Abstract
Cell behavior is guided by the complex interplay of matrix mechanical properties as well as soluble and immobilized biochemical signals. The development of synthetic scaffolds that incorporate key functionalities of the native extracellular matrix (ECM) for support of cell proliferation and tissue regeneration requires that stiffness and immobilized concentrations of ECM signals within these biomaterials be tuned and optimized prior to in vitro and in vivo studies. A detailed experimental sensitivity analysis was conducted to identify the key polymerization conditions that result in significant changes in both elastic modulus and immobilized YRGDS within visible light photopolymerized poly(ethylene glycol) diacrylate (PEGDA) hydrogels. Among the polymerization conditions investigated, single as well as simultaneous variations in N-vinylpyrrolidinone (NVP) and precursor concentrations of Acryl-PEG3400-YRGDS resulted in a broad range of hydrogel elastic modulus (81 – 1178 kPa) and YRGDS surface concentration (0.04 – 1.72 pmol/cm2). Increasing the YRGDS surface concentration enhanced fibroblast cell adhesion and proliferation for a given stiffness, while increases in hydrogel elastic modulus caused decreases in cell adhesion and increases in proliferation. The identification of key polymerization conditions is critical for the tuning and optimization of biomaterial properties and the controlled study of cell-substrate interactions.
Keywords: PEG hydrogels, free-radical photopolymerization, elastic modulus, immobilized YRGDS, cell adhesion and proliferation
Introduction
Native cell function is dictated by a complex interplay of matrix mechanical properties as well as the spatial and temporal presentation of soluble and immobilized biochemical signaling molecules of the native extracellular matrix (ECM). Tissue engineering has attempted to recreate this multifaceted environment using synthetic biomaterials [1-4]. Among the classes of synthetic polymer scaffolds, hydrogels composed of covalently crosslinked poly(ethylene glycol) diacrylate (PEGDA) have been extensively used in tissue engineering due to their inherent biocompatibility and facile tuning of mechanical and physical properties through alterations in polymerization conditions [5,6]. PEGDA hydrogels are resistant to non-specific protein adsorption and cellular adhesion in the absence of cell signaling molecules thus providing a blank slate upon which specific ECM cues can be incorporated within their crosslinked network. By reacting N-hydroxysuccinimidyl ester derivatives of PEG monoacrylates with the cell adhesion peptide RGD, Hern and Hubbell showed that photopolymerizable RGD containing monoacrylated macromers of PEG (MW=3400 Da) could be used to tether RGD to the hydrogel network upon photopolymerization [7]. Studies employing this approach, have shown that PEGDA hydrogels immobilized with RGD support the specific adhesion of fibroblasts [7], endothelial cells [8,9], smooth muscle cells (SMC) [6], pre-osteoblastic cells [10], and neurites [11,12] in vitro. This chemistry has also been adapted to engineer PEGDA scaffolds that support specific cellular interactions through covalent incorporation of growth factors and crosslinked enzymatically degradable peptide domains sensitive to cell-mediated proteolysis [3,13-24].
In order to develop a functional biomaterial scaffold that supports 3D cell behavior, the incorporation and final concentration of matrix biochemical cues as well as mechanical properties should be quantified a priori to in vitro cell culture or in vivo experiments in a systematic and reproducible manner [25]. While extensive studies have investigated the effects of polymerization conditions on the mechanical and/or physical properties of PEGDA hydrogels and how these properties in turn influence cell function [6,8,13,26], limited studies have investigated the effects of polymerization conditions on immobilized concentrations of ECM signals [7,10], which are equally important scaffold design variables that determine the adhesion, migration and proliferation of anchorage-dependent cells. Previous studies using radiolabeling experiments to track the incorporation of the cell adhesion ligand YRGDS have shown that the incorporation of acryl-PEG3400-YRGDS macromers into ultraviolet light (UV) photopolymerized PEGDA hydrogel scaffolds increased from 81 to 97% through individual variations in either the concentration of the acryl-PEG-peptide conjugate and the PEGDA crosslinking agent in the precursor solution [7]. Based on these results, this study demonstrated that cell adhesion and spreading increased with immobilized YRGDS content in a dose-dependent manner. Khatiwala et al. used a fluorescently labeled lysine as an RGD surrogate to track adhesion peptide incorporation [10] in PEGDA hydrogels photopolymerized in the presence of UV for a fixed exposure time of twenty minutes. They showed that peptide incorporation was independent of gel composition with no significant differences in relative fluorescence intensity observed between gels that varied in stiffness by an order of magnitude. However it is not clear how incorporated levels of YRGDS would change under different polymerization times and variations in other polymerization conditions. Other studies have focused on altering RGD functionalized PEGDA hydrogel mechanical properties independent of variations in biochemical composition through the addition of a non-acrylated PEG that does not participate in the crosslinking reaction to the prepolymer solution [6]. This approach was used to show that increasing the fraction of non-acrylated PEG decreased the hydrogel modulus. Furthermore, 2D SMC’s proliferated to a greater extent on stiffer hydrogels whereas increases in RGD precursor composition resulted in hydrogels that promoted increased SMC focal adhesion area [6]. While the above-mentioned studies have focused on controlling PEGDA hydrogel properties and the incorporation of adhesion ligands via alterations in composition of individual precursor components, a variety of polymerization conditions dictate the final biomaterial properties. Limited studies have looked at the simultaneous variation of polymerization conditions as a means for controlling hydrogel mechanical properties and consequently cell behavior. Using visible light photopolymerization, Elbert and Hubbell [8] showed that through simultaneous increases in the fraction of PEG diacrylates containing attached RGD peptides as well as the concentration of N-vinylpyrrolidinone (NVP), substantial increases in 2D endothelial cell projected area could be achieved. In addition, this study also illustrated that small alterations in NVP precursor concentration led to substantial effects on gel mechanical properties and endothelial cell projected area [8]. However, an investigation of the effects of variations in multiple polymerization conditions on both immobilized ECM signals and mechanical properties of PEGDA hydrogels has not been previously conducted.
While the aforementioned studies have provided insight on the role of polymerization conditions in determining the mechanical properties and/or incorporated levels of ECM signals within hydrogels, there has yet to be a comprehensive examination of how polymerization conditions influence the resultant properties of PEGDA scaffolds formed via free-radical photopolymerization in the presence of visible light. In this study we quantify the effects of alterations in multiple polymerization conditions such as exposure time and light intensity, as well as variations in precursor concentrations of the photoinitiator Eosin Y, comonomer and accelerator of free-radical polymerization, NVP, the biofunctional acrylate-PEG-YRGDS macromer, and the concentration and molecular weight of the crosslinking agent PEGDA on the resultant elastic modulus and immobilized YRGDS concentration. Alterations in these conditions were chosen based on the critical role they play during free-radical photopolymerization influencing both the incorporation of pendant functionalities (RGD) into the scaffold as well as hydrogel crosslinking, which can be correlated to the hydrogel mechanical properties. A detailed experimental sensitivity analysis of the effects of polymerization conditions on PEGDA scaffold mechanical properties as well as the final concentration of the cell adhesion ligand YRGDS in hydrogels formed using visible light free-radical photopolymerization is presented. Our studies show that a broad range of elastic modulus and adhesion ligand concentration can be achieved in this photopolymerization system through the simultaneous variation of specific polymerization conditions. Finally, we evaluate the effect of final matrix properties (immobilized YRGDS concentration and elastic modulus) on 2D fibroblast cell adhesion and proliferation in vitro.
Materials and Methods
Materials
PEG (MW = 3400 & 8000 Da), triethylamine 99%, triethanolamine (TEA) >99.5%, 1-vinyl-2-pyrrolidinone (NVP) >99%, Eosin Y 80% and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) sodium salt 99% were purchased from Sigma-Aldrich (St Louis, MO). Acryloyl chloride 96%, potassium carbonate, magnesium sulfate, diethyl ether 99.9%, sodium bicarbonate, anhydrous dichloromethane 99.9%, sodium chloride, 10× phosphate buffered saline (PBS), Dulbecco’s modified eagle’s medium (DMEM), heat inactivated fetal bovine serum (FBS), penicillin(5000 I.U./mL)-streptomycin(5 mg/mL) solution (PenStrep) and 1× trypsin 0.05% were obtained from Fisher Scientific (Hanover Park, IL).
