Abstract
Biofilms formed by nontypeable Haemophilus influenzae (NTHI) are associated with multiple chronic infections of the airway, including otitis media. Extracellular DNA (eDNA) is part of the biofilm matrix and serves as a structural component. Human β-defensin-3 (hBD-3) is a cationic antimicrobial host defense protein (AMP) critical to the protection of the middle ear. We hypothesized that anionic eDNA could interact with and bind hBD-3 and thus shield NTHI in biofilms from its antimicrobial activity. We demonstrated that recombinant hBD-3 [(r)hBD-3] bound eDNA in vitro and that eDNA in biofilms produced by NTHI in the chinchilla middle ear co-localized with the orthologue of this AMP. Incubation of physiological concentrations of (r)hBD-3 with NTHI genomic DNA abrogated the ability of this innate immune effector to prevent NTHI from forming robust biofilms in vitro. Establishment of NTHI biofilms in the presence of both DNase I and (r)hBD-3 resulted in a marked reduction in the overall height and thickness of the biofilms and rescued the antimicrobial activity of the AMP. Our results demonstrated that eDNA in NTHI biofilms sequestered hBD-3 and thus diminished the biological activity of an important effector of innate immunity. Our observations have important implications for chronicity of NTHI-induced diseases.
Key Words: Cationic host defense peptides, Extracellular DNA, Innate immunity, Middle ear, Nontypeable Haemophilus influenzae, Otitis media
Introduction
Nontypeable Haemophilus influenzae (NTHI) is a Gram-negative bacterium that is a member of the normal flora of the human nasopharynx. This opportunistic pathogen is capable of causing a wide range of diseases of both the lower and upper airways, including exacerbations of chronic obstructive pulmonary disease and otitis media (OM) [1, 2, 3, 4]. OM is the most frequently diagnosed illness in children under 15 years of age with total costs of diagnosing and managing OM exceeding USD 5 billion annually in the US alone [5, 6]. In addition to the high prevalence of OM in developed countries, worldwide it is reported that between 65 and 330 million children suffer from chronic secretory OM, 60% of whom have an associated hearing loss from this disease state which is characterized by chronically draining ears, a condition that can extend over years [7, 8, 9]. Current data support the role of bacterial biofilms in chronic OM and the emergence of this paradigm has had significant implications for a greater understanding of the pathogenesis of and the immune response to this highly prevalent pediatric disease [10, 11, 12].
A biofilm is defined as a community of adherent bacteria encased in a self-produced extracellular polymeric matrix [13]. Whereas NTHI biofilms do not include a unique polysaccharide [4], the matrix contains several outer membrane proteins [14], lipooligosaccharide [14, 15], type IV pilin protein [11], integration host factor protein [16], and a dense meshwork of extracellular DNA (eDNA) [11]. eDNA within bacterial biofilms can be supplied by both the host and pathogen [16], and is linked to many processes, including bacterial aggregation [17], quorum sensing [18], nutrition [19], and structural stability [16]. Residence within a biofilm facilitates bacterial evasion of the host innate immune system as well as affords protection from antibiotics and other therapeutic treatments designed to eradicate the resident pathogens [20, 21]. As such, the protective nature of biofilms contributes to the chronic and recalcitrant nature of many respiratory tract diseases, including OM [22].
β-Defensins are a class of cationic antimicrobial host defense peptides (AMPs) that share a structure typified by six cysteine residues that form three internal disulfide bridges [23, 24]. These molecules are secreted by epithelial cells and their potent antimicrobial and immunomodulatory properties serve to maintain homeostasis at mucosal surfaces [23, 25, 26, 27]. Human β-defensin-3 (hBD-3) is an AMP that kills both Gram-positive and -negative bacteria including NTHI [28, 29] and is expressed in the mammalian middle ear [30]. In animal studies using a chinchilla model of OM, we demonstrated that the orthologue of cBD-3, chinchilla β-defensin-1 (cBD-1), modulated the ability of NTHI to colonize the upper airway [29]. Specifically, addition of exogenous recombinant cBD-1 [(r)cBD-1] peptide to the nasal cavity of chinchillas colonized with NTHI resulted in a decreased bacterial load compared to controls. Conversely, neutralization of cBD-1 in the airway by delivery of antibodies directed against this AMP resulted in an augmented concentration of NTHI in nasopharyngeal lavage fluids compared to untreated controls [29]. In a similar approach, Mason et al. [31] recently reported that the ability to resist cBD-1 lethality in the chinchilla middle ear plays a critical role in promoting NTHI survival in this anatomical location. Collectively, these data demonstrated that disruption of the biological activity of this single defensin alters homeostatic control of NTHI in the nasopharynx and influences the disease course of experimental NTHI-induced OM.
Positively charged AMPs interact with a variety of anionic macromolecules such as alginate [32] and DNA [33, 34, 35, 36], but the impact that these associations have on the function(s) of AMPs is not completely understood. As eDNA is an abundant structural component in NTHI-induced biofilms, and hBD-3 is an important effector of innate immunity in the pediatric middle ear, we hypothesized that hBD-3 might associate with eDNA in NTHI biofilms and that this interaction could have important consequences for altering and perhaps diminishing the biological activity of this molecule. In the current study, we provide evidence that recombinant hBD-3 [(r)hBD-3] bound eDNA in a dose-dependent manner. Furthermore, the ability of this innate immunity molecule to effectively alter biofilm formation as well as kill NTHI was dependent upon the removal of eDNA in the extracellular polymeric matrix.
Materials and Methods
Determination of the Relative Abundance of hBD-3 in the Pediatric Middle Ear
To assess the relative concentration of hBD-3 in the pediatric middle ear, aspirates were recovered from children (11–34 months of age) who had documented chronic or recurrent OM and underwent tympanostomy and tube insertion. Prior to the procedure, informed consent and authorization by the parent(s) or a legally authorized representative were obtained and were in concordance with all institutional and federal guidelines for the use of human subjects under protocol 20HSE014. Middle ear fluids were placed into sterile tubes, transported on ice to The Research Institute at Nationwide Children's Hospital and stored at −80°C. Equal volumes of middle ear effusion from children were lyophilized and suspended in 1/10th the original volume in sterile saline. Quantification of hBD-3 was accomplished via an hBD-3 ELISA kit (PeproTech, Rocky Hill, N.J., USA) according to the manufacturer's instructions; the kit has a published limit of detection of 62 pg hBD-3/ml [37]. The Coomassie Plus reagent (Pierce, Rockford, Ill., USA) was used to determine protein concentrations of samples, and equivalent total protein amounts from each patient were included in the assay, which was run in duplicate. A series of graded concentrations of purified (r)hBD-3 diluted in saline was used to generate a standard curve from which to determine the relative concentration of hBD-3 in middle ear samples.
Immunofluorescence Imaging of NTHI Biofilms Formed in vivo
All studies that involved chinchillas were performed under protocol 01304AR, approved by the Institutional Animal Care and Use Committee of The Research Institute at Nationwide Children's Hospital. NTHI-induced biofilms were established in the middle ear of an adult chinchilla (Chinchilla langinera) purchased from Rauscher's Chinchilla Ranch (LaRue, Ohio, USA), as previously described [11]. Briefly, animals were challenged transbullarly with 2,500 CFU NTHI strain 86-028NP [38] bilaterally, and 21 days later, animals were sacrificed and middle ear biofilms embedded in OCT embedding compound (Fisher Scientific, Pittsburgh, Pa., USA). These parameters were chosen as we have extensively characterized the intricate and dense labeling of eDNA in the matrix of robust NTHI biofilms at this time point [11, 16]. Serial sections of 10-µm thickness were cut and labeled as described [16]. Slides were incubated with a 1:200 dilution of rabbit polyclonal anti-cBD-1 [29] for 24 h at 4°C followed by goat anti-rabbit conjugated to AlexaFluor 594 (Invitrogen, Carlsbad, Calif., USA) for 1 h at room temperature. Slides were then washed three times in buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.05% Tween 20) and incubated for 15 min with 1 µg/ml FilmTracer FM1-43 biofilm cell stain (Invitrogen). Slides were washed again three times and counterstained with 4′,6′-diamadino-2-phenylindole (DAPI) to label nucleic acid. Sections were viewed with a Zeiss LSM 510 Meta confocal scanning laser microscope to detect the relative abundance and spatial distribution of both cBD-1 and eDNA.
Quantification of eDNA within an NTHI Biofilm Formed in vitro
Double-stranded eDNA content was quantified in order to determine the amount of eDNA within biofilms formed by NTHI 86-028NP at various time points in vitro. Bacteria were grown on chocolate agar (Fisher Scientific) at 37°C with 5% CO2 overnight. Isolated colonies were suspended in brain heart infusion broth (BD Life Sciences, Sparks, Md., USA) supplemented with 2 µg/ml of heme (Fisher Scientific) and nicotinamide adenine dinucleotide (sBHI; Fisher Scientific) to an optical density of 0.65 at 490 nm. The culture was then diluted 1:6 with pre-warmed sBHI and incubated statically at 37°C with 5% CO2 for 3 h. Cultures were diluted 1:2,500 with sBHI and 200-µl aliquots were inoculated into an 8-well chamber slide (Fisher Scientific). NTHI cultures were incubated at 37°C with 5% CO2 for 8, 16, 24, 36, 48, 64, or 72 h in sBHI to allow for the development of biofilms, as we have described previously [16]. Culture medium was removed from biofilms after the first 16 h of growth and 200 µl of fresh sBHI was added to slides. Culture medium was then changed every 8 h. Following the indicated incubation period, sterile saline was added to the chamber slide and the resultant biofilm was recovered by repeated pipetting. The picogreen dsDNA quantification kit (Invitrogen, Carlsbad, Calif., USA) was then used to quantify the eDNA content according to the protocol supplied by the manufacturer.
