Abstract
MoS2 is a two-dimensional material that is gaining prominence due to its unique electronic and chemical properties. Here, we demonstrate ligand conjugation of chemically exfoliated MoS2 using thiol chemistry. Using this method, we modulate the zeta-potential and colloidal stability of MoS2 sheets through ligand designs, thus enabling its usage as a selective artificial protein receptor for β-galactosidase. The facile thiol functionalization route opens the door for surface modifications of solution processable MoS2 sheets.
The recent excitement over two dimensional atomically-layered materials has been fueled in large part by desires to exploit their low-dimensional properties, which can be distinct from their bulk counter-parts. One of these emerging materials, MoS2, has received a fair share of attention due to its applicability in areas ranging from catalysis1 and electronics2–5 to biomedicine.6 Nevertheless, to fully harness the capabilities of such new materials, ligand conjugations were often necessity. For example, it is typically a crucial step for solution processing, which enables the usage of nanoparticles as functional assembly blocks in bio-imaging,7 electronics,8 sensing,9 and photovoltaics.10 Previously, attempts to functionalize MoS2 have been limited to bulk-like materials characterized by hydrophobic cleavage planes.11,12 Specifically, Tremel and coworkers13 have modified hydrophobic MoS2 particles using nickel-nitrilotriacetic acid chelation.13 However, colloidal surface modification of water dispersible, chemically exfoliated MoS2 (ce-MoS2) sheets has not been demonstrated.
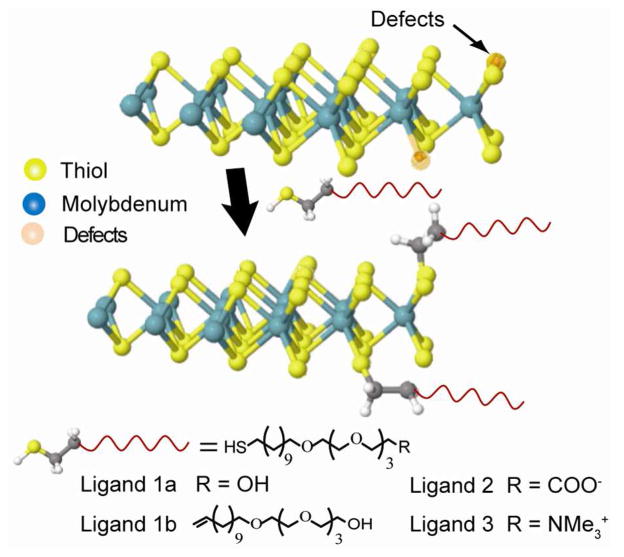
To obtain large quantities of single-layer MoS2 sheets, solution based exfoliation methods are often used. Most typically, this involves chemical exfoliation using lithium intercalation, which can produce single layer MoS2 sheets in scalable quantities.14–16 In this method, lithium is inserted between MoS2 layers and then reacted with water to produce hydrogen gas at the interface. This, in combination with ultrasoncation, then enables the high yield production of water dispersible ce-MoS2 sheets.14 Due to the violent nature of this reaction however, MoS2’s crystal structure becomes deformed,3,17 and internal edges (tears, pinholes and defects) can become visible.18 Previous work has suggested these edge sites to possess higher molecular affinities,19 with theoretical suggestions of thiol edge absorption.20 It is thus possible that ce-MoS2, which pos- sesses defects in both internal edges and perimeter edges, may be amenable to thiol ligand modifications. As this has not been explicitly demonstrated, we show conjugation of ce-MoS2 sheets by thiol-terminated ligands (Figure 1, Supplemental Information S1-5). This was used to conveniently tune ce-MoS2 and its biomolecular interactions. This route opens pathways for making solution processable MoS2 with tunable colloidal properties and can potentially facilitate improvements in its processing.
Figure 1.
Structural models illustrating ligand conjugation of ce-MoS2 sheets.
