Abstract
Basal-like breast cancer (BBC) is an aggressive subtype of breast cancer that has no biologically-targeted therapy. The interactions of BBCs with stromal cells are important determinants of tumor biology, with inflammatory cells playing well-recognized roles in cancer progression. Despite the fact that macrophage-BBC communication is bidirectional, important questions remain about how BBCs affect adjacent immune cells. This study investigated monocyte-to-macrophage differentiation and polarization, and gene expression in response to coculture with basal-like versus luminal breast cancer cells. Changes induced by coculture were compared to changes observed under classical differentiation and polarization conditions. Monocytes (THP-1 cells) exposed to BBC cells in coculture had altered gene expression with upregulation of both M1 and M2 macrophage markers. Two sets of M1 and M2 markers were selected from the PCR profiles and used for dual immunofluorescence staining of BBC versus luminal cocultured THP-1s, and cancer-adjacent, benign tissue sections from patients diagnosed with BBC or luminal breast cancer confirming the differential expression patterns. Relative to luminal breast cancers, BBCs also increased differentiation of monocytes to macrophages and stimulated macrophage migration. Consistent with these changes in cellular phenotype, a distinct pattern of cytokine secretion was evident in macrophage-BBC cocultures, including upregulation of NAP-2, Osteoprotegerin, MIG, MCP-1, MCP-3 and IL-1β. Application of IL-1 receptor antagonist (IL-1RA) to cocultures attenuated BBC-induced macrophage migration. These data contribute to an understanding of the BBC-mediated activation of the stromal immune response, implicating specific cytokines that are differentially expressed in basal-like microenvironments and suggesting plausible targets for modulating immune responses to BBC.
Keywords: breast cancer, basal-like, macrophage, differentiation, polarization
INTRODUCTION
Basal-like breast cancer (BBC) is highly aggressive and disproportionately impacts young African-American women (1, 2). Several studies suggest that breast cancer initiation and progression is linked to inflammation [reviewed in (3)], and investigations have shown a significant increase in the presence of innate and adaptive immune cells, including B cells, T cells, eosinophils, and macrophages in malignant tissues (4–6). In particular, the presence of macrophages or proliferating macrophages appears to be associated with BBC (4, 7). We recently observed that BBC-stromal interactions may result in elevated expression of cytokines, including cytokines that affect macrophage phenotype (8). To date, the signaling pathways that modify the cross-talk between basal-like cells and immune cells in the inflammatory microenvironment of BBCs remain uncertain, with gaps in understanding of how BBCs alter the behavior and phenotype of macrophages.
Macrophages are white blood cells differentiated from monocytes, arising from progenitor cells in the bone marrow (9). As monocytes migrate from peripheral blood into tissue, they differentiate and are referred to as resident macrophages and primarily perform the homeostatic function of debris clearance (10, 11). However, in response to damage-induced inflammation, monocytes migrate into injured tissue where they differentiate and polarize to phagocytize cellular debris, fight infection, and secrete compounds that promote wound healing (10, 12, 13). Macrophages can also promote the development of tumors (14), consistent with observations that wound healing and macrophage responses are correlated with tumor progression (15–18). Tumor-associated macrophages, a subpopulation of M 2-tropic macrophages (M2-macrophages), have been well-documented as the primary class of macrophages in basal cell carcinoma, lung, melanoma, and thyroid cancers (14, 19–22). They enhance tumor growth by stimulating angiogenesis and growth factor secretion (19, 22, 23), suppressing other immune responses (19, 24) or facilitating invasion and metastasis (4, 21).
In light of the important role of macrophages in breast cancer progression, we performed studies focused on the communication between macrophages and BBC. We evaluated whether macrophages have unique interactions with BBCs compared to another, less aggressive breast cancer subtype. Based on our previous findings suggesting elevated cytokine expression due to basal-like interactions with stromal fibroblasts (7), we hypothesized that BBCs recruit and polarize macrophages more effectively than luminal breast cancers. Using the human, monocytic cell line THP-1 as a stable, established source of monocytes, we evaluated the differentiation and polarization responses of monocytes to BBC cells versus luminal breast cancer cells. Our results demonstrate that BBC cells actively drive monocyte-to-macrophage differentiation and elucidate a novel IL-1β-dependent mechanism by which BBCs modulate the microenvironment and promote a pro-tumorigenic milieu.
METHODS
Cell culture
The BBC cell lines used in cocultures were HCC1937, MDA-MB-468, and SUM149. Luminal breast cancer cell lines used included MCF-7, T47D and ZR-75-1. All cell lines were purchased from ATCC and cultured in RPMI media (Gibco/Invitrogen) supplemented with 10% FBS and penicillin/streptomycin. All ATCC cell lines undergo authentication tests during the accessioning process, which is described in the online ATCC brochure Maintaining High Standards in Cell Culture (www.atcc.org). THP-1 cells are an immortalized human cell line cultured from the blood of a patient with acute monocytic leukemia (25). After stringent characterization and validation by Tsuchiya et al., these cells were found to retain all the necessary markers and morphological features to be classified as a monocytic cell population, able to undergo differentiation and polarization into functional, mature macrophages (26). Thus, we have chosen to use them as our model system for ease of handling and high reproducibility of results. THP-1 cells were maintained as a suspension culture at a density of 800,000–1,000,000 cells/mL. Each cell line was individually cocultured with THP-1 cells.
Differentiation of THP-1 monocytes to macrophages
To establish a positive control, THP-1 cells were differentiated and polarized according to established protocols (20, 27), with minor modifications. Briefly, one million cells were plated in 6-well plates in 3 mL RPMI media plus 25 ng/mL phorbol myristate acetate (PMA, Sigma) for 48 hours of treatment to induce differentiation to macrophages. To polarize, THP-1 macrophages were PMA treated for 48 hours, with addition of M1-polarizing cytokines [20 ng/mL interferon-gamma (IFN-γ) and 100 ng/mL lipopolysaccharide (LPS) (Prospec)] or M2-polarizing cytokines [20 ng/mL interleukin-4 (IL-4) and 20 ng/mL IL-13 (Prospec)] during the final 18 hours of treatment. This generated three macrophage populations: PMA-differentiated macrophages, M1-polarized macrophages and M2-polarized macrophages. Changes induced in cocultures were compared against these positive controls. To coculture THP-1 cells with cancer cells, one million basal-like (HCC1937, MDA-MB-468, and SUM149) or luminal (MCF7, T47D and ZR-75-1) breast cancer cell lines were plated into 0.4 μM pore inserts of 6-well transwell plates in 2 mL of media, with 1mL of media added to the bottom of the well. After conditioning the media for 24 hours, one million THP-1 cells were plated into the bottom wells of the transwell plates in 1 mL of additional media. Cocultures were maintained for an additional 48 hours.
Analysis of THP-1 differentiation and polarization
To calculate the average % differentiation of THP-1 cells following cytokine treatment or coculture, four replicates were plated with a set number of cells and after 48 hours the suspended, unattached cells were counted. Differentiated/attached cells were calculated by subtraction assuming no substantial increase in total cell number. Trypan blue was used to evaluate cell viability for all cell counts. To complement these quantitative assessments of percent differentiation, monocyte-to-macrophage differentiation and polarization was confirmed based on characteristic morphological changes and phagocytic function. Increased vacuolization and lysosomal compartmentalization, formation of migratory actin cytoskeletal architecture (i.e. filopodia and lamellipodia), and fibroblast-like changes indicative of polarization were observed and photographed (Fig. 1). Also, polystyrene micro-particles (1μm size, Sigma) were diluted 1:1000, vortexed, and added to treatment media at the end of the 48 hour treatment/coculture. The ability of the THP-1 cells to phagocytize micro-particles was monitored on an Olympus IX-70 microscope by time-lapse capture in two minute intervals at 40X magnification for 4 hours. Movies demonstrating phagocytic function of THP-1 cells in response to coculture can be viewed in the Supplemental material (Supplementary Movies_MCF-7 and _SUM149).