DNA Electrophoretic Mobility Shift Assay
To begin to characterize the ability of (r)hBD-3 (PeproTech) to bind eDNA, we incubated peptide with NTHI genomic DNA (gDNA) and determined the ability of association with this AMP to alter the mobility of NTHI gDNA in an agarose gel. The Invitrogen (Carlsbad, Calif., USA) Purelink mini gDNA extraction kit was used to extract NTHI 86-028NP gDNA according to the manufacturer's protocol. Five hundred nanograms of NTHI gDNA were incubated with 0, 0.5, or 5.0 µg of (r)hBD-3, (r)cBD-1, or bovine serum albumin (BSA) in a total volume of 20 µl ddH2O for 15 min at room temperature. Following this incubation period, samples were separated in an 0.8% w/v agarose gel (Fisher Scientific) at 75 V for 30 min in 1× TAE running buffer (Promega, Madison, Wisc., USA). Gels were stained with GelRed (Phenix Research Products, Candler, N.C., USA) and images were captured using a Syngene U: Genius imaging system (Cambridge, UK).
Atomic Force Microscopy
To visualize the association between AMPs and eDNA, we utilized atomic force microscopy (AFM). Five hundred nanograms of NTHI gDNA were incubated in the absence or presence of 5 µg of (r)hBD-3, (r)cBD-1, or BSA in a total volume of 20 µl ddH2O for 15 min at room temperature. To maximize our ability to view AMP-eDNA complexes, samples were diluted to 5.0 ng AMP and 0.5 ng eDNA in 50 µl ddH2O and each sample was spotted separately onto freshly cleaved mica discs. Mica V-2 optical grade discs (S&S Trading, Floral Park, N.Y., USA) were cleaved with double-sided Scotch tape and samples were incubated on the discs for 10 min at ambient temperature. The mica discs were rinsed with ddH2O, dried at 4°C overnight, and affixed to 15-mm-diameter stubs (Ted Pella, Redding, Calif., USA) prior to visualization. The samples were then imaged using a multimode atomic force microscope (Veeco, Plainview, N.Y., USA) in tapping mode fitted with an NSC15 probe (MikroMasch, San Jose, Calif., USA). Images were analyzed using NanoScope 5.30 software and height data were reported as the mean of three independent images analyzed in triplicate.
Ability of eDNA to Alter (r)hBD-3 Killing of Planktonic NTHI
To determine whether the presence of eDNA mitigated the ability of (r)hBD-3 to kill NTHI 86-028NP, we utilized a previously described microbicidal assay [39]. Here, we modified the protocol such that a selected amount of eDNA (0, 130, 250, 500, or 1,000 ng) was incubated with 5 µg (r)hBD-3/ml in a total volume of 20 µl ddH2O for 15 min at room temperature prior to initiation of the in vitro killing assay.
Ability of (r)hBD-3 to Alter NTHI Biofilm Formation
To determine the impact that (r)hBD-3 had on the prevention of NTHI biofilms, we established NTHI biofilms in 8-well chamber slides (Fisher Scientific) in the presence of selected concentrations of (r)hBD-3. NTHI biofilms were established as described above; however, (r)hBD-3 was added to a final concentration of 0, 0.5, or 5.0 µg peptide/ml sBHI in indicated wells. These concentrations were chosen as they represent a range of (r)hBD-3 concentrations that were detected in effusions recovered from the middle ear of children with OM. Slides were incubated for 16 h at 37°C with 5% CO2 and culture medium was removed. Biofilms were washed with sterile saline and stained with the LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Eugene, Oreg., USA) as per manufacturer's protocol. Biofilms were then fixed with 2% paraformaldehyde in 0.1 M phosphate buffer, pH 7.2, for at least 60 min. Following fixation, biofilms were washed twice with sterile saline, plastic wells were removed from the slide, and a glass coverslip was placed over the slide. The coverslip was sealed in order to minimize dehydration of the sample. Biofilms were imaged with a Zeiss 510 confocal scanning laser microscope (CSLM; Zeiss, Thornwood, N.Y., USA) with a ×63 objective. All biofilm assays were performed in duplicate a minimum of three times.
In order to quantitatively assess the differences in biofilm morphology as a result of the different treatments, we subjected CSLM images to analysis by COMSTAT software. Image files were imported into MATLAB (Albuquerque, N.M., USA) for quantitation of differences in biofilm height, thickness, and biomass. Values presented represent the mean of three experimental replicates conducted on different days.
Impact of eDNA on the Ability of (r)hBD-3 to Alter Biofilms Formed by NTHI
As a means to characterize the effects of an eDNA-(r)hBD-3 complex on the activity of this AMP, NTHI biofilms were established in the absence or presence of 5.0 µg (r)hBD-3/ml. Peptide was incubated with 500 ng eDNA for 15 min at room temperature in sterile water prior to addition to a chamberslide. In addition, wells were also incubated in the presence of 500 ng of gDNA alone purified from NTHI strain 86-028NP. We chose this amount of NTHI gDNA as this concentration represented a conservative estimate of the amount of eDNA in biofilms formed by NTHI between 0 and 16 h based on picogreen assay results. Slides were incubated at 37°C with 5% CO2 for 16 h and then processed for CSLM analysis. All assays were repeated three times, on separate days, and all individual biofilm assays included replicates. Representative images were analyzed by COMSTAT software.
Rescue of (r)hBD-3 Activity by Addition of DNase I to NTHI Biofilms
We next tested whether degradation of eDNA in the biofilm matrix could rescue (r)hBD-3 antimicrobial activity. NTHI biofilms were established in the absence or presence of 0.5 µg (r)hBD-3/ml in combination with either 0.5 µg DNase I or RNase A/ml. A suboptimal dose of DNase I or RNase A was used in order to determine the impact that degradation of deoxy- or ribonucleic acids, respectively, would have on the resultant biofilms. The concentration of the enzymes was chosen such that we could degrade these nucleic acids without observing a significant impact on resultant NTHI biofilm architecture. NTHI biofilms were incubated for 16 h and processed for CSLM. All assays were repeated three times, on separate days, and all individual biofilm assays included replicates. All statistical analyses were performed using Stata V 10.1, Stata (College Station, Tex., USA) and an observed value of p ≤ 0.05 was considered statistically significant.
Results
Determination of the Relative Concentration of hBD-3 in the Pediatric Middle Ear
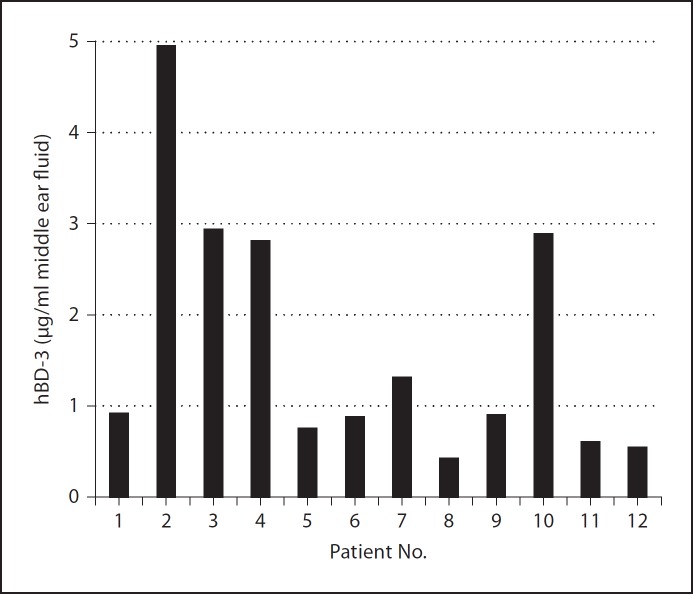
As a foundation to understand how the presence of eDNA within an NTHI biofilm would affect the activity of hBD-3, we first assessed the relative concentration of hBD-3 in the pediatric middle ear during episodic OM. Middle ear effusions that were culture positive for NTHI were obtained from patients and screened via ELISA for the presence of hBD-3. We showed that the mean concentration was 1.7 µg hBD-3/ml effusion with a range of approximately 0.4–4.9 µg hBD-3/ml (fig. 1). These data offered an indication as to the relative amount of hBD-3 within the pediatric middle ear that could be available to bind eDNA and provided a reference for which we could confidently continue with subsequent assays.
Fig. 1.