Cell Culture
NIH 3T3 fibroblasts were maintained in DMEM supplemented with 10% FBS and 1% PenStrep at 37° C and 5% CO2. Cells were grown to confluence and collected by incubation with trypsin for 5 minutes followed by centrifugation for 10 minutes at 200 g.
MTS Cytotoxicity Assay
In this study we make use of visible as opposed to UV light photopolymerization for the fabrication of PEGDA hydrogel scaffolds. To investigate the effect of UV versus visible light photoinitiator on cell viability, an MTS cytotoxicity assay (Promega, Madison, WI) was performed. Fibroblasts were seeded at a density of 5000 cells/well in a 96-well plate and allowed to adhere for 24 hours. The cells were then washed 1× with phosphate buffered saline (PBS) and exposed to either UV or visible wavelength light with and without the corresponding initiator for five minutes. Test conditions included exposure to (i) visible light (λ = 514 nm; 100 mW/cm2), (ii) visible light with 0.05 mM eosin Y and 225 mM TEA (visible light photoinitiator system), (iii) UV light (λ = 365; 11.6 mW/cm2), and (iv) UV light with the photoinitiator 2-hydroxy-2-methylpropiophenone (Irgacure 1173). A five minute exposure time was selected since it is a commonly employed exposure time for UV-based photopolymerized PEGDA hydrogels [27] and since it also represents the longest exposure time used to crosslink the visible light photopolymerized scaffolds in the presented study. Following exposure to above-mentioned test conditions, each well was washed with 1× PBS and fresh culture media added. Control samples were subjected only to the buffer washes. Surviving cells were cultured for 48 hours at 37° C before performing the MTS cytotoxicity assay as outlined in the manufacturer instructions.
Acrylation of poly(ethylene glycol)
PEGDA macromers were produced through the chain-end acrylation of PEG. Briefly, 24 g of PEG (MWPEG = 3400 or 8000 Da) was dissolved in 80 mL anhydrous dichloromethane and combined with triethylamine at a 2:1 mole ratio under argon purged conditions with continuous stirring. Acryloyl chloride was then added at a 4:1 mole ratio to the PEG and reacted overnight in the dark. Aqueous byproducts of the reaction were removed via the addition of 2 M potassium bicarbonate at a 4:1 mole ratio to the PEGDA and collection of the organic phase following a three day separation. The resulting PEGDA solution was washed with magnesium sulfate and precipitated using 4 L diethyl ether. Precipitated PEGDA was dried under vacuum and stored until use at −20° C. Acrylation efficiency of PEGDA was confirmed to be above 90% using nuclear magnetic resonance (1H NMR).
Conjugation of YRGDS
Acryl-PEG3400-YRGDS was synthesized by reacting the terminal amine of the cell adhesion peptide YRGDS (American Peptide, Sunnyvale, CA) with the succinimidyl valerate ester (SVA) of an acryl-PEG3400-SVA (MW = 3400 Da; Laysan Bio Inc, Arab, AL) in 50 mM sodium bicarbonate (pH 8) at a 1:1 mole ratio for two hours. The reaction mixture was then dialyzed for 24 hours (Slide-A-Lyzer® Dialysis Cassette 2000 MWCO; Fisher Scientific, Hanover Park, IL) to remove unreacted peptide, lyophilized and stored at −80° C until use. For simplicity, acryl-PEG3400-YRGDS will be henceforth referred to as YRGDS.
Photopolymerization of PEGDA Hydrogels
PEGDA macromers of varying molecular weight were polymerized in the presence of the chain transfer agent, TEA, the accelerator and co-monomer, NVP, and the visible light photoinitiator, eosin Y. The PEGDA macromer, TEA and NVP were dissolved in deionized water and the pH adjusted to ~8.3. The cell adhesion peptide YRGDS was then dissolved in the precursor solution prior to the addition of eosin Y and subsequent photopolymerization. The standard polymerization condition was defined as a precursor composition consisting of 25% PEGDA, 225 mM TEA, 37 mM NVP, 0.05 mM eosin Y and 5 mg/mL YRGDS with hydrogels polymerized for 1 minute at 100 mW/cm2 using visible light (λ = 514 nm) produced by an Argon Ion laser (Coherent, Inc., Santa Clara, CA).
Quantification of YRGDS Incorporation in PEGDA Hydrogels
The efficiency of YRGDS incorporation into the crosslinked hydrogel network was quantified by radiolabeling the tyrosine residue [7] for a variety of polymerization conditions as identified in Table 1. Briefly, 5 mg unconjugated YRGDS was dissolved in reaction buffer (1× PBS; pH = 6.5), combined with 1 mCi sodium iodine (PerkinElmer Health Sciences Inc., Shelton, CT) in the presence of iodination beads (Pierce, Rockford IL) and allowed to react for 15 minutes. The iodinated YRGDS was dialyzed (Spectra/Por® CE Float-A-Lyzer® 500 Da MWCO, Spectrum Laboratories, Rancho Dominguez, CA) against deionized water for 48 hours with a minimum of four bath water changes to remove any free 125I. The labeled YRGDS was then conjugated to a mono-acrylated PEG as outlined above and stored at −80° C in a lead lined container until use. To track YRGDS incorporation in PEGDA hydrogels, radiolabeled YRGDS was added to the precursor in an amount not exceeding 30 wt % of the total peptide. Prior to polymerization, the activity of the precursor solution for each hydrogel (200 μL) was measured using a Packard Cobra II gamma counter (PerkinElmer Health Sciences Inc., Shelton, CT). Hydrogels were then photopolymerized as outlined above in a 48-well plate followed by washing in deionized water for 36 hours with a minimum of three bath changes to allow for diffusion of unreacted YRGDS. Any unreacted YRGDS that was incapable of diffusing from the gel within this time was assumed to be entrapped within the polymer network. The final activity of each hydrogel was then recorded and compared to the corresponding precursor activity to determine the incorporation efficiency (IE), or fraction of initial activity that remained in the hydrogel for a given polymerization condition. In order to account for changes in YRGDS concentration due to swelling of the matrix, the IE value was used in conjunction with a hydrogel swelling ratio (SR) to calculate the final concentration of YRGDS in the hydrogel according to Equation (1). Briefly, the SR was calculated by dividing the hydrogel mass obtained in the relaxed state immediately following photopolymerization (mr) with the mass of the hydrogel in the swollen state (ms), recorded after the hydrogel achieved equilibrium swelling by incubation in deionized water for at least 24 hours. Average values of IE and SR were determined for each set of polymerization conditions using a sample size of three. Furthermore, IE and SR were found to be independent of initial hydrogel volume (data not shown). The final bulk concentration of YRGDS in hydrogels ([YRGDS]f) was calculated by the following relation,
| (1) |
where [YRGDS]i is the concentration of YRGDS in the precursor solution. The final bulk concentration of YRGDS in the hydrogel was then used to calculate the theoretical surface concentration available for cell adhesion on hydrogel surfaces with the height of the bioavailable surface taken as 10 nm [7]. All radiolabeling experiments were performed in approved areas using designated equipment and waste disposed of in accordance to governing ordinances.
Table 1.
Polymerization conditions used to vary YRGDS incorporation in PEGDA Scaffolds
| Polymerization Condition | Values |
|---|---|
|
| |
| Molecular Weight of PEGDA | 3400 & 8000 Da |
| Exposure Time | 0.5, 1, 2 & 5 mins. |
| Light Intensity | 50, 100 & 200 mW/cm2 |
| PEGDA Concentration | 15, 25 & 40% (w/v) |
| Eosin Y Concentration | 0.025, 0.05 & 0.1 mM |
| Acryl-PEG3400-YRGDS Concentration | 0, 1, 5 & 15 mg/mL |
| NVP Concentration | 37, 74, 111 & 148 mM |
Mechanical Characterization of PEGDA Hydrogels
The mechanical properties of PEGDA hydrogels were quantified using compression experiments. Hydrogels were polymerized using a custom made 10 mm diameter chamber and allowed to reach equilibrium swelling before being loaded onto a TA RSA3 mechanical tester (TA Instruments, New Castle DE) controlled by TA Orchestrator software. Samples were compressed at a rate of −0.5 mm/min to obtain a stress vs. strain curve. The elastic modulus of the hydrogel was taken as the slope of the linear region (<10% strain, r2 > 0.98) of the stress vs. strain curve as has been previously reported [28,29].