Figure 1. THP-1 phenotypes in response to classical stimuli.
THP-1 cells were differentiated and/or polarized with classically-defined stimuli [PMA for differentiation into macrophage and M1-trophic (IFN-γ and LPS) or M2-trophic (IL-4 and IL-13) cytokines for polarization]. (A) THP-1 cells differentiated (78–83%) in treated compared to undifferentiated (UND) conditions (5%), and (B) treated cells undergo characteristic morphological changes, including attachment, increased vacuolization, fibroblast-like changes, and in the M1- or M2-treated cells, formation of lamellipodia and filopodia at their ends. (C) Treated cells were immune-positive for M1-predominant (IL-7R (red), iNOS (blue)) and M2-predominant (CD36 (green), CD163 (magenta)) proteins, with two sets of markers documenting subpopulations of macrophages that are dual positive (yellow) for M1 and M2 markers.
Immunofluorescent staining of differentiated and polarized THP-1 macrophages
Immunofluorescent imaging of cocultures was performed to evaluate classic protein markers of M1 or M2 polarization. Prior to cell plating, 22×22–1.5 glass coverslips (Fisher) were added to the bottom of the wells. Following treatment with chemicals or coculture, these coverslips were fixed and stained for the expression of M1 macrophage markers NOS2a/iNOS (C-11, Santa Cruz Biotechnology) and IL-7R (C-20, Santa Cruz Biotechnology), or M2 macrophage markers CD36 (5–271, BioLegend, Inc.) and CD163 (RM3/1, BioLegend, Inc.), according to standard protocols. Briefly, wells were rinsed with 1X PBS and fixed with cold methanol (−10°C) for 10 minutes. After air drying, cells were washed three times with 1X PBS for 5 minutes, and incubated with 10% normal goat or mouse serum to block for 20 minutes (depending on host antibody). After washing with 1X PBS for 5 minutes, cells were incubated with primary antibody for 1 hour at a concentration of 5.0 μg/mL in PBS plus 1.5% normal goat or mouse serum. Wells were washed three times with 1X PBS for 5 minutes and incubated 45 minutes with Alexa-568 (IL-7R, red), Alexa-488 (CD36, green), Alexa-405 (NOS2a, blue), and Alexa-647 (CD163, magenta) fluorochrome-conjugated secondary antibodies (Molecular Probes) diluted to 2 μg/mL in PBS with 2% normal goat or mouse serum in the dark. Coverslips were washed, removed from the wells and mounted with glycerin for imaging on an Olympus BX-61 microscope at 20X magnification.
RNA isolation and microarray gene expression profiling of control system-differentiated THP-1 cells
RNA was isolated from THP-1 cells using the RNeasy kit (Qiagen). RNA purity and integrity were confirmed by Agilent 2100 Bioanalyzer. THP-1 cells were run in triplicate (three independent experiments) on 4×44K human whole genome microarray slides (Agilent G4112F) according to the Agilent protocol (8). Cy3-labeled reference was Stratagene Universal Human Reference spiked 1:1,000 with MCF-7 RNA and 1:1,000 with ME16C RNA, and Cy-5 labeled cDNAs were from THP-1 total RNA. Labeled cDNAs were hybridized to arrays overnight and washed before scanning on an Agilent G2505C microarray scanner. All array data are available through the Gene Expression Omnibus.
Microarray data analyses
Only genes where more than 70% of microarrays had signal greater than 10 dpi in both channels were included. Data were Lowess normalized and missing data was imputed using k-nearest neighbors’ imputation (with k=10). Data were analyzed by multiclass SAM (Significance Analysis of Microarrays) to identify genes that significantly changed by treatment (4 classes: undifferentiated (monocytes), or PMA-differentiated, M1-polarized, M2-polarized macrophages). Significant genes were evaluated for ontological enrichment using Ingenuity Pathway Analysis (IPA), with Benjamini–Hochberg (B-H) multiple testing correction for the two polarized classes. Significant functions and pathways were defined as those with B-H p-values less than 0.05. Hierarchical clustering with 250 literature-defined differentiation and polarization markers (Supplementary Table 2) was also performed to evaluate concordance between the literature and the expression profiles of our THP-1 positive control (PMA-differentiated, M1- and M2-polarized) macrophages. A set of 43 markers (listed in Table 1) were selected based on concordance between literature and our microarray data and a custom, quantitative RT-PCR array was developed for assaying THP-1 mRNA expression changes in response to breast cancer coculture microenvironments.
Table 1.
Gene targets and controls used to assess macrophage phenotype by RT-PCR.
| Gene Symbol | Gene Name, Entrez Gene ID | Higher in Mo/M1/M2 |
|---|---|---|
| CD11b | Itgam, Integrin, alpha M, 3684 | M2 |
| CD11c | Itgax, Integrin, alpha X, 3687 | M1 |
| CD36 | Leukocyte differentiation antigen, 948 | M2 |
| CD68 | CD68 molecule, 968 | M1 & M2 |
| CD163 | CD163 molecule, 9332 | M2 |
| CD206 | Mannose receptor C type 1, MRC1, 4360 | M2 |
| CCL19 | Chemokine ligand 19, MIP-3b, 6363 | M1 |
| CCL20 | Chemokine ligand 20, MIP-3a, 20297 | M1 |
| CCR2 | Chemokine receptor 2, CD192, MCP-1R, 729230 | Mo |
| CCR7 | Chemokine receptor 7, CD197, 1236 | M1 |
| CXCL1 | Chemokine (C-X-C motif) ligand 1, NAP-3, 2919 | M1 |
| CXCL3 | Chemokine (C-X-C motif) ligand 3, MIP-2b, 2921 | M1 |
| CX3CR1 | Chemokine (C-X3-C motif) receptor 1, 1524 | Mo |
| IL-1b | Interleukin 1, beta; IL1F2, 3553 | M1 |
| IL-6 | Interleukin 6 (interferon, beta 2), 3569 | M1 |
| IL-7R | Interleukin 7 receptor, CD127, IL7RA, 3575 | M1 |
| IL-8 | Interleukin 8, CXCL8, NAP1, 3576 | M1 |
| IL-12 | Interleukin 12A, p35, 3592 | M1 |
| IL-23 | Interleukin 23, alpha, 51561 | M1 |
| Arg 1 | Arginase, liver, 383 | M2 |
| BCL2A1 | BCL2-related protein A1, 597 | M1 |
| COX2 | Prostaglandin-endoperoxide synthase 2, PTGS2, 5743 | M1 |
| BIRC3 | Baculoviral IAP repeat-containing 3, 330 | M1 |
| CSF-1 | Colony stimulating factor 1 (MF), M-CSF, 1435 | M2 |
| CSF-2 | Colony stimulating factor 2 (granulocyte-macrophage), GM-CSF,1437 | M1 |
| EDN1 | Endothelin 1,1906 | M1 |
| F4/80 | Zinc finger protein 808, EMR1, 2015 | M1 & M2 |
| FGL2 | Fibrinogen-like 2, fibroleukin,10875 | M2 |
| FN1 | Fibronectin 1,2335 | M2 |
| HS3ST1 | Heparan sulfate (glucosamine) 3-O-sulfotransferase 1, 9957 | M1 |
| HSD11B1 | Hydroxysteroid (11-beta) dehydrogenase 1, 3290 | M2 |
| INDO | Indoleamine 2,3-dioxygenase 1, IDO1, 3620 | M1 |
