Abstract
A gene regulatory network subcircuit comprising the otx, wnt8, and blimp1 genes accounts for a moving torus of gene expression that sweeps concentrically across the vegetal domain of the sea urchin embryo. Here we confirm by mutation the inputs into the blimp1 cis-regulatory module predicted by network analysis. Its essential design feature is that it includes both activation and autorepression sites. The wnt8 gene is functionally linked into the subcircuit in that cells receiving this ligand generate a β-catenin/Tcf input required for blimp1 expression, while the wnt8 gene in turn requires a Blimp1 input. Their torus-like spatial expression patterns and gene regulatory analysis indicate that the genes even-skipped and hox11/13b are also entrained by this subcircuit. We verify the cis-regulatory inputs of even-skipped predicted by network analysis. These include activation by β-catenin/Tcf and Blimp1, repression within the torus by Hox11/13b, and repression outside the torus by Tcf in the absence of Wnt8 signal input. Thus even-skipped and hox11/13b, along with blimp1 and wnt8, are members of a cohort of torus genes with similar regulatory inputs and similar, though slightly out-of-phase, expression patterns, which reflect differences in cis-regulatory design.
Keywords: Blimp1, Wnt, Hox11/13b, Endomesoderm, Sea urchin, GRN, Dynamic gene expression
Introduction
Vegetal nuclearization of β-catenin is an early input required for specification of endomesoderm in the sea urchin embryo (Logan et al., 1999; Wikramanayake et al., 1998). Initially driven by cell-autonomous processes, the nuclearization of β-catenin marks a permissive condition for installation of all endomesodermal regulatory states. By 7th cleavage (∼11 h post-fertilization, hpf), nuclear β-catenin can be detected in the founder cells of the future skeletogenic and other mesodermal territories, and also in the future endoderm (i.e., respectively, the micromere lineage, the secondary mesenchyme territory, and the veg1 and veg2 territories; Davidson et al., 1998). At a mechanistic level, the permissive state equals the replacement in the transcription factor Tcf of the dominant Groucho repressor moiety by β-catenin (Daniels and Weis, 2005; Range et al., 2005). Multiple regulatory genes essential for defining the territorial specification of all three endomesodermal territories utilize Tcf target sites in their cis-regulatory control apparatus (Davidson, 2006; Davidson et al., 2002a,b; Oliveri and Davidson, 2004), and in the absence of β-catenin all these genes are subject to Groucho/Tcf repression. Transcriptional specification of the endomesodermal territories occurs as a wave of regulatory gene expression moves concentrically outward, beginning in the centrally located skeletogenic domain (5−9 h); then extending to the adjacent ring of mesodermal blastomeres (10−18 h); and finally encompassing the precursors that will generate the gut (>18 h). But, after a few hours of transcription, expression of these same regulatory genes is extinguished, first in the skeletogenic, and then in the mesodermal domain (Fig. 1A; Smith et al., 2007). Thus, the pattern of expression described by these genes is a concentrically moving torus, as illustrated in the following for four different genes. The immediate controller of the moving torus pattern is the wnt8 gene.
Fig. 1.

The blimp1 and wnt8 subcircuit of the gene regulatory network for endomesoderm specification in Strongylocentrotus purpuratus (Smith et al., 2007). (A) Diagrammatic illustration of the dynamic torus pattern of gene expression, as exemplified by wnt8 and blimp1. (B) GRN subcircuit. Regulatory relationships are shown between blimp1, β-catenin/Tcf, GSK-3, and wnt8; nb-Tcf, complex of nuclear β-catenin and Tcf transcription factor. The lines leading from nb-Tcf bifurcate into an arrow and a repression bar, indicating that in the absence of nuclearized β-catenin the Groucho/Tcf complex acts as a dominant repressor at both the wnt8 and blimp1 loci. Positive inputs from both Blimp1 and nb-Tcf thus control wnt8 transcription (Minokawa et al., 2005). The three predicted inputs into the blimp1 cis-regulatory module(s) are indicated: a permissive input from nb-Tcf; activation by Otx; and autorepression.
The architecture of the gene network subcircuit that accounts causally for this dynamic pattern is reproduced in Fig. 1B (Smith et al., 2007). The lynchpin is the blimp1 (formerly krox) regulatory gene. This gene encodes a zinc finger transcription factor of the SET family of proteins (for details and sequence analysis, see Livi and Davidson, 2006, 2007). In their cis-regulatory analysis of the wnt8 gene, Minokawa et al. (2005) proved that, as predicted, Blimp1 is a direct input essential for wnt8 transcription. Indeed, as indicated by whole mount in situ hybridization, the dynamic patterns of expression of these mutual regulatory partners are almost exactly similar (Livi and Davidson, 2007; Minokawa et al., 2005). Morpholino-substituted antisense oligonucleotide (MASO) targeting blimp1 blocks endomesoderm specification (Livi and Davidson, 2007), just as does MASO directed against the wnt8 gene (Minokawa et al., 2005; Wikramanayake et al., 2004). The wnt8 gene also requires the product of its own signal transduction system, β-catenin/Tcf, as a regulatory input, setting up a positive feedback circuit between adjacent cells that both produce and receive Wnt8 (Minokawa et al., 2005). In turn, the blimp1 gene is predicted to utilize a β-catenin/Tcf input as well. There are a number of wnt genes active in the early sea urchin embryo (Croce et al., 2006) and an important additional fact is that it is indeed Wnt8 which generates the essential β-catenin/Tcf input into the blimp1 gene. This was shown directly by the effects of wnt8 MASO on expression of the blimp1 gene (Smith et al., 2007; cf. QPCR data posted at (http://sugp.caltech.edu/endomes/).
The blimp1 gene utilizes two different cis-regulatory modules and two different splice variants in embryogenesis, a late-acting gut specific module and variant, and the early blimp1 vegetal plate module relevant here (Livi and Davidson, 2006, 2007). Gene regulatory network analysis had indicated three inputs into the early blimp1 cis-regulatory system: (i) a β-catenin/Tcf input; (ii) an input from the early embryonic transcriptional activator Otx (see Yuh et al., 2002, 2004; for transcriptional regulatory analysis of otx); and (iii) an autorepressive input. This last was inferred from the observation that MASO directed against the relevant blimp1 transcript leads to a transient but significant early increase in message level (Livi and Davidson, 2006). This autorepressive input was verified by means of a cis-regulatory reconstruction experiment (Smith et al., 2007).
