Abstract
Background
Low levels of selenium have been associated with increased risk of prostate cancer (PCa). Selenoprotein P is the most abundant selenoprotein in serum and delivers ten selenocysteine residues to tissues. Variation in the selenoprotein P gene (SEPP1) may influence PCa development or modify the effects of selenium. We examined the association of SEPP1 single nucleotide polymorphisms (SNPs) with PCa risk and survival, and tested for interactions.
Methods
The Physicians’ Health Study (PHS) is a prospective cohort of 22,071 US physicians; we utilized a nested case-control study of 1,352 PCa cases and 1,382 controls. We assessed four SNPs capturing common variation within the SEPP1 locus. In a subset of men (n=80), we evaluated SEPP1 mRNA expression in tumors.
Results
Two SNPs were significantly associated with PCa risk. For rs11959466, each T allele increased risk (odds ratio (OR)=1.31; 95% confidence interval (CI): 1.02,1.69; ptrend=0.03). For rs13168440, the rare homozygote genotype decreased risk compared to the common homozygote (OR=0.56, 95% CI: 0.33, 0.96). Moreover, there was a significant interaction of rs13168440 with plasma selenium; increasing selenium levels were associated with decreased PCa risk only among men with the minor allele (pinteraction=0.01). SEPP1 expression was significantly lower in men with lethal PCa than long-term survivors.
Conclusions
SEPP1 genetic variation was associated with PCa incidence; replication of these results in an independent dataset is necessary. These findings further support a causal link between selenium and PCa, and suggest that the effect of selenium may differ by genetics.
Keywords: genetic variation, selenium, prostate cancer
Introduction
Several studies have suggested that selenium may act as a chemopreventive agent of PCa (1–4), but the Selenium and Vitamin E Cancer Prevention Trial (SELECT), a large randomized study of selenium supplements, showed no benefit (5). Selenium is incorporated into several antioxidant proteins. Genes that encode selenoproteins contain a Selenocysteine Insertion Sequence (SECIS) structural element in the 3′ UTR region of the mRNA. The SECIS element causes the UGA codon, normally the stop codon, to encode a selenocysteine, which is necessary for the structure and function of the resulting protein and often has redox properties (6). Selenoprotein P (SelP) is the most abundant selenoprotein in serum and delivers selenium to tissues, carrying ten selenocysteine residues per polypeptide (7).
SelP and the gene that encodes it (SEPP1) may themselves be related to PCa. Previously, a study observed lower serum SelP levels in PCa cases than in controls (8). Expression of SEPP1 (9) and SelP (10) was lower in prostate tumor cells than in normal prostate cells. Experimentally, lower SelP expression in tumor prostate cells increased sensitivity to H2O2-induced cytotoxicity, which may lead to higher levels of free radicals, suggesting a mechanism that could promote tumor development (10). The rare AA genotype of SEPP1 rs7579 was associated with increased PCa risk (OR=1.72; 95% CI: 0.99, 2.98) (11). A significant interaction between SEPP1 rs3877899 and a SNP in the manganese super-oxide dismutase (SOD2) gene, rs4880, has also been shown; those with the rs4880 allele encoding alanine had a significantly increased risk of prostate cancer only among those with the CC genotype of rs3877899 (OR=1.43; 95%CI: 1.17, 1.76) (12). We and others have also observed that genetic variation in other selenoproteins is associated with PCa risk or survival (13,14). Based on this evidence, we comprehensively examined the association of SEPP1 genetic variation with PCa risk and survival. We also examined interactions with selenium and selected lifestyle characteristics, as higher BMI and cigarette smoking can lead to an increase in oxidative stress and are additionally associated with PCa mortality.
Materials and Methods
Study Population
The Physicians’ Health Study (PHS) began as a randomized, double-blind trial of aspirin and β-carotene in the prevention of cardiovascular disease and cancer among 22,071 healthy US physicians. Men were excluded if they had any serious medical conditions including all cancers (except non-melanoma skin cancer). Blood samples were collected from 68% of the physicians at baseline in 1982–1984. A detailed description of PHS has been published previously (15). The follow-up rate is 96% for cancer incidence and 100% for mortality. All self-reported PCa cases are verified through medical record and pathology review by the PHS Endpoints Committee. Data were abstracted on PSA at diagnosis, tumor stage, and Gleason score. Death certificates and medical records were reviewed to determine cause of death. Metastases are reported on follow-up questionnaires sent to all men living with PCa and confirmed by additional medical record review. BMI and smoking status were determined from the PHS1 baseline questionnaire; more detailed smoking information was collected on the 60-month follow-up questionnaire to define pack-years.
