Summary
SR proteins are well-characterized RNA binding proteins that promote exon inclusion by binding to exonic splicing enhancers (ESEs). However, it has been unclear whether regulatory rules deduced on model genes apply generally to activities of SR proteins in the cell. Here, we report global analyses of two prototypical SR proteins SRSF1 (SF2/ASF) and SRSF2 (SC35) using splicing-sensitive arrays and CLIP-seq on mouse embryo fibroblasts (MEFs). Unexpectedly, we find that these SR proteins promote both inclusion and skipping of exons in vivo, but their binding patterns do not explain such opposite responses. Further analyses reveal that loss of one SR protein is accompanied by coordinated loss or compensatory gain in the interaction of other SR proteins at the affected exons. Therefore, specific effects on regulated splicing by one SR protein actually depend on a complex set of relationships with multiple other SR proteins in mammalian genomes.
Introduction
SR proteins are among the best characterized splicing regulators in higher eukaryotic cells. They are involved in both constitutive and alternative pre-mRNA splicing; making this family of RNA binding proteins (RBPs) unique compared to other RBPs that function only in regulated splicing (Lin and Fu, 2007; Long and Caceres, 2009; Zhong et al., 2009). SR proteins are RRM (RNA Recognition Motif) containing RBPs and each harbors a signature RS (Arg/Ser-rich) domain(s). Both RRM and RS domains have been implicated in early steps of spliceosome assembly where the RRM is responsible for RNA binding and the RS domain for protein-protein interactions. However, additional biochemical evidence suggests that RRM and RS domain may also respectively participate in protein-protein and protein-RNA interactions (Hertel and Graveley, 2005; Shen et al., 2004).
It has been generally accepted that SR proteins regulate alternative splicing by binding to exonic splicing enhancers (ESEs) to promote exon inclusion, which has been unequivocally demonstrated by tethering SR proteins to an alternative exon (Graveley and Maniatis, 1998). SR protein binding to ESEs is thought to enhance the recognition of weak splice sites by the splicing machinery, an activity equally applicable to both constitutive and regulated exons (Shen and Green, 2006). hnRNP A/B proteins can antagonize the positive effect of SR proteins on splice site selection (Mayeda and Krainer, 1992; Mayeda et al., 1994).
Despite the fact that SR proteins have been extensively characterized on model genes, it has been unclear whether and how various regulatory rules deduced from biochemical studies apply to endogenous transcripts. For example, in vitro binding or in vivo functional SELEX experiments have deduced binding consensus sequences for several SR proteins (Cavaloc et al., 1999; Liu et al., 2000; Liu et al., 1998; Schaal and Maniatis, 1999; Tacke and Manley, 1995). However, different assays appear to reveal distinct consensus motifs, suggesting that SR-RNA interactions may be quite degenerate, context-sensitive, or assay-dependent.
Such uncertainty in SR protein binding consensus makes it difficult to accurately predict SR protein binding sites in mammalian transcriptomes. For example, the ESEfinder program based on functional SELEX predicts SR ESEs on the majority of expressed pre-mRNAs (Cartegni et al., 2003; Lim et al., 2011). However, mapping of in vivo binding sites for SRSF3 (SRp20) and SRSF4 (SRp75) indicates that the two SR proteins only bind small distinct subsets of endogenous transcripts (Anko et al., 2010; Anko et al., 2012). A similar observation has also been made with Drosophila dASF and dSRp55 (Gabut et al., 2007), implying that the interactions of SR proteins with RNAs in vivo may be more selective than previously thought. Alternatively, these initial analyses may not have reached saturation or had the sensitivity to capture most SR proteins binding events on expressed transcripts.
The effect of SR proteins on alternative splicing has also remained a subject of debate. Through binding to ESEs, SR proteins function as positive splicing regulators to promote exon inclusion. However, increasing evidence suggests that they are also involved in exon-skipping events (Gallego et al., 1997; Ghigna et al., 2005; Lemaire et al., 1999; Solis et al., 2008), possibly through competition between the alternative exon and flanking constitutive exons (Sanford et al., 2009). For example, tethering an SR protein to the alternative exon induces its inclusion, but anchoring the SR protein in a flanking exon promotes alternative exon skipping (Han et al., 2011a). These results suggest that SR proteins control alternative exon inclusion in opposite directions, depending on where they bind. The question is how widely do SR proteins use this strategy to regulate splicing in the cell, and if so, whether specific splicing outcomes could be directly linked to the positional effect of SR protein binding in vivo.
Here, we utilized CLIP-seq in combination with splicing-sensitive arrays to address the in vivo RNA binding properties and functions of two classic SR proteins in regulated splicing. We found extensive overlap between SRSF1 and SRSF2 in binding to exons and we detected extensive induction of both exon inclusion and skipping events in response to depletion of either SR protein. Surprisingly, we found little correlation between SR protein binding and induced splicing changes. Further analysis revealed that the loss of RNA binding by one SR protein induces changes in RNA binding by another SR protein, suggesting compensatory or synergistic actions of remaining SR proteins and other splicing regulators in specific SR protein-depleted cells. Mutational analysis revealed that SR protein depletion-induced exon inclusion could be switched to skipping by preventing such compensatory responses, thus suggesting a general regulatory principle that emphasizes the collective contribution of multiple SR proteins to regulated splicing in mammalian transcriptomes.
Results
Preferential and overlapping binding of SRSF1 and SRSF2 on exons
We previously generated two MEF cell lines from conditional SRSF1 and SRSF2 knockout mice (Lin et al., 2005). Each of these MEF lines is functionally complemented with the respective HA-tagged exogenous gene expressed from a Tet-off promoter at the level equivalent to that of the endogenous counterpart, as shown earlier (Lin et al., 2005) and re-confirmed before beginning this study (data not shown). This system permits controlled depletion of SR protein with Doxycycline (Dox) and CLIP-seq analysis for each protein using the same anti-HA antibody, which is efficient in immunoprecipitation (Fig. 1A). Both IPed SR proteins could be labeled with 32P-γ-ATP, likely due to associated SR protein kinases, as noted earlier (Sanford et al., 2008). This provides the location reference for bulk IPed SR proteins. Using T4 kinase, we detected protein-RNA adducts that were sensitive to nuclease trimming (Fig. 1B). We isolated such adducts linked with 30 to 60nt RNA and performed deep sequencing using the established CLIP-seq protocol (Ule et al., 2005; Xiao et al., 2012; Xue et al., 2009; Yeo et al., 2009). We initially obtained 3,694,535 and 4,874,935 non-redundant tags for SRSF1 and SRSF2, respectively, that could be uniquely mapped to the mouse genome (version mm9).
