Abstract
Mitochondrial DNA (mtDNA) is more susceptible than nuclear DNA to helix-distorting damage via exposure to environmental genotoxins, partially due to a lack of nucleotide excision repair (NER). Thus, this damage is irreparable and persistent in mtDNA in the short term. We recently found that helix-distorting mtDNA damage induced by UVC is gradually removed in Caenorhabditis elegans and that removal is dependent upon autophagy and mitochondrial dynamics. We here report the effects of UVC exposure on mitophagy, mitochondrial morphology, and indicators of mitochondrial function in mammalian cells. Exposure to UVC induced autophagy within 24 h; nonetheless, significant mitochondrial degradation was not observed until 72 h post exposure. Mitochondrial mass, morphology and function were not significantly altered. These data further support the idea that persistent mtDNA damage is removed by autophagy, and also suggest a powerful compensatory capacity for dealing with mtDNA damage.
Keywords: Mitochondrial fusion, mitochondrial fission, autophagy, mitophagy, mitochondrial DNA damage, ultraviolet radiation
INTRODUCTION
Mitochondria are the site of most energy production within a cell and are key regulators of apoptosis and calcium homeostasis (1). Mitochondria contain several hundred copies of their own 16.5 kb genome which encodes for a small subset of mitochondrial proteins including 13 polypeptides that are incorporated into four of the five complexes of the OXPHOS system plus 2 rRNAs and 22 tRNAs, with the remaining proteins encoded by nuclear DNA (nDNA) (2). Despite this relatively small contribution, mutations in or depletion of mtDNA are associated with a significant number of inherited mitochondrial diseases which in total are estimated to affect more than 1 in 6,000 people (3), emphasizing the importance of mtDNA integrity to human health. Significant research also implicates mitochondrial dysfunction and mtDNA mutation in the pathogenesis of widespread disorders such as neurodegenerative conditions (4, 5), type 2 diabetes mellitus (6), cancer (7, 8) and aging (9).
There is substantial evidence that mtDNA is uniquely susceptible to damage caused by certain ubiquitous environmental genotoxicants including metabolically-activated polycyclic aromatic hydrocarbons (10-13) and mycotoxins (14, 15), as well as endogenously produced reactive oxygen species (16). Mitochondria house several mechanisms to combat oxidative damage including base excision repair, the repair mechanism responsible for repair of most oxidative DNA lesions. However, mitochondria lack nucleotide excision repair (NER), the mechanism necessary to repair helix-distorting chemical adducts and UVC-induced photodimers. Therefore these lesions persist and have the potential to stall DNA replication (12, 17-20) and transcription (14, 17) potentially leading to depletion of mtDNA and mtDNA-encoded proteins, mutations and subsequently mitochondrial dysfunction.
Mitochondria and mtDNA are degraded by macroautophagy, the lysosomal-driven degradation of cytoplasmic materials (21, 22). It is now well-established that autophagy can selectively degrade dysfunctional mitochondria, referred to as mitophagy, and that this process is dependent in part upon mitochondrial function and morphology and autophagy induction (23-25). Mitochondrial dysfunction resulting from toxicant exposure, mtDNA mutations and ROS can induce autophagy and mitophagy (26-30) and blocking autophagy and mitophagy results in accumulation of dysfunctional mitochondria and damaged mtDNA (31, 32) and increased susceptibility to apoptotic cell death (33-35). Therefore, autophagic mitochondrial degradation may serve as an important mechanism to rid the cell of damaged mitochondria that would otherwise trigger apoptosis.
We recently found that UVC-induced mtDNA damage is gradually removed in vivo in Caenorhabditis elegans and that removal is dependent upon genes involved in autophagy, mitophagy and mitochondrial dynamics (36). Additionally, we demonstrated that UVC exposure induces autophagy in C. elegans. We hypothesize that UVC-induced DNA damage causes mitochondrial dysfunction resulting in changes in mitochondrial morphology and autophagy induction that result in the removal of damaged mtDNAs. In this work, we investigated the effects of UVC exposure on mitophagy, mitochondrial morphology, and indicators of mitochondrial ETC function. We show that UVC-induced mtDNA damage is removed at a similar rate in primary human fibroblasts as in C. elegans and that exposure to UVC induces autophagy within 24 h. Interestingly, significant mitochondrial degradation is not observed until 72 h post exposure. No significant changes in mitochondrial MP, ROS or mitochondrial morphology were observed following UVC exposure. These data further support the idea that persistent mtDNA damage is removed by autophagy; however, future research is needed to elucidate the factors which trigger removal.
MATERIALS AND METHODS
Cell Culture, thymidine block, and UVC/chemical exposures
Primary human skin fibroblasts (CCD-1139sk, ATCC) were maintained in Iscove’s Modified Dulbecco’s Media (IMDM) supplemented with 10% fetal bovine serum, 5% CO2, and 5% penicillin/streptomycin at 37°C. Using a cell culture system to study mitochondrial endpoints is complicated by the tendency of cells cultured in high glucose to utilize glycolysis for energy production rather than OXPHOS, thus rendering mitochondrial function less critical for cell survival (37). We attempted to avoid this by using non-transformed, primary human skin fibroblasts replaced monthly from a low passage frozen stock.
The thymidine block, which was used for all experiments unless otherwise noted, was performed 24 h before analyses on 90% confluent cells by the addition of 3 mM thymidine (final concentration) to IMDM. Thymidine media was replaced every 24 h for the duration of an experiment. For UVC exposure, cells were washed once in PBS and then exposed without medium to 10 J/m2 UVC using an ultraviolet lamp with built-in UVC sensor (CL-1000 Ultraviolet Crosslinker, UVP, Upland, CA, USA) with peak emission at 254 nm; fresh cell culture medium was immediately replaced thereafter. For flow cytometry analyses, cells were seeded at 75K cells/2 ml of normal growth media with or without thymidine in six-well plates, collected by 0.25% trypsin incubation, spun down and resuspended in FACS buffer (1% BSA in PBS). For chemical exposures, 100 nM bafilomycin A1 (Sigma) resuspended to 32 μM in DMSO and further diluted into culture medium was replaced every 24 h for extent of the treatment period.
DNA synthesis and cell cycle analyses
Cells were exposed to UVC with and without 3 mM thymidine in normal growth media and assessed for DNA synthesis by BrdU incorporation at 24, 48 and 72 h post exposure to UVC. Cells were labeled with BrdU, anti-BrdU FITC and PI per the manufacturer’s instructions (BD Biosciences; Cat. No. 347583) and analyzed for FITC and PI fluorescence simultaneously using a FACScan flow cytometer (Becton Dickinson). FlowJo 7.6.4 was used to identify BrdU-positive cells and perform cell cycle analysis. Two biological replicates were analyzed for each treatment at each time point.
