Abstract
Self-complementary AAV (scAAV) vector genomes contain a covalently closed hairpin derived from a mutated inverted terminal repeat that connects the two monomer single-stranded genomes into a head-to-head or tail-to-tail dimer. We found that during quantitative PCR (qPCR) this structure inhibits the amplification of proximal amplicons and causes the systemic underreporting of copy number by as much as 10-fold. We show that cleavage of scAAV vector genomes with restriction endonuclease to liberate amplicons from the covalently closed terminal hairpin restores quantitative amplification, and we implement this procedure in a simple, modified qPCR titration method for scAAV vectors. In addition, we developed and present an AAV genome titration procedure based on gel electrophoresis that requires minimal sample processing and has low interassay variability, and as such is well suited for the rigorous quality control demands of clinical vector production facilities.
Fagone and colleagues address challenges in quantitative PCR titration of self-complementary adeno-associated virus (scAAV). They show that restriction nuclease cleavage of covalently closed hairpins present in scAAV genomes restores quantitative amplification of hairpin-proximal amplicons. An alternative AAV titration procedure that requires minimal sample processing and shows low interassay variability is also presented.
Introduction
Adeno-associated viral (AAV) vectors are achieving striking preclinical and clinical successes and are increasingly being considered for development as human therapeutics for treatment of a variety of human diseases. Concordant with that progress has been an increased focus on the development of rigorous quality control assays to support regulatory applications for new product licensure. One of the most important assays for gene therapy vectors is that of vector genome titration, as the vector genome is the key mediator of therapeutic effect for any genetic therapy, and vector genome quantification is almost always used for dose determination.
AAV vector genome titer has been determined by a wide variety of methods, including Southern blotting (Halbert et al., 1997), dot-blot hybridization (Samulski et al., 1989), ultraviolet (UV) spectrophotometry (Sommer et al., 2003), and quantitative real-time PCR (qPCR) (Clark et al., 1999). UV spectrophotometry is a direct and simple assay, and although it can quantify both capsid protein and genomic DNA, results can be significantly impacted by cellular protein and DNA impurities. Southern blot hybridization involves complex laboratory procedures yet provides high sensitivity and specificity, and additionally confirms the integrity of the vector genome. By omitting the gel electrophoresis step, dot-blot assays provide a simpler method of quantitation by hybridization, but do not provide information about genome integrity and require sample processing to remove protein contaminants, including capsid. qPCR is rapid, convenient to perform, and has a broad dynamic range, but quantifies only the fragment of the vector genome complementary to the PCR primers and probe. The specificity, convenience of use through standardized reagent kits and equipment, and the broad dynamic range have established qPCR as the most frequently used quantitative assay for AAV vector genome titration.
Our self-complementary AAV (scAAV) vector encoding a codon-optimized factor IX cDNA and a small liver-specific promoter has shown robust efficacy in preclinical models of hemophilia B gene therapy (Nathwani et al., 2011), and has to date been administered to six patients with severe hemophilia B in a phase I/II clinical trial (Nathwani et al., 2010). Unlike traditional single-stranded AAV (ssAAV), scAAV vectors use novel terminal repeat mutations and short genome sizes to force packaging of tail-to-tail or head-to-head dimer genomes (Fig. 1a) (McCarty et al., 2003; Wang et al., 2003). As the dimer genomes can anneal immediately on capsid uncoating to form double-stranded, transcriptionally competent DNA templates, they circumvent a significant block to transduction caused by the need for cellular polymerases to copy ssAAV vector genome templates before transcription (McCarty et al., 2001).
FIG. 1.