| iNOS | Nitric oxide synthase 2, inducible; NOS2A, 4843 | M1 |
| LIPA | Lipase A, lysosomal acid, cholesterol esterase, 3988 | M2 |
| MAF | v-Maf musculoaponeurotic fibrosarcoma oncogene homolog, 4094 | M2 |
| MMP14 | Matrix metallopeptidase 14, 4323 | M2 |
| MS4A4A | Membrane-spanning 4-domains, subfamily A-member 4, 51338 | M2 |
| MSR1 | Macrophage scavenger receptor 1, CD204, 4481 | M2 |
| OASL | 2′-5′-oligoadenylate synthetase-like, 8638 | M1 |
| OPN | Osteopontin; SPP1, Secreted phosphoprotein 1, 6696 | M1 |
| PTX3 | Pentraxin 3, long; TNFAIP5, 5806 | M1 |
| TNF-a | Tumor necrosis factor (superfamily member 2), 7124 | M1 |
| VEGF-a | Vascular endothelial growth factor A, 7422 | M2 |
| GAPDH | Gyceraldehyde-3-phosphate dehydrogenase, 2597 | N/A |
| WWP1 | Ubiquitin, protein ligase 1, 11059 | N/A |
| HGDC | Human Genomic DNA Contamination | N/A |
| RTC | Reverse Transcription Control | N/A |
| PPC | Positive PCR Control | N/A |
PCR array expression and analysis of THP-1 cells cocultured in BBC or luminal microenvironments
Quantitative real-time PCR analysis was performed according to manufacturer’s protocol (SA Biosciences) using an Applied Biosystems 7900HT Fast PCR machine. Relative expression values from THP-1 cocultures were obtained by normalizing to GAPDH expression and then normalizing to undifferentiated THP-1 monocytes. Hierarchical cluster analysis was performed based on 38 of 43 markers that showed substantial variation across the BBC and luminal cocultures. CD11c, F4/80, FN1, PTX3, TNF-α data were excluded from further analyses due to low variation.
Immunofluorescent staining of cancer-adjacent, normal tissues from BBC or luminal breast cancer patients
Patient samples were obtained from the University of North Carolina, Lineberger Comprehensive Cancer Center’s Tissue Procurement Facility, with informed consent and under a School of Medicine Institutional Review Board-approved protocol. Histologically normal-appearing sections adjacent to tumors were selected from patients diagnosed with invasive primary tumors and classified as either the BBC or Luminal subtype of breast cancer using the PAM50 algorithm (28) as applied to microarray data collected on adjacent tumors (29). Neither of the patients with BBC had undergone neoadjuvant therapy prior to tissue resection.
Immunofluorescent imaging of tissues adjacent to the tumor microenvironment was performed to evaluate positive in vivo staining for the same M1 or M2 protein markers of macrophage polarization used to evaluate BBC or luminal-cocultured THP-1 cells. Four cancer-adjacent, benign tissue samples were selected based on the patients’ breast cancer subtype (BBC or Luminal A). Paraffin sections 5μM thick were cut and H&E stained by the UNC Histology Core. The additional sections were stained using standard protocols and the same antibody reagents used for in vitro immunofluorescence staining (described above).
Cytokine protein array expression profiling of cocultured THP-1 macrophages
Cytokine protein expression of undiluted culture media was analyzed for 80 cytokines on the Human Cytokine Antibody G Series 5 arrays (RayBiotech, Inc.) according to the manufacturer’s protocol. Cell media (500 μl) was collected into clean tubes, centrifuged at 8000 rpm for 3 min to pellet cellular debris then transferred to clean tubes and stored at −80°C. The glass chip was blocked and incubated overnight at 4°C with the experimental samples. The next day, secondary biotin-conjugated and streptavidin antibody incubations were performed, the slide was washed, air dried, and scanned on a GenePix 4000B scanner at a wavelength of 532 nm using GenePix Pro 4.1 software. Expression for each cytokine was first normalized to the internal control and fold-change was calculated by dividing the normalized expression in coculture by the sum of the normalized expression of corresponding monocultures (i.e. MCF7+THP-1 for MCF7:THP-1 cocultures). Cocultures with a cytokine ratio of at least 1.50 were considered significantly upregulated and those with a ratio less than or equal to 0.65 were significantly downregulated. Expression fold change ratios between 1–1.49 and 0.66–0.99 are classified as non-significantly upregulated and downregulated, respectively. Results are presented for the average of two biological replicates per group (Table 2).
Table 2.
Cytokine expression (ratio and fold change) for THP-1 cocultures with MCF-7 and SUM149 cellsa
| Cytokine | Ratio comparing THP- 1:MCF-7 cocultures to monocultures | Fold-change due to THP-1:MCF-7 coculture | Ratio comparing THP-1:SUM149 cocultures to monocultures | Fold-change due to THP-1:SUM149 coculture | |
|---|---|---|---|---|---|
| More substantially altered in SUM149 cocultures | NAP-2 | 0.74 | −1.35 | 8.95* | 7.95* |
| MCP-1 | 0.54* | −1.85* | 5.06* | 4.06* | |
| Osteoprotegerin | 1.39 | 0.39 | 2.56* | 1.56* | |
| IL-1 b | 0.71 | −1.41 | 1.83* | 0.83* | |
| MCP-3 | 0.62 | −1.61 | 1.83* | 0.83* | |
| MIG | 0.80 | −1.25 | 1.68* | 0.68* | |
| IL-5 | 0.75 | −1.33 | 1.51* | 0.51* | |
| GRO | 0.63* | −1.59* | 0.09* | −11.11* | |
| RANTES | 0.42* | −2.38* | 0.41* | −2.44* | |
| Fractalkine | 0.51* | −1.96* | 0.42* | −2.38* | |
| IL-12 p40p70 | 0.74 | −1.35 | 0.42* | −2.38* | |
| HGF | 0.59* | −1.69* | 0.46* | −2.17* | |
| MIP-1b | 0.61* | −1.64* | 0.52* | −1.92* | |
| PIGF | 0.69 | −1.45 | 0.55* | −1.82* | |
| TGF-b3 | 0.67 | −1.49 | 0.55* | −1.82* | |
| GDNF | 0.64* | −1.56* | 0.58* | −1.72* | |
| MIF | 0.73 | −1.37 | 0.62* | −1.61* | |
| GRO-a | 0.72 | −1.39 | 0.63* | −1.59* | |
| IL-4 | 0.68 | −1.47 | 0.63* | −1.59* | |
| IGFBP-1 | 0.72 | −1.39 | 0.63* | −1.59* | |
| FGF-7 | 0.78 | −1.28 | 0.64* | −1.56* | |
| FGF-4 | 0.70 | −1.43 | 0.64* | −1.56* | |
| PARC | 0.69 | −1.45 | 0.65* | −1.54* | |
|
| |||||
| More substantially altered in MCF-7 cocultures | LIF | 2.13* | 1.13* | 1.88* | 0.88* |
| Osteopontin | 1.90* | 0.90* | 1.70* | 0.70* | |
| IGFBP-2 | 0.38* | −2.63* | 0.39* | −2.57* | |
| ENA-78 | 0.46* | −2.17* | 0.49* | −2.04* | |
| MDC | 0.47* | −2.13* | 0.55* | −1.82* | |
| IL-13 | 0.55* | −1.82* | 0.69 | −1.44 | |
| Eotaxin-2 | 0.59* | −1.69* | 0.60* | −1.66* | |
| IL-1a | 0.61* | −1.64* | 1.23 | 0.23 | |
| IL-2 | 0.62* | −1.61* | 1.09 | 0.09 | |
| GCSF | 0.63* | −1.59* | 0.96 | −1.04 | |
| IL-15 | 0.65* | −1.54* | 1.03 | 0.03 | |
Statistically significant upregulation or downregulation is denoted with an asterisk.