Figure 1. Genomic landscape of SRSF1 and SRSF2 binding.
(A) Western blot of SRSF1-HA and SRSF2-HA using HA antibody, showing efficient immunoprecipitation of both SR proteins in MEFs.
(B) Autoradiograph of 32P-labeled RNA-SR protein adducts with or without crosslinking. The line on the right side of each gel indicates the excised region for CLIP-seq.
(C) The genomic distribution of SRSF1 and SRSF2 tags relative to different proportions of specific genomic regions.
(D) The number of SRSF1 and SRSF2 tags mapped on exon-exon and exon-intron junctions.
(E) The UCSC genome browser view of SRSF1 and SRSF2 binding events on the GPBP1 gene. Control indicates background tags from anti-HA CLIP on a wild-type MEF line that does not carry any HA-tagged protein. Arrows indicate unique binding peaks for a specific SR protein.
(F) Overlap in SR protein binding within exons and introns as a function of gene expression, showing that the overlap is more extensive in exons than introns. See also Figure S1.
The two SR proteins under the investigation bind both exons and introns. However, since exons are much shorter than introns, we detected ~15-fold enrichment in exons after length normalization, suggesting that SR proteins prefer exonic sequences, as expected from their known RNA binding properties (Fig. 1C). This profile largely agrees with data for SRSF1 in HEK293T cells (Sanford et al., 2009), but our data provide more coverage and allow comparison between the two SR proteins. We detected 0.3 to 0.4 million tags for SRSF1 and SRSF2 on intron-exon and exon-exon boundaries, indicating that both SR proteins bind pre-mRNA and spliced mRNA (Fig. 1D). Considering SRSF2 is a non-shuttling SR protein (Caceres et al., 1998), its binding to spliced mRNA suggests it is removed or replaced by other shuttling SR proteins prior to mRNA export.
The most striking observation is an extensive overlap between the two SR proteins in binding to both constitutive and alternative exons, as seen on a representative gene (Fig. 1E). Some binding events are unique to one or the other SR protein, detectable in both intronic and exonic regions (red arrows in Fig. 1E). We observed only background binding on MEFs not expressing any HA-tagged protein (Fig. 1E). The observed binding pattern is also completely different from that of multiple other HA-tagged RNA binding proteins we analyzed by CLIP-seq (data not shown). Thus, the exon-central binding profiles of the two SR proteins are unlikely to result from non-specific binding to the HA tag or IgG beads.
We deduced 50,983 and 56,336 binding clusters (peaks, Benjamini- Hochberg corrected p-value<1e-5) for SRSF1 and SRSF2, respectively, in various classes of RNA, including intron-containing lincRNAs, but by far, the predominant class is intron-containing, protein-coding pre-mRNA (Fig. S1A). The interaction of SR proteins with microRNA is consistent with the reported role of SRSF1 in the regulation of microRNA biogenesis (Wu et al., 2010). SRSF1 and SRSF2 each binds, on an average, ~30% of the exons among 7385 (SFRS1) and 7226 (SFRS2) expressed transcripts that contain introns at the current tag density (Fig. S1B). Gene expression (detected by probes built in Affymetrix exon junction arrays, see Methods) positively correlates with the degree of collective SR binding events on exons, but much less in intronic regions (Fig. 1F, Fig. S1C and S1D). SR protein binding is significantly enriched on the internal exons compared to binding on either the first or the last exon (Fig. S1E and S1F). Reduced binding on the first exon likely reflects a functional interplay of SR proteins with DNA and RNA at the promoter-proximal region (unpublished results). Less frequent SR binding on the last exon may reflect the size of the last exon, which tends to be bigger than internal exons, and thus, while the total number of binding events may be similar, the averaged binding density at individual locations may be lower.
In vivo RNA binding specificity of SRSF1 and SRSF2
The mapped in vivo binding sites for SRSF1 and SRSF2 afford us the opportunity to address their preferred binding motifs in mammalian cells. While it is generally accepted that SRSF1 binds to GA-rich sequences, the RNA binding consensus for SRSF2 has eluded clear description. The in vitro binding SELEX experiments suggest that the RRM of SRSF2 binds to diverse sequences, some of which are also purine-rich (Cavaloc et al., 1999; Schaal and Maniatis, 1999; Tacke and Manley, 1995), but functional selection for SRSF2 responsive elements indicates that this SR protein prefers GC-rich sequences (Liu et al., 2000), which closely resemble the newly proposed consensus SSNG (where S is C or G and N corresponds to any nucleotide) from structural analysis of SRSF2 (Daubner et al., 2012). It has therefore remained unclear, which of these motifs reflect the action of SRSF2 in vivo.
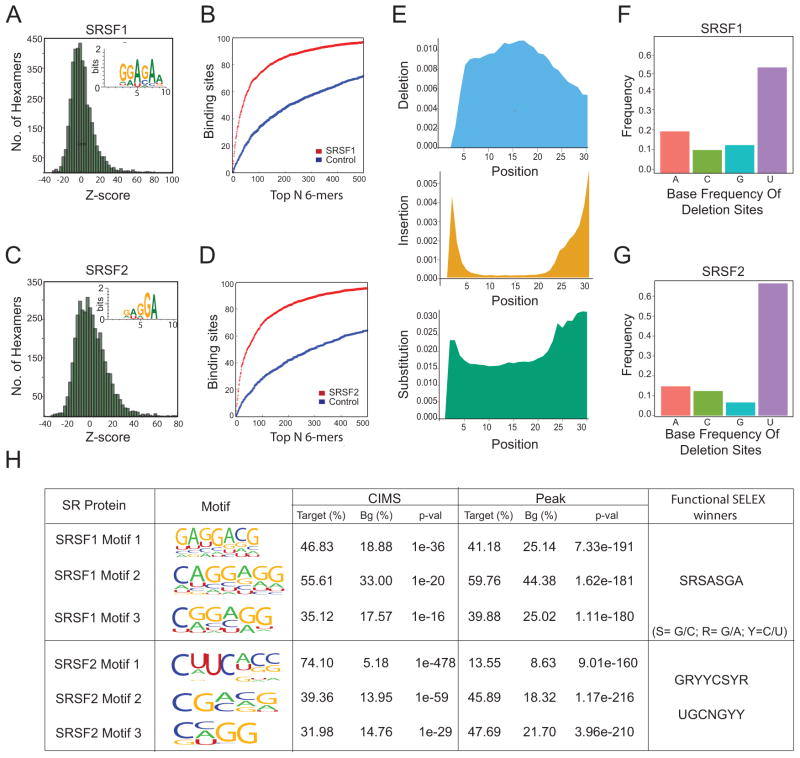
To identify the binding site consensus sequences for SRSF1 and SRSF2 proteins, we first took the standard approach of searching for overrepresented hexamer sequences in detected peaks against randomly sampled genomic sequence as background (Fig. 2A and 2B). Based on Z-scores, we identified the top 143 and 173 hexamers that cover ~80% of all detected peaks for SRSF1 and SRSF2, respectively (Fig. 2C and 2D). We next used these top-ranking hexamers to deduce binding consensus for each SR protein (inserts in Fig. 2A and 2B). We identified the GGAGA motif for SRSF1, which is consistent with the functional SELEX results (Liu et al., 1998) and with the consensus deduced from the initial CLIP-seq study of SRSF1 (Sanford et al., 2009). We could only detected the GA-rich motif for SRSF2, indicating that SRSF2 may bind to RNA with more degenerate motifs (Fig 2C).
