Abstract
Importance of the field
Monitoring cell viability in vitro is critical in many areas of biomedical research, and the ultimate goal in drug discovery is the ability to predict the in vivo toxicology of drug candidates based on their toxicity profile in vitro. Over the last decade, the contribution of high-throughput screening (HTS) toward this goal has been tremendous, providing the ability to screen compounds in parallel against multiple cell types. However, the toxic effects of drug candidates uncovered during clinical trials are by far the main reason for their failure. Over the same period, our understanding of programmed cell death has evolved dramatically with the identification of critical control points in the cell death pathways. As a result, cell viability should no longer be characterized solely on the basis of discrete endpoint measurements such as membrane permeability.
Areas covered in this review/What the reader will gain
This review summarizes the traditional viability assays currently commercially available, focusing on methods amenable to high density format. Assays categorized into the following classes are discussed: dye exclusion assays, DNA condensation-based assays and assays monitoring a metabolic function. We describe each approach, and using case studies, we emphasize their limitations.
Take home message
Current low-content methods based on single parameter readouts are prone to error due to the heterogeneity of cell populations and the multi-faceted nature of cell death. High-content approaches based on continuous, multiplexed readouts are becoming increasingly important for monitoring multiple markers of cell death induction simultaneously, on a cell by cell basis. The use of such content-rich platforms is a necessity to predict the toxicology of drug candidates accurately.
Keywords: apoptosis, cell viability, cyto-toxicity, high-content screening, live cell based assays
1. INTRODUCTION
Our understanding of cell death has evolved from consisting of a binary event – active or passive – to a complex process submitted to multiple regulatory pathways1–5. The assessment of cell viability in vitro is critical in all areas of biomedical research, whether induction of cell death is a desired event such as for the evaluation of anticancer agents, or whether the goal is to identify toxicities associated with the use of a given small molecule or the modulation of a specific pathway. The main reason for the failure of most drug candidates – only 1 out of 10 reaches the market – is their toxicity in patients6. Currently, cytotoxic side effects of failed drug candidates are being discovered late in the drug discovery process, during clinical trials. This failure leads to the loss of large financial investments by pharmaceutical companies. For this reason, the ability to accurately predict the in vivo toxicology of a drug candidate in vitro constitutes the “Holy Grail” in the field of drug discovery. In this article, we review the traditional low-content viability assays that are currently commercially available; we discuss their limitations, and propose new directions toward content-rich platforms allowing multi-parameter analyses that more accurately reflect the status of cell death in cell populations in vitro.
2. LOW CONTENT VIABILITY ASSAYS
Until recently, scientists have relied on single viability assays, with relatively simple endpoint measurements focusing either on the loss of membrane integrity, DNA labeling, or on the assessment of a few discrete metabolic functions. We describe the main categories of commercially available viability assays (Table 1), and we discuss their limitations. We do not intend to provide an extensive review of all commercially available assays, but we rather aim at describing the main general approaches currently employed to assess cell viability.
Table 1.
Summary of commonly employed viability assays.
| Assay | Assay Category | Readout | Compatible with cell by cell analysis |
|---|---|---|---|
| Trypan Blue | Dye exclusion | Fluorometric | Yes |
| Ethidium Bromide | Dye exclusion | Fluorometric | Yes |
| Propidium Iodide | Dye exclusion | Fluorometric | Yes |
| SYTOX | Dye exclusion | Fluorometric | Yes |
| YO-PRO | Dye exclusion | Fluorometric | Yes |
| Hoechst 33258 | DNA condensation | Fluorometric | Yes |
| Acridine Orange | DNA condensation | Fluorometric | Yes |
| MTT and XTT | Redox reaction | Colorimetric | No |
| Alamar Blue | Redox reaction | Fluoro/Colorimetric | No |
| Calcein AM | Esterase substrate | Fluorometric | No |
| Cell Tracker | Esterase substrate | Fluorometric | No |
| CellTiter-Fluor | Protease substrate | Fluorometric | No |
| CellTiter Glo | ATP measurement | Luminometric | No |
| CytoTox-ONE | Enzyme release | Fluorometric | No |
2.1 Dye exclusion assays
Originally used in flow cytometry, a wide variety of fluorescent dyes are available for dye exclusion-based assays. These dyes typically diffuse passively into cells with a compromised plasma membrane, but they are rejected by healthy cells. Dyes that have historically been used for these assays include Trypan Blue, Ethidium/Propidium dyes and cyanine dyes, such as SYTOX and YOPRO. Trypan Blue is a diazol dye that is often used for rapid quantification of viable cells in tissue culture. The red fluorescent Ethidium bromide (EthBr) and Propidium iodide (PI) dyes are widely employed to measure viability in flow cytometry7. While both dyes have been used extensively in cytotoxic assays, PI is usually preferred over EthBr because EthBr carries a single positive charge and will enter into living cells slowly. However, PI carries two charges, which prevents it from entering living cells in a more reliable way7. More recently, PI and EthBr have been replaced with cyanine dyes such as SYTOX and YO-PRO. SYTOX Green Stain is one out of three dyes belonging to the SYTOX family synthesized by Molecular Probes. It is the most widely used by investigators because of its flexibility. SYTOX Green Stain carries three positive charges that allow the dye to be excluded completely from the living eukaryotic and prokaryotic cells7. Its use has been validated for microscopy, fluorometry and flow cytometry readouts8. YO-PRO-1 is one out of a series of monomeric and dimeric cyanine dyes also synthesized by Molecular Probes that are non-fluorescent unless bound to nucleic acids7. These dyes are typically excluded from live cells but stain dead cells that have a compromised membrane. It has been suggested using flow cytometry analysis that YO-PRO-1 specifically labels apoptotic cell populations, and it has subsequently been used for that purpose in flow cytometric assays 9–10 and using fluorescence microscopy10–12.
