Here, a case study of the effects of cryoprotectants on the kinetics of carbonic anhydrase II (CA II) and its inhibition by the clinically used inhibitor acetazolamide (AZM) is presented.
Keywords: carbonic anhydrase, acetazolamide, cryoprotectants, glycerol, sucrose
Abstract
Protein X-ray crystallography has seen a progressive shift from data collection at cool/room temperature (277–298 K) to data collection at cryotemperature (100 K) because of its ease of crystal preparation and the lessening of the detrimental effects of radiation-induced crystal damage, with 20–25%(v/v) glycerol (GOL) being the preferred choice of cryoprotectant. Here, a case study of the effects of cryoprotectants on the kinetics of carbonic anhydrase II (CA II) and its inhibition by the clinically used inhibitor acetazolamide (AZM) is presented. Comparative studies of crystal structure, kinetics, inhibition and thermostability were performed on CA II and its complex with AZM in the presence of either GOL or sucrose. These results suggest that even though the cryoprotectant GOL was previously shown to be directly bound in the active site and to interact with AZM, it affects neither the thermostability of CA II nor the binding of AZM in the crystal structure or in solution. However, addition of GOL does affect the kinetics of CA II, presumably as it displaces the water proton-transfer network in the active site.
1. Introduction
Over the last ∼20 years, there has been a steady shift in data-collection methods for X-ray protein crystallography from cool/room temperature (277–298 K) to cryotemperature (100 K) in order to minimize the detrimental effects of radiation damage (Garman, 2003 ▶). The exact nature of the damage is still uncertain, but it is speculated that X-ray radiation produces free radicals which induce chemical changes on the surface of a protein (Garman & Owen, 2006 ▶). This disordering can lead to the loss of diffraction quality and perturbations in the native structure, which can result in possibly erroneous conclusions about the biological mechanism and function (Holton, 2009 ▶). Strategies to control radiation damage are limited, but the use of cryoprotectants and freeradical scavengers has been implicated in slowing the damage (Kmetko et al., 2011 ▶). The addition of a cryoprotectant is required prior to flash-cooling the crystal in a nitrogen/helium-cooled gas stream to minimize the formation of ice while vitrifying the crystal (Ravelli & Garman, 2006 ▶), which prevents disruption in the order of the crystal lattice (Chapman & Somasundaram, 2010 ▶). The cryoprotectant is administrated prior to flash-cooling via a cocrystallization or pre-treatment soak of the crystal in a solution consisting of the crystallization reservoir solution and a cryoprotectant, with 20–25%(v/v) glycerol (GOL) being the most popularly used cryoprotectant. However, some salts and low-molecular-weight sugars may also be used to fulfill this purpose. Another class of cryoprotectants used in flash-cooling are oils with low scattering and low optical distortion, including Paratone N, paraffin and high-density mineral oils (Garman & Doublié, 2003 ▶; Riboldi-Tunnicliffe & Hilgenfeld, 1999 ▶). Nevertheless, their use is limited and remains overshadowed by GOL. The choice of which cryoprotectant to use is important because it may have deleterious effects on the crystal mosaicity, which may make data reduction and processing more difficult or may affect the active site or a loop conformation of the protein, causing further possible misinterpretation of the structure.
Carbonic anhydrases (CAs) are metalloenzymes that catalyze the reversible hydration/dehydration of CO2/HCO3 −. Humans have 12 active zinc-containing CA isoforms that are expressed in diverse tissues and exhibit various cellular locations, being cytosolic, transmembrane or membrane-bound (Gilmour, 2010 ▶; Aggarwal, Boone et al., 2013 ▶). Human CA II (CA II) is a monomeric ∼29 kDa soluble cytosolic isoform that is found predominantly in red blood cells and has a fast catalytic turnover (∼106 s−1) compared with most other CAs characterized to date (Silverman & McKenna, 2007 ▶). The first step of the ping-pong catalysis in the hydration direction uses a zinc–hydroxide (Zn–OH−) mechanism to reversibly convert CO2 to HCO3 − and a proton, as shown in the reactions below (Silverman & McKenna, 2007 ▶),
The second reaction involves the transfer of a proton from EZnH2O to the bulk solvent (B) in order to regenerate EZnOH−. This transfer is believed to occur via a network of ordered water molecules (conventionally named w1, w2, w3A and w3B) that extends to the side chain of His64 (Fig. 1 ▶ a). Several studies that have previously been conducted to understand proton transfer, including, but not limited to, mutational analyses of the active-site residues (Jackman et al., 1996 ▶; Fisher et al., 2007 ▶; Zheng et al., 2008 ▶), suggest that a disruption in this ordered water network causes a reduction in CA activity.