Figure 2. Motif analysis for SR protein binding sites in the mouse transcriptome.
(A and C) Histogram of Z-scores for hexamers in CLIP-seq peaks for SRSF1 and SRSF2 Insert indicates the deduced consensus based on top hexamers.
(B and D) Percentage of SRSF1 and SRSF2 peaks that contain top hexamers.
(E) Positional profiles of deletion (blue), insertion (yellow) and substitutions (green) in sequenced tags from SRSF2 CLIP-seq tags. A similar pattern was also observed for SRSF1 CLIP-seq.
(F and G) Base frequency at deletion sites for SRSF1 and SRSF2. Deletion of Uracil is predominant in both SR CLIP-seq datasets.
(H) Top three enriched motifs for SRSF1 and SRSF2 deduced by word counting and CIMS analysis and their representations in deletion-containing sequences or peaks relative to general background sequences.
SR protein binding consensus based on induced deletions and genomic distribution
Because the two proteins show extensive overlap in RNA binding, weaker motifs deduced solely from the CLIP sequences may be incidentally derived from binding sites closely occupied by different SR proteins. To address this, we applied a recently published approach that relies on Crosslinking Induced Mutation Sites (CIMS) introduced during the CLIP-seq procedure (Zhang and Darnell, 2011) to more precisely locate the binding site. From all mapped binding events in the mouse transcriptome, we found that 3.5% (SFRS1) and 8% (SFRS2) of tags have one or more internal deletions, the former being somewhat lower than the fractions (8–20%) reported for Nova and Ago2 CLIP tags by Darnell and colleagues. However, these internal deletions are typically distributed within the sequenced tags, distinct from the distribution of insertions and point mutations that were frequently found on the 3′ side of the tags due to sequencing errors (Fig. 2E). Furthermore, U residues were more frequently depleted compared to the other three nucleotides (Fig. 2F and 2G). Because U is most susceptible to UV crosslinking in general, the fact that SFRS1 binding motifs often do not contain Us possibly accounts for the relatively low deletion frequency we observe for this SR protein. This highlights the need to use multiple approaches for deriving motifs from CLIP data, as the crosslinking efficiency of individual nucleotides at variable positions in the motif may influence the numbers and kinds of reads detected.
To complement the analysis of crosslinking-induced deletions, we selected deletion-containing tags that were also mapped within SR binding peaks to deduce consensus motifs by using the HOMER software (Heinz et al., 2010), which is related to the MEME motif identification program (Bailey et al., 2009). This approach again yielded GA-rich consensus motifs for SRSF1, which are well represented in tags with internal deletions and those within mapped SRSF1 binding peaks (Fig. 2H). In contrast, the most enriched motif for SRSF2 among tags containing internal deletions is a CU-rich sequence, followed by two less-well enriched motifs that contain a GC core. However, the top CU-rich motif accounts for only a small fraction (~14%) of the sequences within mapped SRSF2 peaks, while the next two enriched motifs are more representative (close to 50%) (Fig. 2H). We note that the CU and GC-rich sequences agree with the consensus sequences deduced from both binding and functional (in vitro splicing) SELEX experiments (Cavaloc et al., 1999; Liu et al., 2000). Our approach suggests a general approach for motif analysis based on CLIP-seq data by coupling the CIMS strategy with binding frequencies in mammalian transcriptomes.
Compensation for weak splice sites and long introns by the action of SR proteins
The high resolution mapping results permitted us to test a long held theory concerning the ability of SR proteins to compensate for weak 5′ and 3′ splice sites in mammalian cells. We divided the 5′ and 3′ splice sites in the mouse genome into three categories based on the average maximum entropy splice site scores at the 5′ and 3′ splice sites (Yeo and Burge, 2004) and queried CLIP-seq signals for SRSF1 and SRSF2 within each category. We observe an inverse correlation between SR protein binding and the strength of the splice sites (i.e., weaker splice sites show stronger SR protein binding), and this is true for both SRSF1 and SRSF2 (Fig. 3A). We also performed a similar set of analyses on individual exons, with or without dividing them into constitutive or alternative exons, finding a similar trend (Fig. S2). We noted in these analyses that the differences are rather small, implying that the contribution of SR proteins to the selection of weak splice sites might depend not on a single SR protein at each exon, but rather on the collective action of multiple SR proteins on each exon. As a result, the contribution of a given SR protein is less evident from such a metagene analysis.
Figure 3. Correlation between SR protein binding and splicing features.
(A) SR protein binding versus splice site strength. Inserts highlight a reverse correlation between SR protein bindings and splice site strength.
(B) Positive correlation between SR protein binding and the length of both upstream introns (Up intron) and downstream introns (Down intron). Introns bins correspond to (1) <350nt, (2) 350nt-1kb, (Solis et al., 2008) 1kb-2kb, (4) 2kb-4kb, and (5) >4kb.
(C and D) Association of SR protein binding events with decoy or pseudo exons in the mouse genome. See also Figure S2.
As the size of flanking introns has been shown to influence the ability of an internal exon to be recognized in Drosophila (Fox-Walsh et al., 2005), we next tested whether SR protein binding on exons might compensate for the size of introns. For this purpose, we divided exons into 5 bins according to the length of the upstream or downstream intron and compared SRSF1 or SRSF2 binding on exons against the length of flanking introns. Strikingly, we found that binding of both SRSF1 and SRSF2 to exons correlates positively with the length of upstream and downstream introns (Fig. 3B). These observations suggest that exons with longer flanking introns require stronger SR binding to be functionally defined in the mouse transcriptome.
