Abstract
Background
Inborn errors in the Toll like receptor 3 (TLR3)-Interferon (IFN) type I and III pathway have been implicated in susceptibility to herpes simplex virus encephalitis (HSE) in children, but most patients studied do not carry mutations in any of the genes presently associated with HSE-susceptibility. Moreover, many patients do not display any TLR3-related fibroblastic phenotype.
Objective
This suggests the study of other signalling pathways downstream of TLR3 and/or other independent pathways which may contribute to HSE susceptibility.
Methods
We have used the stable isotope labelling of amino acids in cell culture (SILAC) proteomics methodology to measure changes in the human immortalized fibroblast proteome after TLR3 activation.
Results
Cells from healthy controls were compared to cells from a patient with a known genetic aetiology of HSE (UNC93B−/−) and also to cells from an HSE patient with an unknown gene defect. Consistent with known variation in susceptibility of individuals to viral infections, substantial variation in the response level of different healthy controls was observed, but common functional networks could be identified, including upregulation of superoxide dismutase 2 (SOD2). The HSE patients show clear differences in functional response networks when compared to healthy patients and also when compared to each other.
Conclusions
The present study delineates a number of novel proteins, TLR3-related pathways and cellular phenotypes that may help elucidate the genetic basis of childhood HSE. Furthermore, our results reveal SOD2 as a potential therapeutic target for amelioration of the neurological sequelae caused by HSE.
Keywords: Herpes simplex encephalitis (HSE), SILAC, proteomics, mass spectrometry, fibroblast, herpes simplex virus 1 (HSV-1), TLR3, interferon (IFN)
INTRODUCTION
Herpes simplex virus (HSV-1) encephalitis (HSE) is a devastating infection of the central nervous system (CNS)1. HSE is the most common form of sporadic viral encephalitis in Western countries, where it is estimated to occur in approximately 1 in 250,000 individuals per year. It peaks in childhood between the ages of 3 months and 6 years during primary infection with HSV-1, which reaches the CNS via a neurotropic route involving the trigeminal and olfactory nerves2, 3. Treatment with acyclovir decreases the mortality rate in affected children, but significant neurological impairment is observed in most survivors, young children in particular. HSV-1 is widespread and typically innocuous in human populations. Childhood HSE has not been associable with known immunodeficiencies and its pathogenesis remained elusive until we identified the first three genetic etiologies of this condition4, 5. Autosomal recessive UNC-93B deficiency abolishes Toll-like receptor (TLR) 3,7,8, and 9 signaling6, whereas autosomal dominant TLR3 deficiency specifically affects TLR3 signaling7. Recently an autosomal dominant deficiency in TNF receptor-associated factor 3 (TRAF3), an adaptor molecule implicated in the TLR3 pathway, and Toll/IL1R (TIR) domain-containing adaptor inducing IFN-β (TRIF) deficient patients has been described8, 9. These studies suggested that childhood HSE may result from impaired interferon (IFN)-α/β and IFN-λ production in response to the stimulation of TLR3 by dsRNA intermediates of HSV-1 in the CNS. However, only a small fraction of children with HSE carry mutations in UNC93B1, TLR3, TRIF or TRAF3. The study of proteins implicated in the TLR3-IFN pathway for HSE patients enrolled in our laboratory (226 patients) has revealed that only a small proportion of patients bear a genetic defect in these proteins. A larger proportion of patients display an impaired production of IFN type I and III upon TLR3 stimulation of their fibroblasts. Other patients do not show the fibroblastic phenotype associated with impaired IFN production; in these patients the gene defect may affect other pathway(s) that are normally activated in fibroblasts after TLR3 stimulation. In the present study, as an approach to define new candidate genes for HSE, we have used proteomics methods to characterize TLR3-dependent pathways in fibroblasts that might be correlated with HSV-1 infection in the CNS.
METHODS
Subjects and patient P case report
The experimental protocol was approved by the Ethical Committee of Necker-Enfants Malades Hospital (Paris, France) and written informed consent was obtained for this study. P is a French boy of non-consanguineous descent with no family history of encephalitis. At the age of eight years, he suffered fever, convulsions and hemiparesis and he was diagnosed with HSE.
Cell purification
Primary human fibroblasts were obtained from biopsies of an UNC-93B-deficient patient6, three healthy controls, and one HSE patient with unknown gene defect. They were then transformed with an SV-40 vector, as previously described10, to create immortalized fibroblast cell lines: SV40-fibroblasts.
Stable isotope labelling with amino acids in cell culture (SILAC) labelling of human SV40-fibroblast
Cells were grown for six rounds of cell division in DMEM containing 13C6, 15N4 L-arginine, 13C6 L-lysine (Invitrogen, Carlsbad, California, USA) (heavy medium) supplemented with 10% dialysed FCS (Invitrogen) to ensure all the cellular proteins were labelled to saturation. The SILAC technique relies on the intrinsic metabolic machinery of the cell to incorporate “heavy” amino acids into all newly synthesized proteins. Immortalized fibroblast cell lines were used to achieve the full SILAC incorporation without detrimentally affecting the characteristics of the cell line. Our previous work has demonstrated that SV40 immortalised fibroblasts can successfully be used to study TLR3 signalling7–9.
Labelled cells were activated in 24-well plates, at a density of 105 cells/well, with 25 µg/ml polyinosinepolycytidylic acid (poly(I:C)), a TLR3 agonist (InvivoGen, San Diego, CA, USA), for 24 hours. Unlabelled cells were grown in parallel in medium containing normal amino acids (light medium) and were not stimulated. The SILAC labels were shown to be fully incorporated before the experiment was conducted (data not shown).
Protein Separation and In-Gel Enzymatic Digestion
Equal amounts of protein from unlabelled “light” (non-stimulated) and labelled “heavy” (stimulated) cells were mixed and subsequently separated by SDS-PAGE (BioRad Laboratories, Hercules, CA, USA). Proteins were visualized by silver-staining (Sigma, St Louis, MO, USA) and the gel lane was divided into approximately 34 equally sized pieces which were excised from the gel and destained (30mM K3Fe(CN)6; 100mM Na2S2O3) prior to further processing. Gel processing was conducted with a Progest Investigator Instrument (DigiLab, Genomics Solutions, Cambs, UK) for reduction and alkylation according to established protocols11. Briefly, the gel pieces were washed with three cycles of 25 mM NH4HCO3 pH 8.0 and acetonitrile. Finally, gel plugs were rehydrated in 20 µg/mL sequencing grade modified trypsin (Promega, Hamps, UK) and incubated overnight at 37C. Tryptic peptides were eluted, vacuum-dried, and resuspended in 0.1% formic acid.