Bold indicates upregulation.
Migration assays of differentiated THP-1 macrophages
To evaluate the migratory capacity of macrophages under different conditions, undifferentiated THP-1 cells were cultured in ultra-low attachment 6-well plates (Corning) for 48 hours with PMA alone or with breast cancer cells. Breast cancer cell lines were not suspension cultures; these cells were grown on inserts fitted to the 6-well low-attachment plates. Breast cancer cells and THP-1 communication was via soluble factors secreted into media. After brief trypsinization, loosely attached and suspended THP-1 cells were collected and centrifuged at 2000 rpm for 5 minutes. THP-1 cells were resuspended in 1 mL of fresh media, counted and 500,000 of coculture-treated cells were added to 8 μM migration inserts in 1.5 mL of media, with 1 mL of media added to the bottom of the migration chambers. Plates were incubated for 6 hours, then inserts were washed, fixed (10% neutral-buffered formalin, 5 minutes) and stained with 0.2 % crystal violet in 1X PBS. Total number of migrated cells were counted (4 fixed position fields/insert, 20X magnification) using Volocity software on an Olympus IX-81 microscope. For IL-1RA blocking experiments, 500 ng/mL of IL-1RA (R&D Systems, Inc.) was added to THP-1 macrophages in the resuspension media before plating into migration inserts. The average number of migrated cells was evaluated for THP-1 macrophages cocultured with MCF-7 and SUM149 cells, with and without addition of IL-1RA.
RESULTS
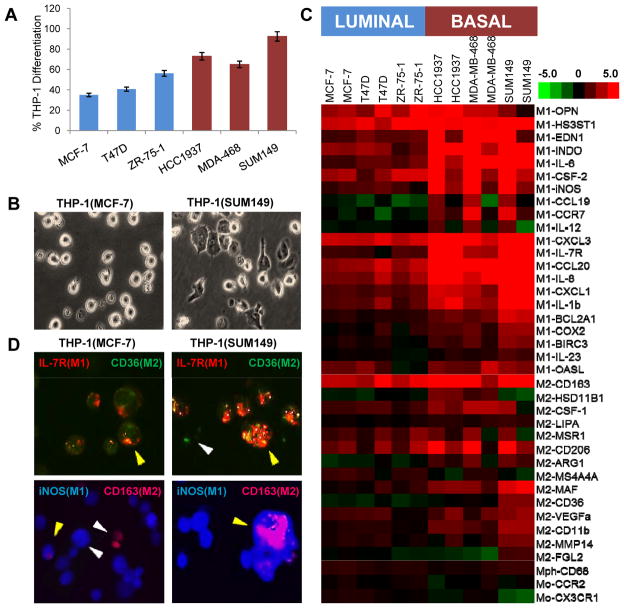
Basal-like breast cancer cells drive differentiation of THP-1 cells
THP-1 cells were treated according to control conditions (PMA to differentiate into macrophages and IFN-γ + LPS or IL-4 + IL-13 to polarize into M1 or M2 macrophages, respectively). Phenotypes of the three populations of macrophages generated by control treatment, including % differentiation, morphological characteristics and immunofluorescent staining of polarization markers, appear in Fig. 1A, B and C, respectively. These populations served as positive controls for comparison of THP-1 macrophage responses to coculture differentiation and polarization. The phenotype of THP-1 cells differed when cocultured with luminal (MCF-7, T47D, ZR-75-1) cells versus BBC (HCC1937, MDA-MB-468, SUM149) cell lines. After 48 hours in coculture, BBCs caused greater differentiation of THP-1 cells into macrophages. BBC cell lines induced approximately 56% differentiation, while luminal cells only produced 28% differentiation (two-tailed t-test p-value = 0.011, Fig. 2A). Similar to morphologic changes in positive control cells (Fig. 1B), morphological changes were observed in cocultured THP-1 macrophages, but were more distinct in BBC cocultures, with representative pictures of SUM149 (basal-like) and MCF-7 (luminal) cells shown in Fig. 2B. SUM149 cocultured THP-1 macrophages are primarily attached, have increased vacuole formation and show characteristics of polarization via cytoskeletal rearrangement, whereas MCF-7 cocultured THP-1 cells still largely resemble undifferentiated cells, though with some increased vacuolization compared to undifferentiated THP-1 monocytes. Both BBC- and luminal-cocultured THP-1 populations demonstrated the ability to phagocytize latex micro-particles (Supplementary Movie files).
Figure 2. Basal-like coculture drives THP-1 differentiation and polarization.
(A) BBC cells significantly differentiated (p-value=0.011) THP-1 cells into macrophages, with percentage differentiation roughly twice that of luminal cells. (B) Morphological differentiation of THP-1 cells is greater after coculture with SUM149 (basal-like) cells compared to MCF-7 (luminal) cells. (C) Similarly, BBC coculturing results in stronger overall q-RT-PCR expression of M1 and M2 markers and decreases in monocytic markers (CCR2 and CX3CR1), while THP-1s cocultured with luminal breast cancer cell lines continue to strongly express monocytic markers. (D) MCF-7 and SUM149 cocultured THP-1 cells were dual-stained for iNOS (blue, M1) and CD163 (magenta, M2) or IL-7R (red, M1) and CD36 (green, M2) using immunofluorescence, suggesting a mixed pattern of M1 and M2-polarized macrophage expression, especially in response to SUM149 cells. THP-1 phagocytic function to engulf latex micro-particles following coculture was also evaluated and time-lapse movies can be viewed in the Supplementary materials.
Gene expression data from cocultured cells mirrored phenotypic changes. Two of the genes assayed were predominantly monocytic markers, CCR2 and CX3CR1 (30). Although THP-1 cells cocultured with all three luminal cell lines increased differentiation as measured by elevated expression of CD68, a macrophage marker, the populations continued to express monocytic markers, indicating the persistence of larger numbers of monocytes in these cocultures (Fig. 2C). These cells may represent an “inflammatory monocyte” phenotype, with high expression of CCR2 and low expression of CX3CR1 (31). This class of monocytes is involved in triggering the immune response and in our coculture data appears to be more abundant in response to luminal cancers. Conversely, THP-1 cells exposed to BBC cells downregulate both CCR2 and CX3CR1, suggesting differentiation to more mature macrophages.