2D Cell Adhesion and Proliferation Assays
Fibroblast adhesion and proliferation on PEGDA hydrogels surfaces with varying stiffness and immobilized YRGDS concentration was visualized by phase contrast imaging and quantified over time using an alamarBlue™ (Trek Diagnostic Systems Inc., Cleveland, OH) proliferation assay [30-32]. Prior to cell seeding, hydrogels were photopolymerized in a 24 well-plate using 300 μL of sterile-filtered precursor solution. Hydrogels were then swollen for 48 hrs in sterile HEPES-buffered saline (HBS; 10 mM HEPES sodium salt and 137 mM NaCl, pH 7.4), punched to fit a 24 well-plate and incubated with culture media for approximately 30 min. Hydrogels were seeded with 50,000 cells/cm2 for adhesion assays and with 10,000 cells/cm2 for quantifying proliferation. In the case of cell adhesion experiments, cells were allowed to adhere for 4 hrs before the hydrogels were rinsed with sodium bicarbonate buffered 1× PBS to remove unadhered cells and transferred to a new well to avoid the inclusion of cells that may have adhered to the polystyrene walls. Hydrogels were then submerged in 600 μL of a 5% alamarBlue™ solution in media and incubated for 6 hrs before the supernatant was read by a Spectrimax Gemini XS fluorescent plate reader (Molecular Devices, Sunnyvale, CA; excitation = 544 nm; emission = 590 nm). The amount of reduced alamarBlue™ was converted to the number of cells present using a standard curve. Fibroblast proliferation was quantified in a similar manner only cells were allowed to adhere for 24 hrs before the first reading was taken. The alamarBlue™ assay was then repeated as described above every other day for two weeks and the number of cells present were normalized to day 1. Following each assay the hydrogels were transferred to fresh media. Cell proliferation studies required a decrease in the incubation period with alamarBlue™ from 6 hrs to 4 hrs and subsequently to 2 hrs to prevent complete conversion of the reagent due to the increases in cell number over time. Additional standard curves were generated to account for the shorter incubation periods.
3D Cell Encapsulation
To confirm that cells would survive visible light photopolymerization and proliferate within PEGDA hydrogels, fibroblasts were encapsulated in an enzymatically degradable PEGDA hydrogel. Briefly, hydrogels were rendered sensitive to cell-mediated proteolysis via the conjugation of a collagenase sensitive peptide, GGL↓GPAGGK (↓ denotes cleavage point by cell-secreted collagenase enzyme between the leucine and glycine residues), to acryl-PEG3400-SVA using a 2:1 PEG to peptide mole ratio. Fibroblasts were then suspended in a 5% (w/v) prepolymer solution at a density of 5,000,000 cells/mL (37 mM NVP, 225 mM TEA, and 0.05 mM eosin Y) and polymerized for one minute in the presence of visible light at an intensity of 100 mW/cm2. Following five days in culture, the cells were formalin fixed and stained for f-actin using Alexa Fluor Phalloidin (5 units/mL in PBS; Invitrogen). The stained cells were then imaged using LSM confocal microscopy (Carl Zeiss).
Statistical Analysis
Comparisons of YRGDS incorporation and elastic modulus in PEGDA hydrogels formed across different polymerization conditions (n = 3), as well for cell adhesion (n = 4) and proliferation (n = 5) studies on PEGDA hydrogel surfaces were expressed as mean ± standard deviation. Statistical significance was evaluated using one-way ANOVA followed by a Tukey’s post-test (SigmaStat 3.5) with p < 0.05 considered significant.
Results
Cytotoxicity of UV and Visible Light Free-Radical Photolymerization
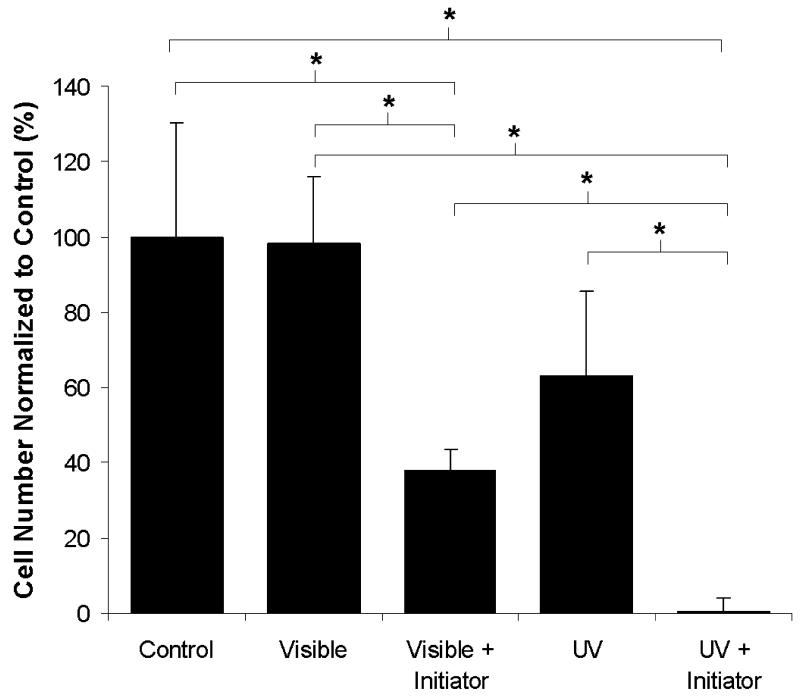
To investigate the effect of UV and visible light on cell viability, fibroblasts were exposed to both wavelengths of light in the absence and presence of the corresponding photoinitiator system. As shown in Figure 1, exposure to visible light (λ = 514 nm) did not decrease in the total number of viable cells as compared to the control. However, exposure to UV wavelength light (λ = 365 nm) resulted in a 37% decrease in total cell number and is likely due to damage or mutations to the cellular DNA. Addition of either type of photoinitiator in combination with light exposure caused a decrease in the total cell number as compared to the control. Specifically, the eosin/TEA photoinitiator system caused a 60% decrease in total cells number as compared to both the control and visible light alone. The combination of UV light exposure and the UV photoinitiator (Irgacure 1173) resulted in no observed cell survival.
Figure 1.
Quantification of cell viability following exposure to UV and visible light in the absence and presence of corresponding initiators (n = 4; * = p < 0.05).
YRGDS Incorporation as a Function of Precursor Composition and Polymerization Conditions
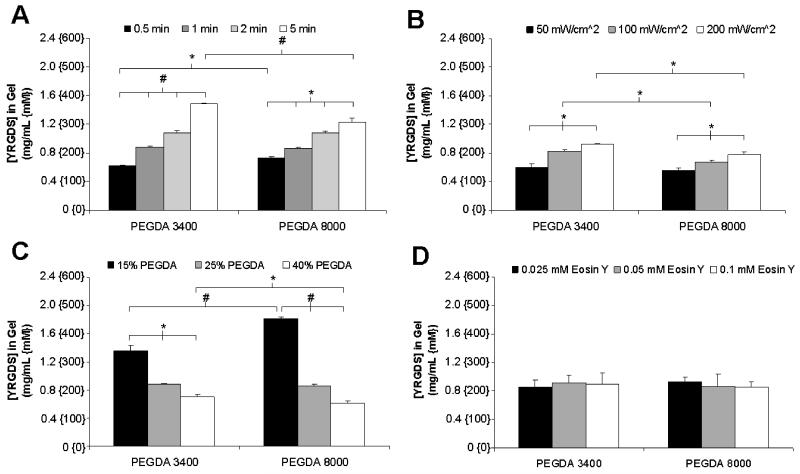
The effect of alterations in polymerization conditions on the incorporation of the cell adhesion ligand YRGDS into crosslinked PEGDA scaffolds was evaluated by radiolabeling and adjusted to account for changes in YRGDS concentration due to hydrogel swelling. In the data presented in Figure 2, YRGDS incorporation was quantified as a function of visible light exposure time, light intensity, PEGDA precursor concentration, eosin Y photoinitiator concentration and PEGDA macromer molecular weight based on the specified ranges presented in Table 1. The PEGDA macromer molecular weights of 3400 and 8000 Da were selected based on the fact that they have been commonly used to support a variety of cell functions for soft tissue engineering [6-8,27,33,34]. Each of the specified polymerization conditions were varied individually while all other conditions were kept constant. The data shown in Figures 2A and 2B demonstrate there is a statistically significant increase in YRGDS incorporation with increased exposure time and light intensity, respectively. Increasing the duration of polymerization from 0.5 to 5 minutes increased the concentration of YRGDS in the resultant hydrogel by 137% (0.63 to 1.49 mg/mL) for hydrogels photopolymerized using a PEGDA macromer molecular weight of 3400 Da as compared to a 68% (0.73 to 1.23 mg/mL) increase in YRGDS concentration when an 8000 Da PEGDA macromer was used. Increases in the intensity of the incident light from 50 to 200 mW/cm2 resulted in a 55% (0.6 to 0.93 mg/mL) and a 39% (0.56 to 0.78 mg/mL) increase in YRGDS concentration for hydrogels photopolymerized using PEGDA macromer molecular weights of 3400 Da and 8000 Da, respectively. As expected, increases in the duration of photopolymerization or the light intensity result in higher monomer conversion and consequently in increased YRGDS incorporation. Decreases in YRGDS incorporation were observed with increasing the PEGDA precursor weight percentage irrespective of the PEGDA macromer molecular weight used to crosslink the hydrogels (Figure 2C). Specifically, increasing the precursor weight percentage of the PEGDA macromer from 15 to 40% (w/vol) resulted in a 49% decrease (1.36 to 0.70 mg/mL) and a 66% decrease (1.82 to 0.61 mg/mL) in YRGDS concentration for hydrogels formed with PEGDA macromer molecular weights of 3400 Da and 8000 Da, respectively. Increases in the PEGDA monomer weight percent results in a decreased mass fraction of YRGDS in the prepolymer solution. It is well established that copolymer composition is only a function of the reactivity ratios which represent the relative reactivity of a monomer to a given radical and the monomer mixture composition. Since the YRGDS monomer mass fraction is reduced in the monomer mixture with increasing PEGDA content, then consequently the YRGDS fraction in the copolymer and resultant hydrogel will decrease. The effect of variations in eosin Y photoinitiator concentration on the final YRGDS concentration within the hydrogels was also investigated. In contrast to the alterations in the polymerization conditions presented in Figure 2 (A-C), increases in the concentration of eosin Y had little effect on the incorporation of YRGDS (Figure 2D). For both PEGDA macromer molecular weights the concentration of YRGDS in the resultant hydrogels remained constant around 0.89 mg/mL for all variations in eosin Y concentration. This suggests that for the given system initiation may be limited by the intensity of the incoming light and not the concentration of eosin Y.