Cell Viability
Cells were collected and resuspended in FACS Buffer (1% BSA in PBS) containing 5 μg/ml (final concentration) Hoechst (Life Technologies) and 1:100 Annexin V APC (Life Technologies) and immediately placed on ice. Fluorescence was measured simultaneously using a FACSVantage Sorter (Becton Dickinson) and acquired data was analyzed using FlowJo 7.6.4. Viable, apoptotic and dead cell populations were defined by unstained and Annexin V APC or Hoechst 33258 individually stained cells and the same quadrants were applied to all samples. Four biological replicates were analyzed for each treatment at each time point. Two-way ANOVA was used to compare the total percentage of apoptotic/dead cells (Q2 + Q3) between treatments and recovery time using Statview 5.0.1.
DNA damage, removal quantification and quantitative PCR
At each recovery time point, cells were scraped, pelleted, flash-frozen, and stored at -80°C until DNA extraction. Genomic DNA was extracted and quantified using a automated extraction procedure and DNA damage analysis was performed using quantitative polymerase chain reaction (QPCR) as described in Furda et al. (2012) (38). At least two time-separated QPCR reactions were performed on each sample and at least two biological replicates were analyzed per treatment and time point. Significant removal/repair at recovery time points 72 and 96 h was determined by one-way ANOVA (effect of time point) and subsequent Fisher’s PLSD (Statview 5.0.1).
Mitochondrial DNA content analysis
Relative mtDNA content was measured by quantitative, real-time PCR as described in Venegas and Halberg (2012) (39) using the QPCR samples described above. For each sample, three technical replicates were averaged from a single real-time PCR run.
Mitochondrial mass, membrane potential, ROS analyses
Cells were stained with 150 nM Mitotracker Green (Life Technologies) for 30 min at 37°C then washed and stained with either 300 nM TMRE (Life Technologies) or 150 nM Mitotracker Red CM-H2XROS (Life Technologies) for 30 min at 37°C. Cells were collected for FACS analysis as described above. Samples were immediately placed on ice and Mitotracker Green and TMRE or Mitotracker Red CM-H2XROS fluorescence were simultaneously measured using a FACScan flow cytometer (Becton Dickinson). Viable cells were gated and geometric mean fluorescence values for MTG, TMRE and Mitotracker Red CM-H2XROS were obtained using FlowJo 7.6.4. TMRE and Mitotracker Red CM-H2XROS were normalized to mitochondrial mass. 2-4 biological replicates were analyzed per treatment per time point for each fluorophore.
LC3II protein quantification by Western blot
Cells were incubated with or without bafilomycin (100 nM) for 3 h then collected and lysed in 100 μl of buffer containing 2% Triton-X and complete protease inhibitor cocktail (Sigma) in PBS for 1 h on ice. Protein concentration was measured by BCA assay (Thermo Scientific). Lysates were resolved by 4-12% LDS-PAGE (NuPage 4-12% Bis-Tris Gels) and electrotransferred onto a PVDF membrane. The membranes were blocked for 1 h in 5% milk then incubated overnight in primary antibodies against LC3B (Novus Biologicals; Cat. No. NB600-1384; 1:2000) and beta-actin (Abcam; Cat. No. ab8224; 1:2000) and for 2 h in secondary antibodies: anti-rabbit-HRP (Pierce; Cat. No. 32460, 1: 200) and anti-mouse-HRP (Immuno Jackson Research, Cat.No. 115-035-174, 1:2000). The blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and developed on film. Densitometry was performed using ImageJ (1.43m). Each sample was analyzed by Western blot at least twice and 2-4 biological replicates were analyzed per treatment per time point.
RNA extraction, reverse transcription and real-time PCR analysis
Total RNA was extracted with Qiagen RNeasy Mini Kit and quantified by NanoDrop 8000 spectrophotometer (Thermo Scientific/NanoDrop). 250ng of isolated RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit (Cat. No. 4368814) using the manufacturer’s instructions and amplified by real time PCR using the 7300 Real Time PCR System (Applied Biosystems), under the following conditions: 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 sec at 95°C and then 60 sec at 60°C for 18S, 36B4, BECN1, COXI, COXIV, FIS1, MAPLC3, MFN1, NRF1, OPA1, and TFAM, at 62°C for PGC1α and at 64°C for DRP1. A dissociation curve was calculated for each sample at the end of each profile. The 25μl PCR reaction contained 12.5μl of SYBR Green PCR Master Mix, 8.5μl H20, 2μl of target-specific primers at 400 nM final concentration, and 2μl of cDNA from the RT reaction already diluted to 2 ng/μl. The ABI PRISM 7300 Sequence Detection System Software, Version 1.1 (Applied Biosystems) was used to carry out data analysis. The average mRNA fold change of each target gene was calculated by comparing the CT (cycle threshold) of the target gene to that of the housekeeping gene 36B4. Two biological replicates were analyzed per treatment and all samples were run in triplicate and were averaged prior to analysis. RT-PCR conditions were optimized for previously published and designed primers; the primer sequences and conditions are listed in Table S1.
Mitochondrial and lysosome colocalization
Cells were plated in 35 mm collagen-coated, glass bottom dishes (MatTek Corporation). Cells were stained with MTG (300 nM) for 30 min, washed and stained with 50 nM Lysotracker Red (LTR; Life Technologies) for 30 min. Cells were incubated in 10 μM leupeptin (Sigma; stock 10 mM in H2O), 7.5 μM pepstatin A (Sigma; stock 2 mM in EtOH) and 50 nM LTR for 1 h. For imaging, media was replaced with Opti-MEM supplemented with 10 μM leupeptin, 7.5 μM pepstatin A and 50 nM LTR.
Fluorescence imaging was performed using a Leica Sp5 laser scanning confocal microscope with 63×/1.20 NA plan apochromat water immersion objective lens at 37°C. Z-stacks were acquired (0.38 μm thickness) at 1024 × 1024 resolution and 3D colocalization of LTR and MTG was performed using Imaris 7.3. Colocalized areas were converted to “surfaces” and the number of surfaces per cell was compared between treatments and time points by two-way ANOVA. Given the significant treatment x time point interaction (P < 0.0001), a treatment effect was analyzed by Fisher’s PLSD at each time point.
Mitochondrial morphology analysis
Cells were plated in 35 mm collagen coated, glass bottom dishes (MatTek Corporation). Cells were stained with MTG (300 nM) for 30 min. For imaging, media was replaced with Opti-MEM. Fluorescence imaging was performed using a Leica Sp5 laser scanning confocal microscope with 63×/1.20 NA plan apochromat water immersion objective lens at 37°C. Z-stacks were acquired (0.38 μm thickness) at 1024 × 1024 resolution. Using Imaris 7.3, MTG fluorescence intensity was used to build a “surface” representing the mitochondria within a cell. The total number of surfaces per cell was quantified as well as the surface area, volume and sphericity of each surface. Mitochondria were separated in the following categories based on volume: 0-10 μm3 (class I), 10-100 μm3 (class II) and ≥100 μm3 (class III) and average inverse sphericity, average surface area/volume and the number of mitochondria were compared between treatments within each class for each time point. At least 20 cells/treatment/time point/experiment were analyzed and two experiments were performed.