Basal-like breast cancer cells drive polarization of THP-1 cells
Because macrophages show plasticity that cannot be captured with a small number of markers, it was important to establish hallmark changes in THP-1 cells across a wide range of genes. It was also important to establish that the reproducible system based on an established cell line (THP-1 cells) reflected M1/M2 changes for primary peripheral blood mononuclear cells that more closely represent resident macrophages. Unsupervised hierarchical clustering of microarray data from PMA and M1- or M2-polarized THP-1 cells revealed reproducible patterns of gene expression (Supplementary Fig. 1A) with expected canonical pathway alterations in response to control conditions (Supplementary Tables 1 and 2). In addition, an extensive review of the literature identified 250 genes indicative of monocyte-to-macrophage differentiation and polarization (Supplementary Table 3). The list of genes was primarily derived from mouse models or investigations using peripheral blood monocytes. Of the 250 genes, 209 (84%) were similarly expressed in THP-1 cells (Supplementary Fig. 1B), demonstrating the usefulness of the optimized THP-1 control system to adequately define the monocyte-to-macrophage differentiation and polarization processes. Using markers that characterize the M1 and M2 polarization states of macrophages (Table 1), we found that relative to luminal breast cancers, basal-like breast cancers induce more widespread phenotypic changes associated with both M1 and M2 macrophage polarization. All three basal-like lines strongly induced THP-1 cells to differentiate and polarize, with the most robust response in SUM149 cells (Fig. 2C), a cell line isolated from inflammatory breast cancer (32), whereas several more of the polarization markers are downregulated in response to MCF-7, T47D or ZR-75-1 luminal breast cancer cells.
Given that BBCs showed increases in both M1 and M2 markers, we evaluated whether this mixed phenotype was due to a dual population of THP-1 cells (i.e. some M1 and some M2 polarized cells) or whether this was evidence of phenotypic plasticity using immunofluorescence. Two sets of M1- and M2-predominant markers were used. In Fig. 2D, both dual positive cells [staining for IL-7R (M1) and CD36 (M2) or iNOS (M1) and CD163 (M2)] and singly positive cells (exclusively expressing either M1 or M2 markers) are shown (33–36). This was similar to the positive controls (Fig. 1D), where a mixed population of M1- and M2-polarized macrophages was also observed. The percentages of positively stained THP-1 cells for CD36, IL-7R, iNOS and CD163 following each culture condition were 3, 0, 0, and 0% for UND, respectively; 39, 87, 62, and 21% for PMA, respectively; 45, 81, 89, and 54% for IFN-γ plus LPS, respectively; and 66, 34, 80 and 40% for IL-4 plus IL-13 treatment, respectively. This phenotypic duality, even in the presence of specific polarization stimuli, has been observed previously with macrophages (37, 38). Consistent with the q-RT-PCR data, the presence of a dual-stained macrophage population was more frequent in THP-1 cells cocultured with basal-like SUM149 cells compared to luminal MCF-7 cells; where the percentages of positively stained THP-1 cells for CD36, IL-7R, iNOS and CD163 were 23, 100, 83, and 29% with MCF-7 cocultured cells, respectively; and 69, 92, 95, and 32% with SUM149 cocultured THP-1 cells, respectively.
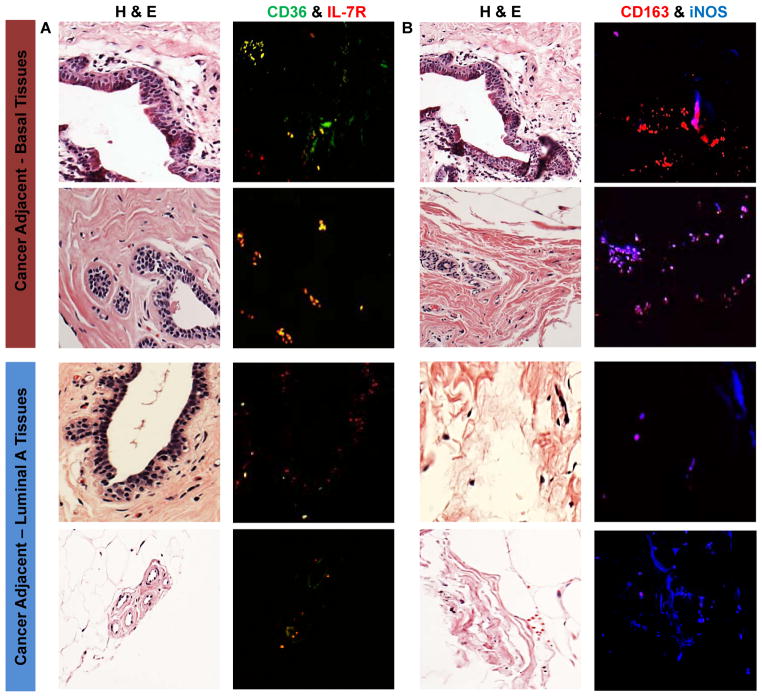
Cancer-adjacent tissue sections from patients with BBC or luminal breast cancer also express differential patterns of macrophage polarization markers
To determine whether the phenotypic patterns observed in THP-1 cocultures were also present within the stromal microenvironments of patients, we used the same M1 and M2 polarization marker pairs (CD36, IL-7R and CD163, iNOS) to dual-stain sections of histologically normal tissues adjacent to tumors. Figure 3A shows the H&E-stained sections for two BBC and two luminal cancer-adjacent tissues, along with the merged images of dual staining for CD36 (M2, green) and IL-7R (M1, red). Dual M1/M2 staining is more pronounced in the basal-adjacent tissues compared to the luminal-adjacent sections. A similar pattern of expression is observed with the CD163 (M2, red) and iNOS (M1, blue) stained tissues (Fig. 3B). Both staining pairs demonstrate there are more M2-positive macrophages in the basal-adjacent normal tissues (CD36-green and CD163-red) and more M1-postive macrophages in the luminal-adjacent normal tissues (IL-7R-red and iNOS-blue). Additionally, there are more dual-staining or mixed-phenotype macrophages in the basal-adjacent stromal compartments compared to the luminal (Fig. 3A-yellow and Fig. 3B-magenta), confirming the PCR expression profiles and recapitulating the in vitro patterns of macrophage expression in vivo.
Figure 3. Cancer-adjacent normal tissues express a similar in vivo pattern of macrophage polarization distinguishing BBC from luminal breast cancer.
(A) H&E stained sections for tissues adjacent to BBC or luminal breast cancer and dual stained immunofluorescent images with staining for M1/IL-7R (red) and M2/CD36 (green). (B) H&E sections for the same specimens, but with dual staining for M1/iNOS (blue) and M2/CD163 (red). More dual stained M1/M2 macrophages and higher numbers of M2 positive macrophages are observed in the BBC-adjacent tissues in this set of samples.
THP-1 cells express differential cytokine profiles in response to basal-like versus luminal coculturing
To identify the soluble cytokines that mediate the communication between BBCs and macrophages, we assayed expression of 80 cytokines using protein antibody arrays. THP-1:MCF-7 coculture resulted in 2 significantly upregulated (≥ 1.5 fold increase in expression) and 13 significantly downregulated (≤ 0.65 fold expressed) cytokines. In contrast, THP-1:SUM149 cocultures significantly upregulated and downregulated 9 or 20 cytokines, respectively (Table 2). The most highly upregulated cytokine in SUM149:THP-1 cocultures was a potent neutrophil chemoattractant NAP-2/CXCL7 (upregulated nearly 8-fold), but MCP-1, Osteoprotegerin, IL-1β, MCP-3, LIF, Ostoepontin, MIG, and IL-5 were also upregulated by at least 50%. Interestingly, G-CSF was significantly downregulated in MCF-7:THP-1 cocultures but not in SUM149:THP-1 cocultures.