Because half of binding sites for SRSF1 and SRSF2 were mapped to intronic regions, we asked whether some intronic binding events might reflect regulatory activities of SR proteins in splicing. A number of studies have implicated decoy exons in alternative splicing (Buratti et al., 2007; Havlioglu et al., 2007). “Decoy exons” only contain one potential 5′ or 3′ splice site whereas “pseudo exons” carry both potential splice sites separated by a sequence of up to 250 nt (Yeo and Burge, 2004). After centering the mapped intronic SR protein binding sites, we examined the frequency of potential 3′ or 5′ splice sites upstream or downstream of the peak. We find a small fraction of potential 5′ splice sites, but not 3′ splice sites, near intronic binding peaks for both SRSF1 and SRSF2 (Fig. 3C and 3D). These observations indicate that only a small fraction of SR protein binding events in introns might represent decoy or pseudo exons. In this context, it is interesting to note that intronic SR protein binding events have been found to inhibit exon inclusion, which do not have to be part of pseudo exons or decoy exons (Erkelenz et al., 2012).
Extensive involvement of SR proteins in exon inclusion and skipping events in vivo
We next investigated how specific SR protein binding events might be linked to the regulation of alternative splicing in the mouse transcriptome. As SRSF1 and SRSF2 were each expressed from a tet-off promoter, we were able to efficiently deplete them by adding Dox to the culture media (Fig. 4A). After Dox-induced depletion, total RNA was isolated for splicing profiling on splicing-sensitive exon junction microarrays (Du et al., 2010). To aid in validation of these candidate events, we divided the data in four bins and selected representative events (colored in Table S1) in each bin for RT-PCR validation. The result indicates that the overall validation rate for events in bin 1 to 3 is 88% (out of 84 events examined, 10 failed). We also complemented the array-based method with the RASL-seq technology (RNA-mediated oligonucleotide Annealing, Selection and Ligation coupled with deep sequencing) we recently developed (Li et al., 2012; Zhou et al., 2012) and validated a large set of candidate events at the rate of 93.5% (46 examined, 3 failed) (Table S1). A representative set of validated events is shown for both SRSF1 (Fig. S3A) and SRSF2 (Fig. S3B).
Figure 4. Induction of exon inclusion and skipping in SR protein-depleted cells.
(A) Western analysis of SR protein depletion using Dox. Actin served as a loading control. “+” and “−” indicate the presence or absence of the SR protein in mock-treated and Dox-treated cells, respectively.
(B) Bar graph presentation of altered alternative events detected by exon junction array and RASL-seq. Depletion of either SR proteins causes both exon inclusion and skipping. Splicing events affected by both are highlighted with similar or opposite responses to depletion of either SR protein on the right.
(C) Validation of SRSF1 and SRSF2 depletion-induced splicing of cassette exons in four different classes. The SR proteins exhibited direct binding in various locations on all of these illustrated genes. See also Figure S3 and Table S1.
In total, we detected 498 SRSF1-dependent and 912 SRSF2-dependent alternative splicing events, consistent with the function of these SR proteins as splicing regulators. Interestingly, among 498 altered splicing events detected in SRSF1-depleted MEFs, 225 events showed increased exon skipping and 276 exhibited increased exon inclusion. Similarly, in response to Dox-induced depletion of SRSF2, 312 exons increased in skipping while 601 exons increased in inclusion (Fig. 4B). Therefore, both SR proteins appear to be more frequently involved in repressing rather than promoting exon inclusion. On the surface, this contradicts the widely perceived roles for SR proteins in promoting exon inclusion. We compared events affected by loss of each protein and found that SRSF1 and SRSF2 jointly regulate 288 alternative splicing events. Of these shared events, 39% are regulated in opposite directions (Fig. 4B), suggesting that the two SR proteins function in a combinatorial fashion to regulate a subset of splicing events in the mouse transcriptome.
Our data show all possible combinations of alternative splicing control by the two SR proteins (Fig. 4C). For example, SRSF1 depletion induced exon skipping (MARCH7 exon 7) or inclusion (CLSTN1 exon 3), while SRSF2 depletion showed no effect on these exons (top left panel). The converse was true on BAP1 exon 5 and TRA2α exon 2 (top right panel), which responded to SFRS2 but not SFRS1 depletion. These exons exhibited restricted responses to only one of the two SR proteins. On FYTTD1 exon 7 and KIFAP3 exon 20, depletion of either SR protein had the same effect (bottom left panel). On RALY exon 4, SRSF1 depletion resulted in exon inclusion, but SRSF2 depletion induced exon skipping, and the converse was true on the MLF1 exon 3 (bottom right panel). These cases exemplify complex functional requirements for different SR proteins at different exons. Although each of these examples deserve detailed molecular dissection to understand the basis for the specific effects detected, the data clearly illustrate more complicated biological functions for SR proteins in regulated splicing than previously appreciated based on studies with model genes.
Functional consequences not linked to simple positional effects of SR proteins
Recent genome-wide studies suggest that many splicing regulators influence splice site selection in a position-sensitive manner (Han et al., 2011b; Witten and Ule, 2011). For SR proteins, the tethering approach has demonstrated that SR protein binding on an internal cassette exon enhances inclusion of that internal exon, whereas tethering to a flanking constitutive exon stimulates skipping of the internal alternative exon (Han et al., 2011a). Our in vivo mapping results coupled with a large number of SR protein-dependent exon inclusion or skipping events presented an opportunity to explore potential positional effects of SR proteins from the global perspective.