Figure 1.

Active sites of (a) CA II, showing ordered waters involved in proton transfer (PDB entry 3ks3; Avvaru et al., 2010 ▶), (b) CA II in complex with AZM (PDB entry 3v2j; this study) and (c) CA II in complex with AZM and GOL (PDB entry 3hs4; Sippel et al., 2009 ▶). Hydrogen bonds are shown as black dotted lines. Waters are colored in gradients of blue from dark (not displaced) to light (AZM displaced) to lighter (GOL displaced). The active-site Zn2+ ion is shown as a magenta sphere. This figure was produced using PyMOL (http://www.pymol.org).
CA II has long been a drug target for the treatment of various diseases such as glaucoma, epilepsy and altitude sickness (Forwand et al., 1968 ▶; Katritzky et al., 1987 ▶; Supuran et al., 2004 ▶; Supuran, 2008 ▶). As such, structures of CA II in complex with various inhibitors have frequently been determined by X-ray crystallography and reported in the PDB (http://www.rcsb.org) and novel techniques of administering already available drugs are being patented (Aggarwal & McKenna, 2012 ▶; Aggarwal, Kondeti et al., 2013 ▶). Since the determination of the first crystal structure of CA II (Liljas et al., 1972 ▶), more than 400 structures of CA II (wild type, variants or in complex with ligands) have been deposited in the PDB (Aggarwal, Kondeti et al., 2013 ▶).
Acetazolamide (AZM) is a classical inhibitor of CA II and has been clinically used in the treatment of glaucoma for decades (Maren, 1967 ▶; Supuran, 2008 ▶; Aggarwal & McKenna, 2012 ▶). It binds and inhibits CA II via the sulfonamide group, which directly interacts with the active-site zinc, displacing the water that is necessary for catalysis. In addition, a further five water molecules (including the proton-transfer water w1) are displaced as the AZM extends out of the active site and makes hydrophobic and polar interactions with the surrounding amino-acid residues of the enzyme (Figs. 1 ▶ a and 1 ▶ b; Sippel et al., 2009 ▶; Avvaru et al., 2010 ▶; Fisher et al., 2012 ▶).
However, many CA II–inhibitor complexes, including CA II–AZM, also show an ordered GOL bound in the active site, a consequence of using GOL as the cryoprotectant during X-ray diffraction data collection. This phenomenon has been observed in more than 40 crystal structures deposited in the PDB. A previously reported high-resolution (1.1 Å) CA II–AZM crystal complex clearly showed the GOL bound adjacent to the AZM ring, displacing a further three water molecules (including the waters w2 and w3B) compared with the uncomplexed CA II structure, with the GOL making seven hydrogen bonds (defined as a proton donor/acceptor distance of between 2.4 and 3.4 Å) to Asn62, Asn67 and Gln92 in the active site and the three remaining water molecules (compare Fig. 1 ▶ c with Figs. 1 ▶ a and 1 ▶ b). As mentioned, a GOL molecule has been observed in this location in the active site in several other high-resolution CA II–inhibitor complexes (Jude et al., 2006 ▶; Srivastava et al., 2007 ▶). Hence, this relatively common observation has raised the question of whether the GOL aids or hinders the inhibitor binding and/or affects the location and interactions that these inhibitors make in the active site of CA II, thereby affecting the interpretation and subsequent drug design (Fig. 1 ▶).