Figure 2.
Quantification of the final concentration of YRGDS in swollen PEGDA hydrogels with varying macromer molecular weights as a function of (A) exposure time, (B) light intensity, (C) PEGDA concentration and (D) eosin Y concentration. Unless noted, hydrogels were formed using a PEGDA concentration of 25% (w/v) and were polymerized for one minute at 100 mW/cm2 with an eosin Y concentration of 0.05 mM. All hydrogels contained a fixed precursor YRGDS concentration of 5 mg/mL (n = 3; * = p < 0.05 and # = p < 0.001).
Figure 2 illustrates that for certain alterations in polymerization conditions statistically significant differences in YRGDS incorporation were observed among hydrogels polymerized under the exact conditions but with different PEGDA macromer molecular weights. These differences in YRGDS incorporation are most evident in the data presented in Figure 2C. Hydrogels formed using a prepolymer solution containing a 15% weight fraction (w/vol) of the PEGDA crosslinking agent exhibited a 34% increase (1.36 to 1.82 mg/mL) in immobilized YRGDS concentration with a PEGDA macromer weight of 8000 Da as compared to hydrogels formed with a macromer molecular weight of 3400 Da. These observed differences in immobilized YRGDS concentration between the two PEGDA macromer molecular weights can be better understood if the respective precursor mole fractions of PEGDA and YRGDS used to crosslink these hydrogels and the molecular weight of the YRGDS macromer conjugate (fixed at 4000 Da) are considered. For example, use of a PEGDA 8000 Da macromer as compared to a PEGDA 3400 Da macromer results in a 31.8% decrease in the precursor PEGDA macromer mole fraction and a 63.6% increase in the precursor mole fraction of YRGDS for a precursor solution consisting of 25% PEGDA, 37 mM NVP, and 5 mg/mL YRGDS. This change alters the kinetics of polymerization resulting in higher YRGDS incorporation. However, these differences do not always appear to be significant and in some cases (Figure 2A and 2B) this effect was negated due to the slower kinetics of the higher macromer molecular weight of the PEGDA crosslinking agent. Therefore, subsequent studies were limited to using a PEGDA macromer of molecular weight equal to 3400 Da.
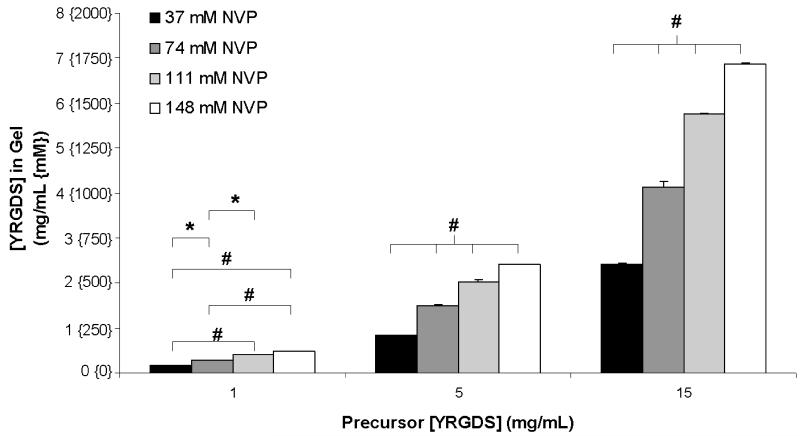
The data presented in Figure 2 illustrates that the selection of polymerization conditions is critical in controlling the final concentrations of immobilized cell adhesion ligands in photopolymerized PEGDA hydrogels. Based on variations in polymerization conditions presented in Figure 2, the range of immobilized YRGDS obtained was between 0.56-1.82 mg/mL. In order to induce more substantial changes in YRGDS incorporation, the effect of NVP, a co-monomer and accelerator of free-radical polymerization, was considered. It has been previously reported that the addition of NVP in the precursor solution accelerates free-radical polymerization by preventing the scavenging of free-radicals by oxygen [35]. Furthermore, previous studies have indicated that variations in precursor concentrations of NVP alter PEG hydrogel mechanical properties [8]. We therefore hypothesized that simultaneous alterations in NVP and YRGDS would also lead to substantial changes in YRGDS incorporation. As indicated in Figure 3, simultaneous variations in NVP and YRGDS precursor concentrations resulted in a broader range of immobilized YRGDS incorporation in hydrogels as compared to those achieved through the altered conditions presented in Figure 2. Specifically, the range of immobilized YRGDS obtained with these simultaneous changes in polymerization conditions was between 0.2-6.9 mg/mL with statistical significant differences observed when comparing groups formed with fixed precursor concentration of YRGDS but with varying NVP concentration. This suggests that NVP plays a critical role in tuning immobilized YRGDS concentration in photopolymerized PEG scaffolds. Figure 3 also demonstrates that increasing the precursor concentration of YRGDS from 1 to 5 to 15 mg/mL for a given concentration of NVP results in significant changes in the concentration of YRGDS in the resultant hydrogels (significance omitted from graph for clarity).
Figure 3.
Quantification of the final concentration of YRGDS in swollen PEGDA 3400 hydrogels as a function of precursor YRGDS concentration for various NVP concentrations. Hydrogels were formed using a 25% (w/v) PEGDA 3400 precursor solution and polymerized for one minute at 100 mW/cm2 (n = 3; * = p < 0.05 and # = p < 0.001).