Quantitation of hBD-3 in effusions collected from the middle ear of children with chronic OM. The concentration of hBD-3 in effusions recovered from 12 children with NTHI-induced OM was determined by ELISA and is shown for each patient.
cBD-1 and eDNA Co-Localize within an NTHI-Induced Biofilm Formed in vivo
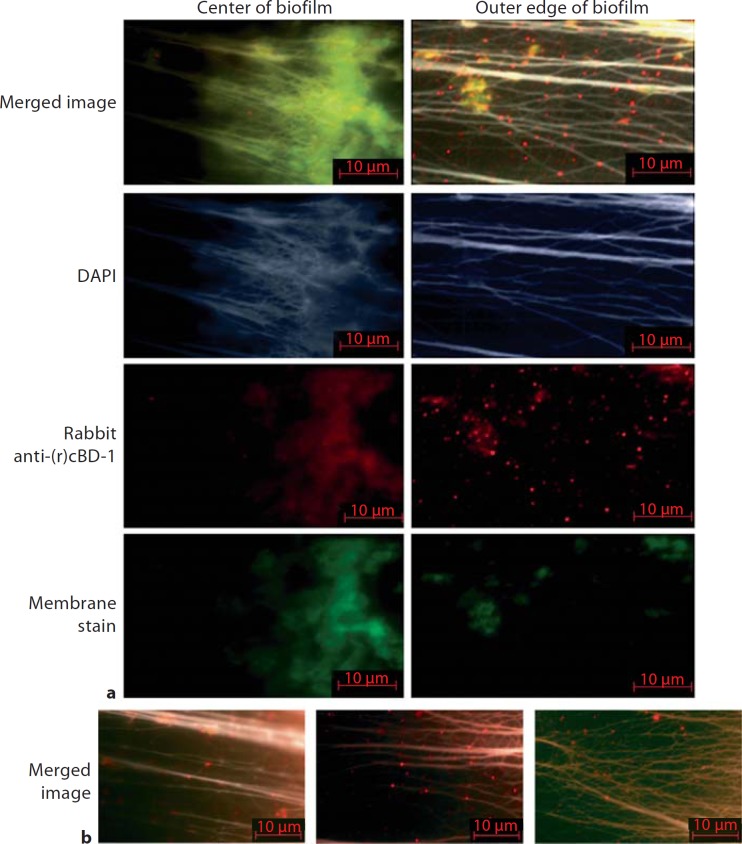
We next wanted to define the relative distribution of cBD-1, the orthologue of hBD-3, and eDNA within an NTHI-induced biofilm recovered from the chinchilla middle ear during experimental OM. We therefore incubated sections derived from an NTHI biofilm recovered from the middle ear of a chinchilla with DAPI, rabbit anti-cBD-1, and a cellular phospholipid membrane stain to label bacteria and host cells. With this approach, we observed positive labeling of DNA and cell membranes, which was co-detected with a weak diffuse signal for cBD-1 near the center of the biofilm (fig. 2a, left panel). No labeling for the AMP was observed after incubation of biofilms with naive rabbit serum, which served as a negative control (data not shown). We detected robust labeling of cBD-1 on the outer edge of the NTHI-induced biofilm relative to interior regions and observed that virtually all cBD-1 appeared to be co-localized in areas that also stained for DNA (fig. 2a, right panel). Furthermore, cBD-1 was observed to decorate long, filamentous strands of eDNA within the biofilm, often seen as punctate red fluorescence in areas that did not show an observable signal with the cellular phospholipid membrane stain (fig. 2a, right panel, 2b). These results showed that cBD-1 co-labeled with bacteria and/or host cells and also co-localized with eDNA in an NTHI biofilm formed in the middle ear. Furthermore, these data demonstrated a spatial distribution of cBD-1 within an NTHI biofilm, whicwh suggested that more cBD-1 was found associated with eDNA in the outer (external) compared to the center (internal) aspects of the biofilm. This distribution of cBD-1 could be due to limited diffusion of the AMP to the inner aspects of the biofilm or attributable to the fact that the exterior of the biofilm is directly exposed to the contents of a middle ear effusion, which includes AMPs.
Fig. 2.
Images of a 21-day biofilm formed in the chinchilla middle ear by NTHI strain 86-028NP. Sections of the biofilm were immunolabeled with rabbit anti-chinchilla β-defensin-1 followed by goat anti-rabbit conjugated to AlexaFluor 594 to label cBD-1 (red). Sections were also stained with a cellular phospholipid membrane stain to label bacteria and host cells (green) and DAPI to label DNA (blue or pseudo-colored white). Individual fluorescent channels (a) and merged images of all three signals (a, b) are shown. The images were derived from the inner aspect of the NTHI biofilm (a, left panel) and the outer edge of the biofilm (a, right panel, b). Note the greater intensity of punctate red labeling, and thereby cBD-1, co-localized with eDNA at the outer periphery of the biofilm. Almost all labeled cBD-1 co-localized with DNA in an NTHI-induced biofilm formed in vivo.
Defensins Interact with eDNA
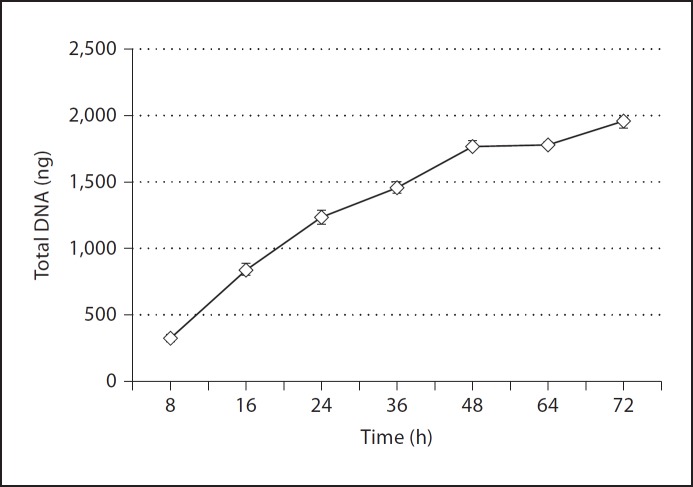
As we had now provided evidence that cBD-1 co-localized with eDNA in an NTHI biofilm established in vivo, we proceeded with in vitro studies to determine the concentration of eDNA within NTHI biofilms and determine whether (r)cBD-1 or (r)hBD-3 indeed bound eDNA. To do so, we first quantified the relative total amount of eDNA in biofilms formed in chamber slides by NTHI over time. We observed that between 8 and 16 h after inoculation, NTHI biofilms contained approximately 300–700 ng of total eDNA and that this value steadily increased over time (fig. 3). We further established that the concentration of eDNA plateaued approximately 64–72 h after inoculation of chamber slides with NTHI and reached approximately 2,000 ng of total eDNA per biofilm when analyzed under these culture conditions.
Fig. 3.
Quantitation of eDNA in an NTHI biofilm formed in vitro. NTHI 86-028NP was incubated in a chamber slide, and at the depicted time points, the established biofilms were recovered and eDNA quantified.
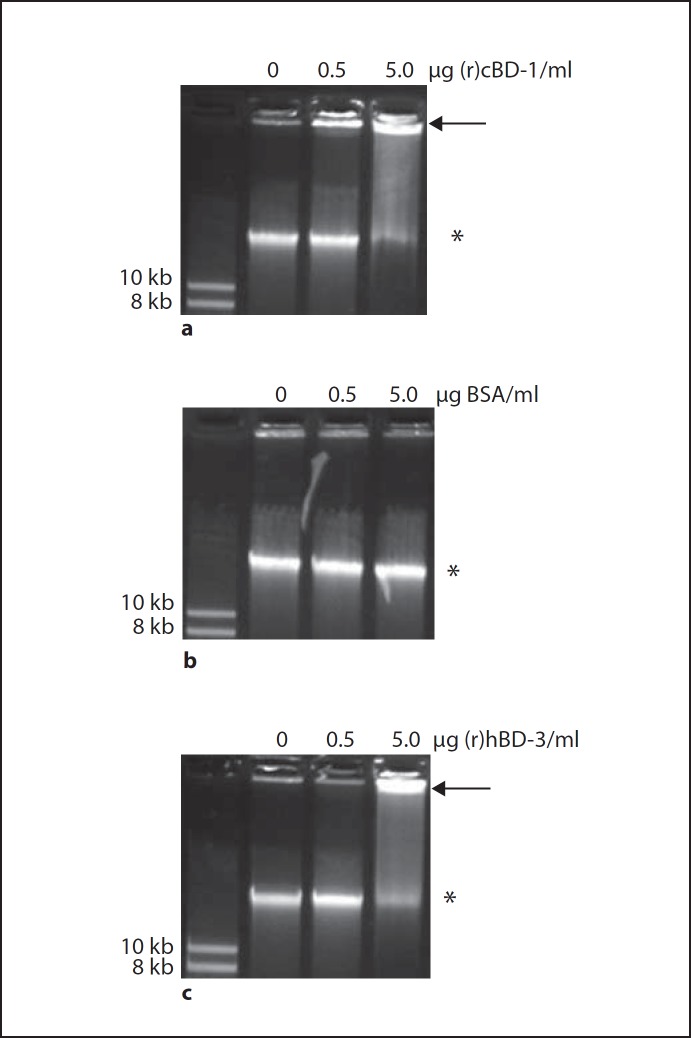
We next utilized an electrophoretic mobility shift assay to determine if our targeted AMPs bound eDNA. We incubated either 0, 0.5, or 5.0 µg (r)cBD-1 with 500 ng NTHI gDNA and subjected the mixture to separation by agarose gel electrophoresis. We found that when in association with (r)cBD-1, the mobility of NTHI gDNA through the agarose gel substantially decreased in a (r)cBD-1 dose-dependent manner compared to NTHI gDNA incubated in the absence of peptide (fig. 4a). We observed that even 0.5 µg/ml (r)cBD-1, which represented the lowest concentration of AMP tested, resulted in the ability to detect a mobility shift for a fraction of the free NTHI gDNA so that the NTHI gDNA-AMP complex migrated slower into the agarose gel (fig. 4a, arrow). The mobility of NTHI gDNA was not altered when incubated with BSA, which served as our negative control for the nonspecific ability of protein to bind NTHI gDNA and cause a commensurate mobility shift (fig. 4b). Our data also demonstrated that (r)hBD-3 directly bound to NTHI gDNA in a dose-dependent manner as shown by the reduced mobility of free NTHI gDNA in an agarose gel (fig. 4c). Of note, incubation of NTHI gDNA with 5.0 µg/ml (r)hBD-3 or (r)cBD-1 resulted in an almost complete shift in the mobility of NTHI gDNA demonstrating that this concentration of AMP essentially bound all free NTHI gDNA.
