Abstract
Background
Human circadian rhythms are regulated by the interplay between circadian genes and environmental stimuli. The influence of altered sleep/wake schedules or light on human circadian gene expression patterns is not well characterized.
Methods
Twenty-one participants were asked to keep to their usual sleep schedules and two blood samples were drawn at the end of the first week for each subject based upon estimated time of dim light melatonin onset (DLMO); the first sample was obtained one and a half hours before the estimated DLMO and the second three hours later, at one and a half hours after the estimated DLMO. During the second week, participants were randomized into two groups, one that received a one hour “blue” light (λmax = 470 nm) exposure in the morning and one that received a comparable morning “dim” light exposure. Two blood samples were obtained at the same clock times as previous week at the end of the second week.
Results
We measured the expression of 10 circadian genes in response to sleep/wake schedule advancement and morning “blue” light stimulation in the peripheral blood of 21 young adults during a two week field study. We found that nine of the 10 circadian genes showed significant expression changes from the first to the second week for participants in both the “blue” and “dim” light groups, likely reflecting significant advances in circadian time.
Conclusions
This wholesale change in circadian clock gene expression may reflect significant advances in circadian time (i.e., advance in DLMO) from the first to the second week resulting from the advanced, daily personal light exposures.
Keywords: Blue light, Circadian gene, Phase shift, Sleep
1. Introduction
The 24 hour circadian clock, the culmination of an ancient adaptation to the light/dark cycles resulting from the earth’s rotation, affects biochemical, physiological, and behavioral processes in almost all living organisms. The mammalian central pacemaker is regulated by the positive and negative transcription translation feedback loops of the so-called circadian genes, the best characterized of which include: CLOCK [1], casein kinase I, epsilon (CSNK1E) [2], cryptochrome 1 (CRY1), cryptochrome 2 (CRY2) [3], period 1 (PER1), period 2 (PER2), period 3 (PER3) [4, 5], neuronal PAS domain protein 2 (NPAS2) [6], TIMELESS [7], and aryl hydrocarbon receptor nuclear translocator-like (ARNTL) (also referred to as brain and muscle Arnt-like protein-1 (BMAL1)) [8, 9]. Polymorphisms in circadian genes have been linked to circadian-related phenotypes, including morning/evening preference and depressive symptoms [10-13]. In particular, polymorphisms in the PER3 gene have been found to be significantly associated with delayed sleep phase syndrome and diurnal preference [10, 13, 14], and a single nucleotide polymorphism located in the 3’ flanking region of the human CLOCK gene was found to be associated with diurnal preference [11].
Light/dark patterns affect both the timing and amplitude of circadian-related processes, including melatonin synthesis and thermoregulation. Mammals have a circadian photoreception pathway that is distinct from those of visual perception. Mice lacking rods and cones, for example, are still able to respond with phase-dependent shifts of behavioral rhythms [15, 16]. Previous studies comparing light spectra indicate that the human circadian system is preferentially sensitive to short-wavelength (~470 nm) light, with greater responses to short wavelength light observed in melatonin suppression [17-19], circadian phase delay [19-21], and circadian phase advance [22-25].
While light dependent effects on traditional markers of circadian rhythmicity, such as melatonin, cortisol, and body temperature, are well documented; the same effects on the expression of circadian genes are generally less well understood. The classic animal experiment conducted by Albrecht et al. provided the first direct evidence that light stimuli can affect core circadian gene expression patterns in mammals [26]. Since then, studies have demonstrated the diurnal time-dependent expression of PER1, PER2, and PER3 in human blood, with less conclusive evidence for BMAL1, CLOCK, TIMELESS, CRY1, and CRY2 [27-29]. Only one study, to our knowledge, has been conducted to measure the effect of sleep entrainment on core circadian gene expression in peripheral blood, which found changes in PER1 and PER2 expression patterns in individuals with altered sleep/wake schedules and those subjected to bright light treatment [30]. Similarly, few studies have been conducted to examine the effect of light stimuli on human circadian gene expression, and only two, restricted to the study of PER3, CRY1, and BMAL1, have examined this effect in human blood [31-34].
In the present study, we test the effects of morning “blue” light exposure and a sleep advance intervention on the expression patterns of 10 circadian genes in the blood of young adults with delayed habitual sleep schedules in a field setting. This represents the first investigation of a near comprehensive panel of well-studied circadian genes in the context of circadian entrainment.