Transmission electron microscopy
Cells were plated in six-well plates for 24 h after which they were dosed with UVC 10 J/m2. Three hours before collection cells were incubated with or without bafilomycin (100 nM). Cells were collected using trypsinization, pelleted then resuspended and placed in the initial fixative, 4% paraformaldehyde, 1% glutaraldehyde (45:1G, pH 7.2-7.4) and stored at 4°C until time of processing. Primarily fixed cells were then pelleted and placed in secondary fixative, (1% osmium tetroxide/0.1 M sodium phosphate buffer, pH 7.2-7.4) for 1 h at room temperature. Resultant secondarily fixed cells were dehydrated in graded ethanol solutions and embedded in Spurr’s resin. Semithin sections (500 nm thick) were cut with glass knives, mounted on glass slides, and stained with 1% toluidine blue O in 1% sodium borate (Figure S4). These sections were used first, to determine presence and relative number of fibroblasts in the block face, and secondly to visualize high resolution light microscopic details of the cells. Ultrathin sections (70-90 nm thick) were cut with a diamond knife, stained with uranyl acetate and lead citrate, and examined using a FEI/Philips 208S transmission electron microscope (TEM) at 80kV accelerating voltage. TEM processing, analysis, and imaging were performed at the Laboratory for Advanced electron and Light Optical Methods (LAELOM), College of Veterinary Medicine, North Carolina State University. Cells were selected at random for imaging. Images were taken at 3300X-5600X to capture the entire cell, at 11000X to capture the majority of the cytoplasmic area and finally at 22000X to investigate the contents of autophagic vacuoles (AVs) within the cell.
RESULTS
We recently reported that removal of otherwise irreparable UVC-induced mtDNA damage is detectable in adult C. elegans within 72 h, and that this removal is dependent on autophagy, mitophagy, and mitochondrial dynamics. The goal of this work was to test if removal would also be detectable in a mammalian cell culture system, and elucidate the effect of UVC exposure on mitophagy and mitochondrial function and dynamics. A cell culture system was utilized to address these goals because of the ease of mitochondrial and autophagy visualization via fluorescence and electron microscopy and availability of high-throughput technologies such as flow cytometry (FACS) to analyze mitochondrial function.
UVC-induced mtDNA damage is removed slowly
First, we sought to determine if removal of UVC-induced photodimers in mtDNA occurred at a similar rate in primary human fibroblasts as it does in C. elegans. Most studies in cell culture have shown the persistence of bulky adducts and UVC-induced photodimers up to 48 h (19, 40, 41). One challenge associated with examining later time points is the potential confounding effect of cell replication on removal quantification due to mtDNA damage dilution. Although mtDNA replication is not dependent on the cell cycle, we wanted to minimize any effect of dilution. Accordingly we blocked nDNA synthesis with excess thymidine (3 mM), a method referred to as a thymidine block (42). Such a block arrests cells in S-phase and has often been used to synchronize cell cultures. BrdU incorporation and propidium iodide-based cell cycle analysis were utilized to test for the effectiveness of the thymidine block. In Figure S1a, the cell cycle profile of 90% confluent cells without the thymidine block displays a typical cell cycle distribution with most cells in G0/G1 (76%) (43). Addition of excess thymidine for 24 h arrested the majority of cells at the beginning of S-phase (Figure S1b). At confluency, nDNA synthesis is very low (~4% of cells positive for BrdU incorporation; Figure S1c) and addition of excess thymidine significantly blocked nDNA synthesis with <1% of cells positive for BrdU incorporation (Figure S1d). At 48 and 72 h after seeding, nDNA synthesis is minimal with or without excess thymidine, presumably due to 100% confluency (Figure S1e).
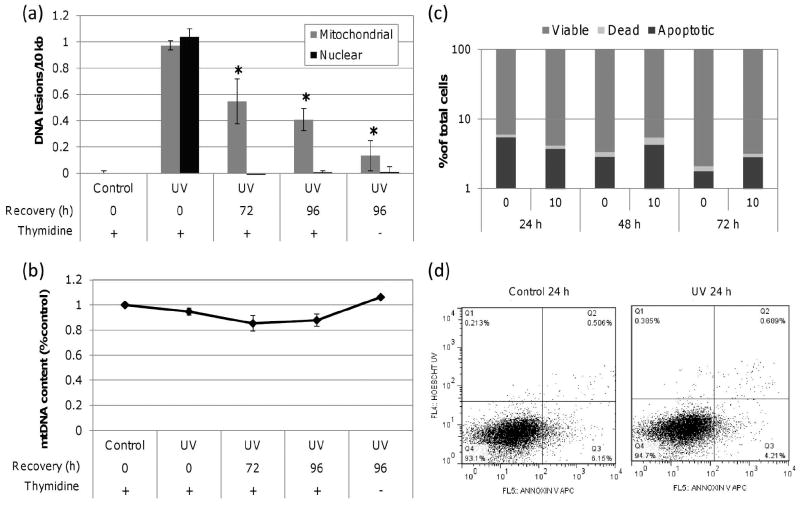
In thymidine blocked cells, we measured UVC-induced DNA lesions in mitochondrial and nuclear DNA at 0, 72 and 96 h post exposure (10 J/m2). As shown in Figure 1a, mtDNA damage was reduced by ~40 % by 72 h and ~60 % by 96 h. There was a nonsignificant trend towards more mtDNA damage removed without the thymidine block. Additionally, the thymidine block did not inhibit nDNA repair (Figure 1a) which was repaired to baseline by 72 h (earlier timepoints were not examined), nor did it inhibit mtDNA replication (Figure 1b). In order to ensure that loss of damage was not the result of cell death, we measured cell viability via FACS analysis of Annexin V APC and Hoescht 33258 cell uptake. Annexin V APC is an indicator of early apoptosis and Hoescht 33258 is an indicator of late apoptotic or necrotic cell death. Lack of a significant increase in the proportion of cells positive for the above markers in control or treated cells led us to conclude that cell viability was not diminished at 24, 48 or 72 h post exposure (Figure 1c-d).
Figure 1. UVC-induced mtDNA damage is removed slowly.
(a) Significant mtDNA damage removal was observed in cells dosed with 10 J/m2 UVC by 72 h compared to initial lesion frequency (one-way ANOVA, effect of time point P = 0.0140; Fisher’s PLSD, P = 0.0316 for 72 h and P = 0.0047 for 96 h). nDNA damage was repaired to baseline by 72 h. (b) mtDNA content was comparable to control throughout the recovery period. (c) No significant decrease in cell viability was observed in cells exposed to 10 J/m2 as measured by Annexin V APC and Hoechst 33258. (d) Representative control and UVC dot plots at 24 h post exposure as measured by FACS with viable (Q4), apoptotic (Q3) and dead (Q2) cell frequencies defined. Bars ± s.e.m.