XCalibur raw files were processed by Quant.exe and Identify.exe of the MaxQuant suite (version 1.0.13.13), in combination with the Mascot search engine (version 2.2, Matrix Science, U.K.), and searched against a concatenated International Protein Index (IPI) human protein database(version 3.54; containing 75,710 entries, including 262 commonly observed contaminants such as porcine trypsin and some human keratins). All proteins were filtered according to a false discovery rate (FDR) of 1% applied at both peptide and protein levels. Proteins were automatically quantified in the MaxQuant software. In the final results files, all Protein Groups with a normalized ratio SigB score of ≤0.05 and a normalised ratio > 1.5 were accepted for downstream analysis. Experiments were done induplicate and all proteins discussed in the text were significant in both replicates for each sample. For details of mass spectrometry analysis, protein inference and quantification, analysis of sample correlations and global functional analysis of protein networks, see Additional Methods at the Online Repository.
Transient transfections
SV40-fibroblasts were transfected with 2 µg of pBI-EGFP-MnSOD (SOD2) or pTRE-Tight-BI-AcGFP1 (Clontech, Palo Alto, CA, USA) as mock vector, in the presence of the FuGENE ®HD Transfection Reagent (Roche Applied Science, Indianapolis, IN, USA), according to manufacturer’s instructions. 24 hours post-transfection, SV40-fibroblasts were activated by incubation with 25 µg/ml poly(I:C) (InvivoGen) for 24 h. pTRE-Tight-BI-AcGFP1 was used as mock vector (pBI-EGFP has been replaced by AcGFP; it has 94% identical amino acid sequence and same biophysical properties).
Western blot
Total cell extracts were prepared from SV40-fibroblasts, both transfected and not transfected. Equal amounts of protein from each sample were separated by SDS-PAGE and blotted onto iBlot™ Gel Transfer Stacks (Invitrogen). Membranes were probed with anti-ICAM-1 rabbit monoclonal (Lifespan Biosciences, Seattle, WA, USA), anti-SOD2 and ITGA2 monoclonals and anti-ITGA5 and PPIF polyclonals (Abcam, Cambridge, MA, USA), anti-ANXA5/7 monoclonals (SantaCruz Biotechnology Inc., SantaCruz, CA), followed by a secondary anti-mouse or rabbit IgG (GEHealthcare, Buckinghamshire, UK). Membranes were stripped and reprobed with anti-GADPH (SantaCruz) to control for protein loading. Antibody binding was detected by enhanced chemiluminescence (ECL; Amersham-Pharmacia-Biotech).
Apoptosis analysis
Levels of SV40-fibroblast apoptosis after poly(I:C) stimulation were assessed by measures of caspase-3 and 7 activity. Cells were plated, in triplicate, in Microtest™ 96 well Assay Plate, Optilux™ Black/Clear Bottom (Falcon, Becton Dickinson, Franklin Lakes, NJ, USA) (1 × 104 cells/well), in DMEM supplemented with 2% FCS; some cells were transfected with 150 ng per well of pBI-EGFP-MnSOD (SOD2) or pTRE-Tight-BI-AcGFP1 as mock vector. 24 hours later, cells were treated with poly(I:C) (25 µg/ml) and incubated for 24h. Caspase-3 and 7 activity was measured 48 hours post-transfection by Caspase-Glo® 3/7 Assay (Promega) as per manufacturer’s instructions.
IFN type I and III determination
SV40-fibroblast cell lines were activated in 24-well plates at a density of 105 cells/well and stimulated with poly(I:C) at a concentration of 25 µg/ml for 24 h. Cells were grown at 37°C under an atmosphere containing 5% CO2. Cell supernatants were recovered and an ELISA was performed for IFN-β (TFB, Fujirebio, Inc., Tokyo, Japan) according to manufacturer’s instructions. The ELISA for IFN-λ was performed as previously described7.
RESULTS
Identification of proteins up-regulated in human SV40 fibroblasts following TLR3 activation
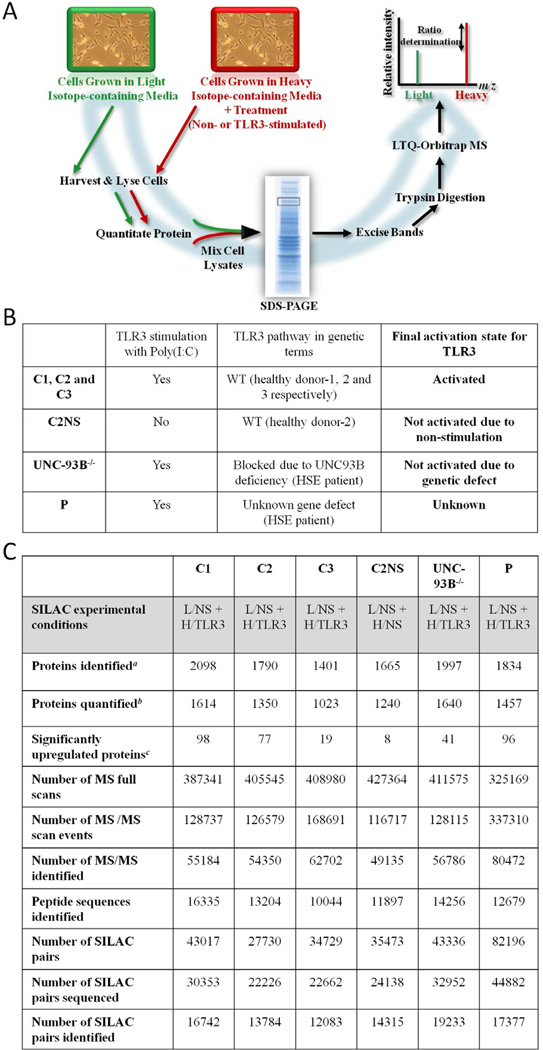
Response to TLR3 activation was defined by altered levels of protein abundance in human SV40-fibroblasts when stimulated with poly(I:C) (used as TLR3 agonist6–9) and was measured by SILAC/MS12–14 monitoring of heavy/light isotope labelling ratios for peptides from proteins of stimulated/unstimulated cells (Figure 1A).
Figure 1.
(A) Illustration of the SILAC workflow. (B) Summary of samples used and TLR3 pathway state for the SILAC experiment. C1–3: healthy controls; C2NS: healthy control-2 non-stimulated; UNC-93B−/−: UNC-93B-deficient patient; P: patient with unknown gene defect. WT: Wild-type HSE: herpes simplex encephalitis. (C) Overview of MS results obtained with MaxQuant software.a Protein groups identified with protein and peptide FDR = 1%.b Protein abundance ratios calculated as median values using a minimum of 3 quantifiable razor peptides.c Protein ratios with SigB ≤ 0.05 and SILAC ratios ≥ 1.5. L: light medium; H: heavy medium; NS: non-stimulated cells; TLR3: Poly(I:C) stimulated cells.