To address this issue, the catalytic activity, melting temperature (T m) and inhibition constant (K i) of AZM for CA II were measured (i) in the presence of GOL, (ii) in the presence of sucrose and (iii) in buffer as a control. In addition, crystal structures of CA II–AZM complex were determined at (i) room temperature (298 K) and (ii) cryotemperature (100 K) using sucrose as a cryoprotectant and compared with previously reported high-resolution cryo-structures of AZM-complexed (Sippel et al., 2009 ▶) and uncomplexed CA II in the presence of GOL (Avvaru et al., 2010 ▶). The results suggest that GOL does not affect either the thermostability or the inhibitor binding of CA II. However, GOL does affect the kinetics of CA II, presumably as it displaces the solvent in the active site.
2. Experimental procedures
2.1. Expression and purification of CA II
The recombinant gene for CA II was cloned into a pET-32(b) plasmid vector with an ampicillin-resistance gene and expressed in Escherichia coli BL21 (DE3) cells. The culture was grown at 310 K in the presence of ampicillin (100 µg ml−1) until it reached an OD600 of 0.6 and was thereafter induced with IPTG (100 µg ml−1) for protein expression. The lysate was purified by affinity chromatography using a p-aminomethylbenzene sulfonamide column. Nonspecifically bound proteins were washed off the column using buffers (200 mM sodium sulfate) at pH 7.0 and 9.0, finally eluting the protein with elution buffer (400 mM sodium azide). The enzyme was then buffer-exchanged to remove sodium azide and concentrated to 15 mg ml−1 (450 µM) using a 10 kDa filter.
2.2. Cocrystallization and X-ray data collection of the CA II–AZM complex
Cocrystals of the CA II–AZM complex were obtained using the hanging-drop vapor-diffusion method. Drops of 10 µl [0.3 mM CA II, 0.7 mM AZM, 0.1%(v/v) DMSO, 0.8 M sodium citrate, 50 mM Tris–HCl pH 8.0] were equilibrated against the precipitant solution (1.6 M sodium citrate, 50 mM Tris–HCl pH 8.0) at room temperature (298 K). Crystals were observed after 5 d. Based on visual selection, two crystals were selected for diffraction studies. For one crystal, data collection was performed at RT (298 K) mounted in a quartz capillary; the other crystal was looped and cryoprotected by quick immersion into 20%(w/v) sucrose precipitant solution and flash-cooled by exposure to a gaseous stream of liquid nitrogen at 100 K. X-ray diffraction data were collected in-house (263 images for the RT crystal and 207 images for the cryoprotected crystal) using an R-AXIS IV++ image-plate system on a Rigaku RU-H3R Cu rotating anode (λ = 1.5418 Å) operating at 50 kV and 22 mA with Osmic Varimax HR optics. The crystal-to-detector distance was set to 80 mm and the oscillation steps were 1° with 5 min exposure per image for both data sets.
2.3. Structure determination
The diffraction data were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). Starting phases were calculated from PDB entry 3ks3 (Avvaru et al., 2010 ▶) with waters removed. The PHENIX package (Adams et al., 2010 ▶) was used for refinement, with 5% of the unique reflections randomly selected and excluded from the refinement data set for the purpose of R free calculations (Brünger, 1992 ▶). Manual refitting of the model was performed in Coot (Emsley & Cowtan, 2004 ▶). Complete refinement statistics and model quality are included in Table 1 ▶.
Table 1. Crystallographic details of CA II–AZM structures.