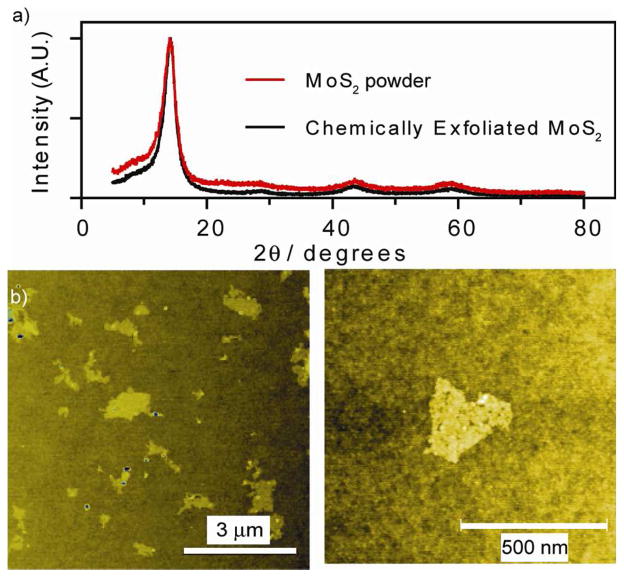
In this report, the as-made ce-MoS2was first purified using exhaustive dialysis (7 days under continuous water flow). Survey of the material using X-Ray Diffraction (XRD) showed primary peak at 2θ = 14, thus indicating absence of mixed lithium phase (Figure 2a).3,21 This was confirmed using Inductive Coupled Plasma-Mass Spectrometry (ICP-MS), which showed molecular ratios of samples obtained through dialysis to contain more than ten-fold less Li than the typical centrifugation purification process (vide infra). Atomic Force Microscopy (AFM) revealed sheets with average diameters of 0.7μm, and average thicknesses between ~0.8 and 1.5 nm, consistent with reported values of monolayers.22 Additionally, the internal edges mentioned previously become are visible (Figure 2b, S6).18 Transmission Electron Microscopy (TEM) Selective Area Diffraction (SAD) showed six-fold symmetrical diffraction spots (2H phase) with additional points in between those diffraction points (1T phase, Figure S7). This is consistent with previous reports by Dungey, et al,17 for ce-MoS2, thus indicating a mixed 1T and 2H material. Separately, Dynamic Light Scattering (DLS) show native ce-MoS2 sheets to have a ζ-potential of −50 mV (Figure 3a). To this end, there are different models in literature regarding the origin of this negative charge. Principally, Divigalpitiya, et al,19 proposed a model with charges at edges. Separately Heising, et al,23 has a model based on partially oxidized Mo atoms on the basal plane due to Li intercalation. However, both agree that the sheets are defective, thus there is the possibility of chemical modification.
Figure 2.
(a) XRD spectra of ce-MoS2. (b) Low and high magnification AFM micrograph of ce-MoS2.
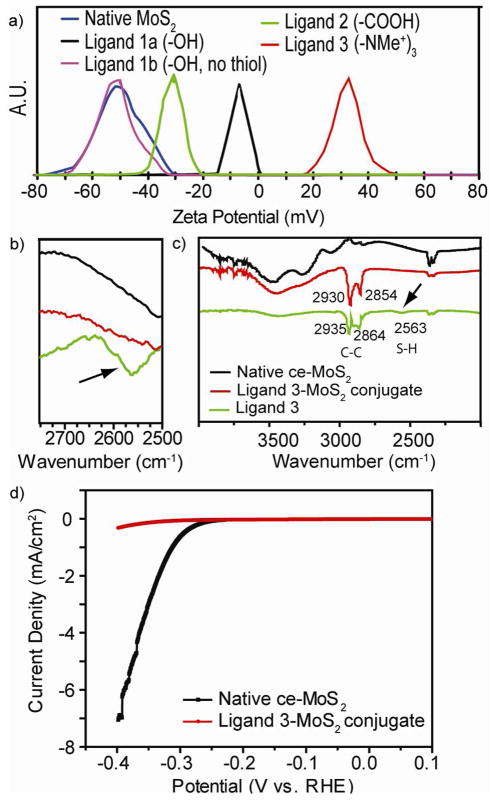
Figure 3.
(a) ζ-potential, (b) FT-IR spectra focused upon the thiol peak, (c) FT-IR spectra showing all peaks and (d) HER polarization curve before and after ligand conjugation
To illustrate this process, we first sought to understand the ligand affinity of chemically exfoliated MoS2. For this, we synthesized a series of polyethylene glycol (PEG) ligands with head-groups of different charge (Figure 1, S1–S5). By including and excising the thiol moiety, we show that the thiol group is responsible for the ce-MoS2 modification (ligand 1a vs. 1b).