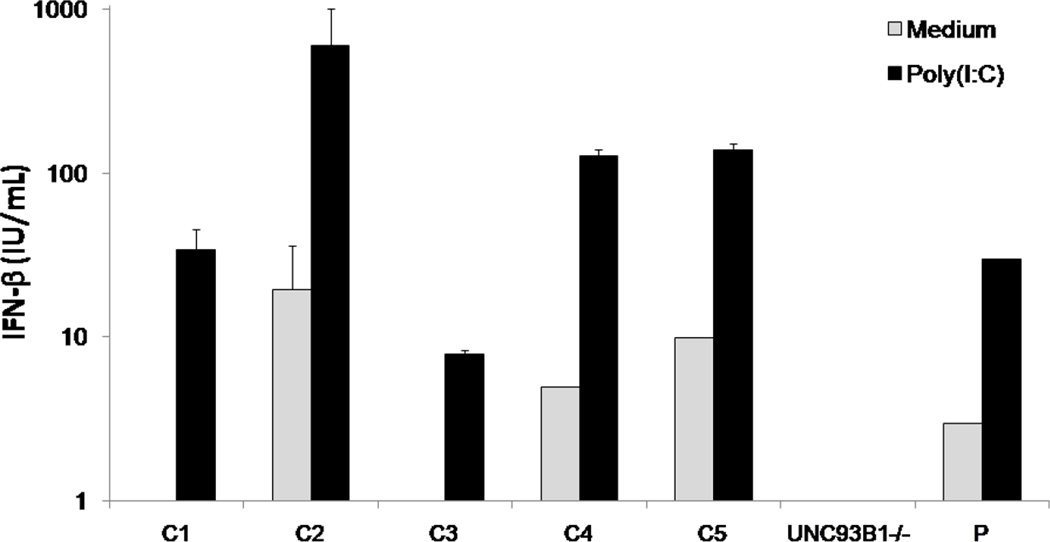
Three Healthy samples (C1, C2 and C3) are positive controls that provide a measure of variability between individuals (Figure 1B). Negative control, C2NS, is a healthy sample with strong TLR3 response, but for which neither cell population (light/heavy) was stimulated (Figure 1B). The UNC-93B-deficiency blocks TLR3, 7, 8 and 9 pathways, so cells from the UNC-93B−/− patient (sample UNC-93B−/−) should not respond to TLR3 agonists6. We used SV40-fibroblasts from a homozygous UNC-93B−/− patient rather than a patient with heterozygous TLR3 mutation due to the complete deficiency of TLR3 pathways observed in UNC93B−/− cells6. Finally, we included cells from an HSE patient with unknown gene defect (P) (Figure 1B), which produce detectable amounts of IFN type I after TLR3 stimulation (Figure 2).
Figure 2.
Production of IFN-β by SV40-fibroblasts after poly(I:C) stimulation for 24 hours as assessed by ELISA. C1–C5: positive healthy controls. UNC93B−/−: UNC-93B-deficient patient. Mean values ± SD were calculated from three independent experiments.
All six biological samples were subjected to duplicate MS analysis15, 16 and protein lists were obtained by combining both datasets prior to analysis of differential protein abundance (E Table 1–6). The MS data is summarized in Figure 1C, and E Figure 1 illustrates the quality of the MS data for a single SILAC peptide pair detected in a key protein of our study.
Figure 1C shows the number of proteins found to be significantly upregulated (normalized H/L (SILAC) ratio ≥ 1.5 and Significance B (SigB)16 < 0.05, see below) in the different cells following activation (except for C2NS) with poly(I:C). We have previously observed variability in response of healthy control fibroblasts upon TLR3 stimulation, particularly in terms of cytokine and interferon production (Figure 2). To take this into account, we included sample C3, from a healthy donor with known weaker response to TLR3 stimulation compared to C1 and C2 (Figure 2). In line with expectation, this control showed a reduced number of upregulated proteins (19 with SILAC H/L ratios between 1.50-fold and 2.15-fold; Figure 1C). Comparison of upregulated proteins between the control samples revealed variation in those proteins most strongly upregulated. This variability might arise from a) the conservative condition that protein SILAC ratios were calculated from at least three separate H/L ratio measurements, and/or b) differences in the extent to which different proteins change in abundance in cells taken from different individuals. A qualitative test of cellular variability amongst healthy controls displays biologically-relevant positive correlation distinguishable from the negative control (C2NS) (see Additional Results, E Figure 2, E Figure 3 and E Table 7 at the Online Repository).
Detection of Functional Networks
Given the observed variability in detection and measurement of specific protein regulation across experiments, we sought common functional networks amongst the most strongly upregulated proteins in order to characterise response to TLR3 stimulation and identify disease-related differences by comparison. We used MetaCore™ version 6.3 (GeneGo Inc.) to investigate pathways and biological functions represented by differentially regulated proteins in all samples (see Additional Results and E Figure 2 at the Online Repository).
The analysis indicated that the most significant GeneGo process networks for C1/C2/C3 are similar and are implicated in immune responses, such as IFN-signalling and antigen presentation pathways (Table 1A), known to be implicated following TLR3 stimulation17, 18. Similar analysis was performed for the non-stimulated C2NS control sample. The majority of proteins in the non-stimulated control C2NS show SILAC ratios ≈ 1 (E Table 4) which represent “measurement noise”. As may be expected, the 8 proteins designated as upregulated (H/L = 1.50 to 3.07-fold) did not associate with canonical pathways or process networks implicated in immune response, and networks inferred from them were of limited statistical significance and scattered over diverse pathways and processes (Table 2A). The UNC-93B−/− sample did not associate with the immune-related networks identified above, further suggesting that the TLR3 pathway is completely abolished by this deficiency (Table 2B). UNC-93B−/− did, however, yield several upregulated proteins (E Table 5), with SILAC ratios of 1.5 to 3.95-fold, which may represent attempts by these cells to compensate for the genetic defect. Canonical pathways associated with these proteins were largely metabolic (glycolysis, gluconeogenesis, fructose metabolism), however, and process networks inferred were particularly diverse (Table 2B).
Table 1.
GeneGo pathway maps and process networks with their proteins associated. (A) Analysis of the individual data sets for C1–3. (B) Enrichment analysis for the union of the C1–3 data sets. Grey panels: pathways/networks known to be implicated in TLR3-IFN signalling. Blue panels: new pathways/networks activated. Threshold: 1.5 (SILAC ratio), SigB p-value < 0.05. Sorting method is statistically significant.