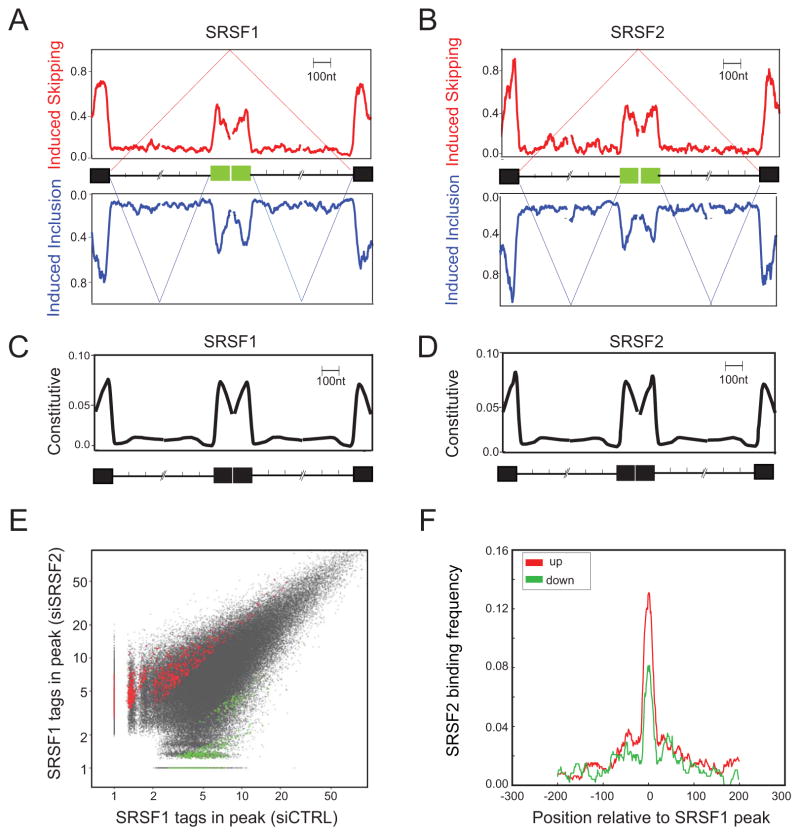
Based on the large number of validated splicing events in addition to those reliably detected by RASL-seq, we constructed cassette exon models separately for induced exon inclusion and skipping events, and mapped SR binding information to the internal alternative and flanking constitutive exons, as well as surrounding intronic regions (for the exon-proximal 300 nt, see Fig. 5A and 5B). For comparison, we constructed a similar cassette exon model from a set of randomly selected constitutively spliced exons (Fig. 5C and 5D). Unexpectedly, we could not see any relationship between SR protein binding and functional outcomes. The only observable feature common for both SR proteins is lower binding on the internal alternative exon compared to the flanking constitutive exons in the regulated cassette exon model, but this trend is not evident on the constitutive exon model (compare Fig. 5A–B and 5C–D). This is consistent with those internal exons being alternative, perhaps due to insufficient SR protein-dependent activities to make them constitutively included. The puzzle then is why depletion of a specific SR protein would induce exon inclusion in one set of these genes but cause exon skipping in another set.
Figure 5. Composite RNA maps for SRSF1 and SRSF2 and interplay between the two SR proteins.
(A and B) Composite maps for SRSF1 (A) and SRSF2 (B) on induced exon inclusion or skipping events based on the most reliable RASL-seq data streamlined by RT-PCR in addition to array-derived and RT-PCR validated events.
(C and D) Composite maps constructed on constitutive exons.
(E) Scatter plot of SRSF1 binding peaks before (siCtrl) and after (siSRSF2) knockdown of SRSF2. X- and Y-axes indicate the number of tags associated with each peak. Red and green indicate peaks that showed significant increase or decrease in SRSF1 binding in response to SRSF2 depletion.
(F) SRSF2 binding frequency on increased (red) and decreased (green) SRSF1 binding peaks.
We hypothesized that specific functional outcomes might depend on the collective contribution of multiple SR proteins acting on individual alternatively spliced exons. As such, depletion of one specific SR protein might alter the competition between the alternative and flanking constitutive splice sites. For example, as illustrated earlier (Han et al., 2011a), skipping of an alternative exon might increase in response to the removal of an SR protein if the SR protein contributes more to the selection of the alternative exon than the flanking exons. Conversely, inclusion of the exon may increase if the depleted SR protein mainly acts on the flanking constitutive exon(s) while other SR proteins bind to the internal alternative exon. This would cause weakening of the flanking exon(s), thereby increasing the competitiveness of the alternative exon, leading to increased inclusion. In other words, the specific outcome after depletion of an SR protein for each exon under consideration depends on the activities of other functionally related SR proteins and their binding sites on the exon itself and on its flanking exons.
Compensatory changes in SR protein binding to RNA
To test the above hypothesis, we mapped SRSF1 binding sites by CLIP-seq in MEFs depleted of endogenous SRSF2 by RNAi. We performed two biological replicates, yielding at total of 9,206,798 (no depletion) and 8,912,433 (SRSF2 depleted) non-redundant and uniquely mapped tags for SRSF1 (see Methods). Because SR protein binding is positively correlated with levels of gene expression, we focused on transcripts with similar overall levels of gene expression and SR binding (<1.5-fold changes in total CLIP-seq tag density per transcript). We normalized each dataset to the same number of total counts (5M) and identified statistically significant changes in SRSF1 binding that were induced by SRSF2 depletion. This identified 786 increased (red) and 479 decreased (green) SRSF1 binding events (Fig. 5E), indicating that binding of SRSF1 is influenced by the action of SRSF2 at many locations. These altered SRSF1 binding events do not seem enriched for alternative exons versus constitutive exons, although such changes on regulated exons might more readily lead to measurable functional outcomes. It is important to emphasize that CLIP-seq tags measure SR-RNA interactions before and after splicing, and therefore, do not strictly reflect final levels of spliced mRNA.
To further understand how SRSF2 binding influences the interaction of SRSF1 with RNA, we analyzed SRSF1 binding peaks that responded to SRSF2 depletion by determining the binding frequency of SRSF2 around SRSF1 binding sites. This analysis revealed that strong SRSF2 binding sites occurred more frequently on and near the sites that showed increased SRSF1 binding after SRSF2 depletion. In contrast, the frequency of strong SRSF2 binding sites was lower on and near sites where SRSF1 binding was reduced in SRSF2 depleted cells (Fig. 5F). We interpret these observations to indicate that SRSF1 and SRSF2 compete for binding at many sites. On the sites that show less binding by both SR proteins, however, reduction in SRSF2 binding might indirectly enhance other RNA binding proteins, such as hnRNP A/B proteins, to compete with adjacent SRSF1 binding events, thus causing a coordinated loss in SR binding.
Both coordinated loss and compensatory gain of SR proteins can be better appreciated on some representative genes (Fig. 6A–C). In the case of HNRNPA2B1 transcripts, for example, SRSF1 and SRSF2 bound similarly to both alternative and flanking exons. Upon SRSF2 knockdown, SRSF1 binding was reduced in the middle alternative exon (arrow in Fig. 6A), while little change was detected on the two flanking exons. This coordinated loss of both SR proteins likely contributes to weakening of the alternative exon, thus causing its efficient skipping in SRSF2-depleted cells (Fig. 6D, left panel with a model below). On CDC45I transcripts, the reduction of SRSF2 binding allowed SRSF1 to bind better on two locations, one on the alternative exon and the other on the downstream constitutive exon (arrows in Fig. 6B). The preferential gain in SRSF1 binding on the internal alternative exon may thus be responsible for the increased inclusion of the exon in response to SRSF2 depletion (Fig. 6D, right panel with a model below). Similarly, on CCNL1 transcripts, the reduction of SRSF2 binding allowed SRSF1 to bind better on the alternative exon (arrows in Fig. 6B), possibly explaining the increased inclusion of the alternatively spliced exon in SRSF2 depleted cells (Fig. 6D, right panel). These observations expose the complex contributions of individual SR proteins to regulation of alternative splicing that arise from competition or collaboration with one another and with other splicing regulators at specific sites in pre-mRNAs within the cell.