Although dye exclusion-based assays are very popular among viability assays, a major drawback of this approach lies in the fact that dying cells may still retain their membrane integrity for a substantial period of time after injury. As a consequence, depending on the time of readout, the cytotoxicity of a treatment may be overlooked7, 13. In addition, most probes used in dye-exclusion based assays are nuclear dyes that are cytotoxic, preventing their use for continuous assays over a long period of time.
2.2 DNA Condensation-based Assays
The fluorescence of nuclear dyes such as Hoechst 33258 and Acridine Orange (AO) is sensitive to DNA conformation and chromatin state. Viability assays based on the use of these dyes take advantage of this property to discriminate between living and apoptotic cells, the latter featuring a higher level of chromatin compaction. Hoechst fluorescent probes are benzimide dyes that can be excited in the UV region of the light spectrum, making them suitable for multiplexing with other fluorophores. Hoechst 33258 is one out of three Hoechst stains available from Molecular Probes that fluoresces blue upon binding to the minor groove of DNA. Hoechst 33258 has been shown to be a useful detector of DNA damage using flow cytometry14 and more recently, it has been used for high-content screening as a nuclei detector and quantifier15–17. Similarly, Acridine Orange is a fluorescent nucleic acid stain which interacts with DNA and RNA by intercalation or electrostatic attraction. It has mostly been employed in flow cytometric applications, but it has also been used to measure apoptosis in embryos18.
Despite their widespread use, Hoechst 33258 and Acridine Orange have their limitations. The major drawback of nuclear dyes, as mentioned earlier, is their toxicity. Also, in condensed chromatin, the bulk of DNA does not allow efficient acridine orange intercalation19, while Hoechst dyes exhibit a wide spectrum of affinity that is sequence-dependent, which can lead to high variability in the assay20. Finally, the use of these dyes requires multiple washing steps which can render the assays unattractive to HTS.
2.3 Assays monitoring a metabolic function
Multiple viability assays rely on the assessment of a metabolic function. A large number of them rely on tetrazolium and resazurin reduction. Tetrazolium salts are water-soluble, colorless compounds widely used for detecting the redox potential. After reduction, these salts form brightly colored formazans that can be quantified using standard spectrophotometric techniques following solubilization. The most common tetrazolium salts used for viability are MTT and XTT. Nevertheless, the MTT assay requires a solubilization step prior to quantification, limiting its use. In addition, a high variability has been described for this assay, based on the exposure time with MTT21. In contrast to MTT, XTT does not need to be solubilized before quantitation, which can reduce the assay time. However, XTT still suffers from the limitations of tetrazolium salts-based assays, namely a high variability resulting from multiple factors affecting the reductive capacity of the assay environment21. In summary, the major limitations of tetrazolium-based assays are the lack of sensitivity and their toxicity.
Alamar Blue has been commonly used as an oxidation-reduction indicator to detect cell viability and cytotoxicity. Alamar Blue, whose active ingredient is resazurin, , is a nonfluorescent dye which is converted to the fluorophore resorufin in response to a chemical reduction believed to occur in the mitochondria22–23. Advantages of this assay include its homogeneous nature and the stability of the signal generated; for these reasons, Alamar Blue is commonly used for HTS24–25. Still, one of the disadvantages of Alamar Blue is optical interference. Some of the compounds tested can give false positive results. However, this can be overcome using a quench test, as described previously by Antczak et al. In the same way, it is important to note that care should be taken to prevent optical/fluorescence interference when selecting probes as well. There are several commercially-available assays based on the use of Alamar Blue including the AlamarBlue Cell Viability Assay Kit from Biotium and CellQuanti-Blue from BioAssay Systems.
As a variation of dye-exclusion-based assays, cell-permeant esterase substrates are able to measure both enzymatic activity and cell-membrane integrity. As nearly neutral molecules, esterase substrates are able to diffuse freely into most cells. Once they reach the cytoplasm, the nonfluorescent substrates are converted by intracellular esterases into fluorescent products that are retained by cells with intact plasma membranes. Calcein AM, which becomes converted into calcein, is the leading esterase substrate for viability assays. Calcein is a polyanionic fluorescein derivative that bears six negative charges and two positive charges at pH 726. As a consequence, Calcein AM has superior cellular retention compared to PI and is unaffected by pH change27. Despite being used in flow cytometric toxicity assays28, Calcein AM has been shown to be toxic to and pumped out of several cell lines29–30, a serious drawback for the widespread assessment of cell viability. In addition, as seen with the DNA dyes, Calcein AM requires various washing steps, which can be a major disadvantage.
Another family of esterase-activated fluorescent probes is 5-chloromethylfluorescein (CMFDA) derivatives, available from Molecular Probes under the label CellTracker. These thio-reactive fluorescent dyes freely diffuse into cells and react with intracellular thiols, followed by the cleavage of their acetate groups which results in a fluorescent product. CMFDA derivatives have been used to detect viable and dead cells in a variety of cell lines31–32, as well as for the imaging of retinal ganglion cell axons33.
Similarly to esterase substrates, protease substrate-based assays rely on the measurement of the conserved and constitutive protease activity that exists in living cells. A peptide substrate such as Gly-Phe-AFC enters intact cells and is cleaved by proteases, generating a fluorescent signal that is proportional to the number of cells. Protease substrates are widely used in multiplexed assays. For example, CellTiter-Fluor, available from Promega, has been used in a multiplexed assay that encompasses the use of live cell protease, dead cell protease and caspase markers to give a detailed cytotoxic profile34.