| A | |||||
|---|---|---|---|---|---|
| C1 SAMPLE | C2 SAMPLE | C3 SAMPLE | |||
| name | pValue | name | pValue | name | pValue |
| Ontology: Top GeneGo Pathway Maps | Ontology: Top GeneGo Pathway Maps | Ontology: Top GeneGo Pathway Maps | |||
| Immune response_Antigen presentation by MHC class I | 0,0004167 | Immune response_Antiviral actions of Interferons | 5,215E-05 | Immune response_MIF-mediated glucocorticoid regulation | 0,02399 |
| Immune response_IFN alpha/beta signaling pathway | 0,007584 | Immune response_Antigen presentation by MHC class I | 0,0001645 | Immune response_IFN alpha/beta signaling pathway | 0,02615 |
| Ontology: Top GeneGo Process Networks | Ontology: Top GeneGo Process Networks | Immune response_IL-27 signaling pathway | 0,02615 | ||
| Inflammation_Interferon signaling | 4,681E-05 | Immune response_Antigen presentation | 0,0004169 | Immune response_Role of integrins in NK cells cytotoxicity | 0,04112 |
| Immune response_Phagosome in antigen presentation | 0,0006502 | Immune response_Phagosome in antigen presentation | 0,007849 | Ontology: Top GeneGo Process Networks | |
| Immune response_Antigen presentation | 0,00111 | Inflammation_Interferon signaling | 0,01796 | Inflammation_Interferon signaling | 0,01668 |
| Immune response_IL-5 signalling | 0,0779 | ||||
| B | |||||
|---|---|---|---|---|---|
| UNION C1, C2 and C3 SAMPLES | |||||
| Ontology: Top GeneGo Pathway Maps | Ontology: Top GeneGo Process Networks | ||||
| name | pValue | Proteins associated | name | pValue | Proteins associated |
| Immune response_IFN alpha/beta signaling pathway | 0,007584 | ISG54, ISG15, PTP-1B | Inflammation_Interferon signaling | 4,68E-05 | TAP1, TAP2, ICAM1, ISG15, ISG54, IFI56 |
| Immune response_MIF-mediated glucocorticoid regulation | 0,02399 | ICAM1 | Immune response_Antigen presentation | 0,000417 | Tapasin, TAP1, TAP2, Calnexin, ICAM1 |
| Immune response_IL-27 signaling pathway | 0,02615 | ICAM1 | Inflammation_NK cell cytotoxicity | 0,04988 | Histone H1, ICAM1, TfR1, MHC class1 |
| Ontology: Top GeneGo Pathway Maps | Ontology: Top GeneGo Process Networks | ||||
| name | pValue | Proteins associated | name | pValue | Proteins associated |
| Transcription_Role of AP-1 in regulation of cellular metabolism | 0,01016 | ITGA2, ITGA5 | Blood coagulation | 0,000239 | TFPI-2, PLAU, ITGA2, AnnexinV |
| Chemotaxis_CCR4-induced leukocyte adhesion | 0,01171 | ICAM1, Talin | Cell adhesion_Platelet-endothelium-leucocyte interactions | 0,000597 | Integrins, TFPI-2, PAI2, ICAM1, PLAU, FGF2 |
| Apoptosis and survival_Endoplasmic reticulum stress response pathway | 0,01918 | ERP5, SOD2 | Transcription_mRNA processing | 0,01563 | hnRNP F, SNRPA, HYPA, KHSRP, SAM68, DDX5, SF3B1, U2AF35 |
| Development_Cross-talk between VEGF and Angiopoietin 1 signaling pathways | 0,0283 | ICAM1 | Cell adhesion_Attractive and repulsive receptors | 0,02102 | ICAM1, Integrins, Cortactin |
| Chemotaxis_Inhibitory action of lipoxins on IL-8- and Leukotriene B4-induced neutrophil migration | 0,0319 | ICAM1, Talin | Proteolysis_Connective tissue degradation | 0,03509 | TFPI-2, IRAP, PAI2, PLAU |
| Inhibitory action of Lipoxins on neutrophil migration | 0,03911 | SOD2 | Cell adhesion_Cell-matrix interactions | 0,05649 | Integrins |
| Apoptosis and survival_Anti-apoptotic action of nuclear ESR1 and ESR2 | 0,04006 | SOD2 | Cell adhesion_Integrin-mediated cell-matrix adhesion | 0,05697 | Integrin, Talin, CD82 |
Table 2.
GeneGo pathway maps and process networks after the analysis of the individual data sets for (A) control non-stimulated (C2NS), (B) UNC-93B deficient patient cells (UNC-93B−/−) and (C) HSE patient cells with unknown gene defect (P). Threshold: 1.5 (SILAC ratio), SigB p-value<0.05 and the sorting method is statistically significant.
| A | B | C | |||
|---|---|---|---|---|---|
| C2NS SAMPLE | UNC-93B−/− SAMPLE | P SAMPLE | |||
| name | pValue | name | pValue | name | pValue |
| Ontology: Top GeneGo Pathway Maps | Ontology: Top GeneGo Pathway Maps | Ontology: Top GeneGo Pathway Maps | |||
| Cell adhesion_Cadherin-mediated cell adhesion | 0,00952 | Glycolysis and gluconeogenesis (short map) | 1,87E-09 | Apoptosis and survival_Granzyme A signaling | 0,00097 |
| Development_Slit-Robo signaling | 0,01098 | Glycolysis and gluconeogenesis p.3 / Human version | 6,96E-05 | Transcription_P53 signaling pathway | 0,0021 |
| Neurophysiological process_EphB receptors in dendritic spine morphogenesis and synaptogenesis | 0,0128 | Glycolysis and gluconeogenesis p.3 | 6,96E-05 | Transport_Macropinocytosis | 0,00271 |
| Cell adhesion_Tight junctions | 0,01317 | Glycolysis and gluconeogenesis p. 2 | 0,000152 | Transport_RAB3 regulation pathway | 0,0037 |
| Transcription_P53 signaling pathway | 0,01426 | Glycolysis and gluconeogenesis p. 2/ Rodent version | 0,000167 | Immune response_Antiviral actions of Interferons | 0,00479 |
| Neurophysiological process_Receptor-mediated axon growth repulsion | 0,01644 | Glycolysis and gluconeogenesis p. 2 / Human version | 0,000281 | Normal wtCFTR traffic / Sorting endosome formation | 0,00612 |
| Ontology: Top GeneGo Process Networks | Fructose metabolism | 0,001911 | DNA damage_NHEJ mechanisms of DSBs repair | 0,00681 | |
| Transcription_Transcription by RNA polymerase II | 0,01626 | Fructose metabolism/ Rodent version | 0,00276 | Delta508-CFTR traffic / Sorting endosome formation in CF | 0,00991 |
| Cytoskeleton_Actin filaments | 0,01972 | Muscle contraction_Delta-type opioid receptor in smooth muscle contraction | 0,0035 | Immune response_IFN alpha/beta signaling pathway | 0,01076 |
| Cell adhesion_Synaptic contact | 0,02144 | Transcription_Role of Akt in hypoxia induced HIF1 activation | 0,003772 | Cell adhesion_Endothelial cell contacts by non-junctional mechanisms | 0,01076 |
| Reproduction_Spermatogenesis, motility and copulation | 0,03227 | Ontology: Top GeneGo Process Networks | Ontology: Top GeneGo Process Networks | ||
| Reproduction_Male sex differentiation | 0,03683 | Muscle contraction | 0,00024 | Cytoskeleton_Cytoplasmic microtubules | 0,00013 |
| Cytoskeleton_Spindle microtubules | 0,1304 | Development_Skeletal muscle development | 0,00127 | Cell cycle_Mitosis | 0,00035 |
| Cytoskeleton_Cytoplasmic microtubules | 0,1371 | Transport_Iron transport | 0,004586 | Cytoskeleton_Spindle microtubules | 0,00068 |
| Development_Neuromuscular junction | 0,1732 | Protein folding_ER and cytoplasm | 0,009973 | Inflammation_Interferon signaling | 0,00072 |
| Cell adhesion_Cell junctions | 0,1881 | Cell adhesion_Cell-matrix interactions | 0,03411 | Cytoskeleton_Macropinocytosis and its regulation | 0,00145 |
| Cell adhesion_Attractive and repulsive receptors | 0,2018 | Cell adhesion_Integrin-mediated cell-matrix adhesion | 0,03452 | Cell adhesion_Integrin-mediated cell-matrix adhesion | 0,00498 |
| Immune response_Phagocytosis | 0,03703 | Translation_Translation initiation | 0,00665 | ||
| Development_Blood vessel morphogenesis | 0,04052 | Cytoskeleton_Actin filaments | 0,00762 | ||
| Cell adhesion_Attractive and repulsive receptors | 0,1177 | Translation_Elongation-Termination | 0,00786 | ||
| Signal transduction_Insulin signaling | 0,1177 | Cytoskeleton_Intermediate filaments | 0,00823 | ||
The same analysis was also performed on the union of proteins upregulated in C1/C2/C3 (152 proteins, excluding 7 proteins in common withC2NS or UNC-93B−/− samples), strengthening the association between TLR3 stimulation and IFN α/β signalling pathways (Table 1B, E Figure 4) and further suggesting a clear proteomic signature for activation of these pathways upon TLR3 stimulation.
