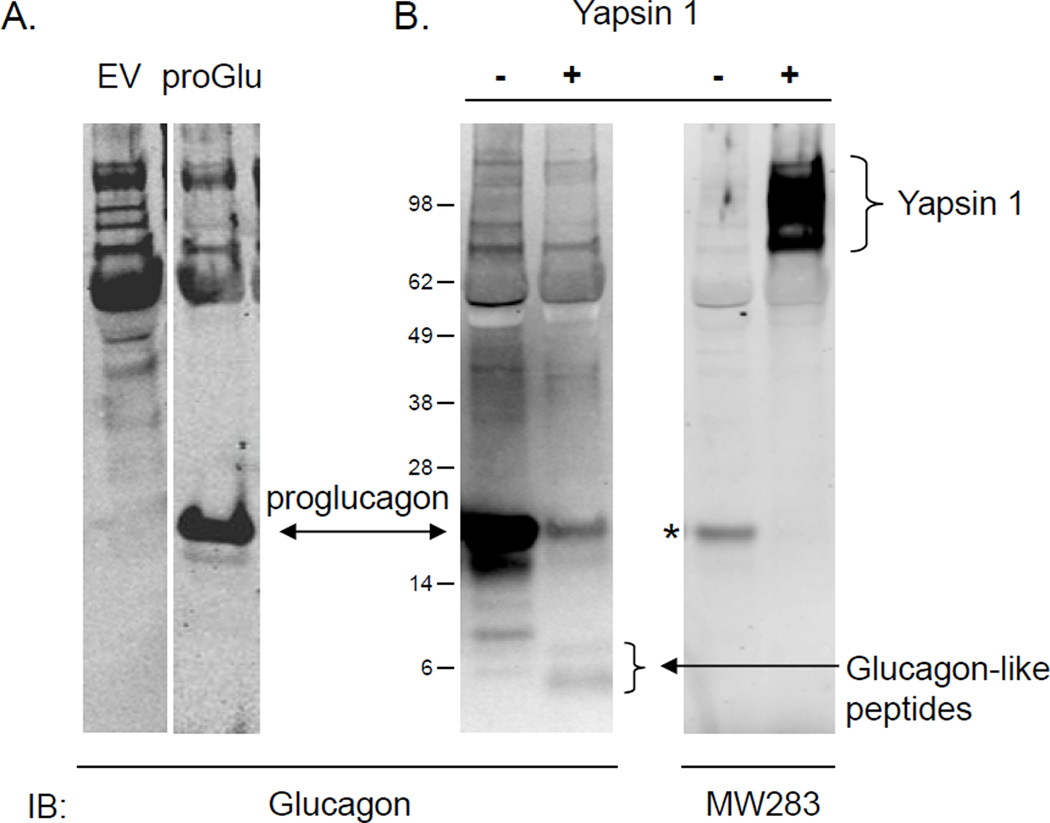
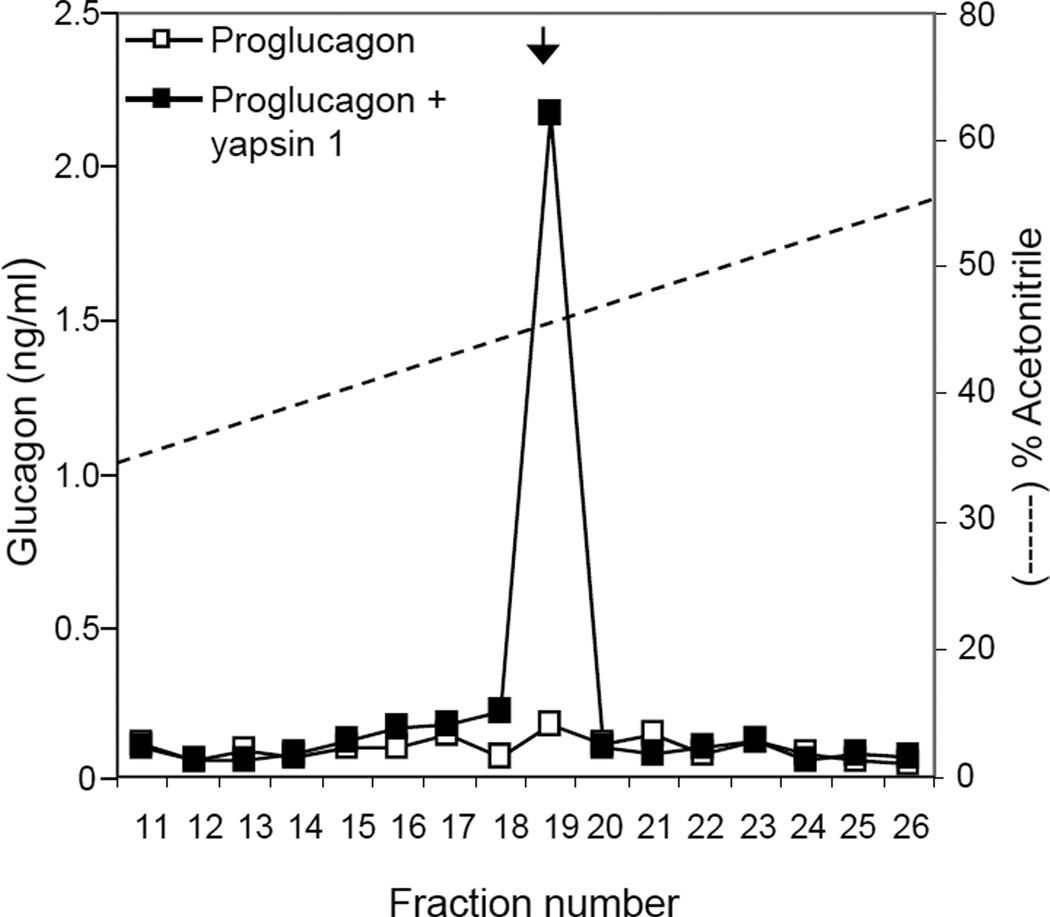
The presence of yapsin 1-IR in glucagon expressing cells prompted us to test if yapsin 1 could process proglucagon in vitro. Proglucagon was found in the culture media of N2a cells transfected with proglucagon cDNA and this was confirmed by Western blot (Fig. 4A). Whereas the N2a cells in general gave non-specific bands in the high molecular mass range, only the cells transfected with proglucagon-pcDNA3.1 showed a strong staining of glucagon-IR at the expected size of ~22 kDa for proglucagon (Fig. 4A). Incubation of the medium containing proglucagon with purified yapsin 1 caused the disappearance of proglucagon and the generation of smaller glucagon-IR products, some consistent with the size of glucagon (Fig. 4B). In this case, the amount of glucagon-IR in the reaction with and without yapsin 1 treatment was 32.1 ng/ml and 28.3 ng/ml, respectively, which was equivalent to the amount in the starting material. Analysis of the peptides generated by yapsin 1 by HPLC followed by EIA identified that authentic glucagon was produced in a yapsin 1 dependent manner (Fig. 5). Of the 1.28 ng of glucagon-IR injected, ~0.24 ng was recovered in fraction #19 representing a ~20 % recovery of authentic glucagon from the digestion of proglucagon by yapsin 1.
Figure 4.
In vitro processing of proglucagon by purified yapsin 1. A. Western blot for proglucagon in conditioned media collected from Neuro2a cells transfected with EV (left) or proglucagon-pcDNA3.1 (right). B. The medium was incubated with (+) or without (−) purified yapsin 1 enzyme and the products analyzed by Western blot. Analysis shows the disappearance of proglucagon and production of glucagon-sized immunoreactive bands (B, left blot, +) produced specifically in the yapsin 1 containing sample as demonstrated by Western blot for yapsin 1 on the same blot (B, right blot, +). * indicates a bleed-through signal from the proglucagon fluorescence channel on the Western blot documentation platform.
Figure 5.
HPLC analysis of glucagon generated by yapsin 1. Aliquots from the in vitro processing reactions were subjected to HPLC to separate the peptides on a reverse-phase C18 column. Enzyme-linked immunoassay of the fractions identified that authentic glucagon was recovered in fraction 19 only from the sample incubated with yapsin 1. The arrow indicates where authentic glucagon standard eluted (Phoenix Pharmaceuticals, Inc.).