A popular method to assess viability is the bioluminescent detection of adenosine triphosphate (ATP). ATP plays a central role in eukaryotic and prokaryotic cell metabolism and can be detected using the firefly luciferase enzyme. After membrane integrity loss, cells lose the ability to synthesize ATP, and ATPases deplete the remaining ATP; thus the amount of viable cells can be quantified through ATP. ATP quantification has become increasing popular in high -throughput chemical screens13 and RNAi screens35. There are a variety of ATP assay kits available including the CellTiter-Glo Luminescent Cell Viability Assay and The ATP Determination Kit available from Promega and Molecular Probes, respectively. One of the limitations of ATP quantification is that ATP levels vary greatly in cells36–37, which is responsible for a great variability for this assay. In addition, cell lysis is required, which limits the use of this method as an endpoint measurement.
Another example of a viability assay relying on the monitoring of a metabolic function is lactate dehydrogenase (LDH) quantification. LDH is released into the surrounding medium when a cell’s membrane is compromised, which is used to indirectly quantify viable cells. LDH catalyzes the conversion of lactate producing NADH. NADH, in the presence of resazurin, is used to drive the reduction of the fluorescent resorufin product. Fluorescence is proportional to the amount of LDH released. LDH-based assays are available from Promega and Molecular Probes. As an example, the CytoTox-ONE Assay (Promega) has been employed to assess the viability of trophoblast cultures38. Although LDH assays can be combined with other enzyme release assays, they are limited by low sensitivity39. Moreover, LDH has a relatively short half-life.
3. LIMITATIONS OF LOW CONTENT CELL VIABILITY ASSAYS
Assays currently used to measure cytotoxicity in vitro may not necessarily accurately reflect in vivo toxicity, which may have dramatic consequences during the progression of a drug candidate into clinical trials. We describe the current approaches for assessing cell viability in vitro and we discuss their limitations. Providing case studies, we explain how traditional viability assays may fail to detect toxicity in cultured cell populations.
3.1 Heterogeneity of cultured cell populations
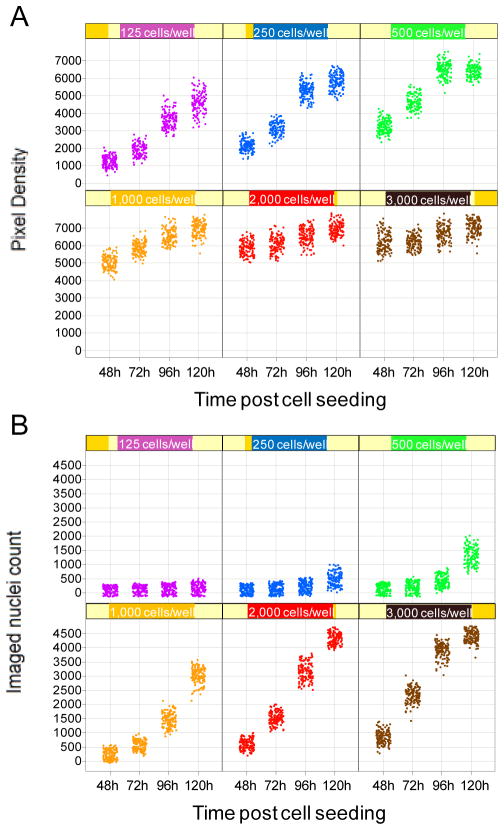
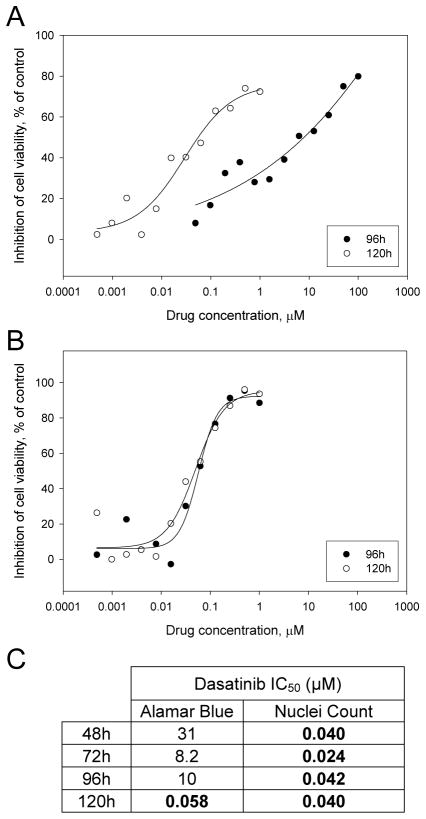
Cell populations studied in vitro are typically asynchronous, resulting in heterogeneity in their cell cycle status. At any given time point, cells will therefore respond differently to treatment. While a fraction of the studied cell population may progress toward cell death, the induced loss in cell viability may be masked by surviving cells. For this reason, any assay measuring the overall response of a cell population to a treatment rather than recording data at the single cell level is prone to error. We illustrate this inherent flaw with an experiment previously described by Antczak et al., using the Alamar Blue assay to assess the growth curves of H1975 human Non Small Cell Lung Cancer cells, compared with direct nuclei count. In this example, H1975 cells were seeded at various densities in 384-well microplates, and Alamar Blue or nuclei count readout was performed at 48, 72, 96 or 120h post-seeding. For nuclei count readout, cells were fixed and stained with Hoechst, and after imaging of four fields per well using the automated microscope INCell Analyzer 1000 (GE Healthcare), automated image analysis was performed to obtain a count for the imaged nuclei. Data for the 128 wells tested for each condition is presented in Figure 1. For Alamar Blue readout (Figure 1A), a cell seeding density of 125 cells per well is optimal, leading to a linear increase of the signal up to 120h. In contrast, at a cell seeding density of 2,000 cells per well, the Alamar blue readout is saturated as early as 48h post-cell seeding. On the other hand, the nuclei count readout shows that at the same cell seeding density of 2,000 cells per well, the growth of H1975 cells is actually linear (Figure 1B). This result demonstrates that while the Alamar Blue assay is sensitive, allowing to measure the growth of as little as 125 cells, signal saturation occurs even though the studied cell population is still in a linear growth phase. As a consequence, when using an Alamar Blue-based assay, unless the cell seeding density is carefully optimized, cell death can easily be overlooked because a few living cells can generate a high signal. In addition, the timing of the assay is important when using Alamar Blue. In our experience, longer incubation periods lead to more accurate measurements. This can be explained by a number of factors. Drugs affecting proliferation require treated cells to complete at least one cell cyle to exert their anti-proliferative properties. However, since cultured cells are typically asynchronous, a larger time window is necessary for all cells to accomplish one cell cyle. As a consequence, an incubation time of as much as two doubling times might be necessary to observe the full effect of a given drug. An assay that measures proliferation directly – such as nuclei count – may be more tolerant regarding this phenomenon. However, an indirect assay such as Alamar Blue, already handicapped by signal saturation at low cell densities, may be particularly sensitive to