Autophagy is induced by UVC exposure within 24 h
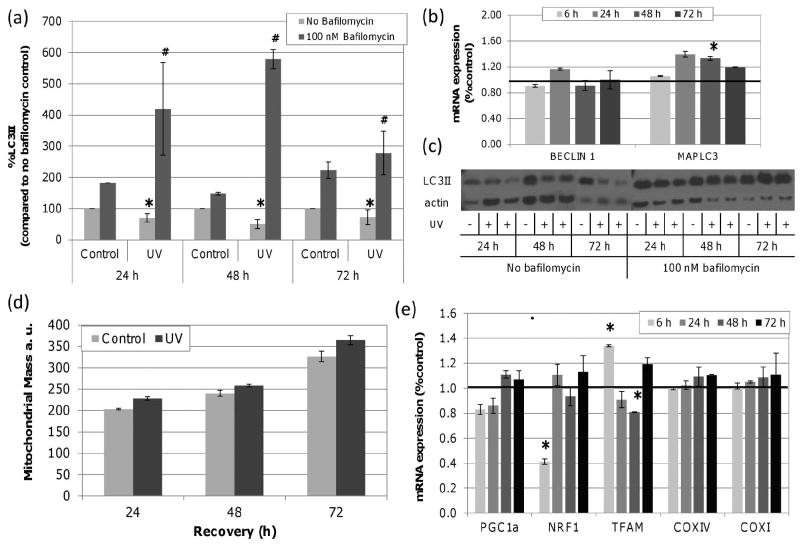
The basal half-life of mtDNA is estimated to be 2-4 days (44) (45, 46). Therefore, the removal of damaged mtDNA observed may reflect natural turnover rather than the result of induced degradation via autophagy. Microtubule-associated protein 1 light chain 3 (MAPLC3) II is incorporated into autophagic membranes and conversion of LC3 to LC3II upon phosphatidylethanolamine conjugation is commonly used as an indicator of autophagy (47). We measured LC3II protein in control and UVC exposed cells. LC3II was significantly lower in UVC exposed cells compared to controls at each time point (Figure 2a); however, these data reflect the steady-state level of LC3II protein which results from both formation and degradation of LC3II. Addition of bafilomycin prevents autophagosomal and lysosomal fusion and allows for the measurement of LC3II production specifically (i.e., autophagic flux) (48). UVC exposed cells had significantly higher LC3II production compared to control samples indicating that lower steady state LC3II in UVC exposed cells was the result of rapid LC3II degradation and that autophagy was induced. In addition, gene expression of MAPLC3, which encodes LC3, and BECN1, which encodes the autophagy protein, BECLIN1 required in the early stages of autophagosomes nucleation, were measured. In accordance with the protein data, we observed a small but significant 1.3 fold induction in MAPLC3 expression (Figure 2b); however, there was no significant change in the expression of BECN1.
Figure 2. UVC exposure increases LC3II formation without changes in mitochondrial mass.
LC3II protein was quantified by Western blot in control and UVC treated cells with and without bafilomycin and normalized to beta-actin. (a) Steady-state (no bafilomycin) levels of LC3II were significantly decreased by UVC exposure at 24, 48 and 72 h when compared to time point controls (asterisks; two-way ANOVA, effect of treatment P = 0.0325). With bafilomycin, UVC exposed cells accumulated more LC3II compared to bafilomycin treated controls at all time points (hash; two-way ANOVA, effect of treatment P = 0.007). (b) MAPLC3 mRNA expression increased by 1.3 fold with UVC exposure 48 h after exposure (asterisks; two-way ANOVA, treatment × time point P = 0.03; Fisher’s PLSD, effect of treatment P < 0.0424 at 48 h). The black line signifies baseline expression level. (c) Representative immunoblots of LC3II in control and UVC (10 J/m2) exposed cells with and without bafilomycin at 24, 48 and 72 h. (d) No significant change in mitochondrial mass was detected after UVC exposure compared to controls (two-way ANOVA, effect of recovery P < 0.0001, treatment × recovery P = 0.4730). (e) mRNA expression of PGC1a, NRF1 and TFAM, COXIV and COXI. NRF1 expression decreased at 6 h (P = 0.0466). TFAM expression was induced 1.3 fold at 6 h but reduced by -1.20 fold by 48 h (two-way ANOVA, treatment × time point P = 0.01; Fisher’s PLSD, effect of treatment P < 0.05 at 6 and 48 h). Bars ± s.e.m.
No change in mitochondrial mass or mtDNA content
Mitochondrial mass can serve as an indicator of mitochondrial biogenesis or degradation. To determine if the increase in autophagy was accompanied by altered mitochondrial mass, we used the mitochondrial selective dye Mitotracker Green (MTG) to quantify mitochondrial mass in control and UVC exposed cells at 0, 48 and 72 h post exposure. MTG selectively accumulates in mitochondria independent of mitochondrial MP. While there was a significant increase in mitochondrial mass with time, there was no significant difference between control and UVC exposed cells at any time point (Figure 2d). As described above, mtDNA content did not change significantly over the course of these experiments (Figure 1b). This suggests that either mitochondrial and mtDNA degradation are not elevated in UVC treated cells, or that they are balanced by mitochondrial biogenesis and mtDNA replication.
To further explore a potential biogenic response, we measured the expression of genes that regulate biogenesis including peroxisome proliferator-activated receptor gamma coactivator 1α (PGC1a), nuclear respirator factor 1 (NRF1) and mitochondrial transcription factor A (TFAM), as well as genes regulated by this pathway: mitochondrial proteins cytochrome c oxidase subunit IV (COXIV) and COX subunit I (COXI). PGC1α regulates both mitochondrial biogenesis and mtDNA replication through enhanced NRF1 expression and transcriptional activity (49, 50). TFAM, the downstream target of NRF1, is essential for mtDNA stability and directly influences mtDNA copy number (51, 52). COXIV and COXI expression are mediated by PGC1 α (53). We observed statistically significant changes in NRF1 and TFAM gene expression over 48 h but these were few and small (Figure 2e). Taken together, our data indicate that mitochondrial biogenesis was not dramatically induced or repressed by 10 J/m2 UVC exposure at least at the level of gene expression.
Increased lysosomal degradation of mitochondrial at 72 h post exposure
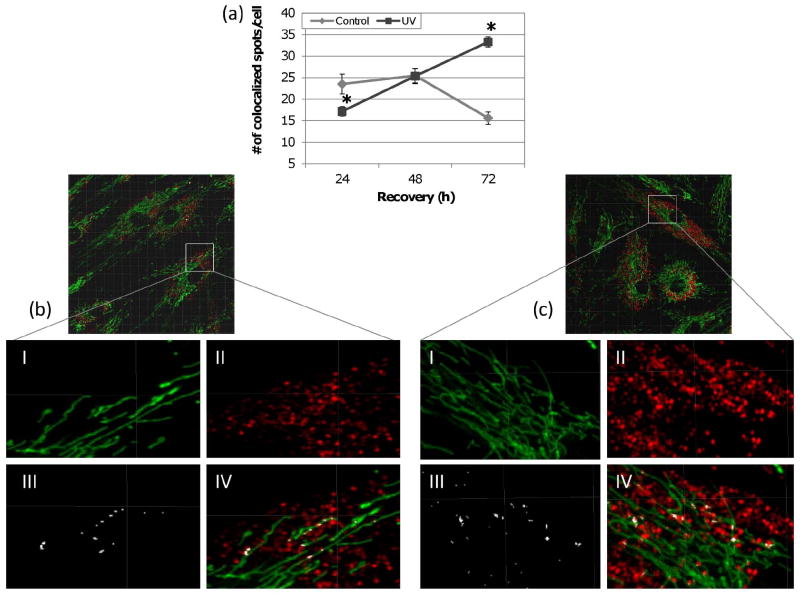
Given that mitochondrial mass and copy number were not affected by UVC exposure, we questioned whether increased autophagy resulted in increased mitochondrial degradation. To evaluate this, lysosomal degradation of mitochondria was captured via live-cell fluorescence microscopy. Lysotracker Red (LTR; 50 nM) and MTG (300 nM) stained cells were incubated with protease inhibitors, leupeptin and pepstatin A, to prevent mitochondrial degradation within the lysosomes. Colocalization of lysosomes and mitochondria was quantified using Imaris Colocalization. In UVC exposed cells, the number of colocalized spots was significantly lower at 24 h, returned to control level by 48 h and was significantly higher at 72 h (Figure 3). This indicates that initially mitochondrial degradation is reduced following UVC exposure but gradually increases and exceeds baseline mitochondrial degradation by 72 h.