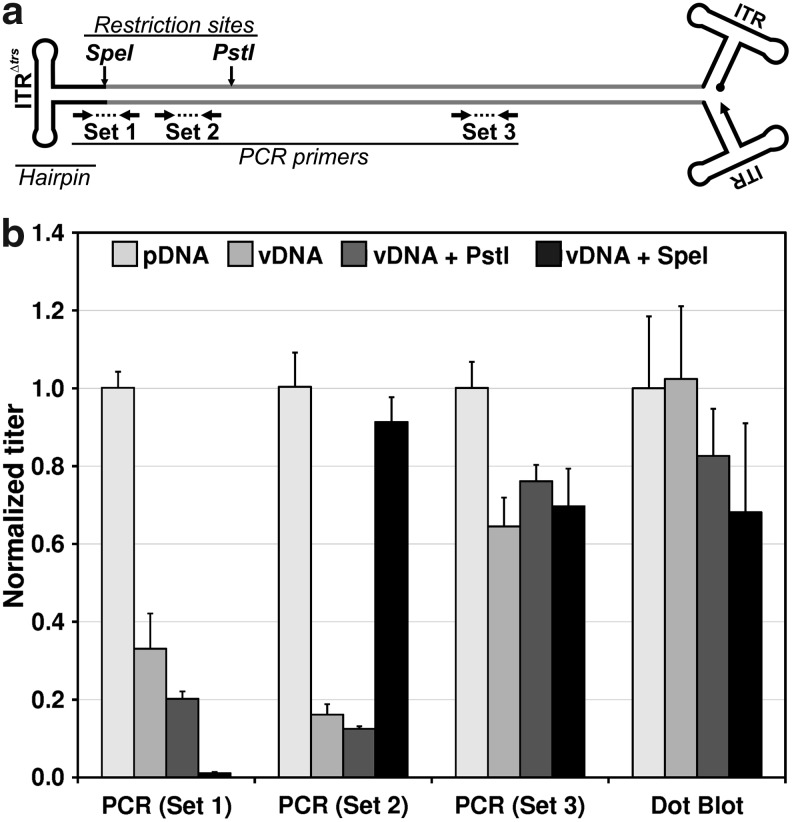
Self-complementary AAV (scAAV) structure and hairpin effect on quantitative PCR (qPCR). (a) Diagram of the self-complementary AAV vector (scAAV-LP1-hFIXco) genome used in the experiment, with primer and restriction endonuclease sites indicated. (b) Measured titer values for each of the prepared templates, using the qPCR or dot-blot method. For each analytical method, the quantity determined for the linearized plasmid was set to a value of one, and the measured values for the other templates at the same input quantity are shown relative to the plasmid value. Error bars indicate the standard deviation of replicates.
During production and quality control of our clinical-grade vector preparation of the scAAV factor IX vector, we noted a discrepancy between the apparent titer of preparations when determined by qPCR relative to other more direct methods such as UV spectrophotometry and the bicinchoninic acid (BCA) assay for total protein (Allay et al., 2011). We now show in this report that the underreporting of titer by qPCR is a systemic error caused by the presence of the covalently closed hairpin at one end of the self-complementary vector genome. In addition, we describe alternative protocols based on both gel electrophoresis and qPCR that circumvent this issue and enable more robust and reliable vector genome titration for self-complementary and single-stranded AAV vectors.
Materials and Methods
Preparation and purification of scAAV2/8-LP1-hFIXco and ssAAV2-hFIX
Preparation, purification, and characterization of scAAV2/8-LP1-hFIXco has been described by Allay and colleagues (2011). The single-stranded AAV vector, AAV2-hFIX, lot number QCR-11002RV, was produced by the vector production facility at Children's Hospital of Philadelphia (Philadelphia, PA).
scAAV-LP1-hFIXco genome purification and endonuclease treatment
Genomic DNA was purified from scAAV2/8-LP1-hFIXco, using a QIAamp MinElute virus vacuum kit (Qiagen, Valencia, CA). Genome fragments with and without hairpin were prepared by incubation with an endonuclease (SpeI, EcoRI, or PstI; New England BioLabs, Ipswich, MA); endonucleases were deactivated by heating at 80°C for 30 min and the restriction buffer was exchanged for Tris–EDTA (TE), using Amicon centrifugal filters (Ultracel-100k; Millipore, Billerica, MA). Treated and untreated DNA samples were quantified by the native agarose gel method. For templates with multiple fragments, an average copy number was calculated, using the fragments with migration and staining intensity within the range of the quantitative standards.
AAV titration by qPCR
Quantitative PCR was done with the EcoRI-linearized hFIXco vector plasmid as reference standard. DNA templates were diluted in 0.05% Pluronic F68 (Sigma-Aldrich, St. Louis, MO) aqueous solution; the 10-fold serial dilution of the reference standard ranged from 2.5×102 to 2.5×108 double-stranded DNA (dsDNA) copies per 25-μl PCR reaction. PCR was done with 1 μM primers (see Table 1 for a list of all qPCR primers used in this work), and the following thermal cycling conditions: one 3-min cycle at 95°C followed by forty 3-step cycles (30 sec at 95°C, 30 sec at 55°C, and 30 sec at 72°C). PCR was done with an iCycler thermal cycler equipped with an MyiQ optical module and both were controlled with MyiQ optical system software 2.0 (version 2.0.1442.0 CR) (Bio-Rad Laboratories, Hercules, CA).