incubation time. To illustrate this issue, we have performed dose response experiments for the drug dasatinib toward dasatinib-sensitive H1975 cells, comparing Alamar Blue and nuclei count readouts. To ensure an accurate measurement of cell death, we used the previously optimized cell seeding densities: 125 cells per well for Alamar Blue readout and 1,000 cells per well for nuclei count readout. The reported IC50 for dasatinib toward H1975 cells is 30–50 nM (Somwar et al., unpublished data). With the Alamar Blue readout, we observe a large discrepancy between the calculated IC50 at the 96h time point (10 μM) and the 120h time point (0.029 μM) (Figure 2A). In contrast, the calculated IC50 using nuclei count readout was very similar for both time points – 0.057 and 0.050 at 96h and 120h – and conform to the reported IC50, as expected (Figure 2B). Further investigation revealed that the nuclei count readout led to a correct IC50 at the 48, 72, 96 or 120h time points, while the calculated IC50 was accurate using the Alamar Blue assay only at the 120h time point (Figure 2C). This result strongly suggests that even after optimizing the cell density in order to ensure being within the linear range of Alamar Blue conversion, the indirect Alamar Blue readout may occasionally be prone to error, due to the heterogeneity of the studied cell population.
Figure 1.
A. Growth curves of human Non Small Cell Lung Carcinoma cells H1975 as assessed using Alamar Blue readout. B. Growth curves of human Non Small Cell Lung Carcinoma cells H1975 as assessed using nuclei count readout.
Figure 2.
A. Dose response of dasatinib toward human Non Small Cell Lung Carcinoma cells H1975 cells as assessed using Alamar Blue readout. Dasatinib IC50 at 96h readout: 10 μM ; IC50 at 120h readout: 0.029 μM. B. Dose response of dasatinib toward human Non Small Cell Lung Carcinoma cells H1975 cells as assessed using nuclei count readout. Dasatinib IC50 at 120h readout: 0.057 μM ; IC50 at 120h readout: 0.050 μM. C. Summary of IC50s for the Alamar Blue and nuclei count readout as a function of time.
In order to avoid erroneous measurements induced by heterogeneous cell populations, several aspects should be considered. A direct measurement of cell proliferation such as the monitoring of thymidine incorporation constitutes the “Gold Standard” of cell viability assays ; unfortunately, its reliance on radiation makes it unamenable to automation on a large scale. Automated imaging and image analysis of nuclei-stained cells provides a powerful and accurate alternative, that can easily be performed even in large screens. Furthermore, the nuclei count readout allows for a direct measurement of the absolute number of surviving cells. This is generally viewed as a great advantage in terms of accuracy, since we and others have previously observed that a few surviving cells may mask toxicity in an indirect assay such as Alamar Blue7,13. In addition, if cell viability is measured using such an assay, then cell seeding density should be carefully optimized, and measurements should be performed at different time points to ensure accuracy. In our study, a longer incubation time allowed to correct for the erroneous IC50 obtained at early time points using the Alamar Blue assay (Figure 2C). Altogether, our examples as well as previous reports illustrate how a drug’s toxicity may easily be overlooked during in vitro cytotoxicity screening due to the limitations of indirect measurements.
3.2 Limitations of single parameter readout
Cell death is increasingly viewed as a dynamic and integrative cellular response whose outcome depends on a net result of signals2, 4–5. Each cellular subtype may respond differently to a specific damage depending on its genetic background and its intrinsic repair ability. Furthermore, each cell integrates signals at critical control points before progressing to a point of no return1, 5. As a consequence, cells respond differently to compounds with different mechanisms of action that affect different cellular pathways6. In this context, it becomes clear that monitoring a single event in the complex process of cell death can easily lead to missing signs of toxicity that will eventually lead to cell death. In addition, a number of regulated but non-apoptotic cell death pathways have been discovered, such as necrosis-like programmed cell death, which occurs in absence of chromatin condensation, or caspase-independent death programmes4. Thus, an assay focused on the induction of apoptosis by monitoring chromatin condensation or caspase activation as a unique readout may be prone to false negatives and may miss compounds activating such signals2. Similarly, concentrating on morphological changes undergone by dying cells may be prone to error, since these features are extremely diverse and may reflect various, continuous modes of death activation3–5. Finally, single parameter readouts are especially prone to false negatives if used – as it is often the case –at single time points. For example, an injured cell may retain its membrane integrity for a relatively long time, and such damage could easily remain undetected using a dye exclusion method at an early time point13. Altogether, these observations highlight the limitations of single parameter readout for monitoring cytotoxicity, and strongly suggest the importance of monitoring multiple parameters when assessing cell death.