Figure 3. Increased lysosomal degradation of mitochondria after exposure to UVC.
(a) The number of colocalized spots between mitochondria and lysosomes was initially lower at 24 h but significantly exceeded that of control by 72 h (two-way ANOVA, treatment × time point P < 0.0001; Fisher’s PLSD, effect of treatment at 24 h P = 0.0068 and 72 h P < 0.0001). Representative images displaying (I) MTG stained mitochondria, (II) LTR stained lysosomes, (III) colocalized spots and (IV) an overlay of all three as detected by Imaris 3D colocalization analysis in (b) control and (c) UVC treated cells. Bars ± s.e.m.
No significant change in mitochondrial ROS production or membrane potential
Others have demonstrated that targeting of mitochondria for degradation can be mediated by mitochondrial MP and ROS. We tested whether UVC exposure affected mitochondrial MP or level of mitochondrial ROS considering that later onset of dysfunction may account for the later onset of mitochondrial degradation at 72 h. FACS analysis of tetramethylrhodamine ethyl ester (TMRE) and Mitotracker Red CMH2XROS fluorescence were employed. TMRE selectively accumulates in mitochondria based on membrane potential and is widely used for mitochondrial labeling and MP quantification (54). Of the available fluorescent ROS indicators, Mitotracker Red CMH2XROS has been shown to have the most mitochondrial specificity and exhibits relatively high sensitivity (55). Compared to control, there was no significant change in MP or mitochondrial ROS in UVC exposed cells at any time point (Figure S2), after normalizing to total mitochondrial mass. These data do not support the hypothesis that autophagy was induced by decreased MP or increased ROS production.
Mitochondrial morphology is not affected by UVC exposure
Slow-onset or low level mitochondrial dysfunction can trigger mitochondrial fusion which restores function (56-59) and in many cases inhibits degradation (60, 61). We considered that increased mitochondrial interconnectivity might mask low level dysfunction caused by UVC treatment and modulate mitochondrial degradation. Therefore, mitochondrial morphology was analyzed in both treatments at all time points via live-cell fluorescence microscopy and MTG staining. Morphometric analysis was based on Koopman et al. (62) with the following modifications to facilitate 3D analysis. To evaluate the effect of UVC on mitochondrial interconnectivity and elongation, the number of surfaces (i.e., mitochondria) per cell as well as the inverse sphericity and surface area/volume ratio of each surface were compared between treatments. Inverse sphericity acts as a measure of both elongation and branching (1=perfect sphere) (62). Surface area/volume ratio decreases with size but, within a specific size range, increases with elongation and branching. Size distribution among the mitochondrial population of each cell was heavily skewed toward small mitochondria in these cells such that such that taking the average of these morphology parameters per cell would fail to accurately capture variation of the mitochondrial population and would reduce the sensitivity to detect slight treatment differences (Figure S3). Therefore, we chose to classify mitochondria by volume (size) into the following groups: 0.1-10 μm3 (Class I), 10-100 μm3 (Class II) and <100 μm3 (Class III) and the number of mitochondria, average surface area/volume and average inverse sphericity were quantified per class for each cell.
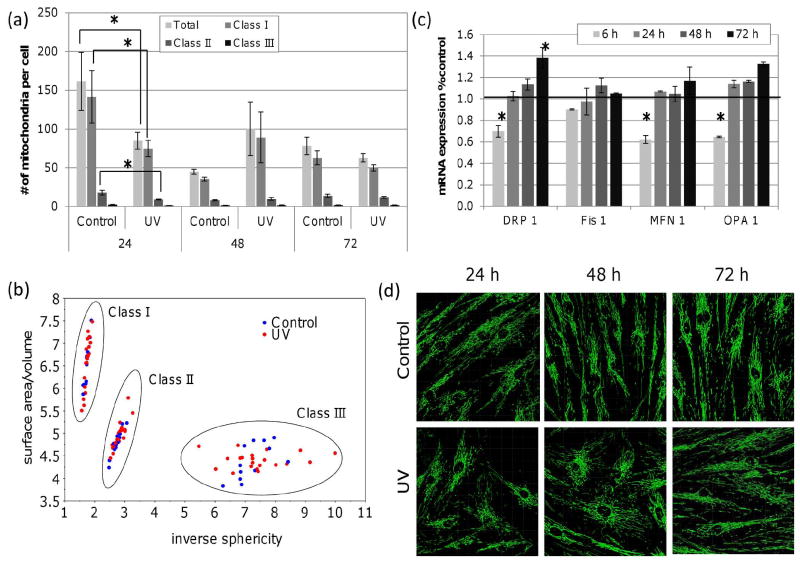
Figure 4a displays the average number of mitochondria per cell within each class for each treatment and time point. There was initially a decrease in the number of mitochondria per cell in UVC treated cells at 24 h. This reflected primarily a decrease in class I and II mitochondria therefore there were less small mitochondria. This could result from an increase in mitochondrial fusion or fewer mitochondria overall. However, as shown above, mitochondrial mass was unaffected by UVC exposure. At later time points, there was no significant effect of UVC on the total number of mitochondria per cell or the size distribution of mitochondria. UVC exposure did not significantly affect mitochondrial elongation or interconnectivity as measured by inverse sphericity or surface area/volume ratio at any time point within any class (Figure 4b). Therefore, we concluded that UVC treatment of cells was not associated with significant alteration in mitochondrial morphology.
Figure 4. No detectable change in mitochondrial number or mitochondrial morphology in UV exposed cells.
(a) The total number of mitochondria per cell was decreased 24 h after UVC exposure (two-way ANOVA, treatment × time point P = 0.0143; Fisher’s PLSD, effect of treatment at 24 h in P = 0.0345), particularly in class I and II sized mitochondria (three-way ANOVA, treatment × time point × class P = 0.0034, two-way ANOVA per class, treatment × time point P < 0.05; Fisher’s PLSD effect of treatment at 24 h P < 0.05). By 48 h mitochondrial number and size distribution had returned to control levels. (b) Scatterplot displaying the average surface area/volume as compared to the average inverse sphericity of mitochondria in each size class. Mitochondrial interconnectivity and elongation (inverse sphericity or surface area/volume) were not affected by UVC exposure at any time point in any class (three-way ANOVA, treatment × time point × class P > 0.05). (c) A small but significant ~40 % reduction in DRP1, MFN1 and OPA1 expression was observed within 6 h of UVC exposure. (d) Representative images of control and UVC exposed mitochondria stained with MTG. Bars ± s.e.m.