Table 1.
qPCR Primers Used in Titration Assays
| Target vector | Primer label | Primer distance from the hairpin (bp) | Primer sequence |
|---|---|---|---|
| scAAV-LP1-hFIXco | Set 1 | 41 | Forward: ATTTTATGTTTCAGGTTCAGGGGGAGGTG Reverse: GCGCAGAGAGGGAGTGGACTAGT |
| Set 2 | 162 | Forward: TGTGAACTGGATCAAGGAGAAG Reverse: AAGCAATAGCATCACAAATTTCA |
|
| Set 3 | 1204 | Forward: CATGGAGGAGAAGTGCAGCTTTGA Reverse: ATGTTGCAGGTCACATCCAGCTCA |
|
| Set 4 | 1154 | Forward: GCAGCTGCAAGGATGACATCAACA Reverse: GCAGAACTGCTCACATCTGCCATT |
|
| Set 5 | 2014 | Forward: GCAAGCAGCAAACAGCAAACACAC Reverse: TGCTCCTCTCCACCGAAATTCCAA |
|
| scAAV-LP1-PPCA | Set 6 | 320 | Forward: TGGAGATGTAGACATGGCCTGCAA Reverse: TGAGAAAGGCGATGTGGGAGAACT |
| Set 7 | 1015 | Forward: ATTTCTTCCGCCTCTTTCCGGAGT Reverse: ACCAGGGAGTTGTCATTCTGCTCA |
|
| Set 5 | 2092 | Forward: GCAAGCAGCAAACAGCAAACACAC Reverse: TGCTCCTCTCCACCGAAATTCCAA |
|
| scAAV-LP1-Cre | Set 8 | 567 | Forward: ACGTACTGACGGTGGGAGAATGTT Reverse: AGCTACACCAGAGACGGAAATCCA |
| Set 9 | 1197 | Forward: TGCCCAAGAAGAAGAGGAAGGTGT Reverse: ATCACTCGTTGCATCGACCGGTAA |
|
| Set 5 | 1702 | Forward: GCAAGCAGCAAACAGCAAACACAC Reverse: TGCTCCTCTCCACCGAAATTCCAA |
|
| scAAV-LP1-GFP | Set 10 | 445 | Forward: TCTTCTTCAAGGACGACGGCAACT Reverse: TTCACCTTGATGCCGTTCTTCTGC |
| Set 5 | 1395 | Forward: GCAAGCAGCAAACAGCAAACACAC Reverse: TGCTCCTCTCCACCGAAATTCCAA |
DNA dot blotting
Reference DNA standard and the viral genome templates in 0.4 M NaOH were transferred onto a Zeta-Probe GT (genomic tested) blotting membrane (Bio-Rad Laboratories), washed with 2×saline–sodium citrate (SSC) buffer, and UV-cross-linked. [α-32P]dCTP (MP Biomedicals, Solon, OH) DNA probe was generated from an EcoRI–PstI DNA fragment excised from the scAAV-LP1-hFIXco plasmid with an NEBlot kit (New England BioLabs) as per the manufacturer's protocol. The membrane was prehybridized for 1 hr at 42°C with Hybrisol I (Millipore) and incubated overnight with the radiolabeled probe in Hybrisol I. After exposure of a phosphorus screen to the membrane, the latent image was recovered with a Personal Molecular Imager system controlled by Quantity One (version 4.6.2, build 070; Bio-Rad Laboratories). Spot intensity was determined with Image Lab software (version 2.0.1, build 18; Bio-Rad Laboratories); regression analysis was done with Excel (Microsoft, Redmond, WA).
scAAV titration by endonuclease digestion qPCR
Five microliters of scAAV was mixed with 5 μl of 1.0% sodium dodecyl sulfate (SDS) and the final volume was adjusted to 50 μl with double-distilled H2O, incubated at 95°C for 10 min, and then cooled to room temperature at a thermal gradient of 3°C/min. The denatured virus was then supplemented with 50 μl of a 1.0% aqueous solution of Nonidet P-40 (NP-40; Roche Applied Science, Indianapolis, IN) and 10 μl was used in a 50-μl endonuclease reaction using 5 units of SpeI (New England BioLabs) for the removal of the hairpin. The cleaved product was diluted 10-fold in 0.05% Pluronic F68 aqueous solution and used as template in accordance with the already described qPCR protocol. For pretreatment with DNase I, 5 μl of scAAV was mixed with 40 U of DNase I (Invitrogen, Carlsbad, CA) in 40 μl of 1×restriction buffer 3 (New England BioLabs) and incubated for 30 min at 37°C; the sample was then supplemented with 10 μl of proteinase K cocktail (0.5% SDS and proteinase K [20 U/ml; Sigma-Aldrich] in nuclease-free water), incubated for 30 min at 56°C and for 20 min at 96°C, and then cooled to room temperature at a thermal gradient of 3°C/min. The denatured virus was then supplemented with NP-40 and treated with SpeI as described previously. The linearity of this assay was confirmed for input scAAV vector preparations with titers in the range of 5×1010 to 1×1013 genome copies (GC)/ml. Below the lower limit the assay underreported, possibly due to incomplete endonuclease digestion (data not shown). It was not tested at input vector titers above 1013 GC/ml.