Here we present data authenticating at the DNA sequence level the predicted β-catenin/Tcf and Otx inputs into blimp1 expression. The endomesoderm gene regulatory network (Davidson, 2006; Davidson et al., 2002a,b; Oliveri and Davidson, 2004) predicts that the subcircuit driving torus expansion entrains the expression of other regulatory genes in addition to blimp1 and wnt8 (Fig. 1), viz. even-skipped (eve) (Ransick et al., 2002) and hox11/13b (Arenas-Mena et al., 2006). We present detailed analyses of the expression patterns of these genes, which shows that they, blimp1 and wnt8, are transcribed in similar, yet subtly distinct ways. If the expanding ring of expression can be thought of as a circular wave-like pattern, then expression of this group of genes can be thought of as “out-of-phase” with the respective peak of expression of each gene slightly offset from the others. This is because their cis-regulatory systems are not identical: while both eve and hox11/13b are predicted to receive Blimp1 and β-catenin/Tcf inputs, they differ in the additional inputs to which they respond, and hox11/13b in turn controls eve spatial expression. In the following, we verify this directly by mutation of target sites in the eve cis-regulatory control system.
Materials and methods
Reporter constructs, microinjection, and QPCR measurement of GFP mRNA in eggs expressing GFP constructs
Reporter constructs were based on a blimp1 BAC GFP recombinant. The recombinant was made in the Strongylocentrotus purpuratus clone 163 O19 BAC, a clone 93,775 bp in length containing the blimp locus (see http://sugp.caltech.edu/resources/annot-seq.psp?seq-163O19). A cassette containing the GFP coding region was inserted into the blimp1 start of translation by homologous recombination following procedures previously described (Lee et al., 2001). Standard PCR and fusion PCR techniques using the High Fidelity PCR Kit (Roche, Indianapolis, IN) were then used to build reporter constructs based on this BAC GFP recombinant. PCR products for all significant reporter constructs were subsequently cloned using the EPICENTRE (Madison, WI) Copy Control Cloning System in the case of large inserts (>5 kb), or the pGEM-T Easy Vector System from Promega, and confirmed by sequencing.
The following consensus sequences were used to identify putative transcription factor binding sites: Tcf, 5′-TTCAAAGG (Oosterwegel et al., 1991); Otx, 5′-(A/G)GATTA (Yuh et al., 2004). Binding site sequences were mutated by PCR and resulting constructs checked by sequencing. Briefly, PCR primers were designed with tailed non-priming sequences including the mutant form of the candidate transcription factor binding sites. In general, mutations were designed by swapping A to C, T to G, and vice versa. Additional non-priming tails were added 15 nucleotides in length for subsequent fusion PCR. In the case of putative Tcf and Blimp binding sites, priming oligonucleotides covering both sites were used (disrupting one or both sites) due to the proximity of each pair of sites. Putative Blimp1 sites were preserved when Tcf site disruptions were introduced and vice versa. This was achieved as follows. Blimp1 site (1): 5′-GAAATTCAAAG to 5′-GAAAGCGAAAG for Tcf disruption; 5′-GAAATTCAAAG TO 5′-TCCCTTCAAAG for Blimp1 disruption. Blimp1 site (2): 5′-TTCAAAGGAAAGG to 5′-GGTAAAGGAAAGG for Tcf disruption; 5′-TTCAAAGGAAAGG to 5′-TTCAAAGGCCCTT for Blimp1 disruption. At the eve locus: 5′-GAAAGTCAAAG to 5′-TCCCTTCAAAG for Blimp1 disruption; 5′-GAAAGTCAAAG to 5′-GAAAGCGAAAG for Tcf disruption.
PCR products or linearized cloned constructs were purified with the Qiagen Qiaquick PCR purification kit and microinjected into fertilized S. purpuratus eggs as described. Approximately 1500 molecules of the desired construct were injected along with a 6-fold molar excess of HindIII-digested carrier sea urchin DNA per egg in a 4 pL volume of 0.12 M KCl. Embryos were collected at different stages for observation by fluorescence microscopy to assess spatial activity qualitatively; or for quantitative analysis of transcript prevalence using real-time PCR (QPCR). All experimental and control constructs were tested in multiple batches of eggs. Microinjection and measurement of GFP mRNA by QPCR were performed as described (Revilla-i-Domingo et al., 2004).
Whole-mount in situ hybridization (WMISH)
The WMISH protocol used here is based on the method of Minokawa et al. (2005), with minor modifications. Embryos were incubated with antibody overnight at 4 °C and transferred into 50% glycerol in a stepwise manner and mounted on slides sealed with clear nail polish.
Results
Identification of the blimp1 cis-regulatory module
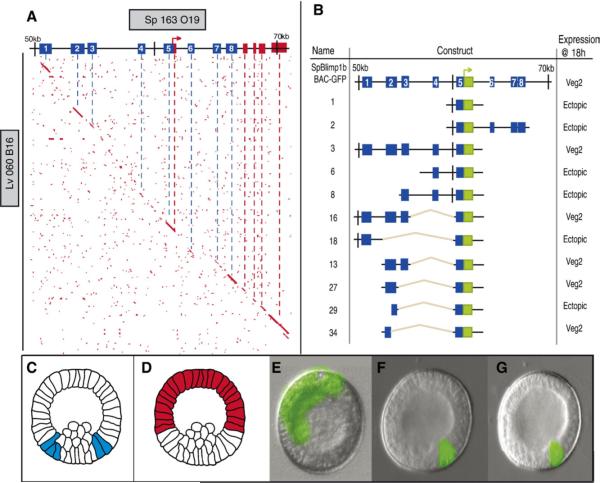
Putative cis-regulatory modules of the blimp1 gene were identified by interspecific sequence comparison, as in many previous studies (Brown et al., 2002; Cameron et al., 2005; Minokawa et al., 2005; Revilla-i-Domingo et al., 2004; Yuh et al., 2002). Conserved patches of genomic non-coding sequence were found by use of the Family Relations II program (Brown et al., 2005). An S. purpuratus BAC sequence (BAC 163 O19, 93,775 bp in length) containing the Spblimp1 gene was thus compared with an Lytechinus variegatus BAC sequence (BAC 060 B16, 55,657 bp) containing the Lvblimp1 gene. The organization of the blimp1 coding sequences is well conserved between the two species, including the exons for the alternately transcribed variants (Livi and Davidson, 2006). The blimp1a variant is not expressed until gastrulation and is not relevant to this study. The early blimp1b transcript begins with exon 1b, located 14.5 kb downstream of the start of transcription of blimp1a. Using 20 bp windows and requiring 85% identity, eight conserved sequence patches were identified. These are located upstream of the exon1b start site and between exon 1b and exon 2 (Fig. 2A). One of these patches, Conserved Region 5 (CR5), partially overlaps the 5′ UTR of the blimp1b transcript and extends ∼500 bp upstream of the first exon. On the basis that this region must contain the basal promoter, it was included in most subsequent reporter constructs.
Fig. 2.