Elastic Modulus of PEGDA Hydrogels as a Function of Precursor Composition
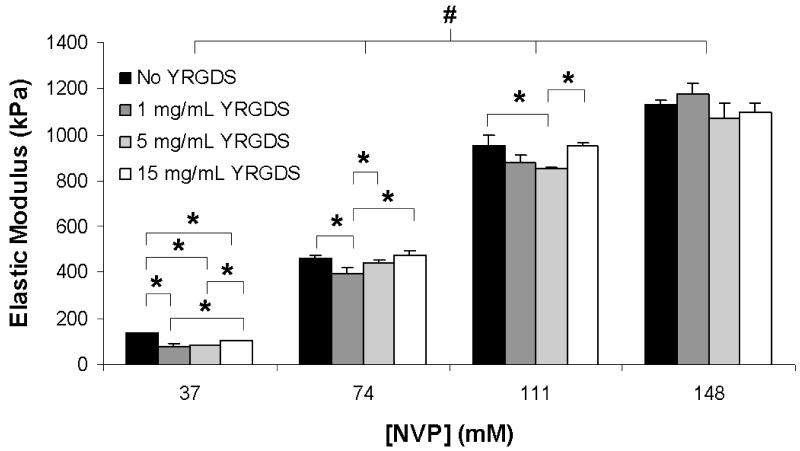
The simultaneous variation in precursor concentrations of NVP and YRGDS also led to a broad range in hydrogel elastic modulus. As shown in Figure 4, increasing the NVP concentration resulted in statistically significant increases in the elastic modulus of the resultant hydrogels. A four-fold increase in NVP concentration increased the elastic modulus by an order of magnitude from 136, 81, 84 and 103 kPa (at 37 mM NVP) to 1131, 1178, 1070 and 1095 kPa (at 148 mM NVP) for hydrogels polymerized with 0, 1, 5 and 15 mg/mL precursor YRGDS concentrations, respectively. While the data demonstrate statistically significant differences in elastic modulus among groups of constant NVP concentration with variable YRGDS precursor concentration, these observed differences were not as substantial as compared to the differences noted in elastic modulus for hydrogels polymerized with varying NVP concentration. For a given concentration of NVP, the elastic modulus varies only on average by 7.9 ± 8.9% from the mean for a mass fraction range of YRGDS between 0-5.4%. In contrast, a four-fold increase in NVP concentration resulted in a 10 fold increase in elastic modulus.
Figure 4.
Elastic modulus of PEGDA 3400 hydrogels as a function of NVP concentration for various precursor concentrations of YRGDS (n = 3; * = p < 0.05 and # = p < 0.001).
Adhesion and Proliferation of Fibroblasts as a Function of Scaffold Properties
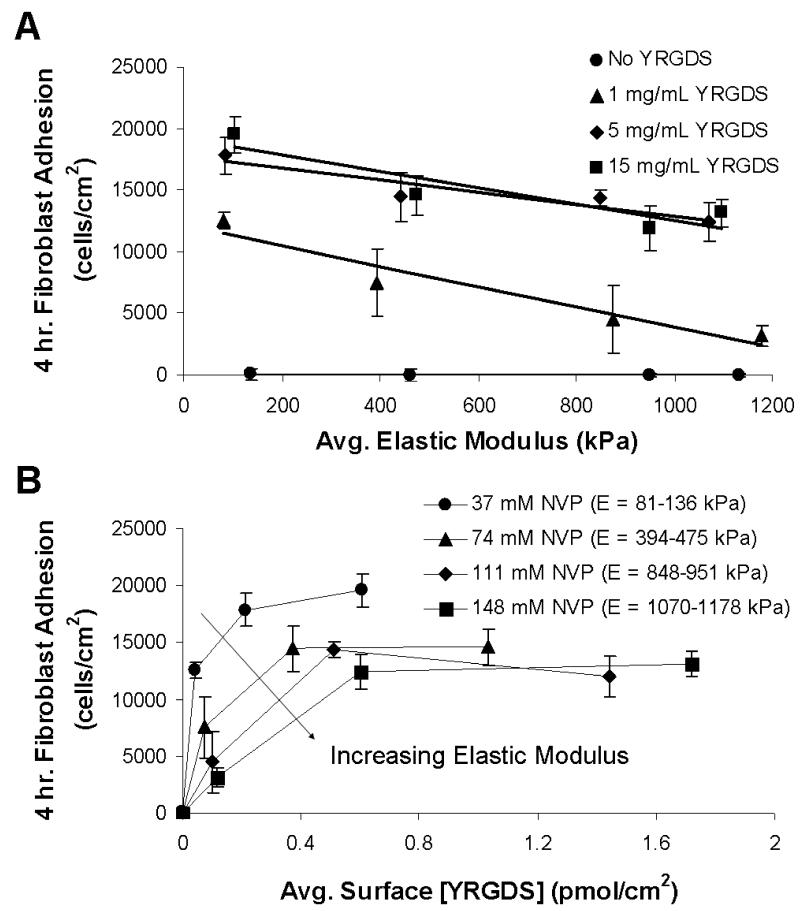
The interplay between scaffold modulus and the concentration of immobilized cell adhesion ligand (YRGDS) on cell adhesion and proliferation was quantified using an alamarBlue™ proliferation assay. Fibroblast adhesion on the surface of PEGDA hydrogels was quantified based on the broad range of matrix properties identified in Figures 3 and 4. Figure 5A shows fibroblast adhesion following four hours of incubation as a function of elastic modulus for various precursor concentrations of YRGDS. The data indicate that fibroblast adhesion increases on hydrogel surfaces with decreasing elastic modulus. The data in Figure 5A also illustrate that cell adhesion is enhanced when these hydrogels are polymerized with increasing precursor concentrations of YRGDS and that cell adhesion eventually remains constant above a certain precursor concentration of the cell adhesion ligand (5 mg/mL) across the range of elastic modulus investigated. Fibroblast adhesion was also quantified in terms of the average immobilized surface concentration of YRGDS for a variable range of elastic modulus (Figure 5B). Higher levels of fibroblast adhesion were achieved with initial increases in immobilized surface concentration of YRGDS and decreasing elastic modulus. Cell adhesion was also found to eventually approach saturation with respect to immobilized surface concentration of YRGDS for all ranges of elastic modulus investigated. Furthermore, more compliant PEGDA hydrogel matrices exhibited saturation in cell adhesion at lower YRGDS surface concentration as compared to stiffer surfaces suggesting that less YRGDS is required for cells to bind to less stiff substrates.
Figure 5.
3T3 fibroblast adhesion to PEGDA 3400 hydrogels following a four-hour incubation as a function of (A) elastic modulus for various precursor concentrations of YRGDS and (B) bioavailable surface concentration of YRGDS for various NVP concentrations (n = 4). The curves shown in (A) are best fit lines generated using linear regression.
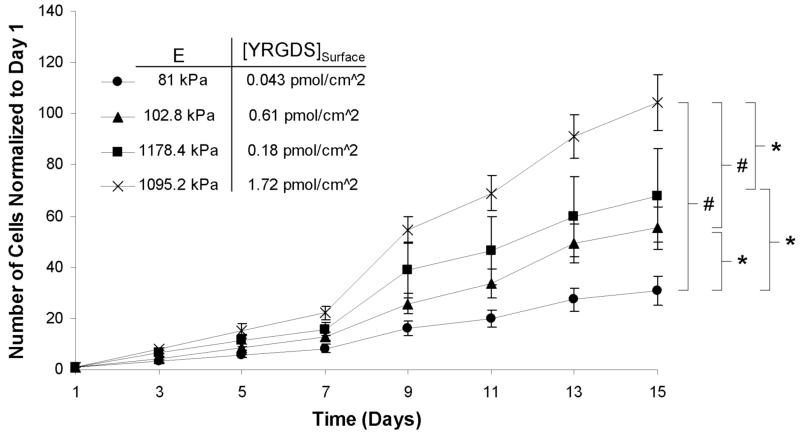
The proliferation of fibroblasts on PEGDA hydrogel surfaces with varying elastic modulus and surface concentration of YRGDS was also quantified. Four polymerization conditions were chosen such that the extremes in elastic modulus and YRGDS surface concentration were investigated. Figure 6 illustrates that fibroblast proliferation increased with increasing hydrogel stiffness over a period of two weeks. Furthermore, fibroblast proliferation was found to increase with increasing YRGDS surface concentration on hydrogels with a similar elastic modulus. Figure 6 also suggests that stiffness had a greater effect on fibroblast proliferation than the bioavailable surface concentration of YRGDS. While not statistically significant, by day 15 fibroblasts had proliferated 23% more on stiffer scaffolds (1178.4 kPa) with a low YRGDS concentration (0.18 pmol/cm2) than on more compliant scaffolds (102.8 kPa) that contained a higher concentration of immobilized YRGDS (0.61 pmol/cm2).
Figure 6.
Proliferation of 3T3 fibroblasts over time on the surface PEGDA 3400 hydrogels with varying stiffness and surface concentration of YRGDS (n = 5; * = p < 0.05 and # = p < 0.001 for day 15 values).