We also measured the mRNA levels of MFN1, OPA1, DRP1 and FIS1 which encode proteins necessary for mitochondrial fusion and fission. At 6 hr post UVC exposure, OPA1, MFN1 and DRP1 gene expression were significantly reduced (-1.5, -1.6 and -1.4 fold, respectively; Figure 4c). A reduction in both fusion and fission gene expression could reflect an overall decrease in mitochondrial dynamics and this would have minimal effects on mitochondrial morphology since it reflects the balance between fusion and fission. At 24 hr, endogenous expression was restored in all cases and by 72 hr post exposure, expression of DRP1 and OPA1 genes was elevated, significantly so in the case of DRP1.
Mitochondrial degradation via autophagy increases during recovery
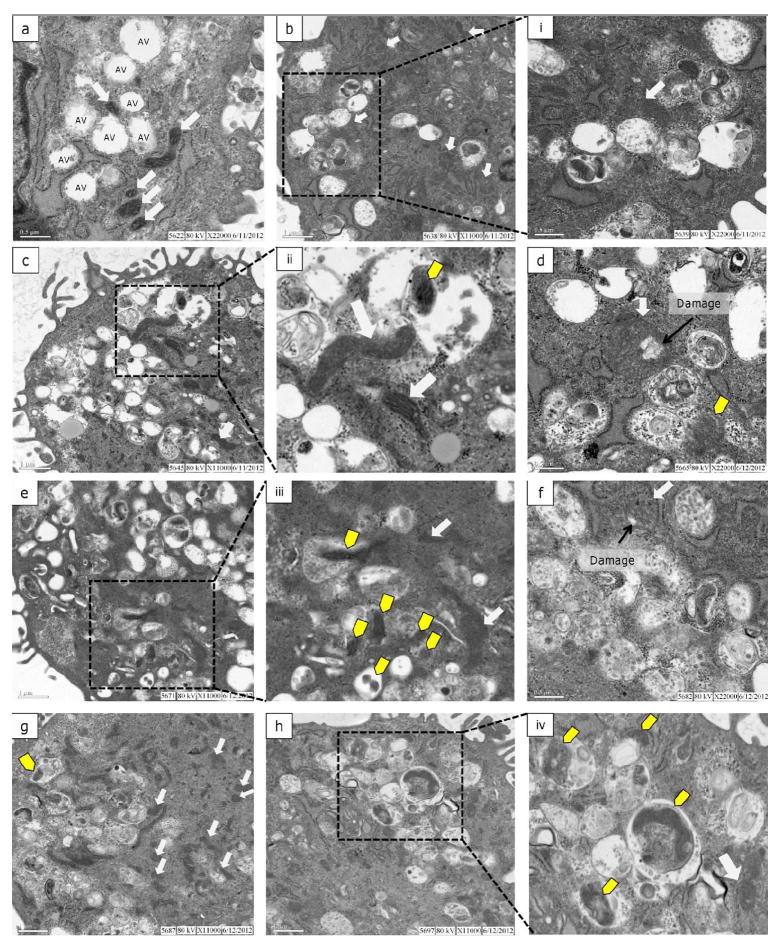
The mitochondrial/lysosomal colocalization data described above suggests that mitochondrial degradation increased at 72 h post exposure. While this approach provides detailed 3D data it does not lend information regarding the specificity of the degradation process (autophagy vs. mitophagy) since other cellular components potentially present in the lysosomes are not visualized. Therefore, we next investigated mitochondrial degradation using TEM which allows for the visualization of autophagic vacuoles and differentiation of their specific contents. In order to capture autophagosomes and their contents prior to degradation, bafilomycin was added three hours before cell fixation to inhibit fusion of autophagosomes and lysosomes.
As shown in Figure 5a, control cells without bafilomycin contained several AVs. Mitochondria in these cells were numerous, well-defined and dense. As expected, AVs representing multiple stages of autophagy including multivesicular endosomes, early and late autophagosomes and lysosomes were present in bafilomycin-treated cells. The occurrence of AVs containing recognizable mitochondria was low in control cells and mitochondria in bafilomycin-treated control cells were less dense and had less defined cristae (Figure 5b). In UVC-exposed cells mitochondrial ultrastructure was variable ranging from long, dense mitochondria with well-defined cristae to rounded, enlarged mitochondria with less well-defined membranes and cristae (Figure 5c-h). In two cases, mitochondria in UVC exposed cells revealed a focal area of low electron density signifying swelling of the matrix (Figure 5d, f). There was a trend toward increasing occurrence of AVs containing mitochondria at 48 and 72 h post exposure (Figure 5e, h). Of these, many contained other cytoplasmic components. Additionally, there were several AVs without mitochondria that contained general cytoplasmic contents such as glycogen and membranous tubules. These observations support a UVC-induced increase in non-specific autophagy leading to increased mitochondrial degradation later in recovery.
Figure 5. Autophagic degradation of mitochondria is enhanced during recovery.
Electron micrographs of control (a), control + Baf (b), UV 24 h + Baf (c-d), UV 48 h + Baf (e-f) and UV 72 h + Baf (g-h). (a) Control cells without bafilomycin had autophagic vacuoles (AV) that corresponded to multivesicular endosomes (MVE) or early autophagosomes, and several mitochondria with defined cristae (solid white arrows). (b) Mitochondria in control cells with bafilomycin were less defined and several AVs representing MVEs, autophagosomes and lysosomes were present. (i) Higher magnification (22,000 ×) of a mitochondrion highlighting a lack of electron density and cristae definition. In UVC-exposed (10 J/m2) cells a range of mitochondrial phenotypes was observed from dense mitochondria with well-defined cristae to enlarged mitochondria with less density and poorly defined cristae. At 24 and 48 h (d,f) large round mitochondria were observed with areas of low electron density suggesting damaged or swollen inner membrane. Autophagosomes containing mitochondria (yellow arrows) were observed at 24, 48 and 72 h with an increasing trend in frequency at 48 and 72 h. Several autophagosomes containing general cytoplasmic materials were also observed throughout the recovery period.
DISCUSSION
We recently found that in adult C. elegans ~40% of UVC-induced mtDNA damage was removed within 72 h of exposure (36). We induced a similar number of mtDNA lesions in non-replicating human fibroblasts and found that damage removal occurred at a similar rate with ~40% removal by 72 h post exposure suggesting that the kinetics of removal are conserved between lower eukaryotes and mammals, at least in non-replicating cells.