2. Methods
2.1 Participant recruitment
Participant characteristics have been described in detail in a previous publication by Sharkey et al [35]. The study was approved by the Institutional Review Boards at Brown University. Briefly, participants were recruited using advertisements targeting “struggling night owls” ages 18-30. Interested individuals called the laboratory to undergo preliminary screening. Inclusion criteria included general good health, lack of medication use with known effects on sleep or circadian rhythms, proficiency in written and spoken English, habitual consumption of less than 360 mg of caffeine and fewer than 10 cigarettes per day, and an average reported total sleep time of ≤ nine hours per night (so that the intervention would not result in significant sleep deprivation). Exclusion criteria included a personal history of a sleep disorder, bipolar disorder, psychosis, seizure disorder, or chronic medical condition (e.g., diabetes, asthma, cancer), and a family history of psychosis or bipolar disorder. Individuals who reported using recreational drugs and most prescription drugs were also excluded. Night shift workers and anyone who had traveled beyond two time zones in the month prior to the study, were also excluded.
Young adults who had misalignment between their actual sleep schedules and daily activities were chosen as subjects as these individuals may represent a useful model of subsyndromal delayed sleep phase. Thus, the definitive inclusion criterion was the presence of a misalignment between the participant’s usual sleep/wake times and his or her routine schedule. This was defined this as the existence of an obligatory morning commitment that required the participant to awaken 1-2.5 hours earlier than his or her average reported wake time at least one day per week. The schedule misalignment formed the basis of the change in the participants’ sleep/wake times during the advanced schedule intervention.
2.2 Study overview
As illustrated in the sample protocol in Figure 1, participants completed one baseline nonscheduled week followed by one week on a phase-advanced sleep/wake schedule. During the second week, participants were exposed to either “blue” or “dim” light shortly after awakening. Participants underwent a laboratory overnight session at the end of each week for blood collection for measuring circadian gene expression. During both weeks of the study, participants wore Daysimeters and wrist actigraphs to measure light exposure and activity/rest patterns. All participants completed the Horne-Östberg morningness-eveningness questionnaire as a measure of circadian phase preference.
Figure 1. Sample protocol diagram.
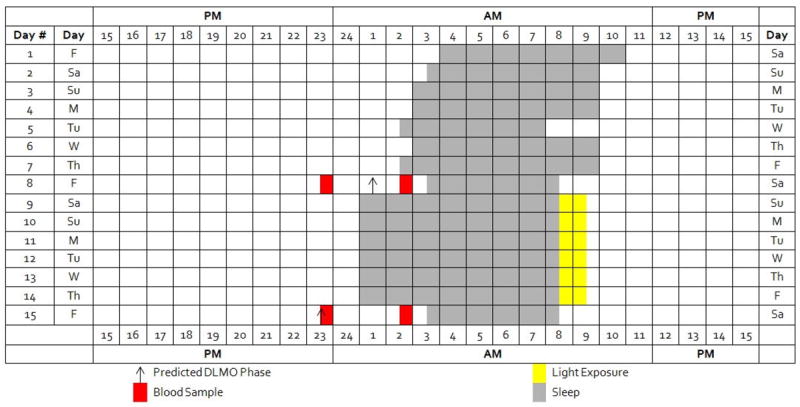
At the end of the first, baseline week, two blood samples were drawn for gene expression assessment at two times; the time of blood draws were individualized based upon each participant’s advanced sleep/wake schedule. The first sample was obtained 1.5 hours before the each participant’s predicted DLMO and the second three hours later, 1.5 hours after the predicted DLMO. Following the advanced sleep/wake schedule of the second week, the two blood samples were obtained at the same clock times as those at the end of the baseline week.
2.3 Home monitoring
The home monitoring protocol has been explained in detail previously [35]. Briefly, participants were instructed to wear Daysimeters and actigraphs, as well as to keep track of sleep activity, throughout the two week study period. The Daysimeter was used to calculate “circadian light” (i.e., spectrally weighted corneal irradiance or circadian illuminance, in units of CLA) based upon the spectral sensitivity of human nocturnal melatonin suppression model by Rea et al [36, 37]. The average log CLA and average log lux for the first 90 minutes after waking and across the entire day while the Daysimeter was worn were calculated for the baseline week and the six days of the phase-advanced sleep schedule. The Daysimeter was also used to collect information on individual participant activity levels to measure compliance with wearing the device. Participants were instructed to wear the Daysimeter during waking hours throughout the two week study period except during bathing, vigorous exercise, and sleep. Participants were also instructed to place the Daysimeter in a safe place in the room where they slept in order to expose the sensor to ambient light. For the Daysimeter data to more accurately reflect corneal light exposures, participants were asked to avoid wearing sunglasses or brimmed hats during the study. Log books were also completed by participants to report times the Daysimeter was worn.