3.3 Limitations of fixed time point assays
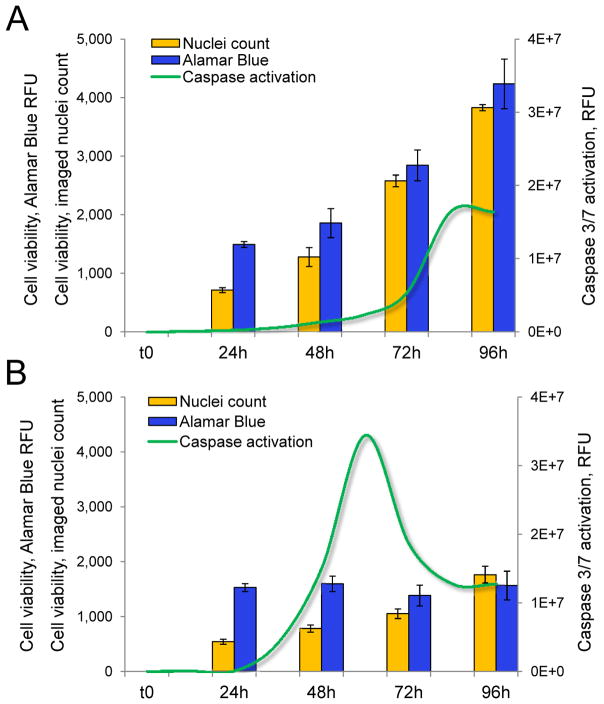
As previously mentioned with the example of a dye exclusion assay, any assay performed at a single time point may fail to correctly assess cytotoxicity. This is true of direct assays such as nuclei count as well, since cellular damages induced by a cytotoxic compound or by the modulation of a specific pathway may yet have to be translated into a decrease in cell number at the time of readout. We illustrate this point with the example of HeLa cells transfected with a Cell Death siRNA pool targeting essential human genes or with a negative control GFP siRNA. We monitored the cytotoxicity induced by both transfections in 384-well microplates over time using a multiplexed readout consisting of Alamar Blue and nuclei count, with assessment of caspase 3/7 activation in parallel. caspase activation was continuously monitored using a live fluorometric readout we have previously optimized and validated for high-content screening (HCS)40. Cells transfected with the negative control siRNA grew linearly over 96h post-transfection as expected, as demonstrated by nuclei count readout(Figure 3A). The increase in cell count was translated into an increase in intensity for the Alamar Blue readout at each time point (Figure 3A). Despite the apparent lack of toxicity of the control transfection as assessed using Alamar Blue and nuclei count readouts, we observed caspase activation at late time points, starting at 84h post-transfection. In contrast, transfection with the Cell Death siRNA pool resulted in a dramatic decrease in cell proliferation, as assessed by nuclei count (Figure 3B). The intensity of Alamar Blue readout was drastically decreased as well for all time points after 24h compared to control transfection. Of note, while the cell number did increase over time (as assessed by nuclei count), the Alamar Blue fluorescence readout reached saturation at 24h and failed to increase at later time points. This observation confirms our previous observation that Alamar Blue-based assays suffer of signal saturation at low cell densities and may fail to pick up variations in cell number at low cell densities. caspase activation in HeLa cells transfected with Cell Death siRNA pool gradually increased after 24h and reached a peak at 60h post-transfection (Figure 3B). The signal then gradually decreased but did not return to the original baseline ; instead it reached a new baseline at a level previously observed with control transfection (Figure 3A). Presumably, the non-specific caspase activation observed at late time points with control transfection was also observed with Cell Death siRNA transfection, which is responsible for the high baseline observed after 72h. Our observations in this case study demonstrate the importance of assessing cytotoxicity at multiple time points. Both the Alamar Blue and nuclei count readouts failed to detect any effect of the knockdown of essential genes until 72h post-transfection. In contrast, the caspase activation readout recorded induction of apoptosis as early as 36h post-transfection. This result is expected because caspase activation precedes any decrease in cell population in the apoptotic process ; however our results highlight the difficulty of assessing an optimal time point for the Alamar Blue and nuclei count readouts. While a late time point is usually better suited for these readouts, non-specific toxicity may occur in vitro at late time points that may interfere and lead to false-positives. Similarly, if the caspase activation assay is performed as a single point read out, it may be prone to error. In our example, identical levels of non-specific caspase activation were observed for both transfections after 80h post-transfection. In addition, caspase activation, which is a transient event in the cell, may occur at different time points depending on the mechanism of action of different cytotoxic compounds or of the signaling pathways triggered by different cytotoxic stimuli40. Overall, our observations in this case study strongly emphasize the importance of performing viability assays at different time points in order to avoid missing early or late cytotoxicity. This implies that continuous assays allowing multiple readouts for the same well are preferred so as to avoid performing multiple experiments for multiple time points40. Likewise, since the cytotoxicity of a compound depends on its concentration and exposure time, a quantitative HTS approach can overcome some of the disadvantages of making a measurement at one time point. For example, the method used by Inglese et al. demonstrates the use of concentration-response curves to eliminate false positives in their kinase screen.
Figure 3.
A. Multiplexed assessment of cytotoxicity induced by the transfection of A. control anti-GFP siRNA or B. Cell Death siRNA pool in HeLa cells. Alamar Blue and nuclei count readouts are performed at 24, 48, 72 and 96h post-transfection ; continuous assessment of Caspase 3/7 activation is performed at 12, 24, 36, 48, 72, 84 and 96h post-transfection.
3.4 Cost/feasibility considerations
An important factor to consider when performing viability assays on a large scale is cost. Since, as we previously demonstrated, a multi-parameter approach is necessary for accuracy – and ideally at multiple time points, the cost of running multiple parallel screens with different assays becomes prohibitive. In addition, to be truly informative, viability assays need to be performed on a panel of cell lines in order to assess the toxicity of a specific treatment across multiple cell types. Some of these cell types may not even be amenable to the production of a large cell population for such screens. Additionally, for compound screening, multiple replicates at multiple concentrations are required. For these reasons, live, continuous assays that are amenable to multiplexing emerge as a practical solution to the limitations of traditional low-content viability assays because they allow the extraction of multiple information from a single well.