Discussion
The cloning of yapsin 1, originally referred to as yap3 (Bourbonnais, et al. 1993; Egel-Mitani et al. 1990), as it was the third aspartic protease to be cloned from Saccharomyces cerevisiae, provided evidence of a new sub-class of aspartic proteases in that it was able to process a yeast prohormone, pro-α-mating factor, correctly at paired-basic residue cleavage sites. It did so in the absence of Kex2, a prohormone processing enzyme of the serine protease class, the first protease to be unequivocally identified both biochemically and genetically, as a prohormone processing enzyme (Julius, et al. 1984; Thomas, et al. 1988). The fact that yapsin 1 could perform this function also introduced the idea, at that time, that aspartic proteases may have more specific roles in endocrinology. Indeed, a mammalian aspartic protease was identified and characterized several years prior to the cloning of yapsin 1 (Loh et al. 1985). That enzyme, now termed yapsin A, to indicate it as the first mammalian yapsin-like enzyme, was shown to process prohormones such as pro-opiomelanocortin (Loh et al. 1985), provasopressin (Loh, et al. 1988; Parish, et al. 1986) and proinsulin (Loh et al. 1985). In addition, its unique high molecular mass relative to other aspartic proteases, of ~68–70 kDa is similar to the mass for deglycosylated yapsin 1 (~65 kDa) (Cawley et al. 1995). In almost all aspects, yapsin A appears to be a homologue of yapsin 1, even immunologically. An antibody was generated against yapsin 1 and used to analyze mammalian tissue by Western blot and immunohistochemistry (Cawley et al. 1996b). Staining of yapsin 1-IR was seen in cells from anterior and intermediate lobes of the pituitary in addition to selected cells in areas of the brain such as mouse arcuate nucleus and hippocampus and the rat supraoptic and paraventricular nuclei, cortex, striatum, and reticular nucleus as well as in extracts from bovine anterior pituitary secretory granules. However, while specific staining was observed in these tissues, characterization of yapsin A at the nucleotide level has not been achieved, hence the amino acid sequence of yapsin A is still unknown.
In the present study we analyzed human islets with the yapsin 1 antiserum. Our results show strong, specific staining of yapsin 1-IR in glucagon producing cells only. The specific nature of the staining, in that it was absent from β- and δ-cells, indicated to us that possibly a yapsin A enzyme was present in human islet α-cells where glucagon is produced and secreted. We tested whether yapsin 1, as the prototypical enzyme for this sub-class of enzymes, could generate glucagon from proglucagon, since in many cases tested, yapsin 1 could process prohomorne substrates in a similar pattern to that of yapsin A. Indeed, we showed that purified yapsin 1 could process proglucagon to generate authentic glucagon. This processing pattern is consistent with yapsin 1 specificity since both the N- and C-termini are flanked by Lys-Arg cleavage sites, sites that are readily recognized by yapsin 1 (Cawley et al. 1996a). Furthermore, an additional Arg present in the P2’ position at the C-terminal cleavage site favorably enhances the processing by yapsin 1 as has been demonstrated previously (Ledgerwood, et al. 1996; Olsen et al. 1998). The selectivity of the enzyme is also observed since two Arg residues (Arg69-Arg70) found within the glucagon sequence (NCBI accession # P55095) were not cut.
These results raise the question as to whether a yapsin-like enzyme contributes to the important physiological function of glucagon production in vivo. One can speculate that similar to the ability of yapsin 1 to process pro-α-mating factor in the absence of Kex2, mammalian yapsin A may be able to participate in the production of glucagon in α-cells in times where PC2, the mammalian homologue of Kex2 involved in glucagon production in vivo (Rouille, et al. 1997), is down-regulated. Efforts at cloning this enzyme are underway.
Highlights.
Yapsin 1, a basic residue specific yeast aspartic protease can generate glucagon from proglucagon.
A yapsin 1 antibody labels glucagon expressing cells of human islets of Langerhans.
The yapsin 1 antibody did not stain insulin or somatostatin producing cells.
Acknowledgements
We would like to thank Dr. Savita Dhanvantari, Lawson Health Research Institute, Canada for the plasmid expressing proglucagon. GPG thanks Professor Lars Grimelius for facilities at the Institute of Genetics and Pathology, Uppsala University, Sweden. This research is supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Author contributions
NXC and HL performed and analyzed the in vitro processing of proglucagon. GPG performed the immunohistochemistry. YPL supported and advised throughout the project and edited the manuscript. NXC wrote the manuscript.