Family Relations II analysis of blimp1 locus and cis-regulatory reporter constructs. (A) Pairwise sequence analysis of two BAC sequences containing blimp1 genes, using the Family Relations II program. The horizontal line at the top represents the sequence of the Strongylocentrotus purpuratus BAC clone 163 O19; the ordinate represents Lytechinus variegatus BAC clone 060 B16. Red dots indicate 20-nucleotide stretches of sequence sharing ≥80% identity between the two species. Red boxes on the horizontal sequence indicate exons with the bent arrow showing the start of transcription in exon 1b of the Spblimp1 gene; numbered blue boxes indicate conserved regions. Conserved sequence patches are revealed by contiguous dots which produce short diagonal segments. (B) GFP reporter constructs for blimp1 cis-regulatory analysis. The top horizontal line represents the SpBlimp1b BAC-GFP recombinant in which the GFP gene has been inserted by homologous recombination at the site of the start of translation of the blimp1 gene, replacing exon1b. The various reporter constructs derived from the SpBlimp1b BAC-GFP recombinant are diagrammed below; construct names are at the left and the patterns of expression generated by these constructs at 18 hpf are indicated at the right. At this time the normal domain of blimp1 expression is the veg2 endoderm. (C, D) Diagrams of mesenchyme blastula stage embryos displaying a normal endodermal domain of expression (blue) or ectopic ectodermal expression (red). (E) Representative fluorescent image of an 18 h embryo bearing construct 2, which is expressed ectopically. (F) Representative 18 h embryo displaying correct expression of the recombinant SpBlimp1b BAC-GFP. (G) Representative 18 h embryo bearing construct 34, which also generates a correct expression pattern.
A BAC knock-in was constructed, in which the coding sequence of GFP and a 3′-trailer and polyadenylation site replace exon1b in the Spblimp1 gene. By monitoring GFP expression by fluorescence microscopy, we found that when microinjected into fertilized sea urchin eggs, the recombinant blimp1 GFP-BAC faithfully recapitulates expression of endogenous blimp1 (quantitative data on this parental control construct are given below). Between 18 and 22 hpf, the reporter is expressed in veg2 endoderm cells, as depicted, for example, in Fig. 2F. The blimp1 GFP BAC knock-in was then used to determine the functional values of the conserved sequence patches. These were isolated from the BAC in various combinations, and combined with the basal promoter and the knock-in GFP reporter of the BAC to produce expression constructs, as described in Materials and methods. The constructs thus generated are shown in Fig. 2B, and each was tested for its activity in the embryo (for schematics of expression patterns, see Figs. 2C, D).
The results showed that when combined with the CR5 basal promoter, module CR2 contains the necessary and sufficient regulatory information for proper spatial expression. Any construct lacking CR2, e.g., constructs 6 or 18 (Fig. 2B), displayed gross ectopic expression, illustrated for construct 2 in Fig. 2E. Construct 27, which contains ∼700 bp of CR2, and construct 34, which contains only the distal ∼300 bp portion of CR2, which is all that is required for accurate expression, were used for the following experiments. A representative embryo illustrating construct 34 expression is shown in Fig. 2G. Although some hours earlier the construct had been expressing in the more centrally located mesodermal domain, enough time had elapsed at the time this observation was made so that the GFP protein is no longer visible in the central region of the torus where gene expression had been extinguished.
Predicted target sites of the blimp1 cis-regulatory module
The GRN analysis predicted that sites for Tcf, Blimp itself, and Otx should be present in the cis-regulatory module controlling pre-gastrular blimp1 expression, and since the distal ∼300 bp of CR2 is evidently sufficient when linked to CR5 (Fig. 2), we searched these regions for the putative target sites. Results are mapped on the CR2 and CR5 sequences in Fig. 3. The site sequence models used for the search are listed in Materials and methods. Within the 300 bp upstream portion of CR2 are two candidate Tcf binding sites, separated by only 2 bp, which retain the core TTCAAAG sequence. Both sites are conserved in L. variegatus, as indicated by the shading in Fig. 3. Furthermore, in the S. purpuratus CR2 sequence, putative Blimp1 target sites immediately flank the pair of Tcf sites. Note that the more downstream candidate Blimp1 target site appears to overlap the 5′AAAG of the neighboring Tcf site. Although the upstream (i.e., distal) of the two candidate Blimp1 sites does not display as strong a convergence with the consensus sequence, this sequence has been found to have low affinity binding to Blimp1 in vitro (Kuo and Calame, 2004) and as noted it is conserved in L. variegatus and overlaps with the 5′TT of the adjacent Tcf site. A further 60 bp downstream of these sites is an apparent Otx target sequence (Otx1). Another strong candidate Otx site is found near the proximal end of CR2 (Otx2) in the region which is missing from construct 34 but included in construct 27. A third possible Otx site (Otx3) is present in CR5, approximately 250 bp upstream of the putative TATA recognition site. The functional significance of each of these candidate transcription factor target sites was tested by mutating the individual sequences; data concerning the Blimp1 target sites utilized for autorepression are reported elsewhere (Smith et al., 2007). Performance of the resulting constructs was assayed by means which differed according to the nature of the inputs. The assay for mutated Otx sites in a blimp1 reporter tests for a quantitative decrease in reporter expression. Tcf binding mediates gene repression via the action of Groucho, thus disruption of Tcf target sites will lead to loss of spatial control: the assay for mutated Tcf sites is hence ectopic reporter activity.
Fig. 3.
Sequences of blimp1 conserved regions 2 and 5. Gray highlight indicates nucleotide sequence that is conserved within 20 bp windows at or above 85% identity between Strongylocentrotus purpuratus and L. variegatus. Bent black arrows indicate primer locations for various reporter constructs (13, 27, 29, and 34) in conserved region 2. Candidate Blimp1 binding sites are in blue characters, putative Tcf target sites in red, Otx target sites in green. The overlapping Blimp1 and Tcf sites are in purple. The translation target site in CR5 is indicated. The actual site of transcription initiation has not been determined. It is at or upstream of the5′ end of our longest cDNA clone, and the possible TATA site indicated may or may not be functional.
Mutational analysis of Otx target sites in the blimp1 cis-regulatory module
Sequences of the three predicted Otx sites in construct 27 were scrambled and the activity of the mutated constructs was compared to that of the wild-type control. Effects were assayed by QPCR measurement of the GFP reporter mRNA, as summarized in Fig. 4. Mutation of Otx1 or Otx3 results in a severe decrease in reporter GFP mRNA. However, disruption of Otx2 did not significantly affect reporter GFP expression, a result consistent with the observation that construct 34 produces normal expression. It is interesting that neither the Otx1 nor Otx3 site is intact in L. variegatus. We visually confirmed proper spatial expression of these mutated constructs by fluorescence microscopy, and noted much dimmer expression when either sites 1 or 3 are disrupted, but not site 2.
Fig. 4.