Fig. 4.
NTHI gDNA was incubated in the absence or presence of increasing concentrations of (r)cBD-1 (a), BSA (b) or (r)hBD-3 (c) and the samples were then separated in a GelRed-stained agarose gel. The arrows indicate the mobility shift of DNA after incubation with peptide compared to free DNA alone (asterisks). A DNA size standard was used in lane 1 of all images with the respective size indicated to the left of the gel. Both (r)cBD-1 and the human orthologue (r)hBD-3 bound eDNA in a dose-dependent manner.
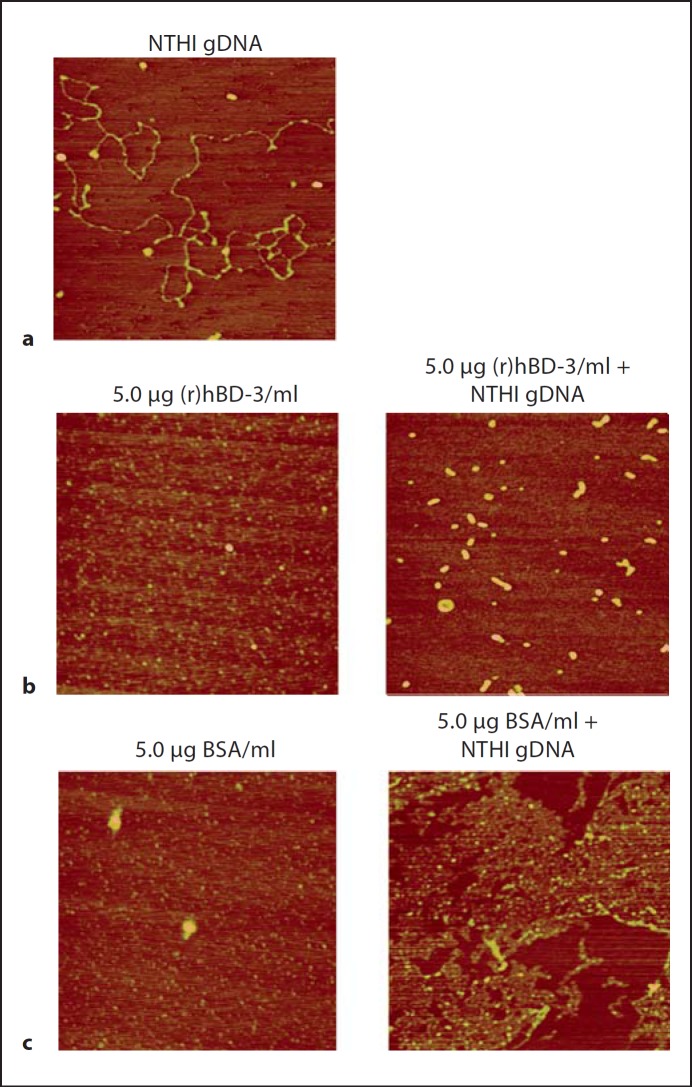
To further demonstrate that (r)hBD-3 physically interacted with DNA, we utilized AFM to capture images of DNA-AMP complexes formed in vitro. In the absence of peptide, NTHI gDNA was easily detectable as filamentous strands on the mica surface with a mean height of 0.62 nm (fig. 5a). Visualization of (r)hBD-3 alone appeared as a diffuse layer of peptide with a mean height of 0.48 nm (fig. 5b, left panel). In contrast, (r)hBD-3 incubated with NTHI gDNA resulted in the complete absence of observable free DNA by AFM and the concomitant appearance of discrete structures with a mean height of 2.27 nm (fig. 5b, right panel). These same structures were not observed when BSA was incubated in the absence (fig. 5c, left panel) or presence of NTHI gDNA (fig. 5c, right panel). These data showed that eDNA could bind (r)hBD-3 likely via charge interactions and further suggested that this association would likely have consequences for the bactericidal activity of these AMPs against NTHI.
Fig. 5.
NTHI gDNA was incubated in the absence (a) or presence of 5.0 µg (r)hBD-3/ml (b, right panel) or 5.0 µg BSA/ml (c, right panel), and images were collected by AFM. Images of 5.0 µg (r)hBD-3/ml alone (b, left panel) and 5.0 µg BSA/ml alone (c, left panel) are also shown. No observable free DNA was detected when NTHI gDNA was incubated in the presence of (r)hBD-3, which demonstrated that this AMP bound eDNA.
eDNA Diminishes the Ability of (r)hBD-3 to Kill Planktonic NTHI
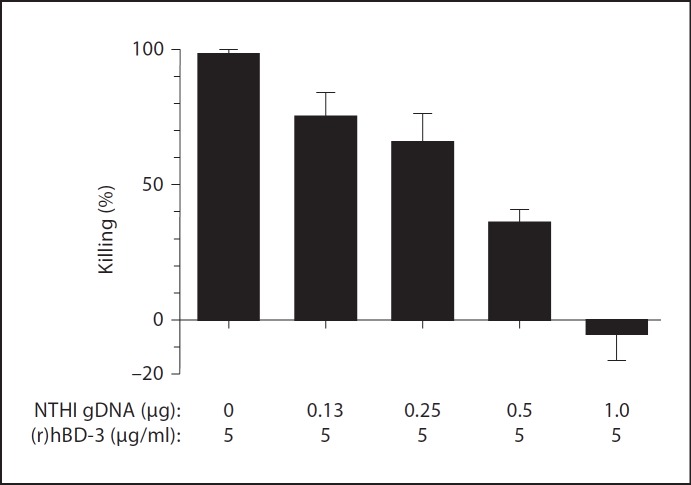
We previously established that incubation of planktonically grown NTHI 86-028NP with 2.5 µg (r)hBD-3/ml for 1 h results in almost 100% killing of bacteria [29]. With this in mind, we therefore next used this in vitro bacterial killing assay to determine if the presence of eDNA could abrogate (r)hBD-3-mediated killing of NTHI. We observed that incubation of NTHI with (r)hBD-3 at a concentration of 5 µg/ml resulted in 100% killing of bacteria in 1 h (fig. 6). In contrast, we demonstrated a dose-dependent inhibition of bacterial killing when 5 µg (r)hBD-3/ml was incubated for 15 min at room temperature with NTHI gDNA prior to initiation of the killing assay (fig. 6). We observed that pre-incubation of 5 µg/ml (r)hBD-3 with 1 µg NTHI gDNA completely abrogated detectable bacterial killing over 60 min. These data demonstrated that eDNA indeed inhibited the ability of (r)hBD-3 to kill planktonic NTHI.
Fig. 6.
Ability of eDNA to alter the killing activity of (r)hBD-3. Planktonic NTHI 86-028NP was incubated with (r)hBD-3 alone or AMP pre-complexed with increasing amounts of eDNA, and the number of surviving colony forming units were determined. Addition of eDNA abrogated the (r)hBD-3-mediated killing of NTHI in a dose-dependent manner.
Physiologically Relevant Concentrations of (r)hBD-3 Alter NTHI Biofilm Structure
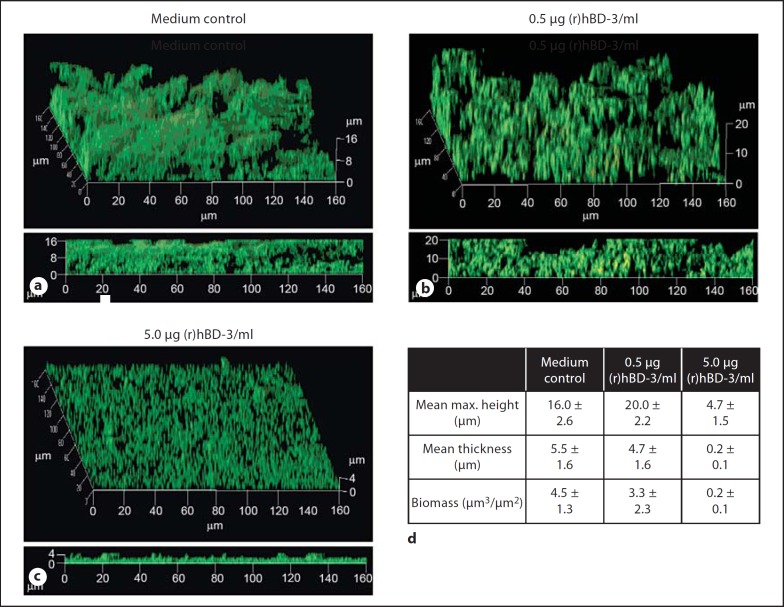
In order to characterize the impact of (r)hBD-3 on the structure of biofilms formed by NTHI, NTHI biofilms were allowed to form in vitro in either medium alone or the presence of 0.5 or 5.0 µg/ml of (r)hBD-3 for 16 h. We observed that 0.5 µg (r)hBD-3/ml sBHI had minimal effect on the resultant biofilms in terms of maximum height, with control and peptide-treated biofilms attaining a mean maximum height of 16 or 20 µm, respectively (fig. 7a, b, d). In addition, the mean biomass and thickness of the biofilms were similar among the control and treatment groups with mean biomass values of 4.5 or 3.3 µm3/µm2 and the thickness observed to be 5.5 or 4.7 µm, respectively (fig. 7d). In contrast, co-incubation of NTHI with 5.0 µg (r)hBD-3/ml, a concentration that resulted in 100% killing of planktonic NTHI (fig. 6), was able to diminish but not eradicate the resultant biofilms in terms of these same metrics of analysis. The mean maximum height of the biofilm formed in the presence of (r)hBD-3 was decreased 70% and mean biomass was reduced from 4.5 to 0.2 µm3/µm2 (fig. 7a, c, d). These data demonstrated that (r)hBD-3 exhibited the capacity to alter NTHI-induced biofilm formation in vitro but that NTHI biofilms were not measurably altered at a lesser concentration of the AMP. Taken together with data that showed direct binding of (r)hBD-3 with eDNA, these data suggested that the presence of eDNA likely significantly dampened the ability of (r)hBD-3 to prevent NTHI-induced biofilms.