4. EXPERT OPINION: TOWARD CONTENT-RICH PLATFORMS FOR CELL VIABILITY ASSESSMENT
Advances in cell biology have led to a better understanding of the complexity of cell death, once seen as a binary process – either active (apoptosis) or passive (necrosis). Increasingly, cell death is being described as a multi-faceted event, with apoptosis and necrosis constituting two extremes of a continuum of death modes, with varying contributions of cellular actors2, 4–5. Recent progress in the study of dying cells suggests that cell death is far more complex than originally envisioned, with the likely implication of intricate pathways yet to be discovered4–5. For example, the discovery that the inhibition of caspase activation does not necessarily protect against cell death stimuli goes against earlier expectations4. This paradigm shift implies that traditional hallmarks of cell death, if monitored independently of each other, may not accurately reflect the death status of a cell population. Rather, the complexity of cell death suggests that studying multiple markers is essential to accurately record this process.
Advances in HTS now allow for the rapid screening of large libraries of small molecules on multiple cell lines in parallel25, 41, thereby allowing to define a cytotoxicity profile for a given small molecule across multiple cell types. This advance is significant, since in some cases specific tissue functions activated only in relevant cell lines may be necessary to identify the toxic effect of a given molecule. While the power of HTS for assessing cytotoxicity has mainly been used for parallel screening of multiple compounds on multiple cell lines, it also allows for multiplexed, high-content assays to be performed at the same time in the same well. In light of the newly-described complexity of cell death, combining assays focused on different markers most likely will enable to record induction of cell death more accurately. Increasingly, drug signatures are obtained using a combination of assays. Pharmaceutical companies already typically employ an array of assays when screening for cytotoxicity, combining the different categories of assays we previously reviewed. For example, the formation of reactive oxygen species can be monitored along with the intracellular glutathione status and the integrity of the cell membrane, to provide an in-depth characterization of the toxic effects of a panel of compounds6, 42. However, the future in cell viability assessment consists in performing multiple assays for characterizing multiple aspects of cell death induction in the same well. This constitutes the main benefit of multiplexed high-content assays. In addition, high-content assays afford the added bonus of overcoming the noise induced by heterogeneous cell populations by providing access to cell by cell analysis. Such an approach has already been validated ; a report demonstrated that a multiplexed high-content assay based on the simultaneous use of four fluorescent dyes reporting on calcium levels, mitochondrial membrane potential, nuclear area, cell number and membrane permeability produced predictions highly concordant with results obtained combining seven conventional, separate assays43. This study, which encompassed 611 drugs, accurately predicted toxicity for 97% of them. Furthermore, the in vitro prediction of toxicity identified in patients was 87%6, 43. This encouraging result demonstrates the potential of multiplexed high-content assays and validates their use for the prediction of in vivo toxicity. We hypothesize that the use of live, continuous assays will further improve the accuracy of toxicity predictions by allowing to record multiple time points for a same well, providing multiple snapshots of cell death induction at different exposure times. Such an assay for live monitoring of caspase activation has already been validated for high-content screening, allowing continuous cell by cell analysis of caspase 3/7 activation40.
We envision that the assessment of cell viability will move away from basic and discrete measurements reflecting late events in the cell death process. Rather, monitoring multiple signaling pathways in real time will provide far more information regarding the mechanism of action of cell death stimuli. Instead of measuring the average response of an heterogeneous population, cell by cell analysis will afford much improved accuracy. Such a transition will be made possible by the discovery of new and improved molecular probes, as well as by progress in bioinformatics to isolate and map significant interactions between multiple cellular actors. As progress in cell engineering allows to combine multiple cell types to closely recapitulate a tissue function, the availability of complex cellular models will dramatically improve the significance of data generated in vitro. As those contributions from different fields converge, detailed in vitro cytotoxicity signatures may well be generated that will predict toxicity in vivo.
References
- 1•.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004 Jan 23;116(2):205–19. doi: 10.1016/s0092-8674(04)00046-7. This review summarizes the critical control points in the cell death pathway. [DOI] [PubMed] [Google Scholar]
- 2••.Degterev A, Yuan J. Expansion and evolution of cell death programmes. Nat Rev Mol Cell Biol. 2008 May;9(5):378–90. doi: 10.1038/nrm2393. This review highlights the diversity of programmed cell death mechanisms. [DOI] [PubMed] [Google Scholar]