UVC exposure induced autophagy, as previously reported in cell culture (33) and in C. elegans (36). Interestingly, our data suggest that mitochondrial degradation is initially inhibited at 24 h post exposure despite elevated autophagy but recovers and exceeds control levels by 72 h. Despite this correlation between the timing of mitochondrial degradation and the timing of detectable damage removal, we cannot know with certainty that autophagy is removing damaged mtDNA without inhibiting autophagy. A lag time between induction of autophagy and mitochondrial degradation has recently been reported by others (63-65) such as during amino acid starvation (63) and has been attributed to enhanced mitochondrial elongation (64, 65). Interestingly, at 24 h when mitochondrial degradation was suppressed, we observed a corresponding decrease in smaller sized mitochondria suggesting increased fusion. However, mitochondrial elongation and interconnectivity at 24 h were not affected at least at the level that we could detect. Thus an exciting future direction is to investigate directly the effect of UVC on fusion and fission events.
Although ~80% of mtDNAs are expected to contain at least one lesion at the level of mtDNA damage induced in the present study, the effects of this damage appear to be within the compensatory range of the mitochondrial population such that mtDNA damage was removed with no significant perturbation of mitochondrial mass, mtDNA content, MP or ROS level. However, it is likely that mtDNAs harboring UVC-induced photodimers are distributed randomly and unequally during fission events resulting in a subset of mitochondria with a higher proportion of damaged DNA and potentially compromised mitochondrial function. MP and ROS analyses performed here reflect an average among all mitochondria within a cell; thus, effects in a subset of mitochondria cannot be ruled out.
We observed an increasing trend in AVs containing mitochondria in UVC treated cells at 48 and 72 h post exposure; however, the contents of AVs overall appeared to be heterogeneous. Therefore, non-specific autophagy rather than more specific mitophagy appears to be mediating mitochondrial degradation following UVC exposure. However, as discussed above (65, 66), even a non-specific autophagic response may be selective at some level for dysfunctional mitochondria so it is possible that dysfunctional mitochondria may be degraded preferentially thus facilitating mtDNA damage removal.
An effect of damage to nDNA on autophagy and mitochondrial fate is also conceivable even though UVC is relatively nucleic acid-specific and nDNA damage is repaired relatively quickly. Autophagy is induced by UVC-induced activation of DNA damage responsive genes ATR, ATM and p53 (67-69) and ATM-deficient cells exhibit defective mitophagy (70). However, it was recently shown that autophagy induction following UVC exposure proved temporally dependent. For example, ATR was dependent up to 6 h but ATR independent at 24 and 48 h (33). Further research is needed to understand the role of DNA damage responsive genes in mediating mitochondrial turnover by autophagy and mitophagy.
Supplementary Material
Acknowledgments
The work was supported by the National Institute of Environmental Health Sciences [P42 ES010356-10A2, 1R01-ES017540-01A2.
References
- 1.Nelson DL, Cox MM. Principles of Biochemistry. Fourth. New York City: W.H. Freeman and Company; 2005. [Google Scholar]
- 2.Scheffler IE. Mitochondria. second. Hoboken: John Wiley and Sons; 2008. [Google Scholar]
- 3.Greaves LC, et al. Mitochondrial DNA and Disease. J Pathol. 2011 doi: 10.1002/path.3028. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18(R2):R169–76. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissman L, et al. DNA repair, mitochondria, and neurodegeneration. Neuroscience. 2007;145(4):1318–29. doi: 10.1016/j.neuroscience.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 6.Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science. 2005;307(5708):384–7. doi: 10.1126/science.1104343. [DOI] [PubMed] [Google Scholar]
- 7.Jin S. Autophagy, mitochondrial quality control, and oncogenesis. Autophagy. 2006;2(2):80–4. doi: 10.4161/auto.2.2.2460. [DOI] [PubMed] [Google Scholar]
- 8.Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25(34):4647–62. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- 9.Terman A, et al. Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxid Redox Signal. 2010;12(4):503–35. doi: 10.1089/ars.2009.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balansky R, et al. Induction by Carcinogens and Chemoprevention by N-Acetylcysteine of Adducts to Mitochondrial DNA in Rat Organs. Cancer Res. 1996;56(7):1642–1647. [PubMed] [Google Scholar]
- 11.Allen JA, Coombs MM. Covalent binding of polycyclic aromatic compounds to mitochondrial and nuclear DNA. Nature. 1980;287:244–245. doi: 10.1038/287244a0. [DOI] [PubMed] [Google Scholar]
- 12.Stairs PW, Guzelian PS, Van Tuyle GC. Benzo[a]pyrene differentially alters mitochondrial and nuclear DNA synthesis in primary hepatocyte cultures. Res Commun Chem Pathol Pharmacol. 1983;42(1):95–106. [PubMed] [Google Scholar]
- 13.Backer JM, Weinstein IB. Mitochondrial DNA is a major cellular target for a dihydrodiol-epoxide derivative of benzo[a]pyrene. Science. 1980;209(4453):297–9. doi: 10.1126/science.6770466. [DOI] [PubMed] [Google Scholar]
- 14.Niranjan BG, Bhat NK, Avadhani NG. Preferential attack of mitochondrial DNA by aflatoxin B1 during hepatocarcinogenesis. Science. 1982;215(4528):73–5. doi: 10.1126/science.6797067. [DOI] [PubMed] [Google Scholar]
- 15.Bedard LL, Massey TE. Aflatoxin B1-induced DNA damage and its repair. Cancer Lett. 2006;241(2):174–83. doi: 10.1016/j.canlet.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 16.Jeong YC, et al. Pyrimido[1,2-a]-purin-10(3H)-one, M1G, is less prone to artifact than base oxidation. Nucleic Acids Res. 2005;33(19):6426–34. doi: 10.1093/nar/gki944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Podratz JL, et al. Cisplatin induced mitochondrial DNA damage in dorsal root ganglion neurons. Neurobiol Dis. 2011;41(3):661–8. doi: 10.1016/j.nbd.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salazar I, et al. The effect of benzo[a]pyrene on DNA synthesis and DNA polymerase activity of rat liver mitochondria. FEBS Lett. 1982;138(1):45–9. doi: 10.1016/0014-5793(82)80391-8. [DOI] [PubMed] [Google Scholar]
- 19.Clayton D, Doda J, Friedberg E. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proceedings of the National Academy of Sciences. 1974;71(7):2777. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasiviswanathan R, et al. Human mitochondrial DNA polymerase gamma exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J Biol Chem. 2012 doi: 10.1074/jbc.M111.306852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462(2):245–53. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemasters JJ, et al. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim Biophys Acta. 1998;1366(1-2):177–96. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 24.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8(1):3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- 25.Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777(9):1092–7. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geisler S, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6(7):871–8. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- 27.Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vives-Bauza C, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010;107(1):378–83. doi: 10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vila M, Ramonet D, Perier C. Mitochondrial alterations in Parkinson’s disease: new clues. J Neurochem. 2008;107(2):317–28. doi: 10.1111/j.1471-4159.2008.05604.x. [DOI] [PubMed] [Google Scholar]