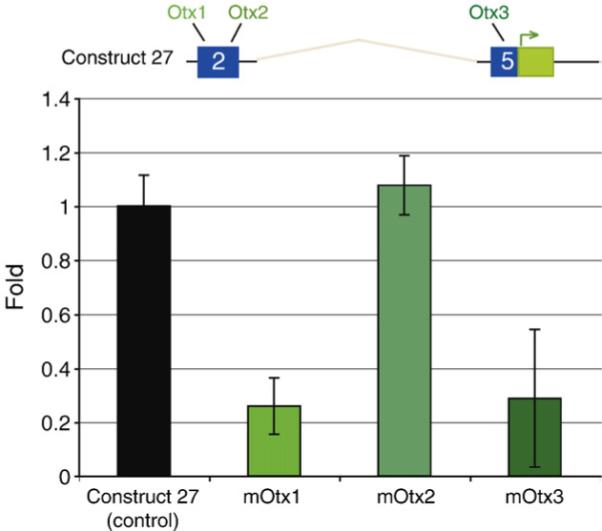
Mutational analysis of cis-regulatory blimp1 Otx sites. The top diagram represents the structure of construct 27 with conserved regions 2 and 5 indicated in blue and the GFP coding sequence in green. Mutations in each of the potential Otx sites were introduced in the construct 27 background and injected into fertilized sea urchin eggs. GFP transcript levels were then assessed by QPCR at 18 hpf. The chart shows fold differences in expression with reference to control construct 27. Disruption of either Otx site 1 or site 3 led to dramatic decreases in GFP message level, while disruption of the Otx site 2 did not.
Mutational analysis of Tcf binding sites in the blimp1b cis-regulatory module
Binding by Tcf at the blimp1 locus is predicted to mediate a logic “toggle switch” between gene repression and activation: Tcf target genes will be dominantly repressed by the Groucho co-factor outside of the endomesodermal domain of β-catenin nuclearization, i.e., in the presumptive ectoderm; while these same genes will be in a condition permissive for activation in the endomesoderm, where the β-catenin/Tcf complex is instead present. Disrupting the Tcf sites that function in this way should therefore cause ectopic ectodermal reporter gene expression, driven by the Otx factor, which at this stage is ubiquitously present (Li et al., 1997; Yuh et al., 2002; there could as well be ancillary ubiquitous activators binding in the blimp1 cis-regulatory module). We found that mutations in either one or both of the Tcf sites in construct 27 indeed produce a dramatic increase in ectopic gene expression (Fig. 5). The same result was obtained when these mutations were made in the Tcf sites of construct 13 or construct 34 (cf. Fig. 2B). Reporter constructs with both Tcf sites disrupted rarely, if ever, reproduced the endogenous pattern of expression; only 5% of embryos bearing these mutated constructs were scored as having the normal 18−24 h veg2 expression pattern. In these experiments, the control construct 27 or the parental blimp1 BAC GFP recombinant produced correct veg2 endoderm expression in 93% and 96% of embryos displaying GFP fluorescence (Fig. 5).
Fig. 5.

Mutational analysis of candidate Tcf sites in the blimp1 cis-regulatory module. The table shows spatial reporter expression of blimp1 BAC-GFP, construct 27, and construct 27 with both Tcf sites mutated. (A, B) Diagrams depicting embryos at early mesenchyme blastula stage, and displaying the wild-type blimp1 pattern of veg2 expression at this stage in blue, ectopic expression in the ectoderm in red. (C–F) Representative fluorescence images of transgenic embryos: (C) correct GFP reporter expression driven by construct 27; (D–F) examples of gross ectopic reporter expression driven by construct 27 in which either one or both Tcf sites had been mutated (D, Tcf site 1 mutated; E, Tcf site 2 mutated; F, both mutated).
Other genes entrained by the torus subcircuit
The endomesoderm GRN indicates that additional genes are linked into the torus subcircuit driven by the wnt8 and blimp1 genes. Two belonging to this cohort are the eve and hox11/13b genes, the relevant network connections of which are depicted in Fig. 6. Network analysis suggests several similarities for the regulatory inputs among these genes as well as some differences. The predicted inputs controlling hox11/13b expression are the same as those found regulating blimp1 transcription (as reported herein and in Smith et al., 2007), though these inputs have not yet been confirmed by cis-regulatory mutation. Meanwhile, even-skipped is predicted to respond to Otx and β-catenin/Tcf inputs as does blimp1, but also to activation by Blimp1, just as does wnt8. In addition, the network predicts that the eve gene is repressed by Hox11/13b. We revisited this last prediction by injection of Hox11/13b MASO; eve transcript levels were increased by 2.4 and 2.4 cycles of QPCR in two trials in embryos harvested at 22 h post-fertilization.
Fig. 6.
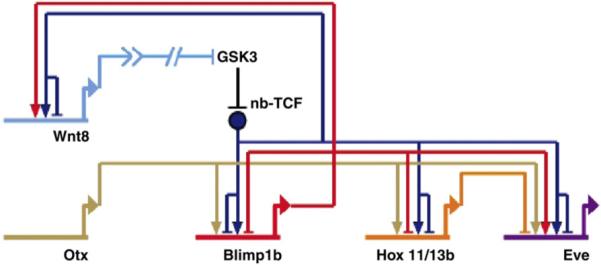
Subnetwork regulatory relationships between blimp1, wnt8, hox11/13b, eve, and otx; abbreviations and notation as in Fig. 1; Eve, even-skipped. The gene regulatory network indicates that hox11/13b responds to an activation input by Otx and the nb-Tcf/Groucho-Tcf toggle switch. The predicted inputs into eve, tested and verified in this work, are activation by Otx and Blimp1b, the nb-Tcf/Groucho-Tcf toggle switch; and repression by Hox11/13b.
Similar yet distinct patterns of expression of entrained genes
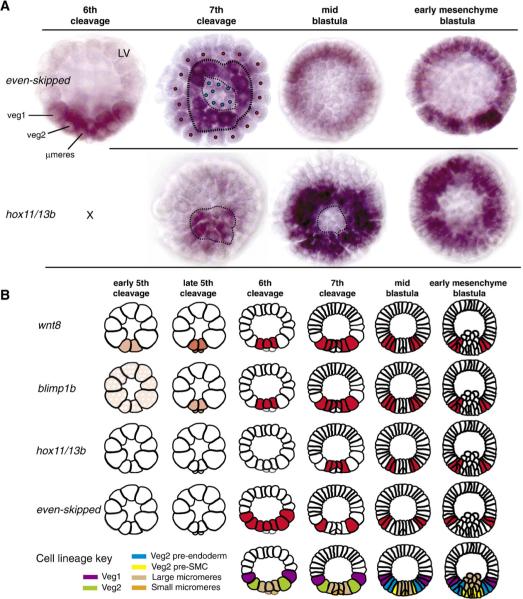
The similar yet distinct cis-regulatory inputs for the various genes entrained by this patterning subcircuit generate similar yet distinct temporal and spatial expression outputs. Fig. 7A shows the spatial expression patterns of eve and hox11/13b: both produce a torus-shaped dynamic pattern of expression, but they differ in that the hox11/13b pattern lies within the eve pattern, and the initial expression of hox11/13b is delayed, displaying only a brief phase of micromere labeling in late 7th cleavage embryos.
Fig. 7.