We utilized a nested case-control study, with controls selected through risk-set sampling and matched to cases on age at baseline (±1 year for cases ≤55 and, if necessary, ±5 years for cases >55 years), smoking status (never, former, current), and follow-up time. We restricted the current study to self-reported Caucasians to reduce potential population stratification. 1352 cases (diagnosed from 1982–2005) and 1382 controls were included in the case-control analyses; additional cases diagnosed after they were matched as controls were included for the outcomes analyses (n=1414). During follow-up through March 9, 2010, 197 men died of PCa or developed bone metastases.
SNPs, DNA, and Genotyping
Using the HapMap database, we selected 5 SNPs to capture variation (with R2>0.80) within SEPP1 and 5 kb up- and downstream. Selection was restricted to SNPs with a minor allele frequency >5% in the International HapMap CEPH samples. DNA was extracted from whole blood. Genotyping was performed with BioTrove OpenArray Technology (16). Four SNPs had high genotyping success (92–97%); rs230819 failed genotyping, but did not substantially reduce the overall gene coverage as it tagged only itself.
Plasma Selenium and SEPP1 Expression Measurement
In a subgroup of participants, plasma levels of selenium were measured as previously described (3) in baseline blood samples from 793 cases (782 in nested case-control study) and 547 controls who also had genotyping data. SEPP1 mRNA expression was measured in a sample of PCa tumors from radical prostatectomy specimens as part of a larger project using the Illumina DASL technology (17); the subset of these cases with expression and genotyping information (N=80) were included in this study.
Statistical Analysis
Analyses were performed with SAS version 9.1 statistical software; all P values are two-sided. Using Pearson’s goodness-of-fit test, none of the SNPs violated Hardy-Weinberg Equilibrium (p>0.05). SNPs were analyzed under a co-dominant genetic model to determine effect estimates, with p-values reported for the additive model. Imputation of non-genotyped, previously published SNPs (rs7579, rs3877899) was performed using the MACH program (18) and genotypes from the CEPH population in HapMap (19,20).
In a nested case-control study, we assessed the risk of incident PCa using unconditional logistic regression models, adjusting for matching factors (age at randomization, smoking status, and duration of follow-up). We also conducted subgroup analyses, comparing aggressive cases (defined as Gleason score ≥8, clinical stage T3, T4, N1, or M1 or fatal/metastatic disease) to controls. Among PCa cases, we performed an analysis of time to lethal PCa outcome using a Kaplan-Meier analysis and Cox regression model, adjusting for age at diagnosis. A lethal PCa outcome was defined as death due to PCa (n=184) or the development of bone metastases (n=13); follow-up began at the time of PCa diagnosis and individuals were censored at the time of death from another cause or the end of follow-up on March 9, 2010.
Pre-diagnostic plasma selenium was normally distributed. We used linear regression to determine if the SEPP1 SNPs were associated with selenium levels among cases and controls combined, adjusting for analytic batch, the matching factors, and case-control status. Quartiles of selenium were created using batch-specific cut points from the controls in the case-control analysis and batch-specific cut points from all cases for the mortality analysis. Interaction terms of ordinal quartiles of selenium and genotype were used in an unconditional logistic regression model for risk and Cox regression model for mortality.
We used ANOVA to determine if the SEPP1 SNPs were associated with SEPP1 mRNA expression levels. Interaction terms were created for SNPs and categories of pack-years of smoking (0, 1–10, 11–20, 21–30, 31–40, >40 years), and for SNPs and two categories of BMI (<25 and ≥25 kg/m2 only, as very few participants were obese) with SNPs modeled as additive.
Results
Characteristics of the cases and controls are presented in Table I. Two of the four SEPP1 SNPs examined were significantly associated with PCa incidence (Table II). For rs11959466, each additional T allele significantly increased the risk of PCa (odds ratio (OR)=1.31; 95% confidence interval (CI): 1.02, 1.69; ptrend=0.03). For rs13168440, while the test for trend was not significant (p=0.41), the minor allele homozygote genotype (CC) was associated with a decreased risk compared to the homozygote TT referent (OR=0.56, 95% CI: 0.33, 0.96; p=0.03). None of the SNPs were associated with PCa survival (Table II) or specifically with aggressive disease (results not shown).
Table I.