We first examined the conjugation results using ligand 1a and ligand 1b. As can be seen in Figure 1, these ligands are nearly identical except for the thiol group on ligand 1a. Despite their similarities however, only the ce-MoS2 samples incubated with ligand 1a showed significant ζ-potential shifts after purification. The resultant ζ-potential of −7 mV represents a 43 mV shift from native ce-MoS2, and is consistent with a neutral PEG functionalized colloid.19 Comparatively, ligand 1b produced no ζ-potential change, indicating no modification (Figure 3a). Due to the attenuated ζ-potential, ligand 1a conjugate was found to be less stable in water.
To expand upon the conjugations, ligand 2 and ligand 3 were synthesized. Here, the conjugates also show appropriate changes in ζ-potential as well, with ligand 2 producing a ζ-potential of −29 mV and ligand 3 a ζ-potential of +36 mV. Overall, ligand 3 conjugates exhibited the greatest colloidal stability. We believe this to be attributed to the pH independent nature of the charged NMe3+group on ligand 3.
Further characterizations were then performed. First, Fourier Transform Infrared spectroscopy (FT-IR, Figure 3b, 3c, S9) revealed the exposed S-H band from free ligands at 2563 cm−1. This peak becomes absent after conjugation with ce-MoS2, giving indication that the thiol moiety becomes buried on to the ce-MoS2 surface (Figure 3b).24,25 Presence of the ligands on the conjugates were also validated by monitoring the C–H aliphatic bands 2854 and 2930 cm−1. Here, these aliphatic bands appear on the conjugates at 2864 and 2935 cm−1 (Figure 3c).26 Unique pegylated ligand signature on conjugated ce-MoS2 surfaces were also verified using X-Ray Photoelectron Spectroscopy (XPS). For example, ether (C-O-C) bonds characteristic of PEG ligands appeared in all conjugated samples (286.6 eV, Figure S8). These results further corroborate presence of PEG ligands on the ce-MoS2, and are consistent with thiol-PEG functionalized materials.27–29
With the conjugation assessed, we then evaluate the effect of residual lithium (Li) from the exfoliation process. Here, ce-MoS2 purified using the 3X centrifugation process described by Heising23 produced a Li:Mo ratio of 0.25, consistent with their report. ce-MoS2 purified using the dialysis process described here reduced the Li:Mo ratio to 0.02, more than tenfold lower (Figure S10). However, the residual Li doesn’t appear to effect conjugation results radically (Figure S11)
We also examined the effect of ligand conjugation on the hydrogen evolution reaction (HER) process. As previous studies suggest edges to dominate HER catalysis,30 we expect that ligand conjugation to suppress HER activity. Indeed, decrease catalytic activity was clearly observed (Figure 3c, S12). It thus appears that the thiol ligands can at minimum functionalize the various edges in ce- MoS2.
We then monitored the changes in colloidal stability with ligand conjugation. Most observably, native ce-MoS2 irreversibly precipitated due to restacking within the first 21 days (Figure 5a).14 Comparably, the conjugates maintained colloidal stability (ligand 3), or can be readily re-dispersed with shaking (ligand 1a, 2) (Figure 4a). This is likely due PEG ligand preventing complete restacking. Due to the pH independent nature of the NMe3+group on ligand 3, its conjugates are also capable of resisting flocculation in acidic media (Figure 4b). To compare, nativece-MoS2 flocculated at low pH.14 Ligand 3 conjugates, on the other hand were able to maintain colloidal stability (Figure 4b).
Figure 5.