The relative expression patterns of wnt8, blimp1, hox11/13b, and eve. (A) Whole mount in situ hybridization (WMISH) of hox11/13b and eve, all panels vegetal view except where indicated lateral view, LV. At 6th cleavage, expression of eve is detectable in veg1, veg2, and micromeres. At 7th cleavage, the expression of eve wanes in veg1 and micromere descendents (red and blue dots, respectively), and eve is seen primarily in veg2 cells (second panel from left; dotted lines circumscribe the veg2 tier of cells). The loss of eve labeling in 7th cleavage micromere descendents corresponds with expression of hox11/13b in that territory at that stage (left panel; dotted lines mark micromere-veg2 boundary). At mid blastula stage, eve labeling is evident in presumptive endoderm cells, while hox11/13b is expressed in the presumptive mesodermal cells. By early mesenchyme blastula stage (right panels), eve expression has moved to veg1 endoderm, while the expression of hox11/13b is confined to veg2 descendents; n.b., the eve image is at a deeper plane of focus than the hox11/13b image. Expression of hox11/13b appears nested within the torus of eve expression. (B) Diagrammatic summary of WMISH results for wnt8, blimp1, eve and hox11/13b (this work, Smith et al., 2007, and Fig. S1). Lateral views are presented, which can be interpreted by the developmental key at bottom. Stippling in early 5th cleavage blimp1 figure indicates inferred maternal Blimp1 factor (Smith et al., 2007).
In Fig. 7B are the results of a high-density time series of WMISH studies on these two genes plus the blimp1 and wnt8 genes, shown diagrammatically from the side, with a key at the bottom indicating lineages and cell fates as seen in this view. The spatial patterns of expression of the four genes can here be compared in detail. Expression of wnt8 occurs first, at early 5th cleavage. As demonstrated by Smith et al. (2007), this is due to the presence of some maternal Blimp1 factor (stippled pink in diagram) plus the β-catenin nuclearized by the maternal system. By late 5th–6th cleavage blimp1 transcription has been activated (Smith et al., 2007). Note that this requires the intensified levels of β-catenin/Tcf which are the result of the wnt8 intercellular autoactivation circuit (Smith et al., 2007). Thereafter wnt8 and blimp1 transcripts are detected together in the four micromeres, as also reported earlier (Minokawa et al., 2005; Livi and Davidson, 2006), and subsequently, as summarized in Fig. 7B, they describe a very similar pattern of expression. By 7th cleavage, they are expressed synonymously (representative WMISH images of 7th cleavage expression of these two genes are shown in Fig. S1). Their expression expands out to the presumptive mesodermal cells in early blastula stage, following which expression in the micromere descendents is extinguished; then expression disappears in mesodermal cells and expands to endodermal cells by early mesenchyme blastula stage (Smith et al., 2007). The later expression of hox11/13b largely overlaps that of blimp1.
Transcription of eve is also evident in the vegetal plate in 6th cleavage (Ransick et al., 2002; Figs. 7A, B). The broad initial domain of expression of eve corresponds to the initial domain of nuclearization of β-catenin (Logan et al., 1999) to which this gene is particularly responsive (Ransick et al., 2002). Its expression extends across the macromeres and their descendents, the veg1 and veg2 cells. Since it extends beyond that of blimp1, the level of β-catenin/Tcf produced by the automomous maternal system suffices for this gene, in contrast to blimp1, which needs the enhanced levels driven by the intercellular wnt8 “pump”. As shown in the 7th cleavage stage embryo in Fig. 7A, expression of eve then fades from the veg1 cells (the “outer” tier of cells when viewed from the vegetal pole as in this figure) just as does β-catenin nuclearized by the autonomous maternal system (Logan et al., 1999). Expression also disappears from the micromeres. Expression of eve continues in veg2 descendents until about the hatched blastula stage, when it fades from these cells and turns on in veg1 descendents. The expression of eve can thus be described as being slightly “out-of-phase” with respect to the rest of the cohort of torus genes, running at the leading edge of the expanding patterning wave.
Verification of predicted inputs controlling eve expression
We sought to verify the cis-regulatory inputs driving this distinct pattern of eve expression relative to the other genes patterned by the torus subcircuit. A BAC clone (079 A02) containing 158 kb of genomic sequence surrounding the eve locus was used to generate a BAC-GFP knock-in by homologous recombination, replacing the first eve exon with a GFP coding sequence. Previously, it had been found that the proximal upstream ∼2 kb and intronic ∼1 kb are sufficient to recapitulate endogenous eve expression (A. Ransick, unpublished data). We narrowed this to ∼1.5 kb of upstream sequence and ∼500 bp of intronic sequence, and used this construct for the ensuing experiments (Fig. 9, 2.5 kb Eve-GFP). By Family Relations analysis with the comparable BAC sequence for L. variegatus, we found most, but not all, of this proximal upstream region highly conserved between the two species (Fig. 8A). Within this conserved patch are overlapping Blimp1 and Tcf sites (Fig. 8B), reminiscent of those found at both the blimp1 and wnt8 loci. Additionally a pair of putative Hox sites is present in the more distal portion of the upstream sequence (Fig. 8B). These sites are perfect 10-mer repeats consisting of 5′ TAATAATAAT separated by 5 bp. Though these sites lay outside the conserved patch of upstream sequence, the previous analysis had indicated that this region is important for proper reporter GFP expression. The functional values of the candidate Blimp1, Tcf, and Hox sites were then tested by mutation.
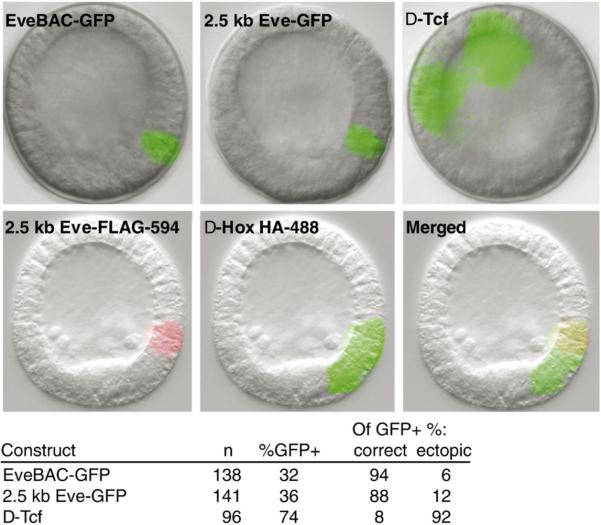
Fig. 9.