Participants were also asked to wear actigraphs to measure activity patterns. Actigraphs were worn on the nondominant wrist continuously throughout the two-weeks study period, except during bathing. Each participant completed a daily sleep diary and phoned the laboratory’s time-stamped voicemail at bedtime and wake time daily. At the end of each monitoring week, the actigraph data were compared to the sleep diary and voicemail information to ensure compliance to the protocol.
2.4 Sleep advance intervention
For the first week of home monitoring, participants were instructed to keep their usual sleep schedule. Bedtimes and rise times were not fixed, and participants were given no instructions with regard to their exposures to light. Participants were instructed to sleep in their own beds, and avoid napping and staying awake all night.
Following the first in laboratory session at the end of the first study week, participants continued to wear the Daysimeter and actigraph for the next six days at home during which they were required to follow their individualized fixed, advanced sleep/wake schedule that included seven and a half hours of time in bed per night and was determined based on each individual participant’s morning schedule requirement. Scheduled wake times were one to two and a half hours earlier than each participant’s average reported wake time during the baseline week, thus representing a phase advance in sleep schedule. Participants were required to be in bed, in the dark, and to try to sleep during their scheduled sleep times. Sleep schedule compliance was confirmed with wrist actigraph data, sleep diaries, and time-stamped phone calls.
2.5 Morning light intervention
We used a novel eye-level light monitor that detects spectrally weighted radiation, the Daysimeter [38], to ensure compliance to the “blue-light” intervention and to quantify light exposures throughout the day and evening, while participants were awake. Participants followed their normal sleep/wake schedules during the first week while wearing the Daysimeter. During the second week, the participants sat in front of an Apollo P2 GoLite light box (Phillips Respironics, Amsterdam, The Netherlands) for one hour immediately after awakening on the six mornings of the advanced sleep/wake schedule. The GoLite P2 is a 6 × 6 inch device with an array of 66 LEDs that emit blue (short wavelength) light with a peak wavelength of 468 nm. Participants were randomly assigned to receive either the “blue” light intervention (treatment group, n=12), where they sat with the GoLite 24 inches from their eye directed at their face with light intensity set at 50 percent maximum brightness (~225 lux), or the “dim” light intervention (control group, n=13), where the participant sat with the GoLite perpendicular to their face, directed across their workspace with light intensity set at 10 percent maximum brightness (< 1 lux). Participants were instructed to wear the Daysimeter device during the light exposure and throughout all waking hours.
2.6 Biosample collection
Participants came in for overnight in-laboratory sessions on the last day of each study week for blood and saliva collection. Laboratory conditions have been described in detail previously [35]. At the end of the first, baseline week, two blood samples were drawn for gene expression assessment at two times; times of blood draws were individualized based upon each participant’s advanced sleep/wake schedule from which dim light melatonin onset (DMLO) phase was estimated using the algorithm of Burgess and Eastman [39]. Blood sampling times were individually scheduled so that the first sample was obtained one and a half hours before the predicted DLMO (pDLMO) and the second was obtained one and a half hours after pDLMO. After imposition of the advanced sleep/wake schedule of the second week and the light treatment, two blood samples were obtained at the same clock times as those from the baseline week. Peripheral whole blood from venipuncture was dispensed into micro-tubes pre-aliquoted with PAXgene™ reagent. The sample was gently inverted and stored at -70°C for later RNA isolation. Viable blood samples were available for eleven subjects in the “dim” light group and ten subjects in the “blue” light intervention group for both weeks of data collection.
Saliva samples were also collected every 30 minutes from three and a half hours before pDLMO to two hours after pDLMO to measure salivary melatonin levels, as detailed elsewhere [35]. Bedtime was scheduled to occur immediately following saliva sample collection at two hours after pDLMO, which required participants to stay awake beyond their usual bedtimes.
2.7 RNA extraction
RNA was extracted from whole blood using the PAXgene™ Blood RNA System Kit (Qiagen, Valencia, CA) according to the manufacturer’s guidelines. Briefly, the samples were removed from -70°C and incubated at room temperature with lysis reagent for two hours to ensure complete lysis. Following lysis, the tubes were centrifuged for 10 min at 5,000 ×g, the supernatant was decanted, and 500 μL of RNase-free water was added to the pellet. The tube was vortexed to thoroughly re-suspend the pellet, centrifuged for 10 minutes at 5000 × g, and the entire supernatant was discarded. The remaining pelleted lysate was re-suspended in 360 μL of buffer BR1 by vortexing and RNA was extracted following the manufacturer’s protocol.