Characteristics of prostate cancer cases and controls in the Physicians’ Health Study.
| Case-control incidence analysis | Cases (n=1352) | Controls (n=1382) |
|---|---|---|
| Age at study onset, mean ± s.d. | 57.8 ± 8.5 | 57.5 ± 8.4 |
| Gleason score, n (%) | ||
| 2–6 | 635 (51.7) | |
| 7 | 409 (33.3) | |
| 8–10 | 185 (15.1) | |
| Clinical stage, n (%) | ||
| T1, T2 | 1137 (88.6) | |
| T3, T4, N1, M1 | 147 (11.4) | |
| Baseline selenium (ug/g), median (10th–90th)* | 0.109 (0.086–0.133) | 0.110 (0.088–0.134) |
|
| ||
| Case-only survival analysis | Cases (n=1414) | |
|
| ||
| Age at diagnosis, mean ± s.d. | 70.4 ± 7.6 | |
| Deaths/metastases due to PCa, n (%) | 197 (13.9) | |
| Diagnosis to PCa death/mets, median years (range) | 5.7 (0.1–21.0) | |
| Follow-up time to censored, median years (range) | 11.0 (0.01–27.2) | |
selenium levels are available for 793 cases and 547controls
PCa=prostate cancer
Table II.
Genotype frequency and association of SEPP1 SNPs with prostate cancer risk and mortality.
| SNP | Risk analysis
|
Survival analysis
|
||||
|---|---|---|---|---|---|---|
| cases, n (%) | controls, n (%) | OR (95% CI) | PCa deaths/mets, n (%) | all other cases, n (%) | HR (95% CI) | |
| rs11959466 | ||||||
| CC | 1100 (88.6) | 1156 (91.0) | 1.00 (ref) | 146 (88.5) | 1005 (88.7) | 1.00 (ref) |
| CT | 135 (10.9) | 114 (9.0) | 1.24 (0.95, 1.61) | 19 (11.5) | 122 (10.8) | 1.05 (0.65, 1.69) |
| TT | 6 (0.5) | 1 (0.1) | 6.54 (0.79, 54.45) | 0 (0.0) | 6 (0.5) | |
| p-value* | 0.03 | 0.82 | ||||
| rs13168440 | ||||||
| TT | 942 (71.7) | 951 (71.3) | 1.00 (ref) | 131 (69.0) | 853 (72.1) | 1.00 (ref) |
| CT | 350 (26.7) | 344 (25.8) | 1.03 (0.86, 1.22) | 57 (30.0) | 310 (26.2) | 1.18 (0.87, 1.61) |
| CC | 21 (1.6) | 38 (2.9) | 0.56 (0.33, 0.96) | 2 (1.1) | 20 (1.7) | 0.66 (0.16, 2.66) |
| p-value | 0.41 | 0.52 | ||||
| rs12517112 | ||||||
| TT | 636 (48.6) | 661 (49.5) | 1.00 (ref) | 86 (45.5) | 574 (48.6) | 1.00 (ref) |
| CT | 558 (42.6) | 576 (43.1) | 1.01 (0.86, 1.19) | 89 (47.1) | 495 (42.0) | 1.16 (0.86, 1.56) |
| CC | 116 (8.9) | 99 (7.4) | 1.22 (0.91, 1.63) | 14 (7.4) | 111 (9.4) | 0.88 (0.50, 1.55) |
| p-value | 0.32 | 0.81 | ||||
| rs230820 | ||||||
| CC | 415 (31.6) | 398 (30.0) | 1.00 (ref) | 61 (32.1) | 381 (32.2) | 1.00 (ref) |
| AC | 646 (49.1) | 655 (49.3) | 0.95 (0.80, 1.13) | 98 (51.6) | 567 (47.9) | 1.07 (0.78, 1.48) |
| AA | 254 (19.3) | 276 (20.8) | 0.88 (0.71, 1.10) | 31 (16.3) | 237 (20.0) | 0.85 (0.55, 1.31) |
| p-value | 0.25 | 0.59 | ||||
p-value for trend test
SNPs=single nucleotide polymorphisms, OR=odds ratio, HR=hazard ratio, mets=metastases
A significant interaction between rs3877899 and a SNP in the manganese super-oxide dismutase (SOD2) gene, rs4880, was shown previously, as described above (12). We had genotyped and examined rs4880 in the past (21), and we imputed rs3877899 (HapMap R2=0.62 with rs13168440), but did not observe a significant interaction when examined in the same manner as reported, with both SNPs modeled as dominant (data not shown). We also attempted to replicate the published finding that the rare AA genotype of rs7579 was associated with increased PCa risk (11); although this SNP was not genotyped in our study, we imputed it (HapMap R2=0.96 with rs12517112). As in the previous study, we observe an increased but nonsignificant association for those with the AA genotype compared with the GG referent), though the magnitude of effect was lower (OR=1.27; 95% CI: 0.96, 1.68).
