Abstract
Two forms of the glutaredoxin (Grx) domain (full length Grx domain and short Grx lacking the N-terminal region) of Mus musculus thioredoxin glutathione reductase (TGR) were isotopically labelled with 15N and 13C isotopes, expressed and purified to homogeneity. We report here the 1H, 13C and 15N NMR assignment for both Grx forms of this mouse TGR. This investigation represents the first NMR analysis of a mammalian TGR.
Keywords: Thioredoxin Reductase, Redox biology, NMR spectroscopy
Introduction
Aerobic life involves metabolism of molecular oxygen. The cytotoxic by-products of this process are commonly known as reactive oxygen species (ROS). The cellular redox environment represents a balance between the production of ROS and their removal by antioxidant enzymes and low molecular-weight antioxidants. Dysbalance in this equilibrium leads to oxidative or reductive stresses. However, organisms evolved sophisticated cellular defence systems that protect them against redox stress [1-6].
Redox regulation of many cellular processes is provided by thioredoxin and glutathione systems. Biochemical and genetic studies have shown that there is an extensive functional overlap between these two reduction systems. Generally, thioredoxin- and glutathione-dependent systems are responsible for maintaining the reducing environment in the cell. These systems are involved in providing reducing equivalents to ribonucleotide reductase and methionine sulfoxide reductase, antioxidant defence and regulation of activities of various transcription factors, kinases, and phosphatases. These systems were also implicated in the redox control of cell growth and death, transcription, cell signalling, and other processes [5-9].
The thioredoxin system is composed of thioredoxin (Trx), thioredoxin reductase (TrxR), and thioredoxin peroxidase, while the glutathione redox system consists of glutathione (GSH), glutathione reductase (GR), glutaredoxin and glutathione peroxidases [5, 10-12]. Thioredoxin reductase (TrxR; homodimer of 55 kDa subunits) is closely related to GR both structurally and functionally [3, 4, 13]. However, in contrast to GR, TrxRs exhibit a broader substrate spectrum (e.g., Trx, glutaredoxin 2, selenite, etc.) [14, 15].
Three separate genes encode TrxR in mammals [16, 17]. In humans, these genes are designated as TXNRD1, TXNRD2 and TXNRD3. The TXNRD3 gene encodes thioredoxin glutathione reductase and corresponding mouse gene is named Txnrd3. This protein was originally known as TR2 and more recently renamed as TGR for thioredoxin-glutathione reductase. The protein is mainly expressed in the male germ cells, but the transcript is also found in other cell types [18-20]. TGR was proposed to function predominantly in disulfide bond formation and isomerisation of sperm proteins during spermatogenesis [18].
TGR consists of a TrxR module and an N-terminal region that includes a glutaredoxin (Grx) domain. The amino acid sequence of the TrxR module of TGR is closely related to other TrxRs, while the Grx domain has an atypical monothiol -CPHS- motif [17-19, 21]. The latter motif usually catalyzes (de)glutathionylation reactions [22]. The Grx domain of TGR may receive electrons from either the TrxR module of TGR or TrxR1 [20]. In addition, the Grx domain is functionally active either as a separate protein or within TGR [20]. This domain allows TGR to participate in both Trx and glutathione redox systems [19]. Prior to TGR discovery, it was believed that these two redox systems work independently, but increasing evidence suggests a crosslink between the systems [23].
Here we report 1H, 13C and 15N NMR resonance assignments of both full-length and short (lacking an N-terminal unfolded region (22 amino acids)) Grx domains of Mus musculus TGR.
Methods and experiments
The N-terminus his-tagged Grx domain of mouse TGR was cloned and purified as described previously [23]. His-tag was subsequently removed using Thrombin kit (Novagen) following the protocol provided by supplier. The amino acids sequence of Grx domain (hereafter Grx) is as follows: MSSPPGRRARLASPGTSRPSSEAREELRRRLRDLIEGNRVMIFSKSYCPHSTRVKELFSSLGVVYNILELDQVDDGASVQEVLTEISNQKTVPNIFVNKVHVGGCDRTFQAHQNGLLQKLLQDD. In addition, a short version of Grx domain lacking the N-terminal 22 residues, from M1 to E22 (hereafter sGrx) was produced using the analogous procedure reported for a full-length Grx protein.
For expression of 15N- and 13C-labeled proteins, E. coli was cultured in M9 minimal medium containing 1 g/l 15NH4Cl and 4 g/l 13C6-glucose at 37°C. The purity of the proteins was checked by SDS-PAGE and MALDI tof-tof mass spectrometry. The final purity of the proteins was above 95%. The yield of both Grx and sGrx was approximately ~20 mg protein per liter of M9 minimal medium.