Effect of binding site disruption of even-skipped GFP reporters. (A) Representative fluorescent images of transgenic embryos harboring eve GFP reporter constructs. The top pair of panels display correct GFP reporter expression driven by the EveBAC-GFP recombinant and by a 2.5 kb Eve-GFP construct; expression is in veg1 endoderm cells in early mesenchyme blastula embryos. By contrast, embryos injected with the 2.5 kb Eve-GFP construct in which the Tcf site is mutated (Δ-Tcf; upper right panel) show gross ectopic expression in the ectoderm; not evident in these images is the relatively weak GFP reporter expression typical for this class of embryos. In the bottom three panels are experiments using the 2.5 kb Eve-GFP fragment bearing either FLAG or HA epitope tags. In these constructs, the Hox sites were, respectively, either left intact or disrupted. The normal (FLAG) construct is expressed the same as the parent 2.5 kb Eve-GFP construct (left panel); in contrast the Δ-Hox (HA) construct shows ectopic reporter expression in the veg2 clearance zone of endogenous eve expression (center panel). The lower right panel shows a merged image. (B) Table showing the quantitative effects of Tcf site disruption on eve GFP reporter activity. Loss of Tcf input leads to a greatly increased number of embryos displaying ectopic expression.
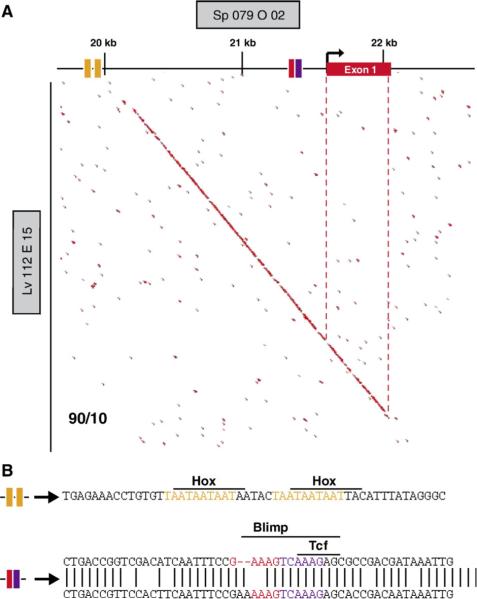
Fig. 8.
Family Relations II analysis of the eve locus and identification of candidate input binding sites. (A) Pairwise sequence analysis of two BAC sequences containing the eve gene using the Family Relations II program. The horizontal line at the top represents the sequence of the Strongylocentrotus purpuratus BAC clone 079 O02; the ordinate represents Lytechinus variegatus BAC clone 112 E15. Red dots indicate 10-nucleotide stretches of sequence sharing ≥90% identity between the two species. Conserved sequence patches are revealed by contiguous dots which produce short diagonal segments. Red box on the horizontal sequence indicates the first exon with the bent arrow showing the start of transcription. Other colored boxes represent candidate factor binding sites: orange, Hox sites; red, Blimp1 site; purple, Tcf site completely overlapped by Blimp1 site. Note the pair of putative Hox sites which lie outside conserved patch. (B) Sequence of candidate input target sites. The pair of Hox sites (orange) consist of perfect 10 bp repeats separated by 5 bp. The candidate Blimp (red) and Tcf (purple) sites lie within a heavily conserved patch of sequence. The top line represents the S. purpuratus sequence and the bottom that of L. variegatus with vertical lines indicating sequence identity. The putative Blimp sites completely overlaps the Tcf site: the core 5′AAAG sequence is conserved in L. variegatus.
Reporter constructs in which the Blimp1 site was mutated were generated from 2.5 kb Eve-GFP, and assayed for GFP expression. As described in Materials and methods, even though the Blimp1 and Tcf sites are partially overlapping, it was possible to produce mutations that destroy each without affecting the other. When the Blimp1 sites were mutated, we found the GFP reporter activity virtually undetectable by eye in the fluorescent microscope (not shown). Consistent with this, QPCR analysis showed GFP mRNA content at 18 h to be only 14 ± 9% that of the control. Thus by this point the accumulation of eve transcripts is strongly dependant on the cis-regulatory Blimp1 input. A similar assay using constructs with a mutated Tcf site displayed a very high frequency of ectopic expression in the ectoderm (Fig. 9, Δ-Tcf). These results indicate that during the dynamic torus phase of its activity the Tcf site restricts eve expression to the vegetal domains where β-catenin is present, while the Blimp1 site is essential for much of its transcriptional activity.
To test the roles of the candidate Hox sites, we generated two new reporter constructs by addition into the C-terminal coding sequence for GFP either a FLAG or HA epitope tag sequence. In the HA-tagged construct, the Hox sites were disrupted by PCR-directed mutagenesis. We then co-injected these constructs and collected embryos at early mesenchyme stage for immunofluorescent analysis. Expression of the wild-type 2.5 kb Eve-FLAG construct was detected by labeling with a secondary antibody conjugated to Alexa-594. This construct displayed correct expression in the veg1 endodermal cells, as evidenced by red channel fluorescence (Fig. 9, bottom left panel). The same embryos were meanwhile labeled with a directly conjugated HA monoclonal antibody tagged with Alexa-488. The clear result was that the green fluorescence produced by the construct lacking intact Hox sites now extends ectopically into the veg2 domain (Fig. 9, bottom central panel). The merged image of a representative embryo is shown in Fig. 9, bottom right panel, illustrating the vegetal extension of reporter expression driven by the Δ-Hox construct.
Discussion
This study adds two further cases to a list that now includes a majority of the more important nodes in the endomesoderm gene regulatory network, at which predicted inputs have been independently authenticated by direct cis-regulatory evidence. The more important nodes can be defined as those which have multiple inputs from elsewhere in the network, and which control production of outputs to other genes in the network. Among genes for which cis-regulatory authentication has been obtained, in addition to the blimp1 and even-skipped genes that are the subject of the present report, are the otx gene (Yuh et al., 2004); the gcm gene (Ransick and Davidson, 2006); the cyclophylin gene (Amore and Davidson, 2006); the gatae gene (P. Y. Lee and E. H. Davidson, unpublished data); the wnt8 gene (Minokawa et al., 2005); the delta gene (Revillai-Domingo et al., 2004 and unpublished data); the brachury gene, E. Chow and A. Cameron, unpublished data); and the endo16 gene, Yuh et al., 2001, 2004). The alx1 gene, the tbr gene, the hox11/13b gene, and the foxa genes are under study, and the list will soon be completed. But it is already clear that the perturbation results underlying the network topology (Davidson et al., 2002a,b; Oliveri and Davidson, 2004) have afforded a sufficient handle on the logic of the system to provide a very high degree of accuracy and predictability.
Cis-regulatory inputs of the blimp1 early transcription unit
Figs. 4 and 5 and data reported elsewhere (Smith et al., 2007) provide direct mutational and other evidence for the functional significance of all three of the regulatory inputs that according to network analysis should account causally for expression of the blimp1 transcript (Minokawa et al., 2005; Livi and Davidson, 2006; and earlier references). Candidate target sites for the Otx, Tcf, and Blimp1 factors were identified in the genomic sequence of CR2, the conserved sequence patch that we show here is the cis-regulatory module which controls blimp1 expression (Figs. 2 and 3). Among these target sites are those with the predicted functions, which can be summarized as follows: (i) The ubiquitous (at the relevant stages) Otx factor provides an essential positive input. (ii) However, because the module operates as an AND logic processor, the positive Otx input affects transcription only in endomesodermal cells receiving the Wnt8 ligand and therefore containing the permissive β-catenin/Tcf complex. (iii) Otherwise, the Tcf sites function as target sites where the alternate Groucho/Tcf complex dominantly represses blimp1 transcription, and this results in an active repression of blimp1 throughout the prospective ectoderm. (iv) The Blimp1 target sites mediate autorepression, accounting for the eventual extinction of blimp1 expression in cells which earlier expressed it (Smith et al., 2007).