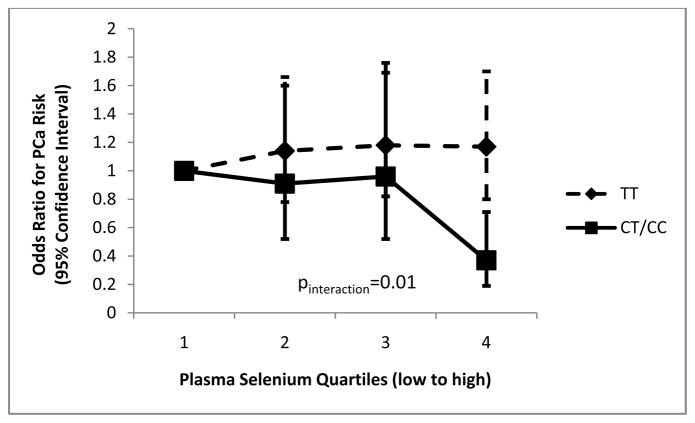
The SEPP1 SNPs were not associated with baseline plasma selenium levels. Selenium quartiles were not associated with case-control status (p=0.22) or PCa survival (p=0.54). However, we observed a significant interaction of rs13168440 with quartiles of plasma selenium for PCa incidence; increasing level of selenium was associated with a decreased risk of PCa only among men carrying the minor C allele, which itself was related to lower risk of PCa -compared to men in the lowest quartile of selenium, those in the highest quartile had an OR of 0.37 (95% CI: 0.19, 0.71; pinteraction=0.01) (Figure 1).
Figure 1.
Association of the interaction of rs13168440 genotypes and plasma selenium quartiles with prostate cancer risk
Although none of the four SNPs was significantly associated with PCa survival, we observed significant interactions of SNPs with pack-years of smoking (rs11959466) and BMI (rs13168440). Increasing category of pack-years of smoking was more strongly associated with an increase in PCa mortality among those carrying the T allele (11.3% of cases) of rs11959466 (HR=1.45; 95% CI: 1.16, 1.81 for each 10-unit pack-year increase) than among those with the CC genotyped (HR=1.09; 95% CI: 0.99, 1.19; pinteraction=0.02). Being overweight or obese was associated with an increased risk of PCa mortality only among those with the common TT genotype of rs13168440 (HR=2.00; 95% CI: 1.41, 2.82) and not among those carrying a C allele (HR=1.01; 95% CI: 0.60, 1.69; pinteraction=0.02).
The mRNA levels of SEPP1 measured in PCa tumors were significantly associated with rs230820 (ptrend=0.05); those with the AA genotype had 3.6% higher expression levels than those with CC. Men with lethal prostate cancer also had significantly lower SEPP1 mRNA expression levels than men with non-lethal disease (p=0.05).
Discussion
We observed that genetic variation in the selenoprotein SEPP1 was associated with PCa risk; one SNP, rs11959466, was associated with an increased risk in an additive manner, while the rare homozygote of another, rs13168440, was associated with a decreased risk. We also observed a significant interaction between rs13168440 and plasma selenium levels where only carriers of the minor allele had reduced PCa risk as selenium levels increased. These findings suggest that this gene may be involved with incidence of PCa, possibly through its function of selenium transport.
Using imputed SNP data, we were unable to replicate a previously reported interaction between an SOD2 rs4880 with SEPP1 rs3877899 (12). However, we did observe a similar borderline significant result for imputed rs7579; the minor allele was associated with increased PCa incidence as previously published (11); this suggests that there may be other SNPs in this gene that are significantly associated with PCa. This SNP is correlated with higher levels of SelP in healthy volunteers, a difference that is no longer significant after selenium supplementation (22), suggesting that supplementation may override the influence of the genetic background.
Although the SNPs themselves were not significantly associated with lethal PCa, one SNP, rs230820, was correlated with gene expression levels of SEPP1 in the tumors of a subset of these men. This SNP itself, or another SNP in linkage disequilibrium with rs230820, could be affecting transcription of this gene. Interestingly, tumor SEPP1 expression levels were significantly lower in men with lethal PCa than among men who did not die from their disease. Additionally, there were significant interactions observed between SEPP1 SNPs and BMI and smoking. All together, these results suggest a potential important antioxidant role of SEPP1 for PCa progression.