Fig. 7.
(r)hBD-3 diminished NTHI biofilms formed in vitro. NTHI 86-028NP was incubated in the absence (a) or presence of increasing concentrations of (r)hBD-3 for 16 h (b, c), and the resultant biofilms were analyzed by CLSM followed by COMSTAT software (d) for differences in biofilm architecture. Incubation of NTHI with 5.0 µg (r)hBD-3/ml altered the resultant biofilm architecture; however, incubation with a lesser concentration of the AMP [0.5 µg (r)hBD-3/ml] resulted in biofilms that more closely resembled those incubated in medium alone.
eDNA Abrogates the Ability of (r)hBD-3 to Diminish NTHI-Induced Biofilms
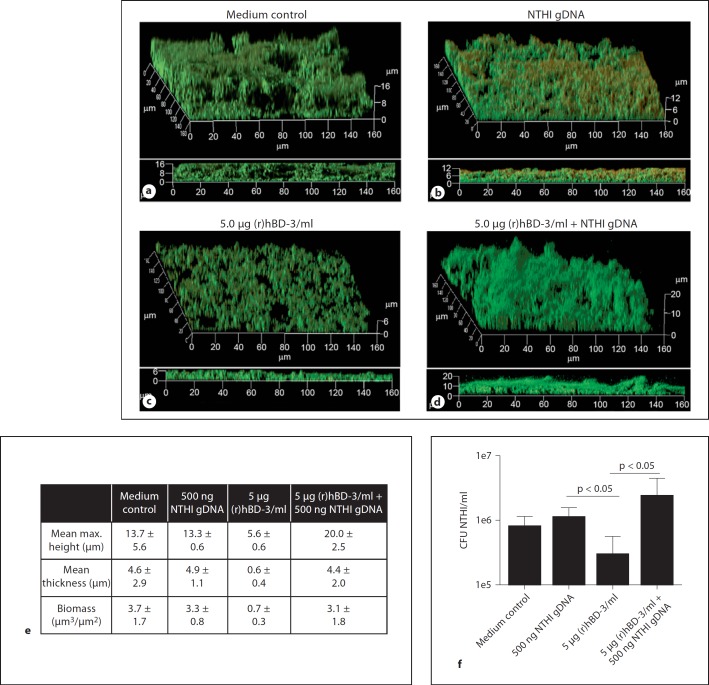
To directly determine the impact that eDNA had on the activity of (r)hBD-3 against NTHI-induced biofilms, NTHI biofilms were allowed to form in the presence of either medium alone, NTHI gDNA, (r)hBD-3, or medium wherein (r)hBD-3 was pre-incubated for 15 min with NTHI gDNA in vitro, as described in the Materials and Methods. Biofilms formed in the presence of medium alone or NTHI gDNA were similar in architecture, mean maximum biofilm height (13.7 vs. 13.3 µm) and thickness (4.6 vs. 4.9 µm; fig. 8a, b, e). As demonstrated previously, incubation of NTHI with 5.0 µg (r)hBD-3/ml reduced the resultant biofilm maximum height, overall biomass, and thickness compared to control biofilms that were established in medium alone (fig. 8a, c, e). In contrast, pre-incubation of 5.0 µg (r)hBD-3/ml with 500 ng NTHI gDNA completely inhibited the ability of the AMP to alter the architecture of an NTHI biofilm. These changes in architecture can be seen qualitatively in CSLM images of the biofilms as well as quantitatively as determined by COMSTAT analysis (fig. 8d, e). We also determined the relative concentration of viable NTHI in these biofilms and showed that incubation of NTHI with 5.0 µg (r)hBD-3/ml resulted in an approximate 40% decrease in the concentration of NTHI compared to medium alone and that this killing was abrogated in the presence of eDNA (fig. 8f). As we demonstrated that (r)hBD-3 interacted with eDNA, our data indicated that eDNA bound (r)hBD-3 and that this interaction inhibited the ability of the AMP to kill NTHI within a biofilm. Furthermore, these data suggested that the maximum concentration of hBD-3 detected in the pediatric middle ear could be rendered ineffective at significantly reducing NTHI biofilms in vivo.
Fig. 8.
Ability of eDNA to mitigate (r)hBD-3 activity in NTHI biofilms. NTHI biofilms were established for 16 h in the presence of either medium alone (a), 500 ng NTHI gDNA (b), 5.0 µg (r)hBD-3/ml (c), or (r)hBD-3 (d) pre-incubated for 15 min with NTHI gDNA. Resultant biofilms were prepared for CLSM and COMSTAT values are shown (e). In addition, the concentration of NTHI in biofilms was determined for each treatment by dilution plating of bacteria on chocolate agar (f). The reddish color seen within the biofilm (b) is due to propidium iodide staining of exogenous NTHI gDNA [11] that was added to this chamber slide and thus is not indicative of bacterial death in this image. Co-incubation of (r)hBD-3 and eDNA resulted in diminished ability of the AMP to alter biofilms and kill NTHI.
DNase I Rescues the Ability of (r)hBD-3 to Alter NTHI Biofilms
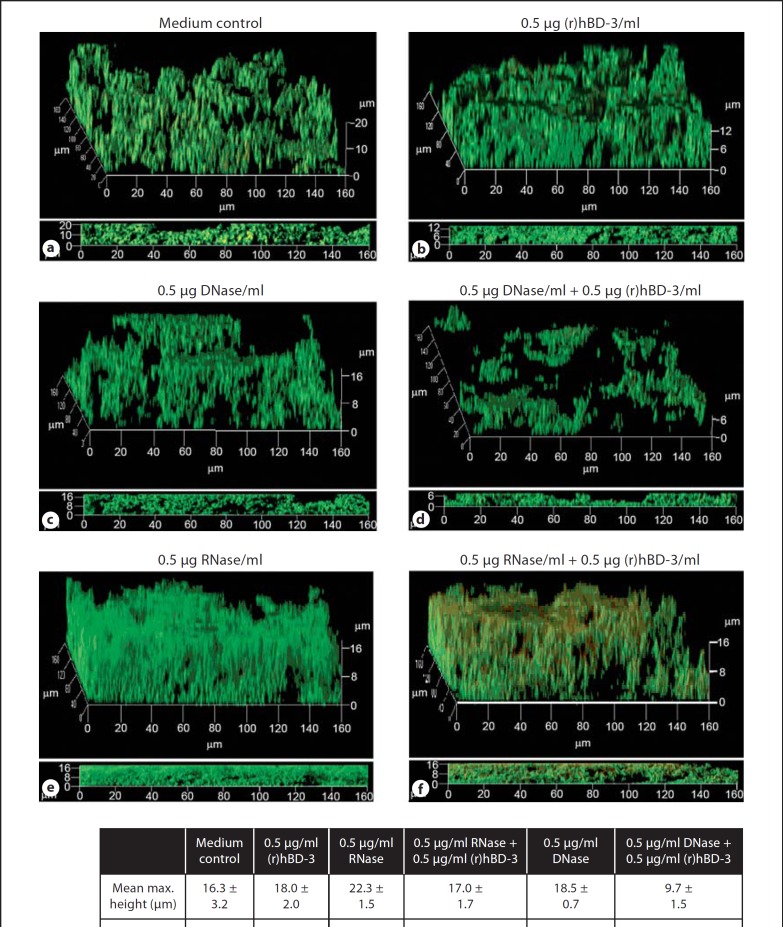
As eDNA abrogated the activity of (r)hBD-3 against NTHI biofilms, we next sought to determine if removal of this macromolecule would render an NTHI biofilm more susceptible to the antimicrobial action of this AMP. To address this question, we established biofilms in either medium alone or in the presence of 0.5 µg (r)hBD-3/ml, the minimum concentration of hBD-3 detected in the middle ear. In addition to these conditions, we also incubated biofilms in the absence or presence of suboptimal doses of DNase I or RNase A (0.5 µg enzyme/ml sBHI) such that we could degrade these nucleic acids without observing a significant impact on resultant NTHI biofilm architecture. Consistent with previous data, incubation of NTHI with 0.5 µg (r)hBD-3/ml had no measurable impact on biofilm height, thickness, and biomass compared to medium alone (fig. 9a, b). In a similar manner, the addition of 0.5 µg DNase I/ml, a suboptimal concentration, resulted in no discernible difference in mean biofilm height (18.5 vs.16.3 µm) or thickness (3.5 vs. 3.7 µm) compared to the control (fig. 9a, c, g). Interestingly, incubation of NTHI with 0.5 µg RNase A/ml did not diminish biofilms but actually resulted in a notable and reproducible increase in mean maximum height (22.3 vs. 16.3 µm) and biomass (5.1 vs. 2.9 µm3/µm2; fig. 9e, g).