Values in parentheses are for the highest resolution bin.
| RT | Cryo-sucrose | |
|---|---|---|
| PDB code | 3v2j | 3v2m |
| Data-collection statistics | ||
| Temperature (K) | 298 | 100 |
| Wavelength (Å) | 1.5418 | 1.5418 |
| Space group | P21 | P21 |
| Unit-cell parameters (Å) | ||
| a (Å) | 42.8 | 42.2 |
| b (Å) | 41.7 | 41.3 |
| c (Å) | 72.9 | 72.1 |
| β (°) | 104.5 | 104.2 |
| Total theoretical reflections | 27697 | 41243 |
| Unique measured reflections | 26188 | 40468 |
| Resolution (Å) | 19.0–1.7 (1.76–1.69) | 19.9–1.5 (1.50–1.47) |
| R sym † (%) | 8.9 (43.2) | 4.5 (19.8‡) |
| 〈I/σ(I)〉 | 12.0 (3.5) | 23.4 (6.5) |
| Completeness (%) | 94.6 (91.1) | 98.1 (96.2) |
| Multiplicity | 4.4 (4.4) | 3.9 (3.7) |
| Final model statistics | ||
| R cryst § (%) | 12.9 (19.0) | 16.3 (41.5‡) |
| R free ¶ (%) | 15.6 (21.5) | 18.7 (45.3‡) |
| Residues | 4–261 | 4–261 |
| No. of atoms†† | ||
| Protein | 2325 | 2261 |
| Drug | 13 | 13 |
| Water | 148 | 242 |
| R.m.s.d. | ||
| Bond lengths (Å) | 0.01 | 0.01 |
| Bond angles (°) | 1.27 | 1.34 |
| Ramachandran statistics (%) | ||
| Most favored | 89.4 | 89.4 |
| Allowed | 10.6 | 10.6 |
| Outliers | 0.0 | 0.0 |
| Average B factors (Å2) | ||
| Main chain | 16.4 | 13.3 |
| Side chain | 24.0 | 17.5 |
| Inhibitor (AZM) | 15.6 | 22.3 |
| Solvent | 32.4 | 25.4 |
R
sym = 
 .
.
Although the cryo-sucrose structure has a low R sym in the highest resolution bin, the R cryst and R free for this bin are high. This is probably owing to the presence of some diffused unassigned density (most likely sucrose) causing under-refinement of the structure.
R
cryst =  −
− 
 × 100.
× 100.
R free is calculated in the same manner as R cryst except that it uses 5% of the reflection data, which were omitted from refinement.
Includes alternate conformations.
2.4. Differential scanning calorimetry (DSC)
DSC experiments were performed using a VP-DSC calorimeter (Microcal Inc., North Hampton, Massachusetts, USA) with a cell volume of approximately 0.5 ml. All CA II–AZM samples were buffered in 50 mM Tris–HCl pH 8.0 at protein concentrations of ∼30 µM. CA II, CA II + 20%(v/v) GOL and CA II + 20%(w/v) sucrose were used as controls. Samples were degassed while stirring for at least 10 min prior to data collection. DSC scans were collected from 293 to 373 K at a scan rate of 60 K h−1. The calorimetric enthalpies of unfolding were calculated by integrating the area under the peaks in the thermograms after adjusting the pre-transition and post-transition baselines. The thermograms were fitted to a two-state reversible unfolding model to obtain van’t Hoff enthalpies of unfolding. The melting-temperature (T m) values of the CA II–AZM samples were obtained from the midpoints of the DSC curves, indicating a two-state transition. All samples were measured in triplicate with the buffer baseline subtracted. The results are shown in Table 3.
2.5. Enzyme kinetics measurements
An 18O-exchange assay was carried out to study the kinetics of the catalyzed HCO3 −–CO2 dehydration–hydration reaction and its AZM inhibition (at 283 K) in the presence of 20%(v/v) GOL, 20%(w/v) sucrose and in the absence of either (representing RT data collection). This method relies on the depletion of 18O from CO2 as measured by membrane inlet mass spectrometry. CO2 passing across the membrane enters a mass spectrometer (Extrel EXM-200), providing a continuous measure of the isotopic content of the CO2. In the first stage of catalysis, the dehydration of labeled bicarbonate has a probability of transiently labeling the active site with 18O. In a subsequent step, protonation of the zinc-bound 18O-labeled hydroxide results in the release of H2 18O into the solvent.