Conclusions
In summary, our findings for SEPP1 common genetic variants, the correlation of a variant with tumor expression, and selected interactions with selenium, BMI, and smoking suggest that this gene may affect risk and progression of PCa. Given the limited sample size of our study, especially for lethal PCa, further studies are necessary to confirm or refute these results. However, a growing body of evidence strongly suggests that the association of selenium with prostate cancer may not be straightforward; selenoproteins and specific genetic alleles may modify the relationship.
Acknowledgments
Funding Source This work was supported by grants from the Department of Defense (PC073618 and PC050569). The PHS was supported by grants CA42182, CA34944, CA40360, CA141298, and CA097193 from the National Cancer Institute and grants HL26490 and HL34595 from the National Heart, Lung, and Blood Institute, Bethesda, Maryland. LAM and KLP are supported by the Prostate Cancer Foundation. KLP was supported by a National Research Service Award (T32 CA009001).
We acknowledge Haiyan Zhang, Natalie DuPre, and Jaquelin Jahn for their assistance with this analysis and manuscript preparation.
Footnotes
Disclosure Statement
None of the authors has a conflict of interest relevant to this study.
References
- 1.Duffield-Lillico AJ, Dalkin BL, Reid ME, Turnbull BW, Slate EH, Jacobs ET, Marshall JR, Clark LC. Selenium supplementation, baseline plasma selenium status and incidence of prostate cancer: an analysis of the complete treatment period of the Nutritional Prevention of Cancer Trial. BJU Int. 2003;91(7):608–612. doi: 10.1046/j.1464-410x.2003.04167.x. [DOI] [PubMed] [Google Scholar]
- 2.Yoshizawa K, Willett WC, Morris SJ, Stampfer MJ, Spiegelman D, Rimm EB, Giovannucci E. Study of prediagnostic selenium level in toenails and the risk of advanced prostate cancer. J Natl Cancer Inst. 1998;90(16):1219–1224. doi: 10.1093/jnci/90.16.1219. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Stampfer MJ, Giovannucci EL, Morris JS, Willett WC, Gaziano JM, Ma J. A prospective study of plasma selenium levels and prostate cancer risk. J Natl Cancer Inst. 2004;96(9):696–703. doi: 10.1093/jnci/djh125. [DOI] [PubMed] [Google Scholar]
- 4.Clark LC, Combs GF, Jr, Turnbull BW, Slate EH, Chalker DK, Chow J, Davis LS, Glover RA, Graham GF, Gross EG, Krongrad A, Lesher JL, Jr, Park HK, Sanders BB, Jr, Smith CL, Taylor JR. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional Prevention of Cancer Study Group. JAMA. 1996;276(24):1957–1963. [PubMed] [Google Scholar]
- 5.Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, Parnes HL, Minasian LM, Gaziano JM, Hartline JA, Parsons JK, Bearden JD, 3rd, Crawford ED, Goodman GE, Claudio J, Winquist E, Cook ED, Karp DD, Walther P, Lieber MM, Kristal AR, Darke AK, Arnold KB, Ganz PA, Santella RM, Albanes D, Taylor PR, Probstfield JL, Jagpal TJ, Crowley JJ, Meyskens FL, Jr, Baker LH, Coltman CA., Jr Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA. 2009;301(1):39–51. doi: 10.1001/jama.2008.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatfield DL, Gladyshev VN. How selenium has altered our understanding of the genetic code. Mol Cell Biol. 2002;22(11):3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hill KE, Lloyd RS, Burk RF. Conserved nucleotide sequences in the open reading frame and 3′ untranslated region of selenoprotein P mRNA. Proc Natl Acad Sci U S A. 1993;90(2):537–541. doi: 10.1073/pnas.90.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer HA, Hollenbach B, Stephan C, Endermann T, Morgenthaler NG, Cammann H, Kohrle J, Jung K, Schomburg L. Reduced serum selenoprotein P concentrations in German prostate cancer patients. Cancer Epidemiol Biomarkers Prev. 2009;18(9):2386–2390. doi: 10.1158/1055-9965.EPI-09-0262. [DOI] [PubMed] [Google Scholar]