Preparation of the internal DNA reference to be used for AAV titration by native and alkaline agarose gel
LITMUS 29 (New England BioLabs) was incubated with ApoI endonuclease (New England BioLabs) to completion of plasmid digestion, as determined by native agarose gel electrophoresis. The treated plasmid was then phenol–chloroform purified, ethanol precipitated, solubilized in TE buffer, and quantified by UV spectrophotometry.
scAAV titration by native gel
AAVs were incubated in 0.5% SDS at 95°C for 10 min and then cooled to room temperature at a thermal gradient of 3°C/min. The viral samples and high DNA mass ladder (Invitrogen) were then mixed with 2×LBR (2×loading buffer with reference: 10% glycerol, 2×Tris–acetate–EDTA [TAE] buffer, 0.6% SDS, 0.02% bromophenol blue, ApoI-treated LITMUS 29 [6 ng/μl]). Samples were loaded onto a 1.0% agarose gel in TAE buffer electrophoresed for 90 min at 5 V/cm of gel length. The gel was then stained for 2 hr in an equal gel volume of DNA-staining solution (2×GelRed in 0.1 M NaCl, prepared from 10,000×GelRed; Biotium, Hayward, CA); the gel image was acquired without destaining under flat-fielding conditions, using the ChemiDoc XRS system and Quantity One (version 4.6.2, build 070; Bio-Rad Laboratories). Band volumes were determined from the gel image with Image Lab software; Microsoft Excel was used to normalize each band volume to the reference band, for the regression analysis, and the subsequent calculation of viral titer. The linearity of this assay was confirmed for input scAAV vector titers as between 2×1011 and 1×1013 GC/ml (data not shown).
ssAAV titration by alkaline gel
The quantification standard (identical in sequence to the test article vector genome) and the intact vector samples were mixed with freshly prepared 2×alkaline loading buffer with reference: 10% glycerol, 2×alkaline running buffer, 0.6% SDS, 0.01% Alizarin yellow R (Alfa Aesar, Ward Hill, MA), and ApoI-treated LITMUS 29 (6 ng/μl). No heating was required because the presence of SDS and the alkaline conditions were sufficient to free the viral genome from the capsid. Samples were loaded onto a 0.8% agarose gel, prepared in TAE buffer, and buffer-exchanged in fresh prepared alkaline running buffer (30 mM NaOH, 1 mM EDTA). After analysis by electrophoresis for 4 hr at 2.5 V/cm of gel length, with recirculating ice-cold alkaline running buffer, the gel was incubated for 1 hr in 3 vol of 100 mM Tris-HCl (pH 8.0) and then stained for 2 hr in an equal gel volume of DNA staining solution (4×GelRed in 0.1 M NaCl). Image acquisition and analysis were done as for the native gel protocol.
AAV titration by UV
ssAAV and scAAV were titrated by UV (Sommer et al., 2003), using constants specific for each virus DNA length: 4300 nucleotides of single-stranded DNA (ssDNA) for ssAAV2-hFIX, 4620 nucleotides of ssDNA for scAAV2/8-LP1-hFIXco, and an average molecular mass per nucleotide of 308 Da; a value of 0.56 was assigned to the A260/A280 ratio for AAV serotype 2 and 8 capsid proteins. Viral samples were diluted in aqueous 0.1% SDS (final concentration), heated at 95°C for 10 min, and then left to cool to room temperature at a thermal gradient of 3°C/min. UV spectra are then acquired with a Beckman Coulter DU 800 UV/visible spectrophotometer and system and application software (version 2.0, build 74; Beckman Coulter, Brea, CA).
Statistical analysis
The mean titers (Table 2) were compared by unpaired t test, using the GraphPad QuickCalcs online calculator (GraphPad Software, La Jolla, CA; www.graphpad.com/quickcalcs/), and considered significant when p<0.05. The coefficient of variation (CV) for each method was calculated as a percentage from the standard deviation (σ) and the absolute value of the mean (μ): (CV=σ÷| μ |×100).
Table 2.
Titers of ssAAV and scAAV Determined by Various Methods
| Meana | Rangea | SDa | n | ||
|---|---|---|---|---|---|
| scAAV2/8-LP1-hFIXco | UV | 6.3×1012 | 4.8–7.5×1012 | 1.4×1012 | 5 |
| ED-qPCR | 4.5×1012 | 3.9–5.2×1012 | 6.8×1011 | 4 | |
| Gelb | 4.5×1012 | 4.1–4.6×1012 | 2.7×1011 | 3 | |
| ssAAV2-hFIX | UV | 1.2×1013 | 9.7–13.6×1012 | 1.4×1012 | 5 |
| qPCR | 7.6×1012 | 6.4–9.7×1012 | 1.3×1012 | 3 | |
| Gelc | 8.9×1012 | 7.8–9.7×1012 | 1.0×1012 | 3 |
ED-qPCR, endonuclease digestion qPCR; scAAV, self-complementary AAV; ssAAV, single-stranded AAV.