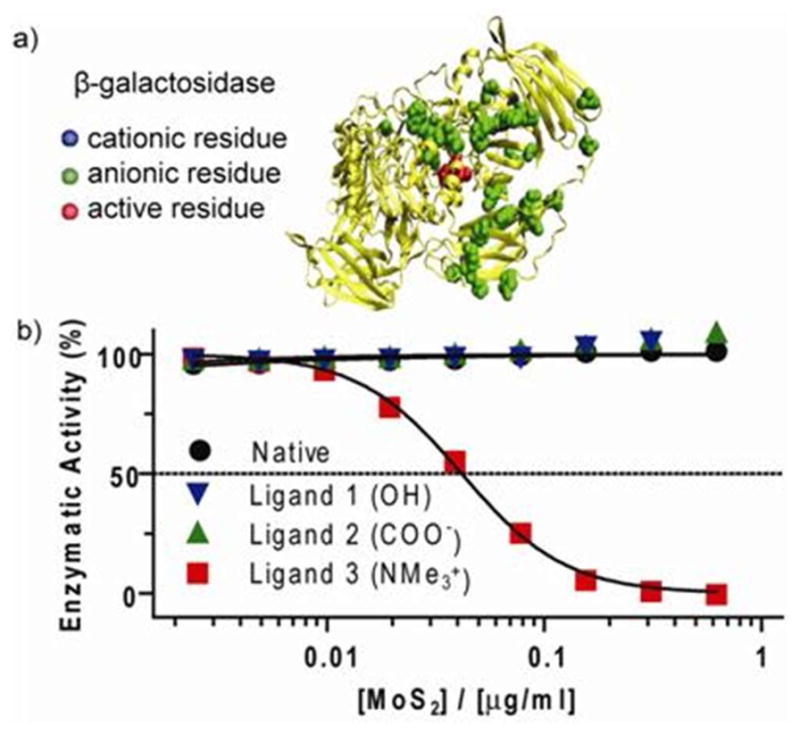
(a) Structure of a single unit of a β-Gal tetramer. Anionic residues (Asp and Glu) surrounding β-gal’s active site are highlighted (bottom) (b) Concentration dependence of β-gal inhibition with various ce-MoS2 conjugate.
Figure 4.
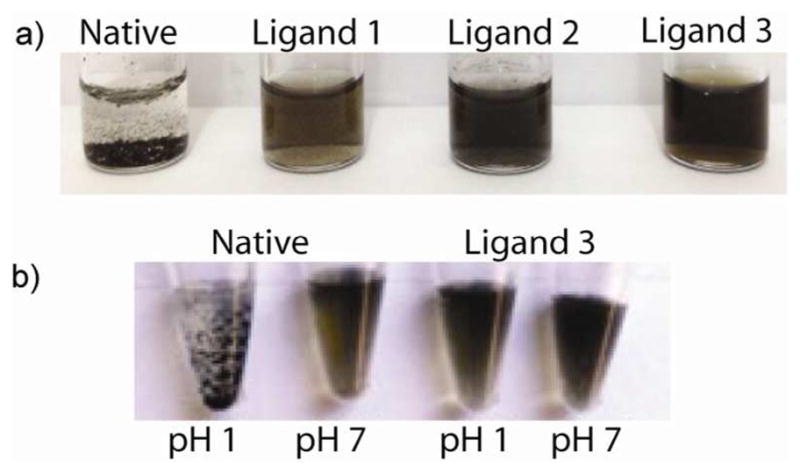
(a) Colloidal stability of ce-MoS2 samples after 21 days in water. (b) Colloidal stability of ce-MoS2 functionalized with NMe3 terminated thiol at various pH.
The combination of ζ-potential tailoring and pegylation also enable the tuning of ce-MoS2 for selective host-guest interactions with biomolecules. As we have recently shown ce-MoS2 to be a promising photothermal agent,31 ligand modifications shown here can have immediate applications. To demonstrate this, we show the ligands ability to change ce-MoS2’s function as an artificial protein receptor. To this end, we use β-galactosidase (β-gal), a hydrolase enzyme. Here, β-gal, due to a ring of anionic residues (Asp and Glu) surrounding its active site does not interact with native ce-MoS2 (Figure 5a).32,33 By tuning ce-MoS2 for the correct electrostatic host-guest interactions we thus enable selective complexation and inhibition of this enzyme.34 For this, 62.5 pM of β-gal were incubated with various concentrations of ce-MoS2-conjugates. After 30 minutes, o-nitrophenyl-β-D-galactopyranoside (ONPG), a chromogenic substrate was added to monitor β-gal activity. From this experiment, it can be seen that only the ligand 3 conjugate, which is cationic, showed ability to modulate β-gal activity. Specifically, 50% of β-gal activity can be inhibited by 0.04 μg/ml of the conjugate (Figure 5b). These results demonstrate the possibility to tune ce-MoS2 for selective host-guest interactions with enzymes. Further development of this demonstrated concept can thus be utilized for biosensor design. 9,35,36
In summary, we have demonstrated ligand conjugation of chemically exfoliated MoS2 sheets through thiol chemistry. In this manner, we were able to tune the ζ-potential and surface functionality of ce-MoS2 sheets to enable its broad usage as artificial receptors for enzymes. Because these aforementioned assemblies have potential applications in sensing,35,37 energy,1,30 etc, and because devices using 2D materials can outperform their nanoparticles,34,38 it is possible that the functionalized ce-MoS2 assemblies may enjoy similar advantages. The ability to functionalize these sheets through facile thiol chemistry suggests that similar chemistry may be applied to other chemically exfoliated transition metal dichalcogenide sheets (Figure S11), and can therefore open the window for further applications and solution processing of these emerging two-dimensional materials.