Fig. 9.
Degradation of eDNA and the subsequent impact on (r)hBD-3 activity. NTHI biofilms were established for 16 h in medium alone (a) or in the presence of 0.5 µg (r)hBD-3/ml (b, d, f). A suboptimal dose of DNase I (c, d) or RNase A (e, f) was also used to determine the impact that degradation of nucleic acid would have on the activity of the cationic AMP without disruption of NTHI biofilm architecture when used alone. Resultant biofilms were prepared for CLSM and COMSTAT values are shown (g). The ability of (r)hBD-3 to alter NTHI biofilms was rescued by degradation of eDNA with DNase I but not degradation of RNA.
In our studies, incubation of NTHI in the presence of both (r)hBD-3 and RNase A (fig. 9f, g) did not affect the formation of biofilms by NTHI as seen qualitatively in CSLM images of the biofilms as well as quantitatively by COMSTAT analysis compared to addition of RNase A alone (fig. 9e, g). In contrast, incubation of NTHI in the presence of a suboptimal dose of (r)hBD-3 and DNase I rescued the activity of the AMP such that the mean biofilm height was reduced from 18 to 9.7 µm, mean thickness from 4.2 to 0.4 µm, and mean biomass from 3.3 to 0.3 µm3/µm2 compared to (r)hBD-3 alone (fig. 9b, d, g). These data represented overall mean decreases of 54, 91, and 90% for the three parameters indicative of successful biofilm establishment, respectively. The data showed that the AMP activity was rescued such that 0.5 µg (r)hBD-3 in combination with DNase I was as effective as 10 times the concentration of AMP alone [5.0 µg (r)hBD-3/ml; compare fig. 8c to fig. 9d). Collectively, these data demonstrated that eDNA mitigated the ability of (r)hBD-3 to alter NTHI biofilm formation and that the inhibitory action of eDNA could be overcome by degradation of the nucleic acid.
Rescue of (r)hBD-3 Activity Results in Increased Killing of Biofilm-Associated NTHI
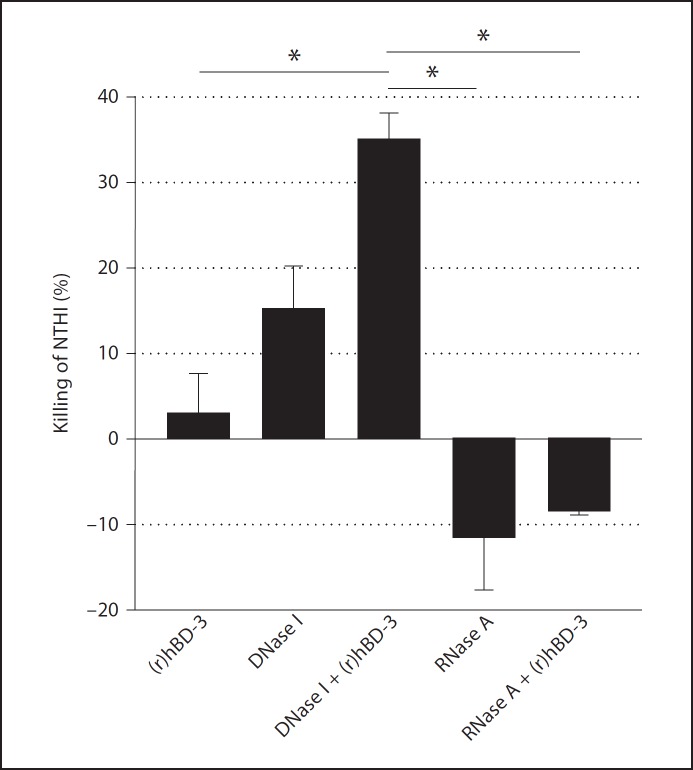
A primary function of AMPs, including hBD-3, in host defense is the ability to kill microorganisms. We therefore next wanted to determine whether incubation of (r)hBD-3 in the presence of DNase I or RNase A would result in increased killing of NTHI within biofilms compared to peptide alone. NTHI biofilms were allowed to form in vitro as above, then the number of viable NTHI in the resultant biofilms was determined. Relative to 0.5 µg (r)hBD-3/ml alone, incubation of NTHI in the presence of DNase I alone, RNase A alone, or RNase A and (r)hBD-3 did not significantly alter the concentration of viable bacteria in biofilms (fig. 10). However, incubation of NTHI in the presence of both 0.5 µg (r)hBD-3/ml and DNase I resulted in 35% NTHI killed, a statistically significant reduction in viable bacteria compared to peptide alone (fig. 10). The concentration of viable NTHI was not significantly altered when NTHI was grown in the presence of 0.5 µg (r)hBD-3/ml and RNase A providing further evidence that eDNA and not ribonucleic acid inhibited the activity of (r)hBD-3. These data demonstrated that removal of eDNA from an NTHI biofilm restores the ability of (r)hBD-3 to alter NTHI biofilm formation through rescue of its antimicrobial activity.
Fig. 10.
Ability of (r)hBD-3 to kill NTHI within a biofilm. NTHI biofilms were established for 16 h in the presence of medium alone or 0.5 µg (r)hBD-3/ml. DNase I or RNase A was also used to determine the impact that degradation of nucleic acid would have on the ability of NTHI to survive under these conditions. The resultant biofilms were recovered and the number of culturable NTHI was determined by dilution plating on chocolate agar. The percent killing of NTHI was determined compared to medium alone. Error bars represent the standard error of the mean. The asterisks indicate a statistically significant change (p < 0.05) in viable NTHI among the treatment groups. Degradation of eDNA by DNase I restored the ability of (r)hBD-3 to kill NTHI.
Discussion
Recently, DNA was established as an important structural component of biofilms formed by NTHI as well as other bacterial species [35, 36] and the role of eDNA in a bacterial biofilm continues to be examined and expounded upon. We hypothesized that one role for eDNA in a bacterial biofilm would be to sequester cationic AMPs, thus inhibiting the ability of these peptides to reach their microbial targets. The current study adds to our understanding of how residence within a biofilm community assists bacteria therein to avoid the action of effectors of innate immunity as others have reported [40]. In addition, our data increase our understanding of how the biofilm component of OM plays a role in chronicity of NTHI diseases and provides tenable methods for prevention of OM development.
Our data demonstrated that the chinchilla host defense peptide cBD-1 and dsDNA were co-detected in biofilms formed during experimental OM and that cBD-1 was frequently co-localized with eDNA. In addition, we showed that (r)cBD-1 and the human orthologue (r)hBD-3 bound to DNA in vitro and that this interaction was not merely an association but a physical interaction between the two molecules. It was recently shown that an additional AMP, the human cathlelicidin LL-37, forms aggregated and condensed complexes with DNA and that this interaction acts as a potent trigger for interferon production by plasmacytoid dendritic cells [41]. Thus, multiple AMP family members exhibit the capacity to bind exogenous DNA and this interaction has an impact on not only the function of AMPs but also cells of the innate immune system. As we detailed that eDNA was abundant in NTHI biofilms formed in the chinchilla middle ear, our data suggested that the interaction between hBD-3 and eDNA would likely have implications for NTHI biofilm development.
We demonstrated that exposure of NTHI to 0.5 µg (r)hBD-3/ml, a concentration of hBD-3 which is typically detected in the pediatric middle ear, did not result in altered biofilm architecture when compared to controls. A limitation to our estimated concentration of hBD-3 in the middle ear is that macromolecules present in effusions, such as mucins, may bind AMPs [42], thus reducing the ability to detect AMP via ELISA which would subsequently result in an underestimation of the actual AMP concentration in vivo. Nevertheless, even when we incubated NTHI with 10 times the concentration of (r)hBD-3, which represented the greatest concentration that was detected in a panel of middle ear effusions of children with active OM, NTHI biofilms were readily detected although at reduced height compared to medium alone. We demonstrated that this concentration of (r)hBD-3 killed 100% of planktonic NTHI in a 1-hour assay, but that killing could be abrogated by the presence of eDNA in vitro. In addition, we showed that incubation with 500 ng exogenous NTHI gDNA plus 5.0 µg (r)hBD-3/ml completely prevented the ability of the peptide to diminish an NTHI biofilm and to reduce the viable concentration of NTHI in these biofilms. Together, these data support the conclusion that the killing activity of the AMP was mitigated in an NTHI-induced biofilm by an interaction with eDNA.
As it is known that eDNA is an integral structural component of biofilms formed by multiple bacterial species, the possibility exists that the altered architecture of NTHI biofilms formed in the presence of (r)hBD-3 may also be due to sequestering of this integral structural component within the biofilm. This hypothesis is supported by our data obtained by AFM wherein we demonstrated that incubation of eDNA with (r)hBD-3 resulted in a dramatic absence of observable free DNA. Interestingly, eDNA has also been implicated in the development of resistance by Pseudomonas aeruginosa to AMPs and traditional aminoglycoside antibiotics through chelation of cationic ions and upregulation of a PhoPQ- and PmrAB-regulated cationic antimicrobial peptide resistance operon [43]. The evidence therefore suggests that eDNA within a biofilm exhibits multiple activities that promote bacterial survival from effectors of innate immunity.