Titers are expressed as genome copies per milliliter.
Native agarose gel electrophoresis.
Alkaline agarose gel electrophoresis.
Results and Discussion
A key assumption for qPCR of AAV genomes is that after heat denaturation, the kinetics of PCR primer annealing to target sites is much faster than the kinetics of annealing of plus and minus strand-complementary vector genomes. Given the discrepancy observed between qPCR titers and other measures of vector concentration for our scAAV vector, our experimental plan was based on the hypothesis that the covalently closed hairpin (derived from the Δtrs inverted terminal repeat [ITR], and hereafter referred to as the “terminal hairpin”) joining the two complementary strands encoding the hFIX expression cassette would enhance reannealing to the exclusion of primer binding. As a result we might see reduced sensitivity with templates containing the hairpin, but not with those from which the hairpin had been removed. We therefore prepared a series of PCR templates derived either from linearized plasmid DNA or self-complementary AAV vector genomic DNA both with and without restriction endonuclease digestion to separate the terminal hairpin from target PCR amplicons (Fig. 1a). The vector genome was purified from clinical-grade scAAV2/8-LP1-hFIXco vector preparations that have been extensively characterized and shown to contain almost exclusively dimer genomes (Allay et al., 2011). After endonuclease digestion all DNAs were quantified by direct visualization of fluorescent dye-stained native agarose gels containing the test article along with known DNA mass standards. These DNA templates were then subjected to four different analyses, in all cases using the plasmid DNA template as a standard. The first three analyses used qPCR with different primer pairs to query amplicons at various distances from the terminal hairpin, whereas the fourth involved dot-blot hybridization using a radioactive probe and quantitative imaging. Figure 1b presents the quantities measured by each of the assays, showing the actual measured value normalized to that determined for the linearized plasmid DNA reference control.
When templates containing a terminal hairpin were analyzed with primers near the hairpin, the values reported were significantly reduced, in some cases by as much as 8-fold. SpeI restriction endonuclease digestion of the genome restored the observed value for primer set 2 to more than 90%, confirming that the terminal hairpin was responsible for the underreporting by qPCR. The primer set 1 amplicon was cleaved between the primers by SpeI, and as expected reported near-zero values on that template. qPCR using primer set 3 and dot-blot hybridization both appeared to be less affected by the molecular form of the amplicon, although small variations were observed.
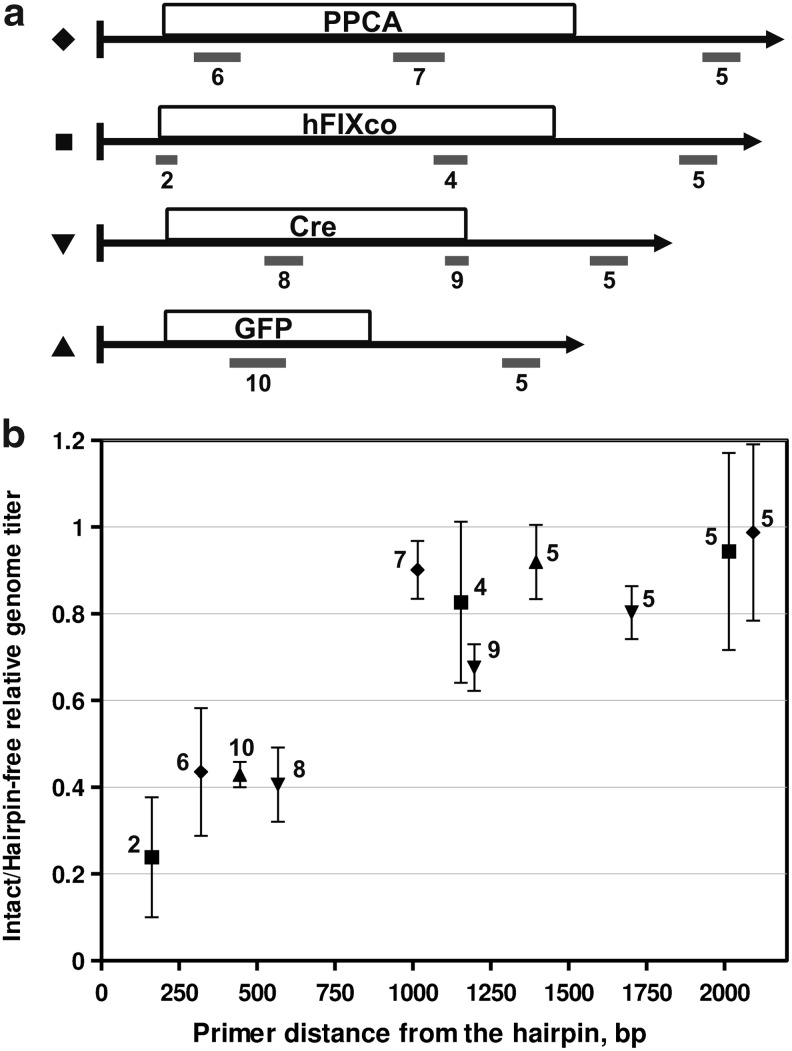
To confirm that the qPCR bias was a general phenomenon and not specific to our particular genome and primer set, we performed qPCR assays on a set of four different scAAV vectors, using eight different primer sets (see Fig. 2a for maps of amplicon locations). In each case, we calculated the ratio of the observed titer without and with digestion with SpeI (which is adjacent to the terminal hairpin in all four vectors). This ratio is shown in Fig. 2b, plotted as a function of the distance of the amplicon from the terminal hairpin. The data clearly show that the magnitude of the bias, interpreted from the ratio of SpeI-untreated versus SpeI-treated titers, is a function of the distance from the terminal hairpin, and not related to any particular primer or template sequence.