Figure 6. Synergistic and compensatory interactions of SR proteins with RNA and associated functional consequences.
(A–C) UCSC genome browser views of the HNRNPA2B1, CDC45I, and CCNL1 genes with binding by SRSF2 (blue), SRSF1 before SRSF2 knockdown (green) and SRSF1 after SRSF2 knockdown (red).
(D) The splicing response of HNRNPA2B1 (left), CDC45I and CCNL1 (right) to SRSF2 depletion. The proposed model on the left indicates potential competition of SR protein binding by other RNA binding proteins (blue bricks), which may quickly occupy the binding site vacated by an SR protein (SR1), thus preventing the compensatory binding by another SR protein (SR2). The proposed model on the right suggests the occupancy of the site vacated by the first SR protein (SR1) by a second SR protein (SR2).
(E) Graphical representation of the CDC45I minigene and specific deletion mutations (labeled X) introduced in SRSF1 binding sites in exon 5 (Ex5.1) and two locations in exon 4 (Ex4.1 and Ex4.2). P1 and P2 indicate PCR primers for splicing analysis. RT-PCR analyses were performed on wt and mutant minigenes before and after SRSF2 knockdown (Bottom). The mutations in exon 4 caused the switch of SRSF2 depletion-induced exon inclusion (highlighted in green boxes) to exon skipping (highlighted in red boxes).
(F) Graphical representation of the CCNL1 minigene and SRSF1 binding site deletion mutations (labeled X) introduced in exon 4 (Ex4.1). P1 and P2 indicate PCR primers for splicing analysis. Splicing of wt and mutant minigene before and after SRSF2 knockdown was analyzed by RT-PCR. The deletion mutation in exon 4 caused the switch of SRSF2 depletion-induced exon inclusion (highlighted in green box) to exon skipping (highlighted in red box).
Conversion from SR protein-dependent exon inclusion to skipping
Loss of SR protein binding on the internal alternative exon explains induced exon skipping after depletion according to the well-known function of SR proteins in promoting exon definition. Here, we wished to further explore the mechanism by which exon inclusion increases upon SR protein depletion. Given the gain in SRSF1 at some sites upon SRSF2 depletion, we wished to test whether an SRSF2-depletion dependent increase in SRSF1 binding could be responsible for increased exon inclusion in SRSF2 depleted cells. We thus constructed two minigenes containing the alternative and flanking exons from the CDC45I and CCNL1 genes, which were spliced similarly to the corresponding regions of their endogenous genes and which showed altered SRSF1 binding after SRSF2 depletion (Fig. 6E and 6F, bottom panel, first two lanes). We next introduced a deletion mutation (indicated by Xs) to the site in the alternative or downstream flanking constitutive exon that showed the alteration in SRSF1 binding after SRSF2 depletion. On CDC45I exon 5, SRSF2 knockdown slightly enhanced SRSF1 binding at the upstream site but reduced its binding at the downstream site. By mutating site 5.1 where SRSF2 depletion induced a minor gain of SRSF1 binding, we found that the mutant had a similar splicing ratio to the wild-type construct before SRSF2 knockdown and caused a small change in the inclusion of the internal alternative exon after SRSF2 knockdown (Fig. 6E, bottom panel). The minor impact by the mutation in this flanking exon might be due to other sequence features that make this exon constitutive; in fact, this construct served as a control for other mutations analyzed.
We next tested mutations in the alternative exon 4 of CDC45I. The prediction is that specific deletion mutations in site 4.1 (the predominant binding site for SRSF2) and 4.2 (the predominant binding site for SRSF1) would each weaken the alternative exon in wild-type cells. A much bigger effect would be predicted in SRSF2-depleted cells, because the Ex4.1 mutation would prevent the gain of SRSF1 binding in SRSF2 depleted cells and the Ex4.2 mutation would weaken SRSF1 binding, which may synergize with the lost of SRSF2 binding. As predicted, we found that both Ex4.1 and Ex4.2 mutants showed a modest effect in wild-type cells. In contrast, both mutants flipped from SRSF2 depletion-induced exon inclusion to skipping (Fig. 6E, bottom panel). We performed a similar analysis on the CCNL1 minigene containing the alternative exon 4, and found that a deletion mutation introduced in the exon also switched SRSF2 depletion-induced exon inclusion to skipping (Fig. 6F, bottom panel). Together, these data demonstrated that the splicing response to changes in function of one SR protein could be profoundly influenced by compensatory actions of another SR protein on regulated exons.
Discussion
The serine/arginine-rich protein family of splicing factors is believed to play a key role in all regulated splicing events studied to date (Lin and Fu, 2007; Long and Caceres, 2009). The well-established concepts for the function of SR protein in regulated splicing include: (i) SR proteins preferentially bind to exonic sequences to promote splice site selection; (ii) Each SR protein has preferred binding site sequences in RNA, although different SR proteins may have shared binding sites; (iii) Individual SR proteins appear to have both unique and redundant functions in splicing; (iv) SR proteins are largely responsible for promoting exon inclusion; and finally (v) The function of SR proteins in splice site selection can be antagonized by heterogeneous nuclear ribonucleoprotein (hnRNP) proteins (Lin and Fu, 2007). By determining in vivo targets for the two classic SR family members and linking their RNA binding activities to functions in regulated splicing, this study has now elucidated several new principles for the function and mechanism of action of SR proteins in mammalian cells.
Most exons are recognized by more than one SR protein
Consistent with their roles in constitutive and regulated splicing by binding exonic splicing enhancers, we found that both SRSF1 and SRSF2 crosslink extensively and preferentially to exons. Interestingly, both of these abundant and ubiquitous SR proteins show extensive overlap in their interactions with RNA, which is quite distinct from the reported binding profiles for another two SR proteins SRSF3 and SRSF4 (Anko et al., 2010; Anko et al., 2012). This raises an intriguing possibility that the SR family members might be composed of those that bind broadly to most exonic sequences, and those that bind to more restricted sets of exons. It is also interesting that SRSF2 binds to both pre-mRNA and spliced mRNA. Because SRSF2 is a non-shuttling SR protein, this observation implies that it has to be removed or exchanged with other shuttling SR proteins prior to mRNA export, as noted earlier (Lin et al., 2005), which may constitute a previously unrecognized step in the nucleus for the regulation of mRNA export.