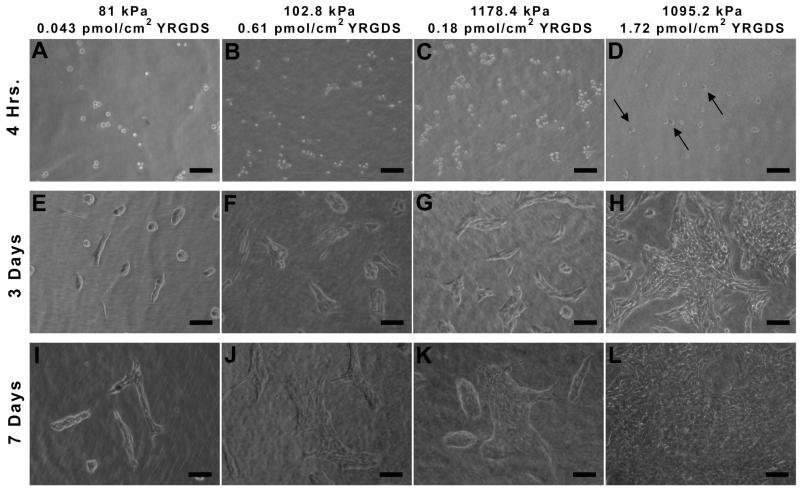
Fibroblast adhesion and proliferation was also visualized by phase contrast imaging. As shown in Figure 7A-C, four hours post-seeding cells are adhered to scaffolds but have a rounded morphology. At this same time point the scaffolds with a higher elastic modulus (1095 kPa) and the highest immobilized concentration of YRGDS, have begun to show spreading and the appearance of a flattened cell morphology (Figure 7D). By day 3 (Figure 7E-H), adhered and fully spread fibroblasts can be seen for all conditions. However, the total number of observed cells increases with both increasing elastic modulus and increasing surface concentration of YRGDS. This trend is also evident seven days post-seeding (Figure 7I-L) with fewer adhered cells present on the more compliant scaffolds with the lowest surface concentration of YRGDS and with increasing number of adhered cells observed with increases in elastic modulus and surface concentration of immobilized YRGDS. The scaffold with the highest elastic modulus and surface YRGDS concentration (Figure 7L) exhibited the largest cell population at Day 7 with cells approaching a confluent monolayer.
Figure 7.
Phase contrast images of 3T3 fibroblast adhesion and proliferation over time on PEGDA scaffolds with varying elastic modulus and immobilized concentrations of the YRGDS adhesion ligand. Arrows in D indicate cells that are fully spread and have taken on a flat morphology (10× magnification; scalebar = 100 μm).
3D Cell Encapsulation
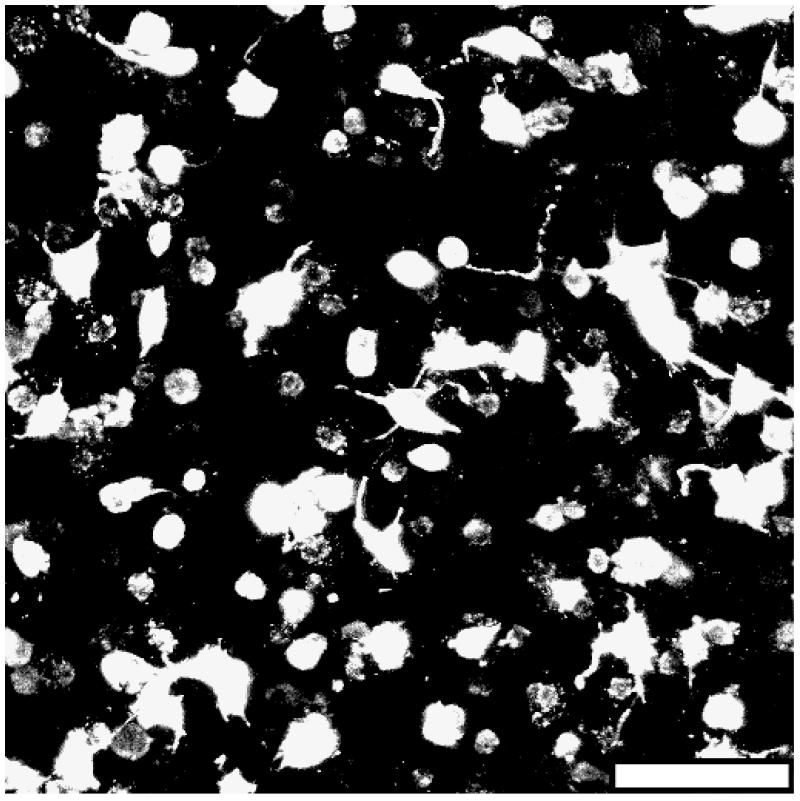
To confirm that cells can survive visible light photopolymerization and that this photopolymerization system is translatable to 3D cellar studies, fibroblasts were suspended in the hydrogel precursor and exposed to visible light during the crosslinking process. Due to the inherent mesh size of crosslinked PEGDA hydrogels being less than the average diameter of a cell [36], the scaffolds were rendered sensitive to cell-mediated proteolysis through the inclusion of a collagenase-sensitive peptide sequence within the polymer backbone to enable cell growth within the crosslinked hydrogel matrix. As shown in Figure 8, after 5 days in culture the fibroblasts have assumed a 3D morphology with extended processes and have begun to form cell-cell connections. This suggests that the fibroblasts not only endured photoencapsulation but that they are proliferating and actively remodeling the polymer matrix.
Figure 8.
Fluorescent image of 3T3 fibroblasts five days post-encapsulation within a collagenase-sensitive PEGDA hydrogel (20× magnification; scalebar = 100 μm).
Discussion
The development of a functional synthetic hydrogel scaffold that supports cell growth and tissue regeneration is highly dependent upon both the mechanical properties and the final concentration of embedded biofunctional signals within the polymeric biomaterial. These biomaterial properties are dictated by the prepolymer composition and selected polymerization conditions whose effects on biomaterial properties should be investigated and fully understood prior to scaffold fabrication for in vitro and in vivo studies. Although significant progress has been made with respect to controlling cell behavior as a function of either gel mechanical properties or immobilized biofunctional signal presentation in PEGDA scaffolds, to our knowledge a quantitative analysis of the effects of multiple polymerization conditions on both the elastic modulus and incorporated levels of an immobilized cell adhesion ligand (YRGDS) has not been previously conducted for visible light photopolymerization.
To investigate UV and visible light cytotoxic effects on cells, fibroblasts were exposed to both wavelengths of light in the absence and presence of the UV and visible light photoinitiators. As indicated by Figure 1, although there was a decrease in cell survival in the presence of both visible light and the visible light photoinitiator as compared to exposure to visible light alone as well as to the control, the exposure of cells to the UV light photoinitiator system resulted in no observed cell survival. In either case, the decrease in total cell number following the addition of the photoinitiator is likely due to the transfer of free-radicals from the initiator to the cells. This could result in cellular damage that could affect their viability proliferation. However, it is important to note that in a polymer system, the free radicals would be transferred from the initiator to the PEG macromers as well as to polymer chains which would serve as additional sinks for free-radicals during polymerization. Therefore, the results from this cytotoxicity study are most likely an overestimation of the toxic effects that the initiators would have on cells during the photopolymerization. Nonetheless, the initiator cytotoxicity studies demonstrate that UV light is more toxic and damaging to cells than longer wavelength visible light. This is an important aspect to consider when cells are polymerized in the presence of light resulting in cellular encapsulation within the hydrogel network. Figure 8 as well as our previously published studies [37,38] using visible light photopolymerization indicate that cells are able to proliferate and form cell-cell contacts within the hydrogel scaffolds indicating that this system is readily translatable to 3D cellular tissue engineering applications.