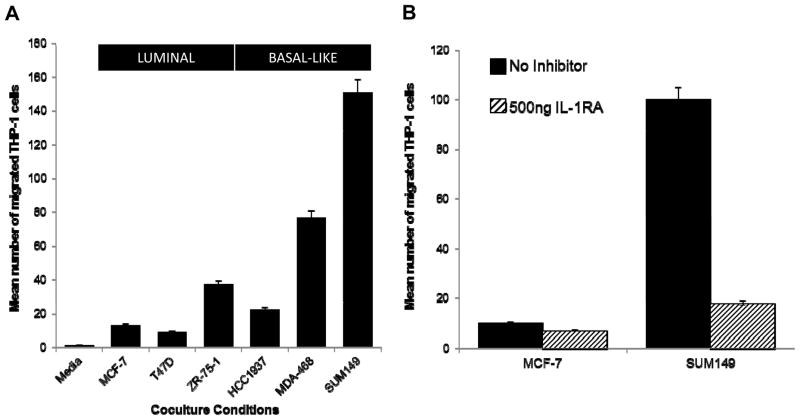
THP-1 cells become more migratory in response to BBC cells in an IL-1 dependent mechanism
Our previous results (8) found that IL-1β was altered in BBC cocultures with fibroblasts, so we were intrigued to observe that this cytokine was also upregulated at RNA and protein levels in BBC:THP-1 cocultures (Fig. 2C (RNA) and Table 2). To evaluate the role of IL-1β in BBC-stromal interactions, the antagonist, IL-1RA was used to block coculture-induced IL-1 signaling. Migratory function was significantly higher in macrophages that were cocultured with BBC cells (two-tailed t-test, p-value=0.039, Fig. 4A) and especially for SUM149-cocultured THP-1 macrophages. Addition of 500 ng of IL-1RA during the 6 hour migration assay dramatically diminished this BBC-induced THP-1 migration (Fig. 4B), but had no significant impact in MCF-7:THP-1 cocultures.
Figure 4. THP-1 migration responses to BBC coculture are reduced by treatment with IL-1RA.
(A) THP-1 cells cocultured with BBCs (HCC1937, MDA-MB-468, SUM149) have significantly greater migratory ability (p-value=0.039) than THP-1s cocultured with luminal cells (MCF-7, T47D, ZR-75-1). The bar chart represents the average of three independent experiments across all three luminal lines or BBC cell lines. (B) THP-1 migration induced by SUM149 coculture is attenuated by treatment with IL-1RA. THP-1 migration levels in BBC cocultures that result after blocking IL-1 receptors are similar to those observed following luminal coculturing.
DISCUSSION
The presence of intratumoral macrophages is associated with tumor progression (19) and with BBC subtype of breast cancer (7). Intravital imaging has demonstrated directly that macrophages intravasate mammary tumors during progression (39). In addition, low grade inflammation without overt clinical consequences, or ‘smoldering inflammation’ (40), has been implicated in cancer etiology. Consistent with this, we and others have recently observed that obese women at a higher risk for BBC and obese cancer patients have a higher prevalence of macrophages in breast adipose tissue (41, 42). Together these results suggest that both etiology and progression of BBCs may be affected by communication with macrophages.
Paracrine loops involving macrophages and breast cancer cells have previously emphasized the role of CSF-1 (39, 43). A CSF-1 signature has also been found in ductal carcinoma in situ (DCIS) (18) and has been shown to predict invasive breast cancer aggressiveness (16). Similarly, inflammatory monocytes with high CCL2 expression play an important role in breast cancer progression (44). Collectively, studies on macrophage-cancer cell interactions have emphasized the diversity of macrophages (19). As explained by Suzuki et al. (45), macrophages have complex transcriptomes designed to control a diversity of gene products for specialized tasks in a temporally and spatially defined manner. We were interested in understanding how BBCs interact with macrophages (similar to our work in fibroblasts (8)), but recognized that a means of characterizing the complex and dynamic changes in macrophages was important. In the current study, we have established and validated a panel of markers that can be used to identify subtypes of macrophages in cancer cell cocultures.
Our findings document that relative to luminal breast cancers, BBCs induce a distinct subpopulation of macrophages both in vitro and in vivo. In vitro, BBCs induced more macrophage differentiation, polarization and migration. Several other coculture systems, including cocultures with endothelial cells (46), adipocytes (47) and mesenchymal stem cells (48) have reported increases in cytokine levels when BBCs interact with stroma. In the case of macrophage-BBC interactions, it appears that the soluble interactions promote a population with features of both types. Mantovani and colleagues have suggested that macrophages in tumors are more similar to alternatively activated M2 macrophages (rather than activated M1 macrophages) (40), with gene profiling experiments on TAMs supporting these findings (49). However, tumor-associated-macrophages have tended not to adhere to these binary definitions (40) and instead often have similarities to both M1 and M2 macrophages, with particular similarities to macrophages present during development (e.g. increased Wnt signaling) (19, 21). Our in vivo findings in a limited number of patient specimens are concordant with our in vitro findings, showing more M1/M2 dual-stained macrophages in basal-like cases than in luminal cases. Future studies should validate our in vitro findings in larger tumor datasets, and should focus on identifying the cytokines that mediate the macrophage behavior in response to BBCs.
Among the cytokines differentially expressed in BBC-macrophage cocultures was IL-1β, which was upregulated by approximately 90%. Using IL-1RA to block the activity of IL-1β, we restricted the macrophage migration to levels observed in luminal cocultures. A previous report has documented increases in IL-1β upregulation in THP-1 cocultures with one basal-like cell line, HCC1937 (50), and demonstrated that IL-1β expression is upstream of COX-2 expression in the cancer cells. Our results extend the observations regarding IL-1β expression from a single cancer cell line to the class of basal-like cell lines and document a phenotypic effect of IL-1RA on macrophages. Furthermore, our results regarding basal-like specificity of the stroma-mediated IL-1β expression may help explain elevated COX-2 in BBCs (51) and in basal-like-stromal interactions (8). Further mechanistic details remain unresolved, but these results suggest that IL-1β may be an important regulator of BBC-stroma interactions. Previous studies have suggested an association between IL-1β expression and progression (52) and genetic variation in IL-1β has been linked with breast cancer survival (53). Likewise, IL-1RA is induced in some mesenchymal stem/stromal cell types (54) as a means of controlling inflammation. If fibroblasts and endothelial cells in the vicinity of the cancer mitigate the effects of IL-1β through upregulation of the decoy receptor, unraveling the role of this cytokine in breast cancer progression will necessitate future studies with intact glands that include several different cell types.
We concluded that BBCs induce distinct macrophage populations. Binary classification of macrophage phenotypes is problematic in our study, due to dual staining of macrophages for both M1 and M2 markers. However this challenge in classifying macrophages based on single markers is consistent with previous literature. For example, Qian and colleagues defined TNF-α as M1-specific (19), in agreement with Mantovani et al., who described TNF-α as upregulated in M1-macrophages and downregulated in M2-macrophages (55). However, Zeyda et al. demonstrated TNF-α expression in both M1- and M2-polarized macrophages (56). Similarly, we originally defined CD163 as an M1 marker based on the work by Fuentes et al. and Qian et al., (19, 57) but ultimately reassigned it to an M2 pattern of expression based on recently published studies (58–60) and our own experimental results with M2-polarizing cytokines. Such discrepancies in “hallmark biomarkers” of macrophage differentiation or polarization make interpretation of polarization markers challenging. Use of a positive control system for polarizing THP-1 cells facilitated our assignment of markers toward M1 or M2 predominance (and therefore represents a database of potential utility to other investigators); but our observation that even chemically polarized macrophages exhibit both M1 and M2 phenotypes simultaneously is a cautionary tale against interpreting macrophage expression based on positivity for only one class of markers.