SR proteins are believed to bind RNA with distinct sequence specificity. However, it has been difficult to obtain well-defined consensus motifs for each SR protein, information important for understanding splicing regulatory networks. For example, early SELEX studies based on in vitro binding or in vitro splicing function provide binding specificities for SR proteins that appear quite degenerate, although some general trends can be deduced (Cavaloc et al., 1999; Liu et al., 2000; Liu et al., 1998; Tacke and Manley, 1995). A degenerate binding mode for SRSF2 has recently been reenforced by the proposed new consensus of the core SSNG motif from structural analysis (Daubner et al., 2012). Our in vivo mapping data largely confirmed the proposed binding specificity for the two SR proteins, suggesting that SRSF1 generally prefers GA-rich sequences while SRSF2 recognizes a GC-rich core. A possible selective advantage for having multiple SR proteins function through degenerate recognition sequences may be to ensure efficient inclusion while still accommodating evolutionary variations in exon sequence due to protein coding, and as levels of individual SR proteins vary in different cell types or during development.
RNA binding of SR proteins compensates for weak splice sites and intron length
Another proposed function of SR proteins is to compensate for weak splice sites in mammalian genomes. Interestingly, the majority of exons in the human genome are short and introns are long (Sakharkar et al., 2004). While we found that SR protein binding (as measured by CLIP peak strength) is inversely correlated with the strength of nearby splice sites, a more striking feature of our data is the correlation between SR protein binding and intron length. Effect of intron length has not been well studied in vitro because substrates with short introns are easier to prepare, yet authentic introns vary considerably with many up to 100 kb in length. As intron size increases, the ability of the cell to identify authentic exons from an increasing mass of intron sequence may demand more efficient SR protein binding.
Genome-wide comparison of constitutive and alternative exons has revealed that alternative exons contain fewer ESEs, which coupled with weaker splice sites, render their inclusion inefficient (Fairbrother et al., 2002; Itoh et al., 2004; Wang et al., 2005; Zhang and Chasin, 2004). This is consistent with our finding that the major SR proteins bind more weakly on alternative exons than flanking constitutive exons. This also agrees with the idea that alternative exons are less efficiently recognized by general splicing factors including the ubiquitous SR proteins so that they can become included only upon the appearance of appropriate cell-type specific or developmentally regulated splicing factors in the cell. Our results reinforce the idea that exon recognition involves a complex interplay of splice site strength, intron length, and binding of splicing enhancing factors, such as SR proteins.
SR proteins create appropriate exon usage by influencing both exon inclusion and skipping in vivo
Perhaps one of the most striking findings here is the involvement of SR proteins in both exon inclusion and skipping events, which contradicts the common assumption that SR proteins mainly function in promoting exon inclusion. Because SRSF1 and SRSF2 often bind to the same exons, we were further surprised by the apparently specialized functions of SR proteins in including discrete sets of exons in the mouse transcriptome. The two major SR proteins exhibit non-redundant functions on some exons, and jointly regulate a smaller fraction of events.
A similar observation was also made recently with the hnRNP family of splicing regulators (Huelga et al., 2012). Again, contrary to expectation that hnRNP proteins antagonize SR proteins to cause exon skipping, many hnRNP proteins appear to function in promoting both exon inclusion and skipping in vivo at different exons. These observations indicate the need to revise the idea that SR proteins are predominantly positive splicing regulators while hnRNP proteins are predominantly negative splicing regulators, given their in vivo effects on both exon inclusion and skipping, as well as our evidence that loss of one SR protein remodels the binding profile of another.
Coordinated action of SR proteins is responsible for complex splicing outcomes
We initially considered the complex effects of SR proteins on regulated splicing to reflect their position-dependent activities, which have been demonstrated on model genes (Han et al., 2011a; Sanford et al., 2009). However, the lack of the anticipated binding site distributions on competing exons from the composite RNA map generated for both SRSF1 and SRSF2 challenges this interpretation. We then reasoned that this simple idea might be confounded by compensatory responses of other SR proteins and splicing regulators upon depletion of an individual member of the family. To test this idea in principle, we profiled SRSF1 binding before and after knocking down SRSF2 and found that depletion of SRSF2 caused both gain and loss of SRSF1 binding at many exons. This means that, to understand SR protein function, we must consider that specific binding events by one SR protein are in competition with binding of other SR proteins and splicing regulators nearby. We further showed by mutational analysis that prevention of such compensatory binding is sufficient to alter the splicing response from SR protein depletion-induced exon inclusion to exon skipping. Therefore, the complex response observed in cells depleted of a specific SR protein likely result from the loss of function of that SR protein in combination with coordinated alteration of binding (and activity in splicing) of other splicing regulators at many exons near the binding sites of the missing protein. These findings provide a conceptual framework to understand complex effects of SR proteins and perhaps other splicing regulators in vivo.
Materials and Methods
Cell culture, RNA extraction, and splicing profiling
The SRSF1 and SRSF2 tet-repressible MEFs were maintained in DMEM plus 10% FBS without tetracycline. SRSF1 or SRSF2 protein was depleted by growing the cells for the indicated period of time in the presence of 10 μl/ml of Dox. RNA isolation, RT-PCR and splicing profiling were carried out according to established standard protocols, which are detailed in the supplement.
Plasmid construction
CDC45I exons 3–5 and CCNL exons 3–5 were PCR amplified and cloned into pcDNA 3.1(+) at the BamH1 and HindIII sites. The mutant plasmids were generated using deletion primers adjacent to the binding site. SRSF2 MEF cells were transfected with the indicated plasmids using Lipofectamine 2000 (Life Technology). Six hours after incubating lipofectamin-plasmid complexes with the cells, the media was changed to serum either containing Dox or without it (to induce SRSF2 depletion). After 48 hrs, the cells were harvested for RNA and protein analyses.
Analysis of genome-wide data
CLIP-seq was carried out as previously described (Xue et al., 2009; Yeo et al., 2009) using anti-HA antibody (Abcam ab9110). For surveying SR protein binding in the absence of another SR protein, we treated SRSF1-HA MEFs with siRNA against SRSF2 (Dharmacon onTargetplus J-044306-05). All analysis of CLIP-seq data was done using custom Python and R scripts, BEDTools (Quinlan and Hall, 2010) and Kent source package (Kent et al., 2002) as detailed in the supplement, which also contains the procedures for correlative analysis of SR protein binding and splicing signals, motif analysis, construction of RNA maps, and in vivo RNA interactions of one SR protein in the absence of another SR proteins.