3. Results and discussion
3.1. X-ray crystallography
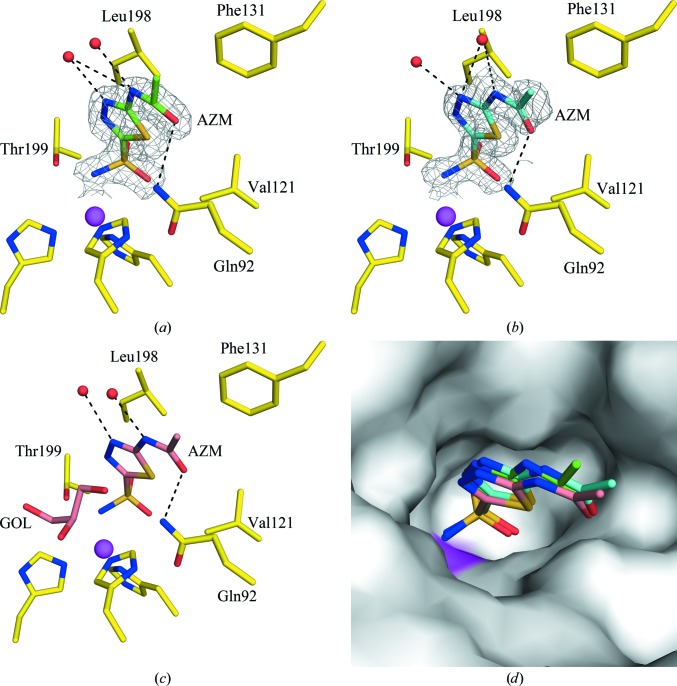
Structures of CA II in complex with AZM were solved using the diffraction data from crystals obtained at 298 K (RT) and 100 K (using sucrose as cryoprotectant; cryo-sucrose) and refined to resolutions of 1.7 and 1.5 Å, respectively (Figs. 2 ▶ a and 2 ▶ b). Although the data could have been collected to higher resolution, the in-house system only permitted collection to a maximal resolution of 1.5 Å. In addition, the analysis of R sym for each image indicated no radiation damage to the crystals during RT or cryo-sucrose data collection. Although the cryo-sucrose structure (PDB entry 3v2m) has a low R sym (19.8%) in the highest resolution bin, the R cryst (41.5%) and R free (45.3%) for this bin are quite high. This is probably owing to the presence of some diffused unassigned density (most likely sucrose) causing under-refinement of the structure. Upon reduction of the data to a resolution of 1.7 Å, the highest resolution bin values of R cryst and R free comply with its R sym.
Figure 2.
Active site of CA II in complex with AZM at (a) RT (298 K), (b) cryo-temperature (100 K) with sucrose and (c) cryo-temperature (100 K) with GOL (PDB entry 3hs4; Sippel et al., 2009 ▶). (d) Surface representation of the three structures superposed. Residues are as labeled. The |2F o − F c| electron density is contoured at 1.0σ. Water molecules are shown as red spheres. Hydrogen bonds are shown as black dotted lines. The active-site Zn2+ ion is shown as a magenta sphere. This figure was produced using PyMOL (http://www.pymol.org).