- 3.Edinger AL, Thompson CB. Death by design: apoptosis, necrosis and autophagy. Curr Opin Cell Biol. 2004 Dec;16(6):663–9. doi: 10.1016/j.ceb.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 4•.Leist M, Jaattela M. Four deaths and a funeral: from caspases to alternative mechanisms. Nat Rev Mol Cell Biol. 2001 Aug;2(8):589–98. doi: 10.1038/35085008. This review summarizes alternative mechanisms of programmed cell death. [DOI] [PubMed] [Google Scholar]
- 5••.Loos B, Engelbrecht AM. Cell death: a dynamic response concept. Autophagy. 2009 Jul;5(5):590–603. doi: 10.4161/auto.5.5.8479. This review emphasizes the importance of integrating new concepts to assess cell death accurately. [DOI] [PubMed] [Google Scholar]
- 6.Schoonen WG, Westerink WM, Horbach GJ. High-throughput screening for analysis of in vitro toxicity. EXS. 2009;99:401–52. doi: 10.1007/978-3-7643-8336-7_14. [DOI] [PubMed] [Google Scholar]
- 7.King MA. Detection of dead cells and measurement of cell killing by flow cytometry. J Immunol Methods. 2000 Sep 21;243(1–2):155–66. doi: 10.1016/s0022-1759(00)00232-5. [DOI] [PubMed] [Google Scholar]
- 8.Roth BL, Poot M, Yue ST, Millard PJ. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl Environ Microbiol. 1997 Jun;63(6):2421–31. doi: 10.1128/aem.63.6.2421-2431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubl W, Iturraspe J, Martinez GA, Hutcheson CE, Roberts CG, Fisk DD, et al. Measurement of absolute concentration and viability of CD34+ cells in cord blood and cord blood products using fluorescent beads and cyanine nucleic acid dyes. Cytometry. 1998 Jun 15;34(3):121–7. doi: 10.1002/(sici)1097-0320(19980615)34:3<121::aid-cyto2>3.3.co;2-u. [DOI] [PubMed] [Google Scholar]
- 10.Idziorek T, Estaquier J, De Bels F, Ameisen JC. YOPRO-1 permits cytofluorometric analysis of programmed cell death (apoptosis) without interfering with cell viability. J Immunol Methods. 1995 Sep 25;185(2):249–58. doi: 10.1016/0022-1759(95)00172-7. [DOI] [PubMed] [Google Scholar]
- 11.Antczak C, Kloepping C, Radu C, Genski T, Muller-Kuhrt L, Siems K, et al. Revisiting old drugs as novel agents for retinoblastoma: in vitro and in vivo antitumor activity of cardenolides. Invest Ophthalmol Vis Sci. 2009 Jul;50(7):3065–73. doi: 10.1167/iovs.08-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briant L, Robert-Hebmann V, Sivan V, Brunet A, Pouyssegur J, Devaux C. Involvement of extracellular signal-regulated kinase module in HIV-mediated CD4 signals controlling activation of nuclear factor-kappa B and AP-1 transcription factors. J Immunol. 1998 Feb 15;160(4):1875–85. [PubMed] [Google Scholar]
- 13.Slater K. Cytotoxicity tests for high-throughput drug discovery. Curr Opin Biotechnol. 2001 Feb;12(1):70–4. doi: 10.1016/s0958-1669(00)00177-4. [DOI] [PubMed] [Google Scholar]
- 14.Kubbies M. Flow cytometric recognition of clastogen induced chromatin damage in G0/G1 lymphocytes by non-stoichiometric Hoechst fluorochrome binding. Cytometry. 1990;11(3):386–94. doi: 10.1002/cyto.990110309. [DOI] [PubMed] [Google Scholar]
- 15.Fennell M, Chan H, Wood A. Multiparameter measurement of caspase 3 activation and apoptotic cell death in NT2 neuronal precursor cells using high-content analysis. J Biomol Screen. 2006 Apr;11(3):296–302. doi: 10.1177/1087057105284618. [DOI] [PubMed] [Google Scholar]
- 16.Morelli JK, Buehrle M, Pognan F, Barone LR, Fieles W, Ciaccio PJ. Validation of an in vitro screen for phospholipidosis using a high-content biology platform. Cell Biol Toxicol. 2006 Jan;22(1):15–27. doi: 10.1007/s10565-006-0176-z. [DOI] [PubMed] [Google Scholar]
- 17.Tonary AM, Pezacki JP. Simultaneous quantitative measurement of luciferase reporter activity and cell number in two- and three-dimensional cultures of hepatitis C virus replicons. Anal Biochem. 2006 Mar 15;350(2):239–48. doi: 10.1016/j.ab.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 18.Tucker B, Lardelli M. A rapid apoptosis assay measuring relative acridine orange fluorescence in zebrafish embryos. Zebrafish. 2007 Summer;4(2):113–6. doi: 10.1089/zeb.2007.0508. [DOI] [PubMed] [Google Scholar]
- 19.Delic J, Coppey J, Magdelenat H, Coppey-Moisan M. Impossibility of acridine orange intercalation in nuclear DNA of the living cell. Exp Cell Res. 1991 May;194(1):147–53. doi: 10.1016/0014-4827(91)90144-j. [DOI] [PubMed] [Google Scholar]
- 20.Loontiens FG, Regenfuss P, Zechel A, Dumortier L, Clegg RM. Binding characteristics of Hoechst 33258 with calf thymus DNA, poly[d(A-T)], and d(CCGGAATTCCGG): multiple stoichiometries and determination of tight binding with a wide spectrum of site affinities. Biochemistry. 1990 Sep 25;29(38):9029–39. doi: 10.1021/bi00490a021. [DOI] [PubMed] [Google Scholar]
- 21.Scudiero DA, Shoemaker RH, Paull KD, Monks A, Tierney S, Nofziger TH, et al. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res. 1988 Sep 1;48(17):4827–33. [PubMed] [Google Scholar]