Even though the sequences of the S. purpuratus and L. variegatus blimp1 cis-regulatory modules are sufficiently similar to have permitted their identification by use of the criterion of evolutionary conservation (Fig. 2), individual sites are often in different places and differently arranged. Given the background level of evolutionary change in site disposition, those cases of tightly conserved site arrangement are likely informative. For example, neither of the two functional Otx sites of the S. purpuratus module are conserved in L. variegatus, though there are three other Otx sites within 500 bp of CR2 in this genome. On the other hand, the unique arrangement of the essential Tcf and Blimp sites in S. purpuratus, in which a pair of Blimp sites surrounds and indeed partially overlaps a pair of Tcf sites separated by only 2 bp, is almost completely conserved in Lytechinus (Fig. 3). About 200 bp downstream in the Lytechinus sequence is another pair of Tcf sites also separated by 2 bp, lacking adjacent Blimp sites, but its functional value is not known. In the wnt8 cis-regulatory module (Minokawa et al., 2005), Tcf and Blimp sites are also contiguous, though there are at least two differences: these sites are not present in pairs; and they function synergistically and positively in the wnt8 gene, while in the blimp1 regulatory module it is these sites which contribute both the autorepression and the Tcf mediated repression of ectoderm expression.
Cis-regulatory inputs of the eve gene
The experiments summarized in Fig. 9 provide direct support, also obtained by specific binding site mutagenesis, for the functional significance of the three chief regulatory inputs which according to network analysis should govern eve gene expression. Candidate binding sites, including overlapping Blimp1/Tcf sites and a pair of Hox sites, were identified in the highly conserved upstream region extending ∼1.5 kb from the start of eve transcription (Fig. 8). The functions encoded in this cis-regulatory apparatus are as follows: (i) Blimp1 provides the predicted activation. (ii) A positive β-catenin/Tcf input is apparently also essential (Ransick et al., 2002), though this remains to be shown directly. (iii) Groucho-Tcf clearly acts as a dominant repressor of eve except in vegetal domains containing the permissive β-catenin/Tcf complex. (iv) Hox11/13b represses transcription within the veg2 domain, allowing expression to continue in veg1 cells (Fig. 9). The combined effect is a distinctive pattern of eve transcription compared with the other genes entrained by the torus subcircuit. To begin, the early expression of hox11/13b at 7th cleavage (Figs. 7A, B) accounts for the early clearance of eve expression from the micromeres, before expression of blimp1 or wnt8 clears from these cells (Fig. 7B, Fig. S1), while at the same time eve is expressed in a broader domain peripherally because of its apparent higher sensitivity to β-catenin/Tcf. Thereafter the repression by Hox11/13b in the more vegetal cell tiers of the dynamic torus pattern confines eve expression to the leading edge of the expanding ring of regulatory gene expression (Fig. 9), while in the center of the region of inactivity the absence of Blimp1, as well as the presence of the Hox11/13b repressor, contributes to eve silencing. But Hox11/13b repression alone suffices, since even if Blimp1 factor is supplied ectopically within the center of the torus, eve expression cannot be restored there (data not shown), while as demonstrated by Smith et al. (2007), expression of blimp1 and wnt8 is restored to the center of the torus under these conditions. Thus the pattern of eve expression is “out-of-phase” with respect to the other genes in this cohort throughout (Fig. 7). The genesis of this pattern is summarized by the cis-regulatory logic diagram shown in Fig. 6.
While the overlapping Blimp1/Tcf sites at the eve locus lie within a highly conserved region and are themselves largely preserved in L. variegatus, the functional twin Hox sites reside outside the conserved proximal patch (Fig. 8A). It is not certain whether this represents an essential difference in eve regulation between the two species, or if there are other functional sites mediating repression by Hox11/13b in L. variegatus. Nonetheless, this is another example of the evolutionary lability of functional repressor sites which lie outside a positively acting cis-regulatory module that is itself conserved.
The Blimp1/Tcf sites of the eve module mark the third locus where Blimp1 and Tcf target sites are close, as reported both in the blimp1 module (this work and Smith et al., 2007), and a wnt8 cis-regulatory module (Minokawa et al., 2005). It is perhaps indicative that overlapping Blimp1/Tcf sites are to be found in a highly conserved proximal upstream region of the hox11/13b gene as well; in fact 9/10 bp of this site are identical to one at the blimp1 locus. However, the Blimp1/Tcf site complexes do not function identically in the three genes. Analysis of their functionality in the hox11/13b cis-regulatory module will help to illuminate the contextual distinctions which predict their activities. More generally, their different roles invite the use of synthetic methods to reveal the determinate design aspects.
A cohort of dynamically expressed regulatory genes
The properties of the blimp1 and the wnt8 cis-regulatory modules explain the function of the subcircuit in which they are linked, which is to drive the dynamic expression of an expanding torus pattern of regulatory gene expression (Smith et al., 2007; Minokawa et al., 2005). Its basic feature is that blimp1 transcriptional autorepression negatively controls the spatial availability of both its own product and of wnt8 transcript (Fig. 1). Thereby, for genes requiring Blimp1 and Tcf/β-catenin inputs, it causes progressive clearance of transcriptional activity within the enlarging center of the torus. The torus meanwhile expands outwards as Wnt8 diffuses to adjacent cells (Smith et al., 2007). We show here that additional regulatory genes are controlled by the wnt8-blimp1 subcircuit. One of these genes, eve is entrained into the dynamic torus expression pattern by means of the encoded target sites of its cis-regulatory module. An elegant aspect of the design of this network subcircuit is the use of another of the downstream cohort of torus genes, hox11/13b, to sculpt the spatial output of eve, a member of the same gene cohort. This too is a direct function of the primary eve cis-regulatory apparatus. The particular kinetics of the cohort expression patterns will depend on transcription and protein turnover rates and DNA–protein interaction constants (cf. Bolouri and Davidson, 2003). But the causal explanation of the linkage of the gene cohort into the torus expression pattern resides directly in the genomic regulatory DNA sequence.
Supplementary Material
Acknowledgments
We thank Veronica Hinman and Ellen Rothenberg for critical reading and helpful suggestions. This work was supported by NIH grant HD37105.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2007.10.042.