In summary, by using multiple markers to assay macrophage polarization phenotypes and by evaluating differentiation with multiple cell-based assays, our results document distinct macrophage behavior in response to BBCs. While BBCs induced in vitro differentiation and migration of macrophages via an IL-1β-dependent mechanism, the migration was repressed in the presence of an IL-1β inhibitor. Given the importance of macrophages in the BBC microenvironment and in BBC-progression, novel in vitro systems for studying the reciprocal communication between BBCs and macrophages are important and could lead to newly identified targets. As pointed out by Ojalvo et al. (21), targeting the macrophages may be more effective than targeting genetically altered, chemotherapy-resistant cancer cells. Future in vivo preclinical studies or epidemiologic studies evaluating this pathway may help to identify novel strategies for targeting aggressive BBCs.
Supplementary Material
(A) Unsupervised hierarchical cluster of Multiclass SAM (FDR = 0) for control treated THP-1 total RNA analyzed on Agilent 4×44K whole genome human microarrays; where 4755 genes were significantly expressed. Of the genes differentially expressed under M1- and M2-specific cytokine stimulation, IPA grouped the two M1 clusters and M2 clusters, totaling 547 and 257 genes listed in Supplementary Table 1, respectively into several canonical pathways related to macrophage maturation and function listed in Supplementary Table 2. (B) Supervised cluster of control system-differentiated THP-1 cells analyzed on 4×44k microarrays to a 250 literature-identified list of genes, already defined as significant in the monocyte-to-macrophage differentiation and polarization processes. In our study, 209 genes were expressed across all samples (84% concordance) from the list of genes (Supplementary Table 3). The scale bar represents a −5.0 to 5.0 fold expression change relative to the median.
Acknowledgments
The authors thank Erick Roman Perez, Alex J. Freemerman, J. Terese Camp, Jessica Rein, Robert Bagnell and the staff of the UNC Pathology and Laboratory Medicine Microscopy Core and Kirk McNaughton of the UNC Histology Research Core Facility for contributions to experiments or data analysis.
GRANT SUPPORT
This publication was made possible by the Breast Cancer and the Environment Research Program (BCERP) award number U01 ES019472 from the National Institute of Environmental Health Sciences (NIEHS), and the National Cancer Institute (NCI), National Institutes of Health (NIH), Department of Health and Human Services (DHHS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS or NCI. The work was also supported by other funding from NIH, DHHS, including an NIEHS Center for Environmental Health and Susceptibility (P30 ES010126), an award from NCI (R01 CA138255), and an NCI Breast SPORE Career Development Award (P50 CA58233-18). Additional funding was provided by the North Carolina State-sponsored University Cancer Research Fund.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS
No potential conflicts of interest were disclosed.
References
- 1.Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295:2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 2.Millikan RC, Newman B, Tse CK, Moorman PG, Conway K, Dressler LG, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109:123–39. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–7. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin EY, Pollard JW. Role of infiltrated leucocytes in tumour growth and spread. Br J Cancer. 2004;90:2053–8. doi: 10.1038/sj.bjc.6601705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujimoto H, Sangai T, Ishii G, Ikehara A, Nagashima T, Miyazaki M, et al. Stromal MCP-1 in mammary tumors induces tumor-associated macrophage infiltration and contributes to tumor progression. Int J Cancer. 2009;125:1276–84. doi: 10.1002/ijc.24378. [DOI] [PubMed] [Google Scholar]
- 7.Campbell MJ, Tonlaar NY, Garwood ER, Huo D, Moore DH, Khramtsov AI, et al. Proliferating macrophages associated with high grade, hormone receptor negative breast cancer and poor clinical outcome. Breast Cancer Res Treat. 2011;128:703–11. doi: 10.1007/s10549-010-1154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camp JT, Elloumi F, Roman-Perez E, Rein J, Stewart DA, Harrell JC, et al. Interactions with fibroblasts are distinct in Basal-like and luminal breast cancers. Mol Cancer Res. 2011;9:3–13. doi: 10.1158/1541-7786.MCR-10-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 10.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8:279–89. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi K. Development and differentiation of macrophages and their related cells. Hum Cell. 1994;7:109–15. [PubMed] [Google Scholar]
- 12.Erwig LP, Henson PM. Immunological consequences of apoptotic cell phagocytosis. Am J Pathol. 2007;171:2–8. doi: 10.2353/ajpath.2007.070135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Mosser DM. Macrophage activation by endogenous danger signals. J Pathol. 2008;214:161–78. doi: 10.1002/path.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J Pathol. 2002;196:254–65. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 15.Troester MA, Lee MH, Carter M, Fan C, Cowan DW, Perez ER, et al. Activation of host wound responses in breast cancer microenvironment. Clin Cancer Res. 2009;15:7020–8. doi: 10.1158/1078-0432.CCR-09-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beck AH, Espinosa I, Edris B, Li R, Montgomery K, Zhu S, et al. The macrophage colony-stimulating factor 1 response signature in breast carcinoma. Clin Cancer Res. 2009;15:778–87. doi: 10.1158/1078-0432.CCR-08-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang HY, Nuyten DS, Sneddon JB, Hastie T, Tibshirani R, Sorlie T, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl AcadSci U S A. 2005;102:3738–43. doi: 10.1073/pnas.0409462102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma M, Beck AH, Webster JA, Espinosa I, Montgomery K, Varma S, et al. Analysis of stromal signatures in the tumor microenvironment of ductal carcinoma in situ. Breast Cancer Res Treat. 2010;123:397–404. doi: 10.1007/s10549-009-0654-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tjiu JW, Chen JS, Shun CT, Lin SJ, Liao YH, Chu CY, et al. Tumor-associated macrophage-induced invasion and angiogenesis of human basal cell carcinoma cells by cyclooxygenase-2 induction. J Invest Dermatol. 2009;129:1016–25. doi: 10.1038/jid.2008.310. [DOI] [PubMed] [Google Scholar]
- 21.Ojalvo LS, Whittaker CA, Condeelis JS, Pollard JW. Gene expression analysis of macrophages that facilitate tumor invasion supports a role for Wnt-signaling in mediating their activity in primary mammary tumors. J Immunol. 2010;184:702–12. doi: 10.4049/jimmunol.0902360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin EY, Pollard JW. Tumor-associated macrophages press the angiogenic switch in breast cancer. Cancer Res. 2007;67:5064–6. doi: 10.1158/0008-5472.CAN-07-0912. [DOI] [PubMed] [Google Scholar]
- 23.Normann SJ. Macrophage infiltration and tumor progression. Cancer Metastasis Rev. 1985;4:277–91. doi: 10.1007/BF00048093. [DOI] [PubMed] [Google Scholar]
- 24.Lechner MG, Megiel C, Russell SM, Bingham B, Arger N, Woo T, et al. Functional characterization of human Cd33+ and Cd11b+ myeloid-derived suppressor cell subsets induced from peripheral blood mononuclear cells co-cultured with a diverse set of human tumor cell lines. J Transl Med. 2011;9:90. doi: 10.1186/1479-5876-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchiya S, Yamabe M, Yamaguchi Y, Kobayashi Y, Konno T, Tada K. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int J Cancer. 1980;26:171–6. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, et al. Induction of maturation in cultured human monocytic leukemia cells by a phorbol diester. Cancer Res. 1982;42:1530–6. [PubMed] [Google Scholar]