- 22.Duarte M, Giordani RB, De Carli GA, Zuanazzi JA, Macedo AJ, Tasca T. A quantitative resazurin assay to determinate the viability of Trichomonas vaginalis and the cytotoxicity of organic solvents and surfactant agents. Exp Parasitol. 2009 Oct;123(2):195–8. doi: 10.1016/j.exppara.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 23.O’Brien J, Wilson I, Orton T, Pognan F. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem. 2000 Sep;267(17):5421–6. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 24.O’Boyle DR, 2nd, Nower PT, Lemm JA, Valera L, Sun JH, Rigat K, et al. Development of a cell-based high-throughput specificity screen using a hepatitis C virus-bovine viral diarrhea virus dual replicon assay. Antimicrob Agents Chemother. 2005 Apr;49(4):1346–53. doi: 10.1128/AAC.49.4.1346-1353.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shum D, Radu C, Kim E, Cajuste M, Shao Y, Seshan VE, et al. A high density assay format for the detection of novel cytotoxic agents in large chemical libraries. J Enzyme Inhib Med Chem. 2008 Dec;23(6):931–45. doi: 10.1080/14756360701810082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiu VC, Haynes DH. High and low affinity Ca2+ binding to the sarcoplasmic reticulum: use of a high-affinity fluorescent calcium indicator. Biophys J. 1977 Apr;18(1):3–22. doi: 10.1016/S0006-3495(77)85592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Clerck LS, Bridts CH, Mertens AM, Moens MM, Stevens WJ. Use of fluorescent dyes in the determination of adherence of human leucocytes to endothelial cells and the effect of fluorochromes on cellular function. J Immunol Methods. 1994 Jun 3;172(1):115–24. doi: 10.1016/0022-1759(94)90384-0. [DOI] [PubMed] [Google Scholar]
- 28.Levesque A, Paquet A, Page M. Measurement of tumor necrosis factor activity by flow cytometry. Cytometry. 1995 Jun 1;20(2):181–4. doi: 10.1002/cyto.990200211. [DOI] [PubMed] [Google Scholar]
- 29.Jonsson B, Liminga G, Csoka K, Fridborg H, Dhar S, Nygren P, et al. Cytotoxic activity of calcein acetoxymethyl ester (Calcein/AM) on primary cultures of human haematological and solid tumours. Eur J Cancer. 1996 May;32A(5):883–7. doi: 10.1016/0959-8049(96)00015-9. [DOI] [PubMed] [Google Scholar]
- 30.Liminga G, Nygren P, Dhar S, Nilsson K, Larsson R. Cytotoxic effect of calcein acetoxymethyl ester on human tumor cell lines: drug delivery by intracellular trapping. Anticancer Drugs. 1995 Aug;6(4):578–85. doi: 10.1097/00001813-199508000-00011. [DOI] [PubMed] [Google Scholar]
- 31.Merrilees MJ, Beaumont BW, Scott LJ. Fluoroprobe quantification of viable and non-viable cells in human coronary and internal thoracic arteries sampled at autopsy. J Vasc Res. 1995 Nov-Dec;32(6):371–7. doi: 10.1159/000159112. [DOI] [PubMed] [Google Scholar]
- 32.Poole CA, Brookes NH, Gilbert RT, Beaumont BW, Crowther A, Scott L, et al. Detection of viable and non-viable cells in connective tissue explants using the fixable fluoroprobes 5-chloromethylfluorescein diacetate and ethidium homodimer-1. Connect Tissue Res. 1996;33(4):233–41. doi: 10.3109/03008209609028880. [DOI] [PubMed] [Google Scholar]
- 33.Kanamori A, Catrinescu MM, Traistaru M, Beaubien R, Levin L. In Vivo Imaging Of Retinal Ganglion Cell Axons Within The Nerve Fiber Layer. Invest Ophthalmol Vis Sci. 2009 Sep 24; doi: 10.1167/iovs.09-4021. [DOI] [PubMed] [Google Scholar]
- 34.Niles AL, Moravec RA, Eric Hesselberth P, Scurria MA, Daily WJ, Riss TL. A homogeneous assay to measure live and dead cells in the same sample by detecting different protease markers. Anal Biochem. 2007 Jul 15;366(2):197–206. doi: 10.1016/j.ab.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004 Feb 6;303(5659):832–5. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 36.Andreotti PE, Cree IA, Kurbacher CM, Hartmann DM, Linder D, Harel G, et al. Chemosensitivity testing of human tumors using a microplate adenosine triphosphate luminescence assay: clinical correlation for cisplatin resistance of ovarian carcinoma. Cancer Res. 1995 Nov 15;55(22):5276–82. [PubMed] [Google Scholar]
- 37.Lundin A, Hasenson M, Persson J, Pousette A. Estimation of biomass in growing cell lines by adenosine triphosphate assay. Methods Enzymol. 1986;133:27–42. doi: 10.1016/0076-6879(86)33053-2. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Douglas GC, Thirkill TL, Lohstroh PN, Bielmeier SR, Narotsky MG, et al. Effect of bromodichloromethane on chorionic gonadotrophin secretion by human placental trophoblast cultures. Toxicol Sci. 2003 Nov;76(1):75–82. doi: 10.1093/toxsci/kfg225. [DOI] [PubMed] [Google Scholar]
- 39.Cho MH, Niles A, Huang R, Inglese J, Austin CP, Riss T, et al. A bioluminescent cytotoxicity assay for assessment of membrane integrity using a proteolytic biomarker. Toxicol In Vitro. 2008 Jun;22(4):1099–106. doi: 10.1016/j.tiv.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antczak C, Takagi T, Ramirez CN, Radu C, Djaballah H. Live-cell imaging of caspase activation for high-content screening. J Biomol Screen. 2009 Sep;14(8):956–69. doi: 10.1177/1087057109343207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckelbarger JD, Wilmot JT, Epperson MT, Thakur CS, Shum D, Antczak C, et al. Synthesis of antiproliferative Cephalotaxus esters and their evaluation against several human hematopoietic and solid tumor cell lines: uncovering differential susceptibilities to multidrug resistance. Chemistry. 2008;14(14):4293–306. doi: 10.1002/chem.200701998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoonen WG, Westerink WM, de Roos JA, Debiton E. Cytotoxic effects of 100 reference compounds on Hep G2 and HeLa cells and of 60 compounds on ECC-1 and CHO cells. I mechanistic assays on ROS, glutathione depletion and calcein uptake. Toxicol In Vitro. 2005 Jun;19(4):505–16. doi: 10.1016/j.tiv.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 43•.O’Brien PJ, Irwin W, Diaz D, Howard-Cofield E, Krejsa CM, Slaughter MR, et al. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch Toxicol. 2006 Sep;80(9):580–604. doi: 10.1007/s00204-006-0091-3. This article validates the use of a multiplexed fluorescent readout for predicting toxicity in vitro and in vivo. [DOI] [PubMed] [Google Scholar]