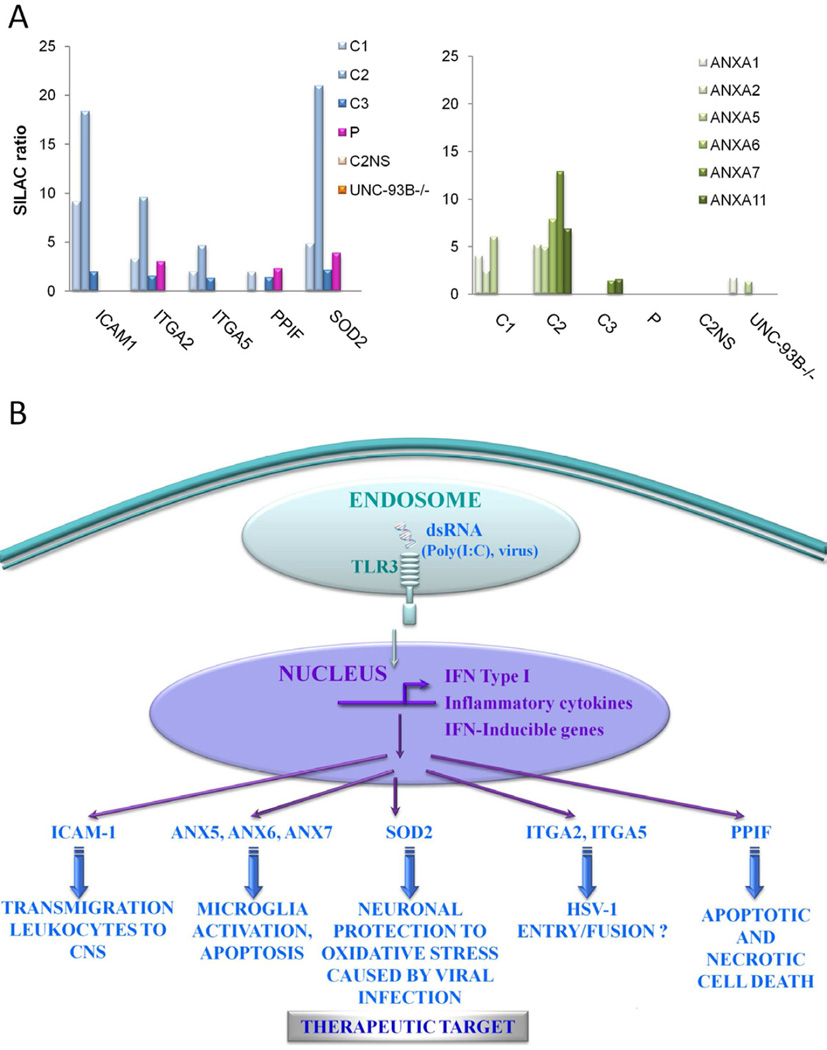
The analysis of healthy controls also revealed other statistically significant proteins/pathways/processes (Table 1B) that do not appear in similar analysis of C2NS or UNC-93B−/− samples (Table 2), and which have not been associated previously with TLR3 pathways. Such proteins included Intercellular adhesion molecule-1 (ICAM-1), integrins ITGA2 and 5, and Superoxide dismutase 2 (SOD2) (Table 1B). Furthermore, some proteins upregulated by TLR3 stimulation in healthy controls were not placed in pathways/networks by GeneGo, but are associated with a role in HSV infection in the CNS by literature evidence. These proteins include several Annexins and Peptidyl-propyl cis-trans isomerise mitochondrial (PPIF) (E Table 1–3), the majority of which were not identity as differentially regulated by analysis of UNC-93B−/− (Figure 3A). E Table 8 shows MS data for individual peptides assigned to these proteins and confirms the quality of their detection and assignment. The expression of these proteins was confirmed by western blot (WB) (E Figure 5) and they are considered further in the discussion.
Figure 3.
(A) For proteins with potential biological significance in HSE, the SILAC ratio detected for healthy controls (C1–3), healthy control 2 non-stimulated (C2NS), UNC-93B-deficient patient (UNC-93B−/−) and patient with unknown gene defect (P). SigB p-value < 0.05. (B) Illustration of potential biological significance in immunity against HSE for proteins up-regulated after TLR3 activation.
Finally, although not the primary focus of our analysis, the union of proteins commonly downregulated upon TLR3 stimulation in healthy controls C1/C2/C3 were subjected to similar pathway analysis. The results are summarised in E Tables 1–3 and reflect strongest association with cytoskeleton remodelling and negative regulation of cell cycle, in accordance with the apoptosis induced upon TLR3 stimulation and in agreement, for example, with upregulation of annexins identified above.
Proteomics of TLR3 stimulated fibroblasts reveals potential new treatments for HSE: SOD2 upregulation
SOD2 is highly upregulated in healthy patient samples C1, C2 and C3 after TLR3 stimulation but not in cells from a patient with known genetic deficiency UNC-93B−/− and neither in unstimulated healthy control sample C2NS (Figure 3A). SOD2 is an antioxidant enzyme strongly upregulated after TLR3 activation in macrophages, protecting these cells from oxidative stress during microbial infection19. SOD2 is increased after HSV-1 infection and SOD2 levels in the CNS are associated with neuronal protection20, 21. Other examples of the protective role of SOD2 have been shown in an animal model of complex I deficiency22, in which SOD2 gene transfer in vivo enhanced cellular resistance to reactive oxygen species and suppressed degeneration of the optic nerve22. This suggests SOD2 as a putative target for future treatment of HSE, in particular to reduce the neurological sequelae that patients suffer after encephalitis.