While our data directly address the question of whether eDNA binds AMPs, evidence in the literature demonstrates that other factors present within the biofilm EPS or in the airway could also interact with cationic host defense peptides, and in turn diminish their anti-biofilm activity. For example, it has been reported that polymyxin B and human neutrophil α-defensin form complexes with capsular polysaccharide from Klebsiella pneumoniae, Streptococcus pneumoniae, and P. aeruginosa thereby reducing the minimum inhibitory concentrations of these host defense peptides [44]. Whereas encapsulated H. influenzae strains may use a similar strategy to inactivate AMPs, this observation would not extend to unencapsulated NTHI strains such as the isolate used in the present studies. Mucins also inhibit the antibacterial activity of AMPs such as LL-37 [42], an AMP expressed in the upper and lower airways. As NTHI is known to upregulate expression of mucins by respiratory epithelial cells [45, 46] and to adhere to mucins in vitro [47, 48], we postulate that the interaction between NTHI and mucins may also provide a niche to evade killing by AMPs produced in the airway. In the dense, complex NTHI biofilm EPS, eDNA is nonetheless clearly one of several compounds likely to interact with innate immune effectors such as AMPs and, in doing so, mitigate their activity.
In the current study, we focused on the ability of eDNA to diminish hBD-3 activity in NTHI-induced biofilms during the initial stages of biofilm formation. One could make the argument that during the earliest stages of biofilm development in the middle ear there is little to no available eDNA to which AMPs such as hBD-3 could bind. We hypothesized that the ability of eDNA to sequester hBD-3 would be minimal when NTHI first encounters mucosal epithelium that would be secreting a basal amount of peptide. At this time, it is likely that NTHI utilizes other mechanisms such as a functional Sap transporter [49, 50] to effectively provide resistance to killing by hBD-3. However, as the concentration of NTHI in the chinchilla middle ear is approximately 1 × 108 CFU/ml within 48 h [51, 52], we predict that the concentration of eDNA in the middle ear would very quickly reach a threshold level to mitigate the activity of hBD-3 in the ear. This conclusion is supported by our observation that the concentration of NTHI in an in vitro 16-hour biofilm is approximately 1 × 108 CFU/ml with an associated eDNA concentration of 500 ng/ml, which was sufficient to completely abrogate the activity of 5.0 µg (r)hBD-3/ml. Our data also suggested that as the biofilm matures, the increase in abundance of eDNA could minimize the ability of newly produced AMPs within the middle ear to effectively eradicate an NTHI biofilm.
Our data provided evidence that degradation of eDNA from a 16-hour NTHI biofilm rescued the activity of (r)hBD-3. When eDNA was removed from the biofilm matrix, (r)hBD-3 significantly reduced the maximum height as well as the mean biomass and thickness of resultant biofilms when compared to controls. In fact, NTHI incubated in the presence of 0.5 µg (r)hBD-3/ml and DNase I formed biofilms that were indistinguishable from when NTHI was incubated with ten times the concentration of peptide, 5.0 µg (r)hBD-3/ml, alone. Moreover, the synergistic activity of 0.5 µg (r)hBD-3/ml when delivered in conjunction with DNase I resulted in a >30% decrease in viable NTHI. The ability to restore AMP activity similar to what is observed in planktonic antimicrobial assays [29] is encouraging given the demonstrated protective nature of the biofilm matrix to antimicrobial treatment. We postulate that addition of DNase I in this experimental approach rescued the activity of (r)hBD-3 due to both disruption of eDNA-(r)hBD-3 complexes, which would release functional peptide, and also due to increased diffusion of the AMP through the now debulked biofilm matrix. An unexpected finding in our studies was that incubation of NTHI with RNase resulted in augmented biofilms and an increase in the concentration of adherent NTHI. The mechanism by which an increased concentration of ribonucleotides results in more robust NTHI biofilms, while not currently understood, is an area of active investigation in our laboratory. Although not presented here, we have demonstrated that the ability to rescue the antimicrobial activity of (r)hBD-3 with the addition of DNase I was not restricted to this AMP but also extended to the closely related molecule recombinant human β-defensin-2, an AMP also expressed in the middle ear (data not shown) [30]. These results suggest that the activity of additional AMPs could be rescued by mechanisms that foster removal of eDNA from biofilms.
Our data have provided evidence to support a novel therapeutic regimen to treat biofilms formed by NTHI during OM. One example of a viable approach could involve the delivery of DNase I in conjunction with an effector of innate immunity such as (r)hBD-3 to the middle ear of a child with chronic, recurrent disease. As tympanostomy tube insertion through the tympanic membrane is common amongst this patient population, access to the middle ear would not be a barrier to this potential treatment regimen. The outcome of this approach would likely be to bolster the ability of the innate immune system to manage NTHI-induced biofilms, subsequently obviating the need for the unnecessary administration of first-line antibiotics typically used to treat patients with OM. In addition, removal of the protective niche provided by the biofilm should also help diminish the recurrent nature of this disease. As overuse of antibiotics leads to the development of newly resistant microorganisms, and is an increasing concern for the medical and scientific communities, our findings could have a wide range of utility.
Collectively, our data showed that the presence of eDNA in a biofilm formed by NTHI abrogated the biological activity of (r)hBD-3, an important effector of innate immunity. These data offer insight into how a molecule that can effectively impact survival of planktonic NTHI in the middle ear is ineffective at protecting this biological niche during chronic disease that includes a biofilm component. As additional diseases with associated bacterial biofilms such as cystic fibrosis contain AMPs bound to eDNA [53], our data have implications for other diseases of the respiratory tract, an observation which warrants future investigation in this regard.
Acknowledgments
We thank Jennifer Neelans for manuscript preparation. This work was supported by grants R01 DC005847 and R01 DC003915 to L.O.B. from the National Institute on Deafness and Other Communication Disorders/National Institutes of Health.
References
- 1.Infante-Rivard C, Fernandez A. Otitis media in children: frequency, risk factors, and research avenues. Epidemiol Rev. 1993;15:444–465. doi: 10.1093/oxfordjournals.epirev.a036129. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal A, Murphy TF. Haemophilus influenzae infections in the H. influenzae type b conjugate vaccine era. J Clin Microbiol. 2011;49:3728–3732. doi: 10.1128/JCM.05476-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foxwell AR, Kyd JM, Cripps AW. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiol Mol Biol Rev. 1998;62:294–308. doi: 10.1128/mmbr.62.2.294-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erwin AL, Smith AL. Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol. 2007;15:355–362. doi: 10.1016/j.tim.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Alsarraf R, Jung CJ, Perkins J, Crowley C, Alsarraf NW, Gates GA. Measuring the indirect and direct costs of acute otitis media. Arch Otolaryngol Head Neck Surg. 1999;125:12–18. doi: 10.1001/archotol.125.1.12. [DOI] [PubMed] [Google Scholar]
- 6.Kaplan B, Wandstrat TL, Cunningham JR. Overall cost in the treatment of otitis media. Pediatr Infect Dis J. 1997;16:S9–S11. doi: 10.1097/00006454-199702001-00003. [DOI] [PubMed] [Google Scholar]
- 7.Woodfield G, Dugdale A. Evidence behind the WHO guidelines: hospital care for children: what is the most effective antibiotic regime for chronic suppurative otitis media in children? J Trop Pediatr. 2008;54:151–156. doi: 10.1093/tropej/fmn042. [DOI] [PubMed] [Google Scholar]
- 8.Leach AJ, Morris PS, Mathews JD. Compared to placebo, long-term antibiotics resolve otitis media with effusion (OME) and prevent acute otitis media with perforation (AOMwiP) in a high-risk population: a randomized controlled trial. BMC Pediatr. 2008;8:23. doi: 10.1186/1471-2431-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization Chronic Suppurative Otitis Media: Burden of Illness and Management Options. Geneva, WHO. 2004 [Google Scholar]
- 10.Murphy TF, Kirkham C. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol. 2002;2:7. doi: 10.1186/1471-2180-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurcisek JA, Bakaletz LO. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–3875. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym PA, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 14.Webster P, Wu S, Gomez G, Apicella M, Plaut AG, St Geme., JW 3rd Distribution of bacterial proteins in biofilms formed by non-typeable Haemophilus influenzae. J Histochem Cytochem. 2006;54:829–842. doi: 10.1369/jhc.6A6922.2006. [DOI] [PubMed] [Google Scholar]