The NMR samples had approximately 1 mM Grx or sGrx uniformly 15N- and 13C-labeled proteins in the buffer containing 10 mM phosphate, 10 mM NaCl and 10 mM β-mercaptoethanol in 95% H2O/5% D2O at pH 7.5. Efforts to decrease solution pH, in order to decrease the exchange rate of amide protons and thus improve detection of NMR signals, resulted in a partial protein precipitation. NMR sample were kept under N2 in order to protect proteins from oxidation.
NMR spectra were acquired at 298 K on a Bruker Avance 600 MHz spectrometer equipped with a 5-mm z-gradient TXI(H/C/N) cryoprobe. Proton chemical shifts were referenced to water, while 15N and 13C chemical shifts were referenced indirectly to a sodium salt of 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) and liquid ammonia, respectively, based on the absolute frequency ratios [24]. The NMR data were processed with the BRUKER XWinNMR version 3.5 software and spectral analysis was performed using CARA version 1.8.4.2 [25].
The backbone and side-chain chemical shift assignments of Grx and sGrx were obtained on the basis of 2D/3D heteronuclear and homonuclear HNCA, HNCACB, CBCA(CO)NH, NHCO, HBHANH, HBHA(CO)NH, HCCH-TOCSY, 15N-1H/13C-1H NOESY, 2D COSY, TOCSY and NOESY spectra.
Assignments and data deposition
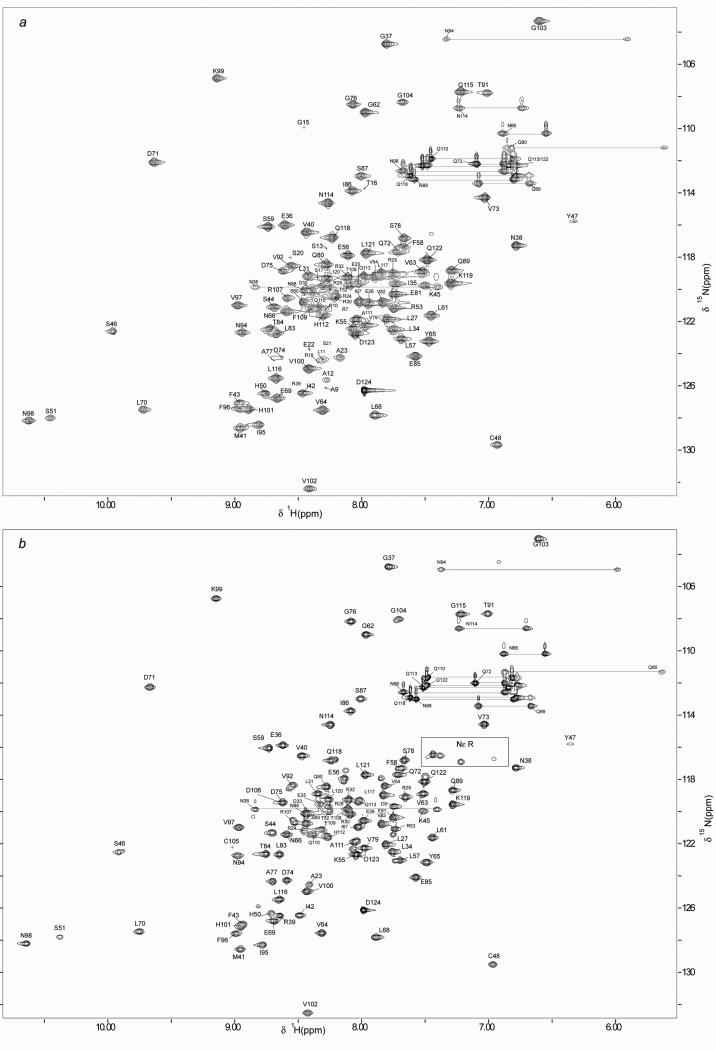
The 2D 1H-15N HSQC spectra of reduced full-length and shortened Grx domains, (Grx and sGrx) of Mus musculus TGR are shown in Fig. 1 (panels a and b, respectively). The peak assignments, indicated in one letter amino acid code and residue number, are also reported in Figure 1. The residues numbering of sGrx domain starts from A23 (Grx sequence consensus). The observed dispersion of chemical shifts for both Grx forms unambiguously indicates that the proteins are either fully or partially folded.
Fig. 1.
2D 1H-15N HSQC spectra of reduced mouse Grx domain (panel a) and sGrx (panel b) in 90:10 H2O:D2O at pH 7,5, 298 K. The assignments are shown with one letter amino acid code and residue numbers. The side-chain resonances of Asn and Gln residues are connected with horizontal lines.