- 9.Calvo A, Xiao N, Kang J, Best CJ, Leiva I, Emmert-Buck MR, Jorcyk C, Green JE. Alterations in gene expression profiles during prostate cancer progression: functional correlations to tumorigenicity and down-regulation of selenoprotein-P in mouse and human tumors. Cancer Res. 2002;62(18):5325–5335. [PubMed] [Google Scholar]
- 10.Gonzalez-Moreno O, Boque N, Redrado M, Milagro F, Campion J, Endermann T, Takahashi K, Saito Y, Catena R, Schomburg L, Calvo A. Selenoprotein-P is down-regulated in prostate cancer, which results in lack of protection against oxidative damage. Prostate. 2011;71(8):824–834. doi: 10.1002/pros.21298. [DOI] [PubMed] [Google Scholar]
- 11.Steinbrecher A, Meplan C, Hesketh J, Schomburg L, Endermann T, Jansen E, Akesson B, Rohrmann S, Linseisen J. Effects of selenium status and polymorphisms in selenoprotein genes on prostate cancer risk in a prospective study of European men. Cancer Epidemiol Biomarkers Prev. 2010;19(11):2958–2968. doi: 10.1158/1055-9965.EPI-10-0364. [DOI] [PubMed] [Google Scholar]
- 12.Cooper ML, Adami HO, Gronberg H, Wiklund F, Green FR, Rayman MP. Interaction between single nucleotide polymorphisms in selenoprotein P and mitochondrial superoxide dismutase determines prostate cancer risk. Cancer Res. 2008;68(24):10171–10177. doi: 10.1158/0008-5472.CAN-08-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penney KL, Schumacher FR, Li H, Kraft P, Morris JS, Kurth T, Mucci LA, Hunter DJ, Kantoff PW, Stampfer MJ, Ma J. A large prospective study of SEP15 genetic variation, interaction with plasma selenium levels, and prostate cancer risk and survival. Cancer Prev Res (Phila) 2010;3(5):604–610. doi: 10.1158/1940-6207.CAPR-09-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arsova-Sarafinovska Z, Matevska N, Eken A, Petrovski D, Banev S, Dzikova S, Georgiev V, Sikole A, Erdem O, Sayal A, Aydin A, Dimovski AJ. Glutathione peroxidase 1 (GPX1) genetic polymorphism, erythrocyte GPX activity, and prostate cancer risk. Int Urol Nephrol. 2009;41(1):63–70. doi: 10.1007/s11255-008-9407-y. [DOI] [PubMed] [Google Scholar]
- 15.Final report on the aspirin component of the ongoing Physicians’ Health Study. Steering Committee of the Physicians’ Health Study Research Group. N Engl J Med. 1989;321(3):129–135. doi: 10.1056/NEJM198907203210301. [DOI] [PubMed] [Google Scholar]
- 16.Roberts DG, Morrison TB, Liu-Cordero SN, Cho J, Garcia J, Kanigan TS, Munnelly K, Brenan CJ. A nanoliter fluidic platform for large-scale single nucleotide polymorphism genotyping. Biotechniques. 2009;46(3 Suppl):ix–xiii. doi: 10.2144/000112887. [DOI] [PubMed] [Google Scholar]
- 17.Setlur SR, Mertz KD, Hoshida Y, Demichelis F, Lupien M, Perner S, Sboner A, Pawitan Y, Andren O, Johnson LA, Tang J, Adami HO, Calza S, Chinnaiyan AM, Rhodes D, Tomlins S, Fall K, Mucci LA, Kantoff PW, Stampfer MJ, Andersson SO, Varenhorst E, Johansson JE, Brown M, Golub TR, Rubin MA. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J Natl Cancer Inst. 2008;100(11):815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorisson GA, Smith AV, Krishnan L, Stein LD. The International HapMap Project Web site. Genome Res. 2005;15(11):1592–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Kantoff PW, Giovannucci E, Leitzmann MF, Gaziano JM, Stampfer MJ, Ma J. Manganese superoxide dismutase polymorphism, prediagnostic antioxidant status, and risk of clinical significant prostate cancer. Cancer Res. 2005;65(6):2498–2504. doi: 10.1158/0008-5472.CAN-04-3535. [DOI] [PubMed] [Google Scholar]
- 22.Meplan C, Nicol F, Burtle BT, Crosley LK, Arthur JR, Mathers JC, Hesketh JE. Relative abundance of selenoprotein P isoforms in human plasma depends on genotype, se intake, and cancer status. Antioxid Redox Signal. 2009;11(11):2631–2640. doi: 10.1089/ARS.2009.2533. [DOI] [PubMed] [Google Scholar]