In this study, a detailed experimental sensitivity analysis of free-radical polymerization conditions was performed to identify conditions that would induce significant changes in hydrogel mechanical properties as well as in incorporated levels of YRGDS and to quantify the effects of these properties on 2D cell behavior. The data indicate that the final concentration of YRGDS is highly dependent on the kinetics of polymerization as well as the precursor composition. YRGDS incorporation increased with increasing YRGDS precursor concentration, exposure time, light intensity, NVP concentration, and with decreasing precursor concentrations of the PEGDA macromer (Figures 2 and 3). Unincorporated YRGDS was found to diffuse from the hydrogels within 24 hours, which could in theory provide a short term signaling pathway to stimulate cells to invade a scaffold. Hydrogel elastic modulus was also found to be highly dependent on the polymerization conditions with the largest changes associated with variations in the NVP monomer concentration (Figure 4). It was also noted that small changes in polymerization conditions could induce significant changes in either elastic modulus or the final concentration of YRGDS (Figures 2-4). Our results demonstrate that simultaneous alterations in NVP and YRGDS precursor concentrations result in wide ranges of hydrogel elastic modulus (81 – 1178 kPa) and final YRGDS surface concentration (0.04 – 1.72 pmol/cm2) (Figures 3 and 4). Furthermore, this wide range of biomaterial properties was capable of inducing statistically significant differences in cell proliferation (Figure 6). Therefore, the simultaneous alteration in YRGDS and NVP precursor concentration play a critical role in controlling the final concentration of RGD as well as the matrix stiffness which in turn provide a wide range of design criteria used for the fabrication of scaffolds that support the growth of multiple cell types and tissues [26].
In order to estimate immobilized surface concentrations of YRGDS, the bioavailable surface was assumed to be the top 10 nm of the hydrogels [7]. This assumption was first utilized by Hern et al in 1998 [7] but was reconsidered in a subsequent study by Elbert and Hubbell suggesting that it overstated the accessible peptide concentration by a factor of 40-100 [8]. In 2003, Behravesh et al. used antibody-based techniques to quantitatively determine that the bioavailable surface of hydrogels composed poly(propylene fumarate-co-ethylene glycol) with a MW of 5000 Da was approximately 4.4 nm [39]. While there is not a general relationship that can be applied to all PEG hydrogels, it is believed that 10 nm is a valid approximation given the fact that the bioavailable surface will vary in thickness across the polymerization conditions investigated.
Previous work on YRGDS incorporation in PEGDA hydrogels formed by UV photopolymerization showed an 81 to 97% incorporation of acrylated YRGDS with incorporation efficiency remaining above 95% except in the case of extremely low precursor YRGDS concentrations [7]. This direct correlation between YRGDS precursor concentration and final YRGDS surface concentration is consistent with the trend observed in the data presented in this study. The use of visible light photopolymerization in the presence of the photoinitiator eosin Y, yielded a wide range in hydrogel elastic modulus (81 – 1178 kPa) as well as final YRGDS concentration (0.17 – 6.86 mg/mL) when coupled with simultaneous variations in initial NVP and YRGDS concentrations. This corresponds to an incorporation efficiency range of 23 to 66%. Although this range of incorporation is lower than that reported in previous studies of PEGDA hydrogels formed using UV photopolymerization, similar surface concentrations of YRGDS were achieved. The quantified values of YRGDS incorporation efficiency in the present study correspond to surface concentrations of YRGDS ranging from 0.04 to 1.7 pmol/cm2 as compared to 0.0087 to 0.71 pmol/cm2 reported previously [7]. In addition, previous studies have indicated that a minimum surface concentration of 1 pmol/cm2 YRGDS, immobilized via a 3400 Da PEG spacer, was required for fibroblast adhesion [7]. In contrast, in the present work cell adhesion was observed for surface concentrations as low as 0.04 pmol/cm2 YRGDS. Although this previous study did not quantify elastic modulus as a function of alterations in polymerization conditions, UV polymerization occurs at a faster rate as compared to visible light induced free-radical polymerization resulting in more crosslinked scaffolds. Since the bioavailable surface of a hydrogel is here defined as the top 10 nm [7], the ability of cells to interact with tethered adhesion sites is a direct function of the crosslink density of the polymer network. Therefore, the observed differences in the minimum YRGDS surface concentration required for fibroblast adhesion is most likely attributed to the lower elastic modulus achieved in our system enabling the cells to more readily interact with pendant YRGDS groups on the hydrogel surface. This hypothesis is also supported in the cell adhesion data presented in Figure 5B, which indicate that a higher surface concentration of YRGDS is required for fibroblasts to bind to stiffer matrices. More crosslinked polymer networks restrict the accessibility of YRGDS to cell surface receptors thus requiring a higher surface concentration to support adhesion.
Among the various parameters evaluated, alterations in the concentration of the NVP monomer had the greatest effect on both hydrogel elastic modulus and immobilized concentration of YRGDS. This result was expected as NVP is an accelerator of free-radical polymerization and has been shown to significantly increase the overall conversion of crosslinked acrylate polymers [35]. White et al. showed that NVP is capable of diffusing through low mobility polymer networks promoting additional reaction diffusion controlled termination and increasing crosslink density through greater double bond conversion [35]. NVP has also been shown to counteract the inhibition of free-radical acrylate polymerization by atmospheric oxygen [40]. Therefore, increases in NVP precursor concentration result in more crosslinked polymer networks while simultaneously increasing the incorporation of pendant YRGDS sites. NVP has also been previously identified as an important parameter in increasing both the storage modulus of PEGDA hydrogels as well as spread area of adherent endothelial cells [8]. Studies have shown that variations in the NVP/acrylate mole ratio from 0.28-0.98 caused an eight-fold increase in hydrogel storage modulus and doubled the spread area of adherent cells for a fixed concentration of RGD for gels polymerized using a PEGDA monomer of molecular weight equal to 3400 Da. We observed similar results using an NVP/PEGDA mole ratio ranging from 0.25-2, which corresponds to an NVP/acrylate mole ratio ranging from 0.125-1. Furthermore, the aforementioned study concluded that the NVP/acrylate ratio in the precursor was more critical in inducing changes in hydrogel storage modulus than the NVP precursor concentration [8]. This fact helps explain the reverse trend in YRGDS incorporation observed as the concentration of PEGDA macromer was increased (Figure 2C). As the concentration of PEGDA in the precursor was increased from 15 to 40%, the mole ratio of NVP/acrylate decreased from 0.42 to 0.16 for a PEGDA macromer molecular weight of 3400 Da and from 0.98 to 0.37 for a PEGDA macromer molecular weight of 8000 Da. These decreases in the NVP/acrylate mole ratio resulted in a reduced PEGDA macromer conversion leading to lower YRGDS incorporation.
Based on the fact that variations in the polymerization conditions have a profound impact on the hydrogel mechanical properties as well as the incorporated levels of biofunctional cues it is important to develop an understanding of how slight variations in these biomaterial properties can in turn influence cell behavior. Previous studies with PEG-based polymer networks have shown that increases in the precursor concentration of RGD caused increases in the projected area of adherent endothelial cells [8], SMC focal adhesion area [6] and more rapid adhesion of human foreskin fibroblasts (HFF) [41]. Although, increases in RGD precursor content are likely to induce increases in its incorporation over certain polymerization conditions, these studies did not quantify cell behavior as a function of incorporated levels of YRGDS presented to cells on the hydrogel surface. In addition, increases in the projected area of adherent endothelial cells [8] and SMC proliferation [6] were observed as hydrogel stiffness increased. Our results indicate that increasing the surface YRGDS concentration resulted in increased adhesion and proliferation for a constant elastic modulus (Figures 5B, 6 and 7). Furthermore, increases in hydrogel elastic modulus caused a significant increase in proliferation while decreasing adhesion (Figures 5A, 6 and 7). This is most likely attributed to the fact that fibroblasts require more YRGDS to bind stiffer matrices in a given time frame. When both YRGDS incorporation and elastic modulus were increased simultaneously, we observed a much greater increases in proliferation (Figure 6). However, failure to supply the cells with an adequate concentration of adhesion ligand could confound these results. Therefore it is important to understand how altering a specific polymerization condition will influence a scaffold’s properties and in turn cell behavior.
Although the current study was limited to fibroblast behavior in a 2D system, the presented techniques can be easily extended to other cell types, for the immobilization of multiple ECM signaling molecules and more complex 3D systems. We present a systematic approach demonstrating that the selection and manipulation of polymerization conditions, such as prepolymer concentrations of NVP and YRGDS, play a critical role in the resultant properties of visible light photopolymerized PEGDA hydrogels, which in turn dictate cell behavior. A fundamental understanding of the effects of these conditions on polymerization and hydrogel crosslinking will ultimately guide the optimization of biomaterial properties and rational design of biofunctional PEGDA scaffolds for numerous tissue engineering applications.