In the case of the full-length Grx domain, 93% of the backbone and side-chain chemical shift assignments were obtained. The assignment procedure of the full-length Grx domain indicated that the majority of the resonances that escaped detection, and thus the assignment, belongs to the N-terminal tail of the protein. This pattern is consistent with the fact that its N-terminal region, M1-E22, is flexible and unstructured. Indeed, while for the most Grx amino acids from the structured protein core the characteristic NOESY patterns in 3D 15N-NOESY spectrum were present, for all N-terminal amino acids, from M1 to E22, NOESY towers were not observed. This observation supports our finding that the N-terminal tail of the Grx domain of mouse TGR is unstructured. In addition, the analysis of secondary structure prediction suggests that the N-terminal region is unfolded (http://bioinf.cs.ucl.ac.uk/). Further inspection of the obtained NMR data for the Grx domain reveals that, in the structured protein core, any NOESY patterns were observed for D74 and A77. Also in the 1H-15N HSQC spectrum, the cross peaks corresponding to these amino acids were rather weak. This indicates that these residues have a higher exchange rate of their amidic protons with water with respect to other amino acids in the protein core. In addition, the signals of three residues: K90, C105 and D106, were not observed in the Grx 1H-15N HSQC spectrum. This observation can be explained by the fact that the resonances of these three residues are heavily overlapped with other protein signals and/or their amidic protons have high exchange rate with water. The assignment for 1H, 13C and 15N chemical shifts of the full-length Grx domain of mouse TGR has been deposited in the BioMagResBank (http://www.bmrb.wisc.edu/) under the BMRB accession number 17636.
To further confirm the hypothesis that the N-terminal region is mobile and unfolded, a new shortened Grx form (sGrx) lacking the N-terminal part of Grx (22 amino acids) was produced and examined with NMR. It should be noted that sGrx was not as stable as the wild-type Grx. Protein precipitation was observed after 1 week, while the full-length protein was stable for months under the same conditions. Comparison of the 1H-15N HSQC spectrum of sGrx and that of the full-length Grx unambiguously reveals a close resemblance of the spectral patterns for both proteins (see Figure 1), thus indicating that their structures are very similar. The NMR assignment procedure for sGrx revealed the lack, in the protein HSQC spectrum, of any resonances assigned to amino acids M1-E22 of the full-length Grx. This observation confirms the correctness of the NMR assignment for the full-length Grx and verifies our proposition that the N-terminal tail of the Grx domain is mobile and unfolded. The residue K90 was not also observed in the sGrx HSQC spectrum. However, in the case of sGrx, it was possible to assign the resonances of residues C105 and D106, which remained unassigned in the case of full-length Grx. The new assignments of sGrx were achieved as a consequence of a less hampered HSQC spectrum in comparison to that of Grx. It is worth mentioning that in the sGrx 1H-15N HSQC spectrum, the cross peaks corresponding to S46, S51, T52, R53, V54, K55, E56, L57, S60, V92, R107, F109, Q110, A111, Q113, Q118 and Q122 residues appear in a doubled form, which could indicate existence of two or more conformations for these amino acids. The latter can correlate with the decreased sGrx stability as compared to full-length Grx domain, mentioned above. In summary, 98% and 90% of backbone and side chain chemical shift assignments were obtained for sGrx. These assignments were deposited in the BMRB data bank under accession number 17637.
The NMR assignments of Grx and sGrx forms pave the way for determination of the three-dimensional solution structures of these proteins that is currently in progress in our laboratories.
Acknowledgements
This research was supported by the Faculty of Natural Sciences and Technology of the Norwegian University of Science and Technology (NTNU) and NIH GM065204. ES acknowledges the NT Faculty, NTNU, for financial support through Post-doctoral fellowship.