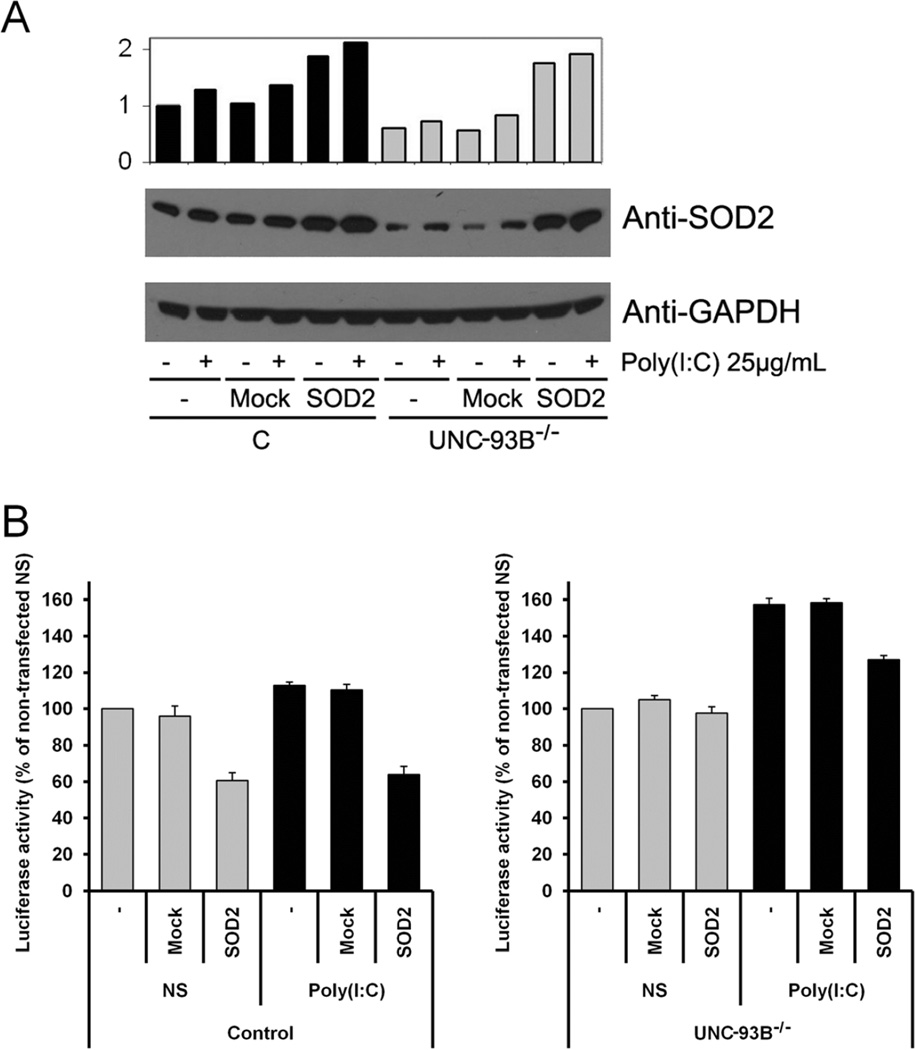
We used gene transfer to further investigate the role of SOD2 in human transformed fibroblasts. WB of healthy cells confirmed upregulation of SOD2 after poly(I:C) stimulation, and very low levels of SOD2 in UNC-93B−/− cells suggested that the defect affects SOD2 expression (Figure 4A). We detected SOD2 overexpression after transient transfections of pBI-EGFP-MnSOD (SOD2), but not with mock transfected vector (Figure 4A). Measurement of apoptosis in SV40-fibroblasts transfected with SOD2 showed that healthy cells overexpressing SOD2 are protected against apoptosis and that this is also the case for stimulation with poly(I:C). Relative apoptosis measured in poly(I:C) stimulated UNC-93B−/− cells against their non-stimulated counterpart was higher than similar measurement for healthy controls, indicating UNC-93B−/− patient cells as more susceptible to apoptosis than healthy cells after TLR3 activation. When transfected with SOD2, however, UNC-93B−/− patient cells were protected against apoptosis (Figure 4B). Together, these results suggest that pharmacological agents that upregulate SOD2 expression or activity could exert a protective anti-oxidant response mechanism to reduce the cell death associated with TLR3 stimulation in HSE patients.
Figure 4.
Human SV40 fibroblast non transfected (−), transfected with SOD2 or Mock vector. (A) WB analysis of SOD2 levels. (−): non stimulated; (+): poly(I:C) stimulated. Densitometry normalized with respect to GADPH, expressed as a fold induction over non stimulated/non transfected control cells. The panel is representative of three experiments. (B) Caspase-3/7 activity in non stimulated (NS) or poly(I:C) stimulated cells. Luciferase activity represented as percentage,100% is healthy control non stimulated. Mean values ± SD were calculated from three independent experiments. C: healthy control C3; UNC-93B−/−: UNC-93B-deficient patient. Two-sided test were used and a p value less than 0.05 was considered statistically significant.
Analysis of a patient with unknown gene defect and without fibroblastic phenotype
30% of HSE patients analyzed do not show the fibroblastic phenotype characterized by reduced IFN type I and III production after TLR3 stimulation (data not shown). In these patients, study of the TLR3 stimulated proteome may help delineate the source(s) of the different cellular phenotype. We conducted a SILAC analysis of a patient whose genetic defect is unknown, but which produces significant amounts of IFN type I and III after TLR3 stimulation (E Figure 6A). The study identified many upregulated proteins shared with stimulated healthy control samples C1, C2 and C3, and which were not observed for UNC93B−/−. The proteins shared those highlighted during earlier analysis of the stimulated healthy controls; with the exception of ICAM-1, which was not detected post-stimulation (Figure 3A and E Table 6) as confirmed by WB analysis (E Figure 6B). ITGA5 (H/L = 1.65) and ANX 11 (H/L = 1.29) are upregulated in patient sample P, but at a ratio significance just outside the threshold of SigB < 0.05 (E Table 6). GeneGo analysis also inferred process networks similar to those related to stimulation of healthy controls, involving immune responses such as IFN-signalling and antigen presentation pathways (Table 2C). This result concurs with previous studies that show cells from patient P to exhibit IFN type I and III production after TLR3 signalling (E Figure 6A). Given the extent of overlap between the analysis of TLR3 stimulation effect in patient P and that in healthy control samples, the non-detectable levels of ICAM-1 upon TLR3 stimulation in P, identified via proteomic analysis, provide an avenue in this patient for a candidate approach centred on ICAM-1 and related genes/proteins. This would be the first such approach in an HSE patient to involve pathways beyond TLR3-IFN signalling.
DISCUSSION
The quantitative SILAC analysis presented here provides important preliminary evidence to suggest that several proteins, pathways and processes could play important and novel roles in TLR3 response and HSE immunity.
ICAM-1 upregulation
After TLR3 stimulation, healthy controls C1, C2 and C3 displayed ICAM-1 upregulation, as described previously23, 24, but there was no corresponding upregulation in UNC-93B−/−, patient P or non-stimulated C2NS (Figure 3A and E Figure 6). ICAM-1 is an intercellular adhesion molecule implicated in HSV-1 infection in CNS25, 26. The present results suggest that lack of ICAM upregulation may be an important factor in susceptibility to HSE, especially for those patients (e.g. P) which show substantial IFN production and lack the fibroblastic phenotype.