- 30.Gil JM, Rego AC. Mechanisms of neurodegeneration in Huntington’s disease. Eur J Neurosci. 2008;27(11):2803–20. doi: 10.1111/j.1460-9568.2008.06310.x. [DOI] [PubMed] [Google Scholar]
- 31.Scheibye-Knudsen M, et al. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J Exp Med. 2012;209(4):855–69. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monick MM, et al. Identification of an autophagy defect in smokers’ alveolar macrophages. J Immunol. 2010;185(9):5425–35. doi: 10.4049/jimmunol.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen LH, et al. Targeting Protective Autophagy Exacerbates UV-Triggered Apoptotic Cell Death. Int J Mol Sci. 2012;13(1):1209–24. doi: 10.3390/ijms13011209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Hernandez A, et al. Coenzyme Q deficiency triggers mitochondria degradation by mitophagy. Autophagy. 2009;5(1):19–32. doi: 10.4161/auto.5.1.7174. [DOI] [PubMed] [Google Scholar]
- 35.Cotan D, et al. Secondary coenzyme Q10 deficiency triggers mitochondria degradation by mitophagy in MELAS fibroblasts. FASEB J. 2011;25(8):2669–87. doi: 10.1096/fj.10-165340. [DOI] [PubMed] [Google Scholar]
- 36.Bess AS, et al. Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dykens JA, Yvonne Will, editors. Drug-Induced Mitochondrial Dysfunction. John Wiley & Sons; New Jersey: 2008. [Google Scholar]
- 38.Furda A, Bess AS, Meyer JN, Van Houten B. Analysis of DNA damage and repair in nuclear and mitochondrial DNA of animal cells using quantitative PCR. In: Bjergbæk L, editor. Methods in Molecular Biology: DNA repair protocols. Springer Science+Business Media; New York, USA: 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venegas V, Halberg MC. Measurement of mitochondrial DNA copy number. Methods Mol Biol. 2012;837:327–35. doi: 10.1007/978-1-61779-504-6_22. [DOI] [PubMed] [Google Scholar]
- 40.Rochette PJ, Brash DE. Human telomeres are hypersensitive to UV-induced DNA Damage and refractory to repair. PLoS Genet. 2010;6(4):e1000926. doi: 10.1371/journal.pgen.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kalinowski DP, Illenye S, Van Houten B. Analysis of DNA damage and repair in murine leukemia L1210 cells using a quantitative polymerase chain reaction assay. Nucleic Acids Res. 1992;20(13):3485–94. doi: 10.1093/nar/20.13.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jackman J, O’Connor PM. Current Protocols in Cell Biology. John Wiley & Sons, Inc; 2001. Methods for Synchronizing Cells at Specific Stages of the Cell Cycle. [DOI] [PubMed] [Google Scholar]
- 43.Ryba T, et al. Genome-scale analysis of replication timing: from bench to bioinformatics. Nat Protocols. 2011;6(6):870–895. doi: 10.1038/nprot.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lipsky NG, Pedersen PL. Mitochondrial turnover in animal cells. Half-lives of mitochondria and mitochondrial subfractions of rat liver based on [14C]bicarbonate incorporation. J Biol Chem. 1981;256(16):8652–7. [PubMed] [Google Scholar]
- 45.Miwa S, Lawless C, von Zglinicki T. Mitochondrial turnover in liver is fast in vivo and is accelerated by dietary restriction: application of a simple dynamic model. Aging Cell. 2008;7(6):920–3. doi: 10.1111/j.1474-9726.2008.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kai Y, et al. Rapid and random turnover of mitochondrial DNA in rat hepatocytes of primary culture. Mitochondrion. 2006;6(6):299–304. doi: 10.1016/j.mito.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 47.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4(2):151–75. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubinsztein DC, et al. In search of an “autophagomometer”. Autophagy. 2009;5(5):585–9. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 49.Lee HC, Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37(4):822–34. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 50.Wu Z, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 51.Kang D, Kim SH, Hamasaki N. Mitochondrial transcription factor A (TFAM): roles in maintenance of mtDNA and cellular functions. Mitochondrion. 2007;7(1-2):39–44. doi: 10.1016/j.mito.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 52.Larsson NG, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18(3):231–6. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- 53.Hock MB, Kralli A. Transcriptional Control of Mitochondrial Biogenesis and Function. Annual Review of Physiology. 2009;71(1):177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 54.Chazotte B. Labeling mitochondria with TMRM or TMRE. Cold Spring Harb Protoc. 2011;2011(7):895–7. doi: 10.1101/pdb.prot5641. [DOI] [PubMed] [Google Scholar]
- 55.Kuznetsov AV, et al. Mitochondrial ROS production under cellular stress: comparison of different detection methods. Anal Bioanal Chem. 2011;400(8):2383–90. doi: 10.1007/s00216-011-4764-2. [DOI] [PubMed] [Google Scholar]
- 56.Ashley N, Poulton J. Anticancer DNA intercalators cause p53-dependent mitochondrial DNA nucleoid re-modelling. Oncogene. 2009;28(44):3880–3891. doi: 10.1038/onc.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tondera D, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. Embo J. 2009;28(11):1589–600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koopman WJH, et al. Inhibition of complex I of the electron transport chain causes O2·-mediated mitochondrial outgrowth. American Journal of Physiology - Cell Physiology. 2005;288(6):C1440–C1450. doi: 10.1152/ajpcell.00607.2004. [DOI] [PubMed] [Google Scholar]
- 59.Zanchetta LM, et al. Mitophagy and mitochondrial morphology in human melanoma-derived cells post exposure to simulated sunlight. Int J Radiat Biol. 2011;87(5):506–17. doi: 10.3109/09553002.2011.556175. [DOI] [PubMed] [Google Scholar]
- 60.Parone PA, et al. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS ONE. 2008;3(9):e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Twig G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27(2):433–46. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koopman WJ, et al. Mitochondrial network complexity and pathological decrease in complex I activity are tightly correlated in isolated human complex I deficiency. Am J Physiol Cell Physiol. 2005;289(4):C881–90. doi: 10.1152/ajpcell.00104.2005. [DOI] [PubMed] [Google Scholar]
- 63.Kristensen AR, et al. Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics. 2008;7(12):2419–28. doi: 10.1074/mcp.M800184-MCP200. [DOI] [PubMed] [Google Scholar]
- 64.Gomes LC, Benedetto GD, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13(5):589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rambold AS, et al. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A. 2011;108(25):10190–5. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim I, Lemasters JJ. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am J Physiol Cell Physiol. 2011;300(2):C308–17. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Unsal-Kacmaz K, et al. Preferential binding of ATR protein to UV-damaged DNA. Proc Natl Acad Sci U S A. 2002;99(10):6673–8. doi: 10.1073/pnas.102167799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hurley PJ, Bunz F. ATM and ATR: components of an integrated circuit. Cell Cycle. 2007;6(4):414–7. doi: 10.4161/cc.6.4.3886. [DOI] [PubMed] [Google Scholar]
- 69.Suzuki K, Kodama S, Watanabe M. Recruitment of ATM protein to double strand DNA irradiated with ionizing radiation. J Biol Chem. 1999;274(36):25571–5. doi: 10.1074/jbc.274.36.25571. [DOI] [PubMed] [Google Scholar]
- 70.Valentin-Vega YA, et al. Mitochondrial dysfunction in ataxia-telangiectasia. Blood. 2012;119(6):1490–500. doi: 10.1182/blood-2011-08-373639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.