Conclusion
A comprehensive experimental sensitivity analysis was performed to evaluate the effects of alterations in polymerization conditions on the mechanical properties and immobilized YRGDS concentration in visible light photopolymerized PEGDA hydrogels and to quantify the effects of these scaffold properties on 2D cell behavior. The results identify the critical polymerization parameters and their impact on both immobilized concentrations of the YRGDS adhesion ligand as well as on the hydrogel elastic modulus. The results show that a broad range of elastic modulus and adhesion ligand concentration can be achieved by simultaneously varying the concentration of NVP and YRGDS in the prepolymer solution. Knowledge of relationships such as these are critical for the rational design of PEGDA scaffolds formed by free-radical photopolymerization to support growth of numerous cell types and the regeneration of various tissues.
Acknowledgements
The authors are grateful to Dr. Eric Brey in the Biomedical Engineering Department at the Illinois Institute of Technology (IIT) for his guidance in developing the YRGDS incorporation studies. The authors would also like to acknowledge Dr. David Venerus and the members of his lab for their help with the compression experiments. This research was supported by funding from the Pritzker Institute of Biomedical Science and Engineering at IIT and the National Institutes of Health (Grant No. R21HL094916).
References
- 1.Hoffman AS. Ann. NY Acad. Sci. 2001;944:62–73. doi: 10.1111/j.1749-6632.2001.tb03823.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee KY, Mooney DJ. Chem. Rev. 2001;101:1869–1879. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 3.Peppas NA, Hilt JZ, Khademhosseini A, Langer R. Adv. Mater. 2006;18:1345–1360. [Google Scholar]
- 4.Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA. Adv. Mater. 2009;21:3307–3329. doi: 10.1002/adma.200802106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon JJ, Hahn MS, Kim I, Nsiah BA, West JL. Tissue Eng. Part A. 2009;15:579–585. doi: 10.1089/ten.tea.2008.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peyton SR, Raub CB, Keschrumrus VP, Putnam AJ. Biomaterials. 2006;27:4881–4893. doi: 10.1016/j.biomaterials.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 7.Hern DL, Hubbell JA. J. Biomed. Mater. Res. 1998;39:266–276. doi: 10.1002/(sici)1097-4636(199802)39:2<266::aid-jbm14>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 8.Elbert DL, Hubbell JA. Biomacromol. 2001;2:430–441. doi: 10.1021/bm0056299. [DOI] [PubMed] [Google Scholar]
- 9.Porter AM, Klinge CM, Gobin AS. Biomacromol. 2011;12:242–246. doi: 10.1021/bm101220b. [DOI] [PubMed] [Google Scholar]
- 10.Khatiwala CB, Peyton SR, Metzke M, Putnam AJ. J. Cell. Physiol. 2007;211:661–672. doi: 10.1002/jcp.20974. [DOI] [PubMed] [Google Scholar]
- 11.Blewitt MJ, Willits RK. Ann. Biomed. Eng. 2007;35:2159–2167. doi: 10.1007/s10439-007-9389-4. [DOI] [PubMed] [Google Scholar]
- 12.Gunn JW, Turner SD, Mann BK. J. Biomed. Mater. Res. A. 2005;72:91–97. doi: 10.1002/jbm.a.30203. [DOI] [PubMed] [Google Scholar]
- 13.Bott K, Upton Z, Schrobback K, Ehrbar M, Hubbell JA, Lutolf MP, Rizzi SC. Biomaterials. 2010;31:8454–8464. doi: 10.1016/j.biomaterials.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 14.Gobin AS, West JL. FASEB J. 2002;16:751–753. doi: 10.1096/fj.01-0759fje. [DOI] [PubMed] [Google Scholar]
- 15.Lee SH, Moon JJ, Miller JS, West JL. Biomaterials. 2007;28:3163–3170. doi: 10.1016/j.biomaterials.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Lee SH, Moon JJ, West JL. Biomaterials. 2008;29:2962–2968. doi: 10.1016/j.biomaterials.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leslie-Barbick JE, Moon JJ, West JL. J. Biomater. Sci. Polym. Ed. 2009;20:1763–1779. doi: 10.1163/156856208X386381. [DOI] [PubMed] [Google Scholar]
- 18.Mann BK, Gobin AS, Tsai AT, Schmedlen RH, West JL. Biomaterials. 2001;22:3045–3051. doi: 10.1016/s0142-9612(01)00051-5. [DOI] [PubMed] [Google Scholar]
- 19.Mann BK, Schmedlen RH, West JL. Biomaterials. 2001;22:439–444. doi: 10.1016/s0142-9612(00)00196-4. [DOI] [PubMed] [Google Scholar]
- 20.Moon JJ, West JL. Curr. Top Med. Chem. 2008;8:300–310. doi: 10.2174/156802608783790983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pratt AB, Weber FE, Schmoekel HG, Muller R, Hubbell JA. Biotechnol. Bioeng. 2004;86:27–36. doi: 10.1002/bit.10897. [DOI] [PubMed] [Google Scholar]
- 22.Seliktar D, Zisch AH, Lutolf MP, Wrana JL, Hubbell JA. J. Biomed. Mater. Res. A. 2004;68:704–716. doi: 10.1002/jbm.a.20091. [DOI] [PubMed] [Google Scholar]
- 23.West JL, Hubbell JA. Macromol. 1999;32:241–244. [Google Scholar]
- 24.Patterson J, Hubbell JA. Biomaterials. 2010;31:7836–7845. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 25.Tibbitt MW, Anseth KS. Biotechnol. Bioeng. 2009;103:655–663. doi: 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemir S, West JL. Ann. Biomed. Eng. 2010;38:2–20. doi: 10.1007/s10439-009-9811-1. [DOI] [PubMed] [Google Scholar]
- 27.Chiu YC, Cheng MH, Engel H, Kao SW, Larson JC, Gupta S, Brey EM. Biomaterials. 2011;32:6045–6051. doi: 10.1016/j.biomaterials.2011.04.066. [DOI] [PubMed] [Google Scholar]
- 28.Bryant SJ, Chowdhury TT, Lee DA, Bader DL, Anseth KS. Ann. Biomed. Eng. 2004;32:407–417. doi: 10.1023/b:abme.0000017535.00602.ca. [DOI] [PubMed] [Google Scholar]
- 29.DeKosky BJ, Dormer NH, Ingavle GC, Roatch CH, Lomakin J, Detamore MS, Gehrke SH. Tissue Eng. Part C Methods. 2010;16:1533–1542. doi: 10.1089/ten.tec.2009.0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fittkau MH, Zilla P, Bezuidenhout D, Lutolf MP, Human P, Hubbell JA, Davies N. Biomaterials. 2005;26:167–174. doi: 10.1016/j.biomaterials.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 31.Larson EM, Doughman DJ, Gregerson DS, Obritsch WF. Invest. Ophthalmol. Vis. Sci. 1997;38:1929–1933. [PubMed] [Google Scholar]
- 32.Voytik-Harbin SL, Brightman AO, Waisner B, Lamar CH, Badylak SF. Vitro Cell Dev. Biol. Anim. 1998;34:239–246. doi: 10.1007/s11626-998-0130-x. [DOI] [PubMed] [Google Scholar]
- 33.Nemir S, Hayenga HN, West JL. Biotechnol. Bioeng. 2010;105:636–644. doi: 10.1002/bit.22574. [DOI] [PubMed] [Google Scholar]
- 34.Guarnieri D, De Capua A, Ventre M, Borzacchiello A, Pedone C, Marasco D, Ruvo M, Netti PA. Acta Biomater. 2010;6:2532–2539. doi: 10.1016/j.actbio.2009.12.050. [DOI] [PubMed] [Google Scholar]
- 35.White TJ, Liechty WB, Guymon CA. J. Appl. Poly. Sci. Part A. 2007;45:4062–4073. [Google Scholar]
- 36.Raeber GP, Lutolf MP, Hubbell JA. Biophys. J. 2005;89:1374–1388. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokic S, Papavasiliou G. Acta. Biomater. 2012;8:2213–2222. doi: 10.1016/j.actbio.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokic S, Papavasiliou G. Tissue Eng. Part A. 2012 doi: 10.1089/ten.tea.2012.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behravesh E, Sikavitsas VI, Mikos AG. Biomaterials. 2003;24:4365–4374. doi: 10.1016/s0142-9612(03)00338-7. [DOI] [PubMed] [Google Scholar]
- 40.Lorenz DH, Azorlosa JL, Tu RS. Radiation Phys. and Chem. 1977;9:843–849. [Google Scholar]
- 41.Drumheller PD, Hubbell JA. Anal. Biochem. 1994;222:380–388. doi: 10.1006/abio.1994.1506. [DOI] [PubMed] [Google Scholar]