Upregulation of integrins ITGA2 and ITGA5
We detected ITGA2 and 5 upregulation after TLR3 stimulation in healthy cells but not in C2NS or UNC-93B−/− (Figure 3A). Cells from patient P showed significant upregulation of ITGA2 (Figure 3A) and upregulation of ITGA5 (H/L = 1.65) just beyond the significance threshold employed (E Table 6). Some studies have demonstrated that integrins are necessary for HSV-1 entry/fusion in cells27 and ITGA2 is upregulated in chronic hepatitis virus infection28. The present results identify specific integrins upregulated following TLR3 activation and whose further investigation might offer new insights into HSE susceptibility.
Upregulation of Annexins
After TLR3 stimulation in healthy cells, we detected upregulation of several annexins (Figure 3A). Microglial cells, after activation in response to harmful stimuli, are able to induce neuronal cell death by ANX129 and ANX5 upregulation30. It has been shown that TLR3 activation with poly(I:C) and HSV-1 infection induce apoptosis by enhancing ANX5 levels31–33. The present data suggests possible roles for ANX5, 6, 7 in the activation and subsequent apoptosis of immune cells in the CNS upon viral infection.
PPIF upregulation
PPIF protein was upregulated after TLR3 stimulation in healthy and patient P cells, but not in those with UNC-93B−/− defect (Figure 3A). PPIF, also called Cyclophilin D, is part of the mitochondrial permeability transition pore in the inner mitochondrial membrane. Activation of this pore is thought to be involved in the induction of apoptotic and necrotic cell death. HSV-1 infection of neurons is known to induce cell death and PPIF has been previously described as a key protein in neuronal cell death34–36. This is the first report of a relationship between PPIF and TLR3, suggesting a possible role for PPIF in HSE immunity.
Previous studies have shown that the TLR3 pathway is essential for susceptibility to HSE6–8. However the analysis of many proteins downstream of TLR3 implicated in the TLR3-IFN type I pathway has failed to reveal a genetic defect in most patients analyzed. In 226 patients enrolled in our laboratory, we have detected mutations in UNC93B1, TLR3 or genes downstream of the TLR3 pathway in a very low percentage. Conversely, study of IFN type I and III production after TLR3 activation in SV40-fibroblasts of HSE patients has shown that only 30%, of a total of 89 patients analyzed, have normal IFN type I and III production (data not shown). This suggests that despite the importance of the TLR3 pathway in HSE immunity, genetic defect(s) responsible for susceptibility to HSV-1 in the CNS could be due to TLR3-independent pathways, or other TLR3, IFN-dependent pathways that are activated after initial TLR3 activation. Overall, the present results suggest a rich diversity of pathways downstream of TLR3 activation that may be relevant for HSE susceptibility and which invite further investigation.
Proteomic analysis of a patient (P) with unknown gene defect but without the fibroblastic phenotype has revealed a lack of ICAM-1, delineating a cellular phenotype which may assist dissection of this genetic aetiology. Finally, an important problem in this pathology is post-infection neurological sequelae, for which the current treatment is aciclovir, an antiviral drug. However, this treatment is largely uneffective2. The present study has now revealed SOD2 as a possible therapeutic target for prevention of the neurological sequelae suffered by HSE patients.
Proteomics has reached the stage where its methods are well developed and capable of generating large, robustly identified, high resolution quantitative datasets that provide a wealth of knowledge at the systems level. Even for a disease such as HSE, of apparent aetiological diversity, coupled with the cellular diversity evident amongst individuals, proteomics has provided an integrated overview of cellular responses that may be crucial for understanding the dynamics of HSE susceptibility.
Supplementary Material
CLINICAL IMPLICATIONS.
Quantitative SILAC-based proteome analysis of TLR3-stimutated human immortalized fibroblasts can delineate cellular phenotypes and may be used to dissect the genetic basis of childhood herpes simplex encephalitis (HSE), as well as new potential therapeutics targets.
ACKNOWLEDGMENTS
We thank the members of the Laboratory of Human Genetics of Infectious Diseases, especially Marjorie Hubeau, Dr C. Guerrera and Prof. L.R. Brown for helpful advice. We thank Mariana Díaz Almiron (Biostatistical Unit, IdiPaz-Hospital La Paz) for the biostatistical analysis. We thank the patient and her family for their participation in this study, which was supported by the AXA Research Fund, the Groupement d’Intérêt Scientifique Maladies Rares, the Action Concertée Incitative de Microbiologie, the March of Dimes, the Agence Nationale pour la Recherche, the Eppley Foundation, the National Institute of Allergy and Infectious Diseases grant number R01AI088364, the Thrasher Research Fund, the Jeffrey Modell Foundation, Talecris Biotherapeutics, the St. Giles Foundation, the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH) grant number 8UL1TR000043, and the Rockefeller University. P.R. is supported by a European Union FP6 grant. J.-L.C. was an international scholar of the Howard Hughes Medical Institute until 2008. The work was supported by The Wellcome Trust Grant to J. G-Z.
Abbreviations
- HSE
Herpes simplex virus encephalitis
- SILAC
Stable isotope labelling of amino acids in cell culture
- HSV-1
Herpes simplex virus
- CNS
Central nervous system
- TLR
Toll-like receptor
- TRAF3
TNF receptor-associated factor 3
- TRIF
Toll/IL1R (TIR) domain-containing adaptor inducing IFN-β
- IFN
Interferon
- poly(I:C)
Polyinosinepolycytidylic acid
- SigB
Significance B
- ICAM-1
Intercellular adhesion molecule-1
- SOD2
Superoxide dismutase 2
- PPIF
Peptidyl-propyl cis-trans isomerise mitochondrial
- MS
Mass Spectrometry
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Since initial involvement in the work, M.W.B.T. has become an employee of Celgene Research SLU, part of Celgene Corporation, and declares no conflicts of interest.