Accession Numbers
Microarray data and CLIP-seq data for SRSF1 and SRSF2 are available at the Gene Expression Omnibus under the accession number GSE44583 and GSE44591, respectively.
Supplementary Material
Highlights.
The two SR proteins under study overlap extensively in binding to exons
SR proteins are prevalently involved in both exon inclusion and skipping
SR protein binding to RNA are both positively and negatively coordinated
Individual SR proteins regulate splicing in synergy with other SR proteins
Acknowledgments
We thank Dr. Chris Brenner for advice on motif analysis, and members of the Fu lab for ideas and extensive discussion during the course of this investigation. This work was supported by NIH grants (GM049369 and HG004659), the China 973 program (2011CB811300), the China 111 program grant (B06018) to X-D.F and NIH grant (GM084317) to G.Y. and M.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anko ML, Morales L, Henry I, Beyer A, Neugebauer KM. Global analysis reveals SRp20- and SRp75-specific mRNPs in cycling and neural cells. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1862. [DOI] [PubMed] [Google Scholar]
- Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome biology. 2012;13:R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Stuani C, De Prato G, Baralle FE. SR protein-mediated inhibition of CFTR exon 9 inclusion: molecular characterization of the intronic splicing silencer. Nucleic acids research. 2007;35:4359–4368. doi: 10.1093/nar/gkm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes & development. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic acids research. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA (New York, NY) 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubner GM, Clery A, Jayne S, Stevenin J, Allain FH. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. The EMBO journal. 2012;31:162–174. doi: 10.1038/emboj.2011.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nature structural & molecular biology. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkelenz S, Mueller WF, Evans MS, Busch A, Schoneweis K, Hertel KJ, Schaal H. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2012 doi: 10.1261/rna.037044.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeh RF, Sharp PA, Burge CB. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Dejardin J, Tazi J, Soret J. The SR family proteins B52 and dASF/SF2 modulate development of the Drosophila visual system by regulating specific RNA targets. Molecular and cellular biology. 2007;27:3087–3097. doi: 10.1128/MCB.01876-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego ME, Gattoni R, Stevenin J, Marie J, Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. The EMBO journal. 1997;16:1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell. 2005;20:881–890. doi: 10.1016/j.molcel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1:765–771. doi: 10.1016/s1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- Han J, Ding JH, Byeon CW, Kim JH, Hertel KJ, Jeong S, Fu XD. SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons. Molecular and cellular biology. 2011a;31:793–802. doi: 10.1128/MCB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Xiong J, Wang D, Fu XD. Pre-mRNA splicing: where and when in the nucleus. Trends in cell biology. 2011b;21:336–343. doi: 10.1016/j.tcb.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlioglu N, Wang J, Fushimi K, Vibranovski MD, Kan Z, Gish W, Fedorov A, Long M, Wu JY. An intronic signal for alternative splicing in the human genome. PLoS One. 2007;2:e1246. doi: 10.1371/journal.pone.0001246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ, Graveley BR. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends in biochemical sciences. 2005;30:115–118. doi: 10.1016/j.tibs.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell reports. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh H, Washio T, Tomita M. Computational comparative analyses of alternative splicing regulation using full-length cDNA of various eukaryotes. RNA (New York, NY) 2004;10:1005–1018. doi: 10.1261/rna.5221604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire R, Winne A, Sarkissian M, Lafyatis R. SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation. European journal of immunology. 1999;29:823–837. doi: 10.1002/(SICI)1521-4141(199903)29:03<823::AID-IMMU823>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Li H, Qiu J, Fu XD. RASL-seq for massively parallel and quantitative analysis of gene expression. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. Unit 4. Chapter 4. 2012. pp. 13pp. 11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Fu XD. SR proteins and related factors in alternative splicing. Advances in experimental medicine and biology. 2007;623:107–122. doi: 10.1007/978-0-387-77374-2_7. [DOI] [PubMed] [Google Scholar]
- Lin S, Xiao R, Sun P, Xu X, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Molecular cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Liu HX, Chew SL, Cartegni L, Zhang MQ, Krainer AR. Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Molecular and cellular biology. 2000;20:1063–1071. doi: 10.1128/mcb.20.3.1063-1071.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Zhang M, Krainer AR. Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. The Biochemical journal. 2009;417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Mayeda A, Munroe SH, Caceres JF, Krainer AR. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. The EMBO journal. 1994;13:5483–5495. doi: 10.1002/j.1460-2075.1994.tb06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakharkar MK, Chow VT, Kangueane P. Distributions of exons and introns in the human genome. In silico biology. 2004;4:387–393. [PubMed] [Google Scholar]
- Sanford JR, Coutinho P, Hackett JA, Wang X, Ranahan W, Caceres JF. Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One. 2008;3:e3369. doi: 10.1371/journal.pone.0003369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y. Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome research. 2009;19:381–394. doi: 10.1101/gr.082503.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaal TD, Maniatis T. Selection and characterization of pre-mRNA splicing enhancers: identification of novel SR protein-specific enhancer sequences. Molecular and cellular biology. 1999;19:1705–1719. doi: 10.1128/mcb.19.3.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20:1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Molecular cell. 2004;13:367–376. doi: 10.1016/s1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- Solis AS, Peng R, Crawford JB, Phillips JA, 3rd, Patton JG. Growth hormone deficiency and splicing fidelity: two serine/arginine-rich proteins, ASF/SF2 and SC35, act antagonistically. The Journal of biological chemistry. 2008;283:23619–23626. doi: 10.1074/jbc.M710175200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. The EMBO journal. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods (San Diego, Calif) 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic acids research. 2005;33:5053–5062. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J. A splicing-independent function of SF2/ASF in microRNA processing. Molecular cell. 2010;38:67–77. doi: 10.1016/j.molcel.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Tang P, Yang B, Huang J, Zhou Y, Shao C, Li H, Sun H, Zhang Y, Fu XD. Nuclear matrix factor hnRNP U/SAF-A exerts a global control of alternative splicing by regulating U2 snRNP maturation. Molecular cell. 2012;45:656–668. doi: 10.1016/j.molcel.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Molecular cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nature structural & molecular biology. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Darnell RB. Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nature biotechnology. 2011;29:607–614. doi: 10.1038/nbt.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes & development. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Molecular cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG, et al. The Akt-SRPK-SR Axis Constitutes a Major Pathway in Transducing EGF Signaling to Regulate Alternative Splicing in the Nucleus. Molecular cell. 2012;47:422–433. doi: 10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.