- 27.Park EK, Jung HS, Yang HI, Yoo MC, Kim C, Kim KS. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm Res. 2007;56:45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 28.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elloumi F, Hu Z, Li Y, Parker JS, Gulley ML, Amos KD, et al. Systematic bias in genomic classification due to contaminating non-neoplastic tissue in breast tumor samples. BMC Med Genomics. 2011;4:54. doi: 10.1186/1755-8794-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–86. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 31.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 32.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomsen LL, Miles DW. Role of nitric oxide in tumour progression: lessons from human tumours. Cancer Metastasis Rev. 1998;17:107–18. doi: 10.1023/a:1005912906436. [DOI] [PubMed] [Google Scholar]
- 34.Al-Rawi MA, Mansel RE, Jiang WG. Interleukin-7 (IL-7) and IL-7 receptor (IL-7R) signalling complex in human solid tumours. Histol Histopathol. 2003;18:911–23. doi: 10.14670/HH-18.911. [DOI] [PubMed] [Google Scholar]
- 35.Helming L, Winter J, Gordon S. The scavenger receptor CD36 plays a role in cytokine-induced macrophage fusion. J Cell Sci. 2009;122:453–9. doi: 10.1242/jcs.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shabo I, Stal O, Olsson H, Dore S, Svanvik J. Breast cancer expression of CD163, a macrophage scavenger receptor, is related to early distant recurrence and reduced patient survival. Int J Cancer. 2008;123:780–6. doi: 10.1002/ijc.23527. [DOI] [PubMed] [Google Scholar]
- 37.Biswas SK, Gangi L, Paul S, Schioppa T, Saccani A, Sironi M, et al. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation) Blood. 2006;107:2112–22. doi: 10.1182/blood-2005-01-0428. [DOI] [PubMed] [Google Scholar]
- 38.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–56. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 40.Mantovani A, Sica A. Macrophages, innate immunity and cancer: balance, tolerance, and diversity. Curr Opin Immunol. 2010;22:231–7. doi: 10.1016/j.coi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Sun X, Casbas-Hernandez P, Bigelow C, Makowski L, Joseph Jerry D, Smith Schneider S, et al. Normal breast tissue of obese women is enriched for macrophage markers and macrophage-associated gene expression. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1789-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris PG, Hudis CA, Giri D, Morrow M, Falcone DJ, Zhou XK, et al. Inflammation and increased aromatase expression occur in the breast tissue of obese women with breast cancer. Cancer Prev Res (Phila) 2011;4:1021–9. doi: 10.1158/1940-6207.CAPR-11-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, et al. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–83. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 44.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–5. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–62. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buess M, Rajski M, Vogel-Durrer BM, Herrmann R, Rochlitz C. Tumor-endothelial interaction links the CD44(+)/CD24(−) phenotype with poor prognosis in early-stage breast cancer. Neoplasia. 2009;11:987–1002. doi: 10.1593/neo.09670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dirat B, Bochet L, Dabek M, Daviaud D, Dauvillier S, Majed B, et al. Cancer-associated adipocytes exhibit an activated phenotype and contribute to breast cancer invasion. Cancer Res. 2011;71:2455–65. doi: 10.1158/0008-5472.CAN-10-3323. [DOI] [PubMed] [Google Scholar]
- 48.Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, Monville F, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer Res. 2011;71:614–24. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ojalvo LS, King W, Cox D, Pollard JW. High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. Am J Pathol. 2009;174:1048–64. doi: 10.2353/ajpath.2009.080676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hou Z, Falcone DJ, Subbaramaiah K, Dannenberg AJ. Macrophages induce COX-2 expression in breast cancer cells: role of IL-1beta autoamplification. Carcinogenesis. 2011;32:695–702. doi: 10.1093/carcin/bgr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, et al. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12:479–91. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soria G, Ofri-Shahak M, Haas I, Yaal-Hahoshen N, Leider-Trejo L, Leibovich-Rivkin T, et al. Inflammatory mediators in breast cancer: coordinated expression of TNFalpha & IL-1beta with CCL2 & CCL5 and effects on epithelial-to-mesenchymal transition. BMC Cancer. 2011;11:130. doi: 10.1186/1471-2407-11-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snoussi K, Strosberg AD, Bouaouina N, Ben Ahmed S, Chouchane L. Genetic variation in pro-inflammatory cytokines (interleukin-1beta, interleukin-1alpha and interleukin-6) associated with the aggressive forms, survival, and relapse prediction of breast carcinoma. Eur Cytokine Netw. 2005;16:253–60. [PubMed] [Google Scholar]
- 54.Prockop DJ, Youn Oh J. Mesenchymal Stem/Stromal Cells (MSCs): Role as Guardians of Inflammation. Mol Ther. 2011 doi: 10.1038/mt.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–11. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 56.Zeyda M, Gollinger K, Kriehuber E, Kiefer FW, Neuhofer A, Stulnig TM. Newly identified adipose tissue macrophage populations in obesity with distinct chemokine and chemokine receptor expression. Int J Obes (Lond) 2010;34:1684–94. doi: 10.1038/ijo.2010.103. [DOI] [PubMed] [Google Scholar]
- 57.Fuentes-Duculan J, Suarez-Farinas M, Zaba LC, Nograles KE, Pierson KC, Mitsui H, et al. A subpopulation of CD163-positive macrophages is classically activated in psoriasis. J Invest Dermatol. 2010;130:2412–22. doi: 10.1038/jid.2010.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burdo TH, Lentz MR, Autissier P, Krishnan A, Halpern E, Letendre S, et al. Soluble CD163 made by monocyte/macrophages is a novel marker of HIV activity in early and chronic infection prior to and after anti-retroviral therapy. J Infect Dis. 2011;204:154–63. doi: 10.1093/infdis/jir214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weaver LK, Hintz-Goldstein KA, Pioli PA, Wardwell K, Qureshi N, Vogel SN, et al. Pivotal advance: activation of cell surface Toll-like receptors causes shedding of the hemoglobin scavenger receptor CD163. J Leukoc Biol. 2006;80:26–35. doi: 10.1189/jlb.1205756. [DOI] [PubMed] [Google Scholar]
- 60.Backe E, Schwarting R, Gerdes J, Ernst M, Stein H. Ber-MAC3: new monoclonal antibody that defines human monocyte/macrophage differentiation antigen. J Clin Pathol. 1991;44:936–45. doi: 10.1136/jcp.44.11.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Unsupervised hierarchical cluster of Multiclass SAM (FDR = 0) for control treated THP-1 total RNA analyzed on Agilent 4×44K whole genome human microarrays; where 4755 genes were significantly expressed. Of the genes differentially expressed under M1- and M2-specific cytokine stimulation, IPA grouped the two M1 clusters and M2 clusters, totaling 547 and 257 genes listed in Supplementary Table 1, respectively into several canonical pathways related to macrophage maturation and function listed in Supplementary Table 2. (B) Supervised cluster of control system-differentiated THP-1 cells analyzed on 4×44k microarrays to a 250 literature-identified list of genes, already defined as significant in the monocyte-to-macrophage differentiation and polarization processes. In our study, 209 genes were expressed across all samples (84% concordance) from the list of genes (Supplementary Table 3). The scale bar represents a −5.0 to 5.0 fold expression change relative to the median.