Fig. 2.
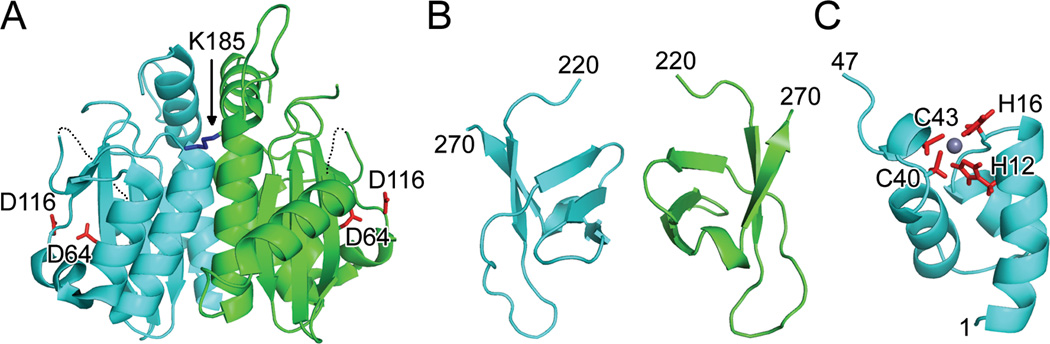
Structures of individual HIV-1 IN domains. (A) X-ray crystal structure (PDB code 1THG) highlighting the common CCD dimer and active site residues Asp64 and Asp116 (red sticks); the third member of the DDE catalytic triad, Glu152, was undecipherable in this initial structure. Only one of two Lys185 solubility-enhancing substitutions (blue stick) is visible in this orientation. Dashed lines indicate disordered gaps in polypeptide chains. (B) CTD NMR structure (PDB code 1IHV) revealed a dimer, with each monomer folded into a five-stranded beta barrel. (C) HIV-1 NTD monomer from PDB code 1WJC. Highlighted are Zn-coordinating residues His12, His16, Cys 40, and Cys43 as well as the bound Zn2+ ion (grey sphere). Visualized IN residue termini are indicated in panels B and C.