Supplementary Material
Acknowledgments
This work was partly funded by the National Cancer Institute Center for Cancer Nanotechnology Excellence (CCNE) initiative at Northwestern University Award No. U54CA119341. JH acknowledges support from National Science Foundation (DMR CAREER 0955612) and the Alfred P. Sloan Research Foundation. S. B. was supported by Basic Science Research Program through the National Research Foundation of Korea funded by the MEST (Project no. 2012-000181). We thank NU-HTA and NU NUANCE and NU-QBIC for characterization equipment availability and expertise. We also thank A. Yan and Y. Huang for assistance with EM microscopy and M. Rotz and Dr. K. MacRenaris assistance with ICP-MS. SC thanks US DHS for a graduate fellowship.
Footnotes
Author Contributions
All authors have approved the final version of the manuscript.
Synthetic protocols, experimental procedures, and additional characterizations are included in the supporting information. This material is available free of charge at http://pubs.acs.org.
References
- 1.Li Y, Wang H, Xie L, Liang Y, Hong G, Dai HJ. Am Chem Soc. 2011;133:7296. doi: 10.1021/ja201269b. [DOI] [PubMed] [Google Scholar]
- 2.Radisavljevic B, Radenovic A, Brivio J, Giacometti V, Kis A. Nat Nano. 2011;6:147. doi: 10.1038/nnano.2010.279. [DOI] [PubMed] [Google Scholar]
- 3.Eda G, Yamaguchi H, Voiry D, Fujita T, Chen M, Chhowalla M. Nano Lett. 2011;11:5111. doi: 10.1021/nl201874w. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, Loh KP, Sow CH, Gu H, Su X, Huang C, Chen ZK. Langmuir. 2004;20:6914. doi: 10.1021/la049887t. [DOI] [PubMed] [Google Scholar]
- 5.Liu KK, Zhang W, Lee YH, Lin YC, Chang MT, Su CY, Chang CS, Li H, Shi Y, Zhang H, Lai CS, Li LJ. Nano Lett. 2012;12:1538. doi: 10.1021/nl2043612. [DOI] [PubMed] [Google Scholar]
- 6.Wu H, Yang R, Song B, Han Q, Li J, Zhang Y, Fang Y, Tenne R, Wang C. ACS Nano. 2011;5:1276. doi: 10.1021/nn102941b. [DOI] [PubMed] [Google Scholar]
- 7.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat Mater. 2005;4:435. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 8.Nakanishi H, Walker DA, Bishop KJM, Wesson PJ, Yan Y, Soh S, Swaminathan S, Grzybowski BA. Nat Nano. 2011;6:740. doi: 10.1038/nnano.2011.165. [DOI] [PubMed] [Google Scholar]
- 9.De M, Rana S, Akpinar H, Miranda OR, Arvizo RR, Bunz UHF, Rotello VM. Nat Chem. 2009;1:461. doi: 10.1038/nchem.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yaacobi-Gross N, Soreni-Harari M, Zimin M, Kababya S, Schmidt A, Tessler N. Nat Mater. 2011;10:974. doi: 10.1038/nmat3133. [DOI] [PubMed] [Google Scholar]
- 11.Rapoport L, Fleischer N, Tenne RJ. Mater Chem. 2005;15:1782. [Google Scholar]
- 12.Yarar B, Kaoma J. Colloids Surf. 1984;11:429. [Google Scholar]
- 13.Tahir MN, Zink N, Eberhardt M, Therese HA, Kolb U, Theato P, Tremel W. Angew Chem, Int Ed. 2006;45:4809. doi: 10.1002/anie.200504211. [DOI] [PubMed] [Google Scholar]
- 14.Joensen P, Frindt RF, Morrison SR. Mater Res Bull. 1986;21:457. [Google Scholar]
- 15.Schumacher A, Scandella L, Kruse N, Prins R. Surf Sci. 1993;289:L595. [Google Scholar]