References
- Amore G, Davidson EH. Cis-regulatory control of cyclophilin, a member of the ETS-DRI skeletogenic gene battery in the sea urchin embryo. Dev. Biol. 2006;293:555–564. doi: 10.1016/j.ydbio.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Arenas-Mena C, Cameron RA, Davidson EH. Hindgut specification and cell-adhesion functions of Sphox11/13b in the endoderm of the sea urchin embryo. Dev. Growth Differ. 2006;48:463–472. doi: 10.1111/j.1440-169X.2006.00883.x. [DOI] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. Transcriptional regulatory cascades in development: initial rates, not steady state, determine network kinetics. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CT, Rust AG, Clarke PJ, Pan Z, Schilstra MJ, De Buysscher T, Griffin G, Wold BJ, Cameron RA, Davidson EH, Bolouri H. New computational approaches for analysis of cis-regulatory networks. Dev. Biol. 2002;246:86–102. doi: 10.1006/dbio.2002.0619. [DOI] [PubMed] [Google Scholar]
- Brown CT, Xie Y, Davidson EH, Cameron RA. Paircomp, Family Relations II and Cartwheel: tools for interspecific sequence comparison. BMC Bioinformatics. 2005;6:70. doi: 10.1186/1471-2105-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RA, Chow SH, Berney K, Chiu TY, Yuan QA, Kramer A, Helguero A, Ransick A, Yun M, Davidson EH. An evolutionary constraint: strongly disfavored class of change in DNA sequence during divergence of cis-regulatory modules. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11769–11774. doi: 10.1073/pnas.0505291102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat. Struct. Mol. Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- Croce JC, Wu SY, Byrum C, Xu R, Duloquin L, Wikramanayake AH, Gache C, McClay DR. A genome-wide survey of the evolutionarily conserved Wnt pathways in the sea urchin Strongylocentrotus purpuratus. Dev. Biol. 2006;300:121–131. doi: 10.1016/j.ydbio.2006.08.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Gene Regulatory Networks in Development and Evolution. Academic Press; San-Diego: 2006. The Regulatory Genome. [Google Scholar]
- Davidson EH, Cameron RA, Ransick A. Specification of cell fate in the sea urchin embryo: summary and some proposed mechanisms. Development. 1998;125:3269–3290. doi: 10.1242/dev.125.17.3269. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science. 2002a;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Schilstra MJ, Clarke PJ, Rust AG, Pan Z, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A provisional regulatory gene network for specification of endomesoderm in the sea urchin embryo. Dev. Biol. 2002b;246:162–190. doi: 10.1006/dbio.2002.0635. [DOI] [PubMed] [Google Scholar]
- Kuo TC, Calame KL. B lymphocyte-induced maturation protein (Blimp)-1, IFN regulatory factor (IRF)-1, and IRF-2 can bind to the same regulatory sites. J. Immunol. 2004;173:5556–5563. doi: 10.4049/jimmunol.173.9.5556. [DOI] [PubMed] [Google Scholar]
- Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- Li X, Chuang CK, Mao CA, Angerer LM, Klein WH. Two Otx proteins generated from multiple transcripts of a single gene in Strongylocentrotus purpuratus. Dev. Biol. 1997;187:253–266. doi: 10.1006/dbio.1997.8610. [DOI] [PubMed] [Google Scholar]
- Livi CB, Davidson EH. Expression and function of blimp1/krox, an alternatively transcribed regulatory gene of the sea urchin endomesoderm network. Dev. Biol. 2006;293:513–525. doi: 10.1016/j.ydbio.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Livi CB, Davidson EH. Regulation of spblimp1/krox1a, an alternatively transcribed isoform expressed in midgut and hindgut of the sea urchin gastrula. Gene Expression Patterns. 2007;7:1–7. doi: 10.1016/j.modgep.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- Minokawa T, Wikramanayake AH, Davidson EH. cis-regulatory inputs of the wnt8 gene in the sea urchin endomesoderm network. Dev. Biol. 2005;288:545–558. doi: 10.1016/j.ydbio.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH. Gene regulatory network controlling embryonic specification in the sea urchin. Curr. Opin. Genet. Dev. 2004;14:351–360. doi: 10.1016/j.gde.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Oosterwegel MA, van de Wetering ML, Holstege FC, Prosser HM, Owen MJ, Clevers HC. TCF-1, a T cell-specific transcription factor of the HMG box family, interacts with sequence motifs in the TCR beta and TCR delta enhancers. Int. Immunol. 1991;3:1189–1192. doi: 10.1093/intimm/3.11.1189. [DOI] [PubMed] [Google Scholar]
- Range RC, Venuti JM, McClay DR. LvGroucho and nuclear beta-catenin functionally compete for Tcf binding to influence activation of the endomesoderm gene regulatory network in the sea urchin embryo. Dev. Biol. 2005;279:252–267. doi: 10.1016/j.ydbio.2004.12.023. [DOI] [PubMed] [Google Scholar]
- Ransick A, Davidson EH. Cis-regulation of Notch signaling input in the sea urchin gcm gene. Dev. Biol. 2006;297:587–602. doi: 10.1016/j.ydbio.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH. New early zygotic regulators expressed in endomesoderm of sea urchin embryos discovered by differential array hybridization. Dev. Biol. 2002;246:132–147. doi: 10.1006/dbio.2002.0607. [DOI] [PubMed] [Google Scholar]
- Revilla-i-Domingo R, Minokawa T, Davidson EH. R11: a cis-regulatory node of the sea urchin embryo gene network that controls early expression of SpDelta in micromeres. Dev. Biol. 2004;274:438–451. doi: 10.1016/j.ydbio.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Smith J, Theodoris C, Davidson EH. A gene regulatory network subcircuit drives a dynamic pattern of gene expression. Science. 318:794–797. doi: 10.1126/science.1146524. in press. [DOI] [PubMed] [Google Scholar]
- Wikramanayake AH, Huang L, Klein WH. Beta-catenin is essential for patterning the maternally specified animal-vegetal axis in the sea urchin embryo. Proc. Natl. Acad. Sci. U. S. A. 1998;95:9343–9348. doi: 10.1073/pnas.95.16.9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikramanayake AH, Peterson R, Chen J, Huang L, Bince JM, McClay DR, Klein WH. Nuclear beta-catenin-dependent Wnt8 signaling in vegetal cells of the early sea urchin embryo regulates gastrulation and differentiation of endoderm and mesodermal cell lineages. Genesis. 2004;39:194–205. doi: 10.1002/gene.20045. [DOI] [PubMed] [Google Scholar]
- Yuh C-H, Brown CT, Livi CB, Rowen L, Clarke PJ, Davidson EH. Patchy interspecific sequence similarities efficiently identify positive cis-regulatory elements in the sea urchin. Dev. Biol. 2002;246:148–161. doi: 10.1006/dbio.2002.0618. [DOI] [PubMed] [Google Scholar]
- Yuh C-H, Dorman ER, Howard ML, Davidson EH. An otx cis-regulatory module: a key node in the sea urchin endomesoderm gene regulatory network. Dev. Biol. 2004;269:536–551. doi: 10.1016/j.ydbio.2004.02.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.