As reported previously (Sippel et al., 2009 ▶; Fisher et al., 2012 ▶), AZM was observed to bind in the active site of CA II with its sulfonamide N atom directly interacting with the zinc; the pentameric ring is stabilized by hydrophobic interactions with Val121 and Leu198 and by polar interactions with Thr199 and Thr200; the molecule also makes hydrogen bonds to two water molecules and the side chain of Gln92 (Figs. 2 ▶ a, 2 ▶ b and 2 ▶ c). The RT and sucrose structures were superposed with a high-resolution structure (PDB entry 3hs4; Sippel et al., 2009 ▶) of the same complex for which GOL was used as the cryoprotectant (main-chain r.m.s.d. = 0.2 Å). Although there were no significant structural differences between them, there was a small reduction in the unit-cell parameters (Δa = 0.6, Δb = 0.4, Δc = 0.8 Å) in the cryo-structure compared with the RT structure (Table 1 ▶). This phenomenon of shortening of unit-cell edges, as explained by Fraser and coworkers, has been partially attributed to the expulsion of water molecules from inside the unit cells because of the addition of cryoprotectants (Fraser et al., 2011 ▶). In addition, a shift (1.2 Å) in the CH3 group at the tail of AZM was observed in the RT structure compared with the cryo-structures (Fig. 2 ▶ d), but it does not seem to affect the hydrogen-bonding pattern or hydrophobic interactions. It is interesting that this bound GOL is absent when CA II crystals are cooled using GOL without the addition of an inhibitor (Fig. 1 ▶ a; Avvaru et al., 2010 ▶). Therefore, we hypothesize that the binding of GOL is stabilized upon the binding of a CA inhibitor such as AZM.
Hence, this opportunistic GOL-binding site provides a possible ‘fragment’ for structure-aided drug design. This opportunistic GOL-binding site in CA II (lined by Asn62, Asn67 and Gln92) differs by at least one amino-acid residue in the other catalytic CAs (Table 2 ▶). Thus, this binding site has potential for the design of isoform-specific CA inhibitors. GOL could be chemically incorporated into an inhibitor to impart additional specific hydrogen-bonding interactions with the hydrophilic side of the active site (Fig. 2 ▶ c). However, if a CA inhibitor has a tail that binds in the GOL-binding pocket, the GOL in the cryoprotectant could compete for this site. This suggests that a cryoprotectant should be chosen wisely when studying CA–ligand interactions using X-ray crystallography and this phenomenon may be of general concern for other proteins with hydrophilic active-site pockets.
Table 2. Conservation of CA II active-site amino acids.
| Position No. | |||
|---|---|---|---|
| CA | 62 | 67 | 92 |
| I | Val | His | Gln |
| II | Asn | Asn | Gln |
| III | Asn | Arg | Gln |
| IV | Asn | Glu | Gln |
| V (murine) | Thr | Gln | Gln |
| VI | Asn | Gln | Gln |
| VII | Asn | Gln | Gln |
| IX | Asn | Gln | Gln |
| XII | Asn | Lys | Gln |
| XIII | Ser | Asn | Gln |
| XIV | Asn | Gln | Gln |
3.2. Differential scanning calorimetry (DSC)
The thermal stabilities of CA II and the CA II–AZM complex in the presence and absence of cryoprotectants were measured using DSC. A single major unfolding transition (T m) was observed in the thermograms (Table 3 ▶). The CA II–AZM complex (with no cryoprotectant) had a T m of 338.4 K, which is an ∼7 K increase in stability compared with uncomplexed CA II (T m = 331.4 K; Avvaru et al., 2009 ▶). This can be attributed to the increased enthalpic contributions resulting from the binding energy of AZM in the active site of CA II. The addition of GOL or sucrose did not have any significant effect on the melting temperature of the CA II–AZM complex (ΔT m = −1.4 and 2.0 K, respectively).
Table 3. The major unfolding transition (T m) of CA II in complex with AZM.
DSC experiments were carried out in triplicate.
| Sample | T m (K) |
|---|---|
| 20 µM CA II | 331.4 ± 0.1 |
| 20 µM CA II + 20%(v/v) GOL | 332.2 ± 0.1 |
| 20 µM CA II + 20%(w/v) sucrose | 333.5 ± 0.1 |
| 20 µM CA II + 40 µM AZM | 338.5 ± 0.1 |
| 20 µM CA II + 20%(v/v) GOL + 40 µM AZM | 337.0 ± 0.7 |
| 20 µM CA II + 20%(w/v) sucrose + 40 µM AZM | 340.4 ± 0.2 |
3.3. Enzyme-kinetics measurements
18O exchange was used to measure the K i values of AZM for CA II inhibition in the presence of buffer, 20%(v/v) GOL and 20%(w/v) sucrose. As shown in Table 4 ▶, there was a significant 30% reduction in enzyme activity in the presence of GOL and a moderate 15% reduction in the presence of sucrose. These solution kinetic data suggest there is a weak inhibition effect on the enzyme activity at high concentrations of GOL and this could be attributed to the displacement of waters w2 and w3B, which have previously been shown to be important for proton transfer (reaction 2) during CA catalysis, as explained in the context of Fig. 1 ▶ (Silverman & McKenna, 2007 ▶). However, the presence of GOL or sucrose did not affect the inhibition of CA II by AZM (which has a nanomolar affinity for the enzyme).