- 16.Zeng Z, Yin Z, Huang X, Li H, He Q, Lu G, Boey F, Zhang H. Angew Chem, Int Ed. 2011;50:11093. doi: 10.1002/anie.201106004. [DOI] [PubMed] [Google Scholar]
- 17.Dungey KE, Curtis MD, Penner-Hahn JE. Chem Mater. 1998;10:2152. [Google Scholar]
- 18.Eda G, Fujita T, Yamaguchi H, Voiry D, Chen M, Chhowalla M. ACS Nano. 2012;6:7311. doi: 10.1021/nn302422x. [DOI] [PubMed] [Google Scholar]
- 19.Divigalpitiya WMR, Frindt RF, Morrison SR. Science. 1989;246:369. doi: 10.1126/science.246.4928.369. [DOI] [PubMed] [Google Scholar]
- 20.Raybaud P, Hafner J, Kresse G, Toulhoat H. Phys Rev Lett. 1998;80:1481. [Google Scholar]
- 21.Joensen P, Crozier ED, Alberding N, Frindt RF. J Phys C. 1987;20:4043. [Google Scholar]
- 22.Late DJ, Liu B, Matte HSSR, Rao CNR, Dravid VP. Adv Funct Mater. 2012;22:1894. [Google Scholar]
- 23.Heising J, Kanatzidis MG. J Am Chem Soc. 1999;121:11720. [Google Scholar]
- 24.Shi W, Sahoo Y, Swihart MT. Colloid Surfaces A. 2004;246:109. [Google Scholar]
- 25.Thangadurai P, Balaji S, Manoharan PT. Nanotechnology. 2008;19:435708. doi: 10.1088/0957-4484/19/43/435708. [DOI] [PubMed] [Google Scholar]
- 26.Porter MD, Bright TB, Allara DL, Chidsey CED. J Am Chem Soc. 1987;109:3559. [Google Scholar]
- 27.Harder P, Grunze M, Dahint R, Whitesides GM, Laibinis PE. J Phys Chem B. 1998;102:426. [Google Scholar]
- 28.Wagner CD, Muilenberg GE. Handbook of x-ray photoelectron spectroscopy: a reference book of standard data for use in x-ray photoelectron spectroscopy. Perkin-Elmer Corp., Physical Electronics Division; 1979. [Google Scholar]
- 29.Weisbecker CS, Merritt MV, Whitesides GM. Langmuir. 1996;12:3763. [Google Scholar]
- 30.Jaramillo TF, Jørgensen KP, Bonde J, Nielsen JH, Horch S, Chorkendorff I. Science. 2007;317:100. doi: 10.1126/science.1141483. [DOI] [PubMed] [Google Scholar]
- 31.Chou SS, Kaehr B, Kim J, Foley BM, De M, Hopkins PE, Huang J, Brinker CJ, Dravid VP. Angew Chem, Int Ed. 2013 doi: 10.1002/anie.201209229. n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verma A, Simard JM, Worrall JWE, Rotello VM. J Am Chem Soc. 2004;126:13987. doi: 10.1021/ja046572r. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson RH, Zhang XJ, DuBose RF, Matthews BW. Nature. 1994;369:761. doi: 10.1038/369761a0. [DOI] [PubMed] [Google Scholar]
- 34.De M, Chou SS, Dravid VP. J Am Chem Soc. 2011;133:17524. doi: 10.1021/ja208427j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miranda OR, Chen HT, You CC, Mortenson DE, Yang XC, Bunz UHF, Rotello VM. J Am Chem Soc. 2010;132:5285. doi: 10.1021/ja1006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miranda OR, You CC, Phillips R, Kim IB, Ghosh PS, Bunz UHF, Rotello VM. J Am Chem Soc. 2007;129:9856. doi: 10.1021/ja0737927. [DOI] [PubMed] [Google Scholar]
- 37.Lee JS, Ulmann PA, Han MS, Mirkin CA. Nano Lett. 2008;8:529. doi: 10.1021/nl0727563. [DOI] [PubMed] [Google Scholar]
- 38.Chou SS, De M, Luo J, Rotello VM, Huang J, Dravid VP. J Am Chem Soc. 2012;134:16725. doi: 10.1021/ja306767y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.