- 15.Jurcisek J, Greiner L, Watanabe H, Zaleski A, Apicella MA, Bakaletz LO. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect Immun. 2005;73:3210–3218. doi: 10.1128/IAI.73.6.3210-3218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, Tjokro N, Li B, Justice SS, Bakaletz LO. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011;4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- 17.Das T, Sharma PK, Busscher HJ, van der Mei HC, Krom BP. Role of extracellular DNA in initial bacterial adhesion and surface aggregation. Appl Environ Microbiol. 2010;76:3405–3408. doi: 10.1128/AEM.03119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spoering AL, Gilmore MS. Quorum sensing and DNA release in bacterial biofilms. Curr Opin Microbiol. 2006;9:133–137. doi: 10.1016/j.mib.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 19.Palchevskiy V, Finkel SE. Escherichia coli competence gene homologs are essential for competitive fitness and the use of DNA as a nutrient. J Bacteriol. 2006;188:3902–3910. doi: 10.1128/JB.01974-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slinger R, Chan F, Ferris W, Yeung SW, St Denis M, Gaboury I, Aaron SD. Multiple combination antibiotic susceptibility testing of nontypeable Haemophilus influenzae biofilms. Diagn Microbiol Infect Dis. 2006;56:247–253. doi: 10.1016/j.diagmicrobio.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 22.Foreman A, Wormald PJ. Different biofilms, different disease? A clinical outcomes study. Laryngoscope. 2010;120:1701–1706. doi: 10.1002/lary.21024. [DOI] [PubMed] [Google Scholar]
- 23.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–720. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 24.Ganz T. Antimicrobial polypeptides in host defense of the respiratory tract. J Clin Invest. 2002;109:693–697. doi: 10.1172/JCI15218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh PK, Jia HP, Wiles K, Hesselberth J, Liu L, Conway BA, Greenberg EP, Valore EV, Welsh MJ, Ganz T, Tack BF, McCray PB., Jr Production of beta-defensins by human airway epithelia. Proc Natl Acad Sci USA. 1998;95:14961–14966. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang D, Chertov O, Bykovskaia SN, Chen Q, Buffo MJ, Shogan J, Anderson M, Schroder JM, Wang JM, Howard OM, Oppenheim JJ. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- 27.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB, Bos NA. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhople V, Krukemeyer A, Ramamoorthy A. The human beta-defensin-3, an antibacterial peptide with multiple biological functions. Biochim Biophys Acta. 2006;1758:1499–1512. doi: 10.1016/j.bbamem.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 29.McGillivary G, Mason KM, Jurcisek JA, Peeples ME, Bakaletz LO. Respiratory syncytial virus-induced dysregulation of expression of a mucosal beta-defensin augments colonization of the upper airway by non-typeable Haemophilus influenzae. Cell Microbiol. 2009;11:1399–1408. doi: 10.1111/j.1462-5822.2009.01339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song JJ, Chae SW, Woo JS, Lee HM, Jung HH, Hwang SJ. Differential expression of human beta defensin 2 and human beta defensin 3 in human middle ear cholesteatoma. Ann Otol Rhinol Laryngol. 2007;116:235–240. doi: 10.1177/000348940711600312. [DOI] [PubMed] [Google Scholar]
- 31.Mason KM, Raffel FK, Ray WC, Bakaletz LO. Heme utilization by nontypeable Haemophilus influenzae is essential and dependent on Sap transporter function. J Bacteriol. 2011;193:2527–2535. doi: 10.1128/JB.01313-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo HH, Chan C, Burrows LL, Deber CM. Hydrophobic interactions in complexes of antimicrobial peptides with bacterial polysaccharides. Chem Biol Drug Des. 2007;69:405–412. doi: 10.1111/j.1747-0285.2007.00518.x. [DOI] [PubMed] [Google Scholar]
- 33.Remijsen Q, Kuijpers TW, Wirawan E, Lippens S, Vandenabeele P, Vanden Berghe., T Dying for a cause: NETosis, mechanisms behind an antimicrobial cell death modality. Cell Death Differ. 2011;18:581–588. doi: 10.1038/cdd.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juneau RA, Pang B, Weimer KE, Armbruster CE, Swords WE. Nontypeable Haemophilus influenzae initiates formation of neutrophil extracellular traps. Infect Immun. 2011;79:431–438. doi: 10.1128/IAI.00660-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beckloff N, Laube D, Castro T, Furgang D, Park S, Perlin D, Clements D, Tang H, Scott RW, Tew GN, Diamond G. Activity of an antimicrobial peptide mimetic against planktonic and biofilm cultures of oral pathogens. Antimicrob Agents Chemother. 2007;51:4125–4132. doi: 10.1128/AAC.00208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiner DJ, Bucki R, Janmey PA. The antimicrobial activity of the cathelicidin LL37 is inhibited by F-actin bundles and restored by gelsolin. Am J Respir Cell Mol Biol. 2003;28:738–745. doi: 10.1165/rcmb.2002-0191OC. [DOI] [PubMed] [Google Scholar]
- 37.Yin L, Chino T, Horst OV, Hacker BM, Clark EA, Dale BA, Chung WO. Differential and coordinated expression of defensins and cytokines by gingival epithelial cells and dendritic cells in response to oral bacteria. BMC Immunol. 2010;11:37. doi: 10.1186/1471-2172-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakaletz LO, Baker BD, Jurcisek JA, Harrison A, Novotny LA, Bookwalter JE, Mungur R, Munson RS., Jr Demonstration of type IV pilus expression and a twitching phenotype by Haemophilus influenzae. Infect Immun. 2005;73:1635–1643. doi: 10.1128/IAI.73.3.1635-1643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris RH, Wilk D, Bevins CL, Munson RS, Jr, Bakaletz LO. Identification and characterization of a mucosal antimicrobial peptide expressed by the chinchilla (Chinchilla lanigera) airway. J Biol Chem. 2004;279:20250–20256. doi: 10.1074/jbc.M400499200. [DOI] [PubMed] [Google Scholar]
- 40.Martinez LR, Casadevall A. Cryptococcus neoformans cells in biofilms are less susceptible than planktonic cells to antimicrobial molecules produced by the innate immune system. Infect Immun. 2006;74:6118–6123. doi: 10.1128/IAI.00995-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Su B, Nestle FO, Zal T, Mellman I, Schroder JM, Liu YJ, Gilliet M. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 42.Bucki R, Namiot DB, Namiot Z, Savage PB, Janmey PA. Salivary mucins inhibit antibacterial activity of the cathelicidin-derived LL-37 peptide but not the cationic steroid CSA-13. J Antimicrob Chemother. 2008;62:329–335. doi: 10.1093/jac/dkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008;4:e1000213. doi: 10.1371/journal.ppat.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Llobet E, Tomas JM, Bengoechea JA. Capsule polysaccharide is a bacterial decoy for antimicrobial peptides. Microbiology. 2008;154:3877–3886. doi: 10.1099/mic.0.2008/022301-0. [DOI] [PubMed] [Google Scholar]
- 45.Jono H, Xu H, Kai H, Lim DJ, Kim YS, Feng XH, Li JD. Transforming growth factor-beta-Smad signaling pathway negatively regulates nontypeable Haemophilus influenzae-induced MUC5AC mucin transcription via mitogen-activated protein kinase (MAPK) phosphatase-1-dependent inhibition of p38 MAPK. J Biol Chem. 2003;278:27811–27819. doi: 10.1074/jbc.M301773200. [DOI] [PubMed] [Google Scholar]
- 46.Chen R, Lim JH, Jono H, Gu XX, Kim YS, Basbaum CB, Murphy TF, Li JD. Nontypeable Haemophilus influenzae lipoprotein P6 induces MUC5AC mucin transcription via TLR2-TAK1-dependent p38 MAPK-AP1 and IKKβ-IĸBα-NF-ĸB signaling pathways. Biochem Biophys Res Commun. 2004;324:1087–1094. doi: 10.1016/j.bbrc.2004.09.157. [DOI] [PubMed] [Google Scholar]
- 47.Kubiet M, Ramphal R, Weber A, Smith A. Pilus-mediated adherence of Haemophilus influenzae to human respiratory mucins. Infect Immun. 2000;68:3362–3367. doi: 10.1128/iai.68.6.3362-3367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davies J, Carlstedt I, Nilsson AK, Hakansson A, Sabharwal H, van Alphen L, van Ham M, Svanborg C. Binding of Haemophilus influenzae to purified mucins from the human respiratory tract. Infect Immun. 1995;63:2485–2492. doi: 10.1128/iai.63.7.2485-2492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mason KM, Bruggeman ME, Munson RS, Bakaletz LO. The non-typeable Haemophilus influenzae Sap transporter provides a mechanism of antimicrobial peptide resistance and SapD-dependent potassium acquisition. Mol Microbiol. 2006;62:1357–1372. doi: 10.1111/j.1365-2958.2006.05460.x. [DOI] [PubMed] [Google Scholar]
- 50.Mason KM, Munson RS, Jr, Bakaletz LO. A mutation in the Sap operon attenuates survival of nontypeable Haemophilus influenzae in a chinchilla model of otitis media. Infect Immun. 2005;73:599–608. doi: 10.1128/IAI.73.1.599-608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGillivary G, Bakaletz LO. The multifunctional host defense peptide SPLUNC1 is critical for homeostasis of the mammalian upper airway. PLoS One. 2010;5:e13224. doi: 10.1371/journal.pone.0013224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson RW, McGillivary G, Denoel P, Poolman J, Bakaletz LO. Abrogation of nontypeable Haemophilus influenzae protein D function reduces phosphorylcholine decoration, adherence to airway epithelial cells, and fitness in a chinchilla model of otitis media. Vaccine. 2011;29:1211–1221. doi: 10.1016/j.vaccine.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 53.Bucki R, Byfield FJ, Janmey PA. Release of the antimicrobial peptide LL-37 from DNA/F-actin bundles in cystic fibrosis sputum. Eur Respir J. 2007;29:624–632. doi: 10.1183/09031936.00080806. [DOI] [PubMed] [Google Scholar]