References
- Azaryan AV, Schiller M, Mende-Mueller L, Hook VY. Characteristics of the chromaffin granule aspartic proteinase involved in proenkephalin processing. J Neurochem. 1995;65:1771–1779. doi: 10.1046/j.1471-4159.1995.65041771.x. [DOI] [PubMed] [Google Scholar]
- Azaryan AV, Wong M, Friedman TC, Cawley NX, Estivariz FE, Chen HC, Loh YP. Purification and characterization of a paired basic residue-specific yeast aspartic protease encoded by the YAP3 gene. Similarity to the mammalian pro-opiomelanocortin-converting enzyme. J. Biol. Chem. 1993;268:11968–11975. [PubMed] [Google Scholar]
- Bourbonnais Y, Ash J, Daigle M, Thomas DY. Isolation and characterization of S.cerevisiae mutants defective in somatostatin expression: cloning and functional role of a yeast gene encoding an aspartyl protease in precursor processing at monobasic cleavage sites. EMBO. J. 1993;12:285–294. doi: 10.1002/j.1460-2075.1993.tb05655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley NX, Chen H-C, Beinfeld MC, Loh YP. Specificity and kinetic studies on the cleavage of various prohormone mono- and paired-basic residue sites by yeast aspartic protease 3. J. Biol. Chem. 1996a;271:4168–4176. doi: 10.1074/jbc.271.8.4168. [DOI] [PubMed] [Google Scholar]
- Cawley NX, Loh YP. Yapsin 1. In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes. edn 3rd. London: Elesevier Academic Press; 2011. (in press) [Google Scholar]
- Cawley NX, Olsen V, Zhang CF, Chen HC, Tan M, Loh YP. Activation and processing of non-anchored yapsin 1 (yap3p) J. Biol. Chem. 1998;273:584–591. doi: 10.1074/jbc.273.1.584. [DOI] [PubMed] [Google Scholar]
- Cawley NX, Pu L-P, Loh YP. Immunological identification and localization of yeast aspartic protease 3-like prohormone processing enzymes in mammalian brain and pituitary. Endocrinology. 1996b;137:5135–5143. doi: 10.1210/endo.137.11.8895388. [DOI] [PubMed] [Google Scholar]
- Cawley NX, Wong M, Pu LP, Tam W, Loh YP. Secretion of yeast aspartic protease 3 is regulated by its carboxy-terminal tail: characterization of secreted YAP3p. Biochemistry. 1995;34:7430–7437. doi: 10.1021/bi00022a016. [DOI] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- Egel-Mitani M, Flygenring HP, Hansen MT. A novel aspartyl protease allowing KEX2-independent MFa propheromone processing in yeast. Yeast. 1990;6:127–137. doi: 10.1002/yea.320060206. [DOI] [PubMed] [Google Scholar]
- Erickson AH, Blobel G. Early events in the biosynthesis of the lysosomal enzyme cathepsin D. J Biol Chem. 1979;254:11771–11774. [PubMed] [Google Scholar]
- Foltmann B. Chymosin: a short review on foetal and neonatal gastric proteases. Scand J Clin Lab Invest Suppl. 1992;210:65–79. [PubMed] [Google Scholar]
- Fukamizu A, Murakami K. New aspects of the renin-angiotensin system in blood pressure regulation. Trends Endocrinol Metab. 1995;6:279–284. doi: 10.1016/1043-2760(95)00156-5. [DOI] [PubMed] [Google Scholar]
- Gagnon-Arsenault I, Parise L, Tremblay J, Bourbonnais Y. Activation mechanism, functional role and shedding of glycosylphosphatidylinositol-anchored Yps1p at the Saccharomyces cerevisiae cell surface. Mol Microbiol. 2008;69:982–993. doi: 10.1111/j.1365-2958.2008.06339.x. [DOI] [PubMed] [Google Scholar]
- Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-a-factor. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot Cell. 2005;4:1364–1374. doi: 10.1128/EC.4.8.1364-1374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunihiro S, Kawanishi Y, Sano M, Naito K, Matsuura Y, Tateno Y, Gojobori T, Yamagata Y, Abe K, Machida M. A polymerase chain reaction-based method for cloning novel members of a gene family using a combination of degenerate and inhibitory primers. Gene. 2002;289:177–184. doi: 10.1016/s0378-1119(02)00547-4. [DOI] [PubMed] [Google Scholar]
- Ladds G, Davey J. Identification of proteases with shared functions to the proprotein processing protease Krp1 in the fission yeast Schizosaccharomyces pombe. Mol Microbiol. 2000;38:839–853. doi: 10.1046/j.1365-2958.2000.02180.x. [DOI] [PubMed] [Google Scholar]