Table 4. Binding affinities (inhibition constants) of AZM for CA II calculated at 283 K in triplicate.
| Buffer | Initial enzyme activity (%) | K i (nM) |
|---|---|---|
| 50 mM Tris–HCl | 100 ± 11 | 0.48 ± 0.08 |
| 50 mM Tris–HCl + 20%(w/v) sucrose | 85 ± 8 | 0.43 ± 0.05 |
| 50 mM Tris–HCl + 20%(v/v) GOL | 69 ± 7 | 0.56 ± 0.07 |
4. Conclusion
The binding of GOL in the active site of CA II only in the presence of the inhibitor AZM has provided an opportunistic lead for CA isoform-specific drug design, as the incorporation of a GOL-like substituent onto the ‘tail’ region of AZM would presumably be involved in the same hydrogen-bond donor/acceptor interactions as those observed for GOL in this crystallographic study. As these GOL-binding residues vary among the human CA isoforms, we postulate a potential therapeutic CA pharmacophore that could impart isoform-specific inhibition.
This study also reports a cautionary tale, however, when selecting a suitable cryoprotectant for enzyme–ligand crystallography. Interactions of the cryoprotectant with the protein could have unforeseen effects on ligand binding, ranging from a commensalistic relationship as observed between GOL and AZM in CA II to a direct competitive inhibition with the ligand as proposed with the aforementioned drug. One is therefore forewarned in the careful consideration of an otherwise prosaic cryoprotectant.
Supplementary Material
PDB reference: carbonic anhydrase II, complex with AZM, 3v2j
PDB reference: in presence of sucrose, 3v2m
Acknowledgments
The work has been supported by an Alumni Fellowship Award, Grinter Award and Medical Guild Research Incentive Award from the University of Florida (MA), an HHMI Science for Life (BK) and National Institutes of Health grant GM25154 (RM and DNS).
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Aggarwal, M., Boone, C. D., Kondeti, B. & McKenna, R. (2013). J. Enzyme Inhib. Med. Chem. 28, 267–277. [DOI] [PubMed]
- Aggarwal, M., Kondeti, B. & McKenna, R. (2013). Bioorg. Med. Chem. 15, 1526–1533. [DOI] [PMC free article] [PubMed]
- Aggarwal, M. & McKenna, R. (2012). Expert Opin. Ther. Pat. 22, 903–915. [DOI] [PubMed]
- Avvaru, B. S., Busby, S. A., Chalmers, M. J., Griffin, P. R., Venkatakrishnan, B., Agbandje-McKenna, M., Silverman, D. N. & McKenna, R. (2009). Biochemistry, 48, 7365–7372. [DOI] [PMC free article] [PubMed]
- Avvaru, B. S., Kim, C. U., Sippel, K. H., Gruner, S. M., Agbandje-McKenna, M., Silverman, D. N. & McKenna, R. (2010). Biochemistry, 49, 249–251. [DOI] [PMC free article] [PubMed]
- Brünger, A. T. (1992). Nature (London), 355, 472–475. [DOI] [PubMed]
- Chapman, M. S. & Somasundaram, T. (2010). Acta Cryst. D66, 741–744. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Fisher, S. Z., Aggarwal, M., Kovalevsky, A. Y., Silverman, D. N. & McKenna, R. (2012). J. Am. Chem. Soc. 134, 14726–14729. [DOI] [PMC free article] [PubMed]