REFERENCES
- 1.Whitley RJ. Herpes simplex encephalitis: adolescents and adults. Antiviral Res. 2006;71:141–148. doi: 10.1016/j.antiviral.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 2.De Tiege X, Rozenberg F, Heron B. The spectrum of herpes simplex encephalitis in children. Eur J Paediatr Neurol. 2008;12:72–81. doi: 10.1016/j.ejpn.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Abel L, Plancoulaine S, Jouanguy E, Zhang SY, Mahfoufi N, Nicolas N, et al. Age-Dependent Mendelian Predisposition to Herpes Simplex Virus Type 1 Encephalitis in Childhood. J Pediatr. 2010 doi: 10.1016/j.jpeds.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Casanova JL, Abel L. Primary immunodeficiencies: a field in its infancy. Science. 2007;317:617–619. doi: 10.1126/science.1142963. [DOI] [PubMed] [Google Scholar]
- 5.Alcais A, Abel L, Casanova JL. Human genetics of infectious diseases: between proof of principle and paradigm. J Clin Invest. 2009;119:2506–2514. doi: 10.1172/JCI38111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casrouge A, Zhang SY, Eidenschenk C, Jouanguy E, Puel A, Yang K, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314:308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 7.Zhang SY, Jouanguy E, Ugolini S, Smahi A, Elain G, Romero P, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 8.Perez de Diego R, Sancho-Shimizu V, Lorenzo L, Puel A, Plancoulaine S, Picard C, et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor-3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33:400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sancho-Shimizu V, Perez de Diego R, Lorenzo L, Halwani R, Alangari A, Israelsson E, et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J Clin Invest. 2011;121:4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapgier A, Wynn RF, Jouanguy E, Filipe-Santos O, Zhang S, Feinberg J, et al. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol. 2006;176:5078–5083. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- 11.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 12.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Ong SE, Foster LJ, Mann M. Mass spectrometric-based approaches in quantitative proteomics. Methods. 2003;29:124–130. doi: 10.1016/s1046-2023(02)00303-1. [DOI] [PubMed] [Google Scholar]
- 14.Mulvey C, Tudzarova S, Crawford M, Williams GH, Stoeber K, Godovac-Zimmermann J. Quantitative proteomics reveals a "poised quiescence" cellular state after triggering the DNA replication origin activation checkpoint. J Proteome Res. 2010;9:5445–5460. doi: 10.1021/pr100678k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 16.Cox J, Matic I, Hilger M, Nagaraj N, Selbach M, Olsen JV, et al. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat Protoc. 2009;4:698–705. doi: 10.1038/nprot.2009.36. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto M, Seya T. TLR3: interferon induction by double-stranded RNA including poly(I:C) Adv Drug Deliv Rev. 2008;60:805–812. doi: 10.1016/j.addr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- 19.Rakkola R, Matikainen S, Nyman TA. Proteome analysis of human macrophages reveals the upregulation of manganese-containing superoxide dismutase after toll-like receptor activation. Proteomics. 2007;7:378–384. doi: 10.1002/pmic.200600582. [DOI] [PubMed] [Google Scholar]
- 20.Hill JM, Lukiw WJ, Gebhardt BM, Higaki S, Loutsch JM, Myles ME, et al. Gene expression analyzed by microarrays in HSV-1 latent mouse trigeminal ganglion following heat stress. Virus Genes. 2001;23:273–280. doi: 10.1023/a:1012517221937. [DOI] [PubMed] [Google Scholar]
- 21.Saha RN, Pahan K. Differential regulation of Mn-superoxide dismutase in neurons and astroglia by HIV-1 gp120: Implications for HIV-associated dementia. Free Radic Biol Med. 2007;42:1866–1878. doi: 10.1016/j.freeradbiomed.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi X, Lewin AS, Sun L, Hauswirth WW, Guy J. SOD2 gene transfer protects against optic neuropathy induced by deficiency of complex I. Ann Neurol. 2004;56:182–191. doi: 10.1002/ana.20175. [DOI] [PubMed] [Google Scholar]
- 23.Rydberg C, Mansson A, Uddman R, Riesbeck K, Cardell LO. Toll-like receptor agonists induce inflammation and cell death in a model of head and neck squamous cell carcinomas. Immunology. 2009;128:e600–e611. doi: 10.1111/j.1365-2567.2008.03041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Starace D, Galli R, Paone A, De Cesaris P, Filippini A, Ziparo E, et al. Toll-like receptor 3 activation induces antiviral immune responses in mouse sertoli cells. Biol Reprod. 2008;79:766–775. doi: 10.1095/biolreprod.108.068619. [DOI] [PubMed] [Google Scholar]
- 25.Brankin B, Hart MN, Cosby SL, Fabry Z, Allen IV. Adhesion molecule expression and lymphocyte adhesion to cerebral endothelium: effects of measles virus and herpes simplex 1 virus. J Neuroimmunol. 1995;56:1–8. doi: 10.1016/0165-5728(94)00110-a. [DOI] [PubMed] [Google Scholar]
- 26.Noisakran S, Harle P, Carr DJ. ICAM-1 is required for resistance to herpes simplex virus type 1 but not interferon-alpha1 transgene efficacy. Virology. 2001;283:69–77. doi: 10.1006/viro.2001.0858. [DOI] [PubMed] [Google Scholar]
- 27.Parry C, Bell S, Minson T, Browne H. Herpes simplex virus type 1 glycoprotein H binds to alphavbeta3 integrins. J Gen Virol. 2005;86:7–10. doi: 10.1099/vir.0.80567-0. [DOI] [PubMed] [Google Scholar]
- 28.Bieche I, Asselah T, Laurendeau I, Vidaud D, Degot C, Paradis V, et al. Molecular profiling of early stage liver fibrosis in patients with chronic hepatitis C virus infection. Virology. 2005;332:130–144. doi: 10.1016/j.virol.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Parente L, Solito E. Annexin 1: more than an anti-phospholipase protein. Inflamm Res. 2004;53:125–132. doi: 10.1007/s00011-003-1235-z. [DOI] [PubMed] [Google Scholar]
- 30.Li L, Lu J, Tay SS, Moochhala SM, He BP. The function of microglia, either neuroprotection or neurotoxicity, is determined by the equilibrium among factors released from activated microglia in vitro. Brain Res. 2007;1159:8–17. doi: 10.1016/j.brainres.2007.04.066. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Q, Wei H, Tian Z. Poly I:C enhances cycloheximide-induced apoptosis of tumor cells through TLR3 pathway. BMC Cancer. 2008;8:12. doi: 10.1186/1471-2407-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillis PA, Okagaki LH, Rice SA. Herpes simplex virus type 1 ICP27 induces p38 mitogen-activated protein kinase signaling and apoptosis in HeLa cells. J Virol. 2009;83:1767–1777. doi: 10.1128/JVI.01944-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swarup V, Ghosh J, Duseja R, Ghosh S, Basu A. Japanese encephalitis virus infection decrease endogenous IL-10 production: correlation with microglial activation and neuronal death. Neurosci Lett. 2007;420:144–149. doi: 10.1016/j.neulet.2007.04.071. [DOI] [PubMed] [Google Scholar]
- 34.Baines CP, Kaiser RA, Purcell NH, Blair NS, Osinska H, Hambleton MA, et al. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 2005;434:658–662. doi: 10.1038/nature03434. [DOI] [PubMed] [Google Scholar]
- 35.Li V, Brustovetsky T, Brustovetsky N. Role of cyclophilin D-dependent mitochondrial permeability transition in glutamate-induced calcium deregulation and excitotoxic neuronal death. Exp Neurol. 2009;218:171–182. doi: 10.1016/j.expneurol.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du H, Guo L, Fang F, Chen D, Sosunov AA, McKhann GM, et al. Cyclophilin D deficiency attenuates mitochondrial and neuronal perturbation and ameliorates learning and memory in Alzheimer's disease. Nat Med. 2008;14:1097–1105. doi: 10.1038/nm.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.