FIG. 2.
Relationship between amplicon distance and qPCR bias. (a) Schematic maps of the four vectors and eight qPCR amplicons that were used in the experiment. Each vector genome map is preceded at the left by the symbol used in the graph below for the observed titer ratio. All vectors are drawn with the covalently closed terminal hairpin (left). Beneath each vector gray bars indicate the location of each numbered amplicon. “hFIXco” corresponds to the clinical vector used in Fig. 1. (b) Plot of relative qPCR genome titer for eight separate qPCR primer sets used in four different vector preparations. Each data point is labeled with the amplicon number and represents the average calculated ratio of observed titer between undigested and digested vector genomes. Error bars indicate the standard deviation of at least three independent quantifications.
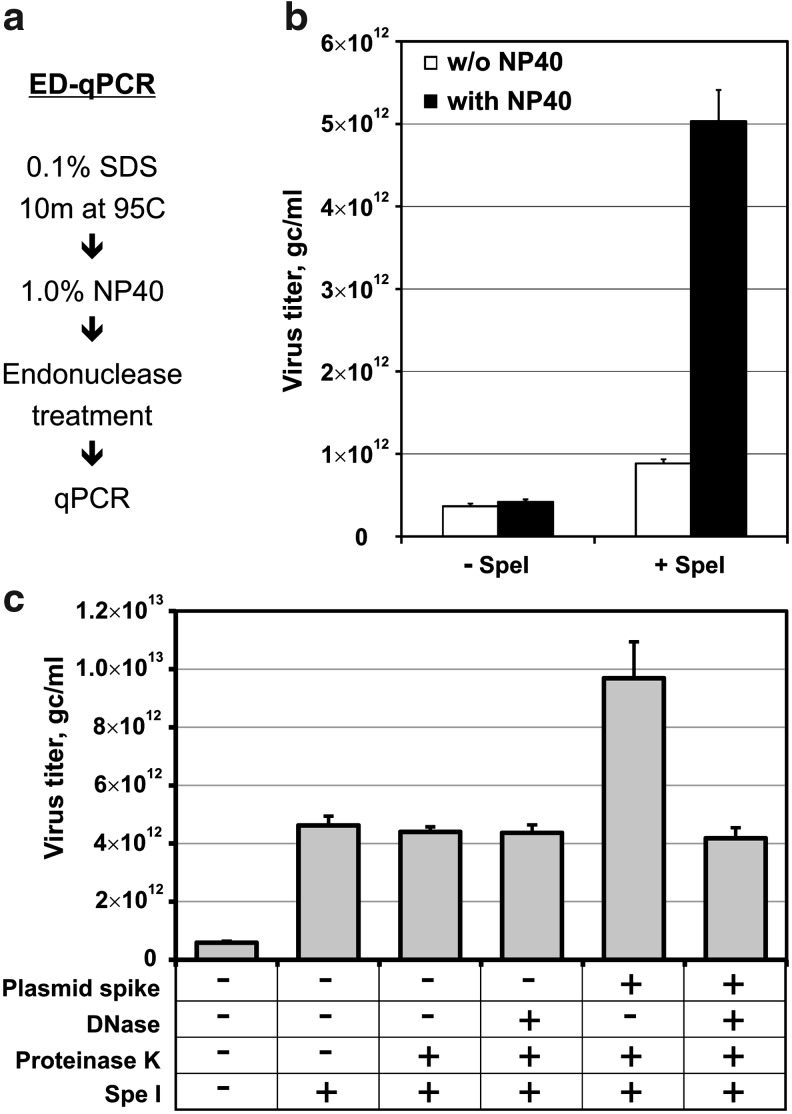
The ability of restriction endonuclease digestion to restore quantitative template behavior led us to develop a rapid, modified qPCR protocol (termed endonuclease digestion qPCR, or ED-qPCR) that could be used on self-complementary vector preparations without the need for DNA purification (Fig. 3a). Particles are first denatured in 0.1% SDS by heating at 95°C for 10 min, and samples are then treated with 1% NP-40 in order to sequester the SDS (Panyukov et al., 2008) and prevent it from interfering with subsequent steps. Preparations can then be directly digested with restriction endonuclease before quantitation by qPCR using primer set 2 (Table 1). Figure 3b shows the affect of these pretreatment protocols on the observed titer from a clinical-grade preparation of an scAAV vector that had a previously determined qPCR titer of ∼4×1011 VG/ml. Only when the dual detergent treatment is followed by restriction endonuclease digestion does the observed titer increase by approximately 10-fold, which is consistent with the 8-fold effect attributable to the hairpin observed previously (Fig. 1). To extend the ED-qPCR technique to samples potentially contaminated with unencapsidated DNA, we additionally show in Fig. 3c that the addition of proteinase K and DNase I treatments before the SpeI digestion step allows the elimination of spiked plasmid DNA without decreasing the signal derived from vector genomic DNA.
FIG. 3.
Endonuclease digestion qPCR. (a) Schematic diagram of the technique. (b) Graph of measured values for clinical-grade scAAV2/8-LP1-hFIXco preparation, when technique is performed under the conditions indicated (see Materials and Methods). Error bars represent standard deviation of replicates. (c) Quantitative PCR results for a similar scAAV preparation, when performed with the additional treatments indicated. “Plasmid spike” indicates addition of 6.8×1012 GC/ml of linearized plasmid standard to vector test article before the procedures.