- Fisher, S. Z., Tu, C., Bhatt, D., Govindasamy, L., Agbandje-McKenna, M., McKenna, R. & Silverman, D. N. (2007). Biochemistry, 46, 3803–3813. [DOI] [PubMed]
- Forwand, S. A., Landowne, M., Follansbee, J. N. & Hansen, J. E. (1968). N. Engl. J. Med. 279, 839–845. [DOI] [PubMed]
- Fraser, J. S., van den Bedem, H., Samelson, A. J., Lang, P. T., Holton, J. M., Echols, N. & Alber, T. (2011). Proc. Natl Acad. Sci. USA, 108, 16247–16252. [DOI] [PMC free article] [PubMed]
- Garman, E. (2003). Curr. Opin. Struct. Biol. 13, 545–551. [DOI] [PubMed]
- Garman, E. F. & Doublié, S. (2003). Methods Enzymol. 368, 188–216. [DOI] [PubMed]
- Garman, E. F. & Owen, R. L. (2006). Acta Cryst. D62, 32–47. [DOI] [PubMed]
- Gilmour, K. M. (2010). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 157, 193–197. [DOI] [PubMed]
- Holton, J. M. (2009). J. Synchrotron Rad. 16, 133–142.
- Jackman, J. E., Merz, K. M. & Fierke, C. A. (1996). Biochemistry, 35, 16421–16428. [DOI] [PubMed]
- Jude, K. M., Banerjee, A. L., Haldar, M. K., Manokaran, S., Roy, B., Mallik, S., Srivastava, D. K. & Christianson, D. W. (2006). J. Am. Chem. Soc. 128, 3011–3018. [DOI] [PMC free article] [PubMed]
- Katritzky, A. R., Caster, K. C., Maren, T. H., Conroy, C. W. & Bar-Ilan, A. (1987). J. Med. Chem. 30, 2058–2062. [DOI] [PubMed]
- Kmetko, J., Warkentin, M., Englich, U. & Thorne, R. E. (2011). Acta Cryst. D67, 881–893. [DOI] [PMC free article] [PubMed]
- Liljas, A., Kannan, K. K., Bergstén, P. C., Waara, I., Fridborg, K., Strandberg, B., Carlbom, U., Järup, L., Lövgren, S. & Petef, M. (1972). Nature New Biol. 235, 131–137. [DOI] [PubMed]
- Maren, T. H. (1967). Physiol. Rev. 47, 595–781. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Ravelli, R. B. & Garman, E. F. (2006). Curr. Opin. Struct. Biol. 16, 624–629. [DOI] [PubMed]
- Riboldi-Tunnicliffe, A. & Hilgenfeld, R. (1999). J. Appl. Cryst. 32, 1003–1005.
- Silverman, D. N. & McKenna, R. (2007). Acc. Chem. Res. 40, 669–675. [DOI] [PubMed]
- Sippel, K. H., Robbins, A. H., Domsic, J., Genis, C., Agbandje-McKenna, M. & McKenna, R. (2009). Acta Cryst. F65, 992–995. [DOI] [PMC free article] [PubMed]
- Srivastava, D. K., Jude, K. M., Banerjee, A. L., Haldar, M., Manokaran, S., Kooren, J., Mallik, S. & Christianson, D. W. (2007). J. Am. Chem. Soc. 129, 5528–5537. [DOI] [PMC free article] [PubMed]
- Supuran, C. T. (2008). Nature Rev. Drug Discov. 7, 168–181. [DOI] [PubMed]
- Supuran, C. T., Scozzafava, A. & Conway, J. (2004). Carbonic Anhydrase: Its Inhibitors and Activators Boca Raton: CRC Press.
- Zheng, J., Avvaru, B. S., Tu, C., McKenna, R. & Silverman, D. N. (2008). Biochemistry, 47, 12028–12036. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: carbonic anhydrase II, complex with AZM, 3v2j
PDB reference: in presence of sucrose, 3v2m