References
- 1.Arner ES, Zhong L, Holmgren A. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods in enzymology. 1999;300:226–39. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 2.Mustacich D, Powis G. Thioredoxin reductase. The Biochemical journal. 2000;346(Pt 1):1–8. [PMC free article] [PubMed] [Google Scholar]
- 3.Becker K, et al. Thioredoxin reductase as a pathophysiological factor and drug target. European journal of biochemistry / FEBS. 2000;267(20):6118–25. doi: 10.1046/j.1432-1327.2000.01703.x. [DOI] [PubMed] [Google Scholar]
- 4.Williams CH, et al. Thioredoxin reductase two modes of catalysis have evolved. European journal of biochemistry / FEBS. 2000;267(20):6110–7. doi: 10.1046/j.1432-1327.2000.01702.x. [DOI] [PubMed] [Google Scholar]
- 5.Holmgren A. Antioxidant function of thioredoxin and glutaredoxin systems. Antioxidants & redox signaling. 2000;2(4):811–20. doi: 10.1089/ars.2000.2.4-811. [DOI] [PubMed] [Google Scholar]
- 6.Halliwell B. Antioxidant defence mechanisms: from the beginning to the end (of the beginning). Free radical research. 1999;31(4):261–72. doi: 10.1080/10715769900300841. [DOI] [PubMed] [Google Scholar]
- 7.Sies H. Glutathione and its role in cellular functions. Free radical biology & medicine. 1999;27(9-10):916–21. doi: 10.1016/s0891-5849(99)00177-x. [DOI] [PubMed] [Google Scholar]
- 8.Rhee SG. Redox signaling: hydrogen peroxide as intracellular messenger. Experimental & molecular medicine. 1999;31(2):53–9. doi: 10.1038/emm.1999.9. [DOI] [PubMed] [Google Scholar]
- 9.Finkel T. Redox-dependent signal transduction. FEBS letters. 2000;476(1-2):52–4. doi: 10.1016/s0014-5793(00)01669-0. [DOI] [PubMed] [Google Scholar]
- 10.Williams CH., Jr. Thioredoxin-thioredoxin reductase--a system that has come of age. European journal of biochemistry / FEBS. 2000;267(20):6101. doi: 10.1046/j.1432-1327.2000.01700.x. [DOI] [PubMed] [Google Scholar]
- 11.Pedrajas JR, et al. Identification and functional characterization of a novel mitochondrial thioredoxin system in Saccharomyces cerevisiae. The Journal of biological chemistry. 1999;274(10):6366–73. doi: 10.1074/jbc.274.10.6366. [DOI] [PubMed] [Google Scholar]
- 12.Seo MS, et al. Identification of a new type of mammalian peroxiredoxin that forms an intramolecular disulfide as a reaction intermediate. The Journal of biological chemistry. 2000;275(27):20346–54. doi: 10.1074/jbc.M001943200. [DOI] [PubMed] [Google Scholar]
- 13.Dolphin D, Poulson R, Avramovic O. Glutathione: Coenzymes and Cofactors. Wiley; New York: 1989. [Google Scholar]
- 14.Johansson C, Lillig CH, Holmgren A. Human mitochondrial glutaredoxin reduces S-glutathionylated proteins with high affinity accepting electrons from either glutathione or thioredoxin reductase. The Journal of biological chemistry. 2004;279(9):7537–43. doi: 10.1074/jbc.M312719200. [DOI] [PubMed] [Google Scholar]
- 15.Jimenez A, et al. Characterization of human thioredoxin-like-1: potential involvement in the cellular response against glucose deprivation. FEBS letters. 2006;580(3):960–7. doi: 10.1016/j.febslet.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 16.Osborne SA, Tonissen KF. Genomic organisation and alternative splicing of mouse and human thioredoxin reductase 1 genes. BMC genomics. 2001;2(1):10. doi: 10.1186/1471-2164-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun QA, et al. Heterogeneity within animal thioredoxin reductases. Evidence for alternative first exon splicing. The Journal of biological chemistry. 2001;276(5):3106–14. doi: 10.1074/jbc.M004750200. [DOI] [PubMed] [Google Scholar]
- 18.Su D, et al. Mammalian selenoprotein thioredoxin-glutathione reductase. Roles in disulfide bond formation and sperm maturation. The Journal of biological chemistry. 2005;280(28):26491–8. doi: 10.1074/jbc.M503638200. [DOI] [PubMed] [Google Scholar]
- 19.Sun QA, et al. Selenoprotein oxidoreductase with specificity for thioredoxin and glutathione systems. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(7):3673–8. doi: 10.1073/pnas.051454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun QA, et al. Reaction mechanism and regulation of mammalian thioredoxin/glutathione reductase. Biochemistry. 2005;44(44):14528–37. doi: 10.1021/bi051321w. [DOI] [PubMed] [Google Scholar]
- 21.Sun QA, et al. Redox regulation of cell signaling by selenocysteine in mammalian thioredoxin reductases. The Journal of biological chemistry. 1999;274(35):24522–30. doi: 10.1074/jbc.274.35.24522. [DOI] [PubMed] [Google Scholar]
- 22.Fernandes AP, Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxidants & redox signaling. 2004;6(1):63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- 23.Gerashchenko MV, Su D, Gladyshev VN. CUG start codon generates thioredoxin/glutathione reductase isoforms in mouse testes. The Journal of biological chemistry. 2010;285(7):4595–602. doi: 10.1074/jbc.M109.070532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Neal S, Wishart DS. RefDB: a database of uniformly referenced protein chemical shifts. Journal of biomolecular NMR. 2003;25(3):173–95. doi: 10.1023/a:1022836027055. [DOI] [PubMed] [Google Scholar]
- 25.Keller RLJ. Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment. ETH; Zurich: 2004. [Google Scholar]