- Ledgerwood EC, Brennan SO, Cawley NX, Loh YP, George PM. Yeast aspartic protease 3 (Yap3) prefers substrates with basic residues in the P2, P1 and P2' positions. FEBS Lett. 1996;383:67–71. doi: 10.1016/0014-5793(96)00219-0. [DOI] [PubMed] [Google Scholar]
- Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc Natl Acad Sci U S A. 2000;97:1456–1460. doi: 10.1073/pnas.97.4.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YP, Birch NP, Castro MG. Pro-opiomelanocortin and pro-vasopressin converting enzyme in pituitary secretory vesicles. Biochimie. 1988;70:11–16. doi: 10.1016/0300-9084(88)90153-8. [DOI] [PubMed] [Google Scholar]
- Loh YP, Parish DC, Tuteja R. Purification and characterization of a paired basic residue-specific pro-opiomelanocortin converting enzyme from bovine pituitary intermediate lobe secretory vesicles. J Biol Chem. 1985;260:7194–7205. [PubMed] [Google Scholar]
- Mackin RB, Noe BD, Spiess J. The anglerfish somatostatin-28-generating propeptide converting enzyme is an aspartyl protease. Endocrinology. 1991;129:1951–1957. doi: 10.1210/endo-129-4-1951. [DOI] [PubMed] [Google Scholar]
- Monod M, Hube B, Hess D, Sanglard D. Differential regulation of SAP8 and SAP9, which encode two new members of the secreted aspartic proteinase family in Candida albicans. Microbiology. 1998;144(Pt 10):2731–2737. doi: 10.1099/00221287-144-10-2731. [DOI] [PubMed] [Google Scholar]
- Olsen V, Guruprasad K, Cawley NX, Chen HC, Blundell TL, Loh YP. Cleavage efficiency of the novel aspartic protease yapsin 1 (Yap3p) enhanced for substrates with arginine residues flanking the P1 site: correlation with electronegative active-site pockets predicted by molecular modeling. Biochemistry. 1998;37:2768–2777. doi: 10.1021/bi9724826. [DOI] [PubMed] [Google Scholar]
- Parish DC, Tujeta R, Alstein M, Gainer H, Loh YP. Purification and characterization of a paired basic residue-specific prohormone-converting enzyme from bovine pituitary neural lobe secretory vesicles. J. Biol. Chem. 1986;261:14392–14397. [PubMed] [Google Scholar]
- Portela-Gomes GM, Lukinius A, Grimelius L. Synaptic vesicle protein 2, A new neuroendocrine cell marker. Am J Pathol. 2000;157:1299–1309. doi: 10.1016/S0002-9440(10)64645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouille Y, Bianchi M, Irminger JC, Halban PA. Role of the prohormone convertase PC2 in the processing of proglucagon to glucagon. FEBS Lett. 1997;413:119–123. doi: 10.1016/s0014-5793(97)00892-2. [DOI] [PubMed] [Google Scholar]
- Ryle AP, Porter RR. Parapepsins: two proteolytic enzymes associated with porcine pepsin. Biochem J. 1959;73:75–86. doi: 10.1042/bj0730075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabattini E, Bisgaard K, Ascani S, Poggi S, Piccioli M, Ceccarelli C, Pieri F, Fraternali-Orcioni G, Pileri SA. The EnVision++ system: a new immunohistochemical method for diagnostics and research. Critical comparison with the APAAP, ChemMate, CSA, LABC, and SABC techniques. J Clin Pathol. 1998;51:506–511. doi: 10.1136/jcp.51.7.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. Specificity of Pepsin and Its Dependence on a Possible 'Hydrophobic binding Site'. Nature. 1963;199:1094–1095. doi: 10.1038/1991094a0. [DOI] [PubMed] [Google Scholar]
- Thomas G, Thorne BA, Thomas L, Allen RG, Hruby DE, Fuller R, Thorner J. Yeast KEX2 endopeptidase correctly cleaves a neuroendocrine prohormone in mammalian cells. Science. 1988;241:226–230. doi: 10.1126/science.3291117. [DOI] [PubMed] [Google Scholar]
- Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol. 2008;181:1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Wolfe MS, Xia W, Moore CL, Leatherwood DD, Ostaszewski B, Rahmati T, Donkor IO, Selkoe DJ. Peptidomimetic probes and molecular modeling suggest that Alzheimer's gamma-secretase is an intramembrane-cleaving aspartyl protease. Biochemistry. 1999;38:4720–4727. doi: 10.1021/bi982562p. [DOI] [PubMed] [Google Scholar]