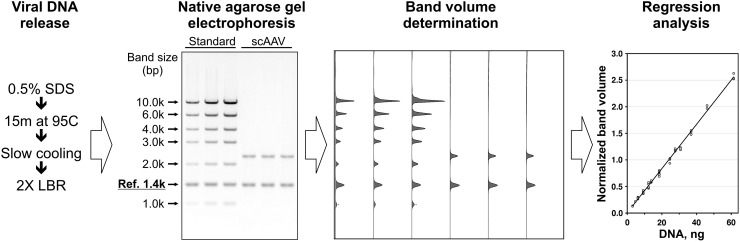
Although qPCR is a sensitive technique that provides a broad dynamic range, interassay variation is typically significant (Lock et al., 2010) and a preferred method would provide robust assay linearity without a requirement for extensive dilution of the test article. We therefore implemented an agarose gel analysis protocol to directly quantify self-complementary AAV genomes within particles. Figure 4 outlines this technique, wherein vector particles are minimally processed to release double-stranded DNA, mixed with loading buffer containing a known concentration of a reference DNA, and loaded on a native agarose gel alongside commercially available quantitative standards. After electrophoresis and staining with fluorescent dye, a digital image can be quantified with commonly available software and the total signal of each DNA band calculated relative to the in-lane reference band, which corrects for uneven gel loading or imaging artifacts. As illustrated by the standard curve shown in Fig. 4, the assay is linear (r2=0.996) and replicates show low variation. Although the native gels provide more reliable quantitation of double-stranded vectors, an alternative, similar protocol using denaturing alkaline gels was also developed for single-stranded vectors, which do not migrate uniformly on native gels. Although this technique performed comparably to the native gel assay (see Materials and Methods; and data not shown), sequence-dependent variability of staining intensities for single-stranded vector genomes necessitated the use of quantitative DNA standards with sequence identical to the vector test article.
FIG. 4.
Agarose gel titration method. Shown are the sample preparation process, a representative gel image used for analysis, pseudo-chromatograms generated by image processing, and the resulting standard curve used for quantitation. LBR, loading buffer with Reference; Ref. 1.4k, internal Reference DNA band.
To corroborate the various titration methods that we have developed, we performed UV spectrophotometry, ED-qPCR, and agarose gel titration on two independent AAV vector preparations. Both were of clinical-grade purity but one was single stranded and one was self-complementary. Each assay method was performed at least three times on separate days, with each assay containing at least three replicates (Table 2). Remarkably, measured values for each preparation were mostly statistically indistinguishable (p>0.05), although the UV-determined values tended to be higher and in the case of the ssAAV vector were significantly different from the other methods. Regarding interassay variation, for the native gel titration method the coefficient of variation was only 6%, and on alkaline gels the variation was still low but slightly higher at 11%.
Our results demonstrate that standard qPCR methods for AAV vector titration can be highly influenced by the molecular structure of the DNA genome; with self-complementary vectors the measured titer can be as much as 10-fold below the actual titer as determined by other methods. Although early publications comparing scAAV and ssAAV vectors used Southern or dot blotting (McCarty et al., 2001, 2003; Wang et al., 2003), procedures that are apparently not affected by the terminal hairpin (Fig. 1b), the qPCR artifact described herein might cause overestimation of the benefit in potency gained by the use of self-complementary vector genome. We note that any sequence generating secondary structure in AAV genomes may also affect the accuracy of titration by qPCR, further emphasizing the benefit of secondary assays to confirm dosing titer accurately. In the case of scAAV vector artifacts, our results also show that the qPCR titers are highly dependent on the specific primer set used, with amplicons closer to the hairpin being significantly underreported. This bias can be corrected by incorporating relatively simple endonuclease digestion steps into the assay protocol. The novel gel titration assay methodology described herein provides an additional robust and reliable quantitative assay for purified vector preparations, one that is well suited for further development into a validated quality control assay for clinical-grade vector preparations.
Acknowledgments
The authors acknowledge Katherine High, Bernd Hauck, and Olga Zelenaia for scientific discussions. This work was supported by grants from the Assisi Foundation of Memphis, the American Lebanese Syrian Associated Charities (ALSAC), the National Heart, Lung, and Blood Institute (grant HL073838), and the National Cancer Institute (Cancer Center Support grant CA027165).
Author Disclosure Statement
The authors have no competing interests to disclose.
References
- Allay J.A. Sleep S.E. Long S., et al. Good manufacturing practice production of self-complementary serotype 8 adeno-associated viral vector for a hemophilia B clinical trial. Hum. Gene Ther. 2011;22:595–604. doi: 10.1089/hum.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K.R. Liu X. McGrath J.P. Johnson P.R. Highly purified recombinant adeno-associated virus vectors are biologically active and free of detectable helper and wild-type viruses. Hum. Gene Ther. 1999;10:1031–1039. doi: 10.1089/10430349950018427. [DOI] [PubMed] [Google Scholar]
- Halbert C.L. Standaert T.A. Aitken M.L., et al. Transduction by adeno-associated virus vectors in the rabbit airway: Efficiency, persistence, and readministration. J. Virol. 1997;71:5932–5941. doi: 10.1128/jvi.71.8.5932-5941.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock M. McGorray S. Auricchio A., et al. Characterization of a recombinant adeno-associated virus type 2 Reference Standard Material. Hum. Gene Ther. 2010;21:1273–1285. doi: 10.1089/hum.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty D.M. Monahan P.E. Samulski R.J. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- McCarty D.M. Fu H. Monahan P.E., et al. Adeno-associated virus terminal repeat (TR) mutant generates self-complementary vectors to overcome the rate-limiting step to transduction in vivo. Gene Ther. 2003;10:2112–2118. doi: 10.1038/sj.gt.3302134. [DOI] [PubMed] [Google Scholar]
- Nathwani A. Tuddenham E. Rosales C., et al. Early clinical trial results following administration of a low dose of a novel self complementary adeno associated viral vector encoding human factor IX in two subjects with severe hemophilia B. Blood. 2010;116:114–114. [Google Scholar]
- Nathwani A.C. Rosales C. McIntosh J., et al. Long-term safety and efficacy following systemic administration of a self-complementary AAV vector encoding human FIX pseudotyped with serotype 5 and 8 capsid proteins. Mol. Ther. 2011;19:876–885. doi: 10.1038/mt.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyukov Y.V. Nemykh M.A. Dobrov E.N. Drachev V.A. Surfactant-induced amorphous aggregation of tobacco mosaic virus coat protein: A physical methods approach. Macromol. Biosci. 2008;8:199–209. doi: 10.1002/mabi.200700145. [DOI] [PubMed] [Google Scholar]
- Samulski R.J. Chang L.S. Shenk T. Helper-free stocks of recombinant adeno-associated viruses: Normal integration does not require viral gene expression. J. Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer J.J. Smith P.H. Parthasarathy S., et al. Quantification of adeno-associated virus particles and empty capsids by optical density measurement. Mol. Ther. 2003;7:122–128. doi: 10.1016/s1525-0016(02)00019-9. [DOI] [PubMed] [Google Scholar]
- Wang Z. Ma H.I. Li J., et al. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 2003;10:2105–2111. doi: 10.1038/sj.gt.3302133. [DOI] [PubMed] [Google Scholar]