2.8 Circadian gene expression measurement by quantitative RT-PCR
Total RNA was first reverse-transcribed in a 20 μl reaction system, which contained MMLV reverse transcriptase (Invitrogen., Carlsbad, CA), 1 mmol/l dNTPs, 40 U RNase inhibitor (Amersham Pharmacia Biotech, Freiburg, Germany), and 300 ng oligo d(T) 12–18 (InVitroGen., Carlsbad, CA). The reaction was performed at 37 °C for 50 minutes, 70 °C for 15 minutes, and 4 °C for five minutes. Forward and reverse primers for the 10 circadian genes and the HPRT1 control gene were designed in-house and chemically synthesized by IDT (Supplemental Table 1). At least one primer in each pair was designed to cross an exon-exon junction in order to reduce the potential for unintended DNA amplification. SYBR Green RT-PCR was performed on a Stratagene MX3000P instrument (Stratagene) using Power SYBR® Green PCR Master Mix (Applied Biosystems, Carlsbad, California) with the following conditions: 30 minutes at 50°C and 15 minutes at 95°C followed by 40 cycles of: 15s at 94°C, 30 seconds at 57°C, and 30 seconds at 72°C. Each gene-specific reaction was run in duplicate, along with a no template negative control for each primer pair in order to identify potential primer-dimer formation. Expression levels were obtained for each circadian gene by calculating a delta cycle threshold (dCt) value for each gene relative to levels of the HPRT1 control gene (dCt = CtHPRT1 − Ctcircadian gene). Pre-pDLMO 2-dCt values were subtracted from post-pDLMO 2-dCt values for each of the two weeks of sample collection, and the values of these differences served as the basis for comparison for our analytical model. Having the comparison between the differences between pre- and post-pDLMO expression rather than raw values of expression allowed for the control of baseline differences in expression between study subjects.
2.9 Mood and stress questionnaires
As described previously [35], participants completed questionnaires to measure their baseline mood and stress at the start of the study and on the evenings of each laboratory session.
2.10 Statistical analyses
All analyses were conducted using the SAS® software package, version 9.1 (SAS Institute). A repeated-measures, mixed-design analysis of variance (ANOVA) was used to compare differences between pre- and post-pDLMO expression between study weeks and light groups (“blue” light and “dim” light).
3. Results
3.1 Participant characteristics
Participants ranged in age from 18 to 30 years (mean ± SD = 21.8 ± 3.0) and included 12 men (48%) and 13 women (52%). Fourteen participants (56%) were non-Hispanic Caucasian; five (20%) were Asian; four (16%) were Hispanic; one (4%) was African American; and one (4%) was multiracial. Horne-Östberg morningness-eveningness scores ranged from 28-54; two participants were definite evening types, 10 were moderate evening types, and 13 were neither types. The two light treatment groups did not differ in age, sex, morningness-eveningness score, habitual bedtime and rise times, or baseline mood scores (refer to previous publication by Sharkey, Carskadon et al.) [35].
3.2 Melatonin circadian phase estimates and relative blood collection times
As described in our previous study [35], the sleep advance intervention resulted in significantly earlier times of salivary DLMO for both light groups. The average DLMO time was 23:24 ± 1:16 at the end of the baseline unscheduled week and 21:59 ± 1:03 at the end of the week on the fixed, advanced schedule, yielding an average DLMO phase advance of 1.4 ± 0.9 hours. In contrast, no significant differences in salivary DLMO were found between light treatment groups. The average predicted DLMO time (23:07 ± 0:40) closely matched the average salivary DLMO time of week 1. Average pre-pDLMO and post-pDLMO blood collection times for week 1 were 1.8 hours before salivary DLMO and 1.2 hours after salivary DLMO, respectively. The average blood collection times for week 2 were 0.4 hours before salivary DLMO and 2.6 hours after salivary DLMO.
3.3 Effects of advanced sleep schedules on circadian gene expression
We tested the impact of our advanced sleep/wake schedule intervention on circadian gene expression patterns by using the repeated measures, mixed design ANOVA model. The mean pre- and post-pDLMO expression levels in week 1 differed significantly from the mean expression levels in week 2 for CRY1, CRY2, and TIMELESS (P < 0.05), but not for the other circadian genes between (see Table 1). In contrast, a significant effect of study week × time of blood collection was found for expression of nine of the ten circadian genes (CLOCK, CRY1, CRY2, CSNK1e, NPAS2, PER1, PER2, PER3, and TIMELESS; P < 0.01). Post-pDLMO expression of all ten circadian genes was diminished relative to pre-pDLMO expression in week 1, while the opposite was true in week 2. As indicated by the week × time interaction term, these differences between pre- and post-pDLMO expression were significantly different between week 1 and week 2 for all circadian genes, with the exception of BMAL. No main effect for blood collection times was found for expression of any circadian gene (Table 1).
Table 1.
Phase advance and blue light treatment effects on circadian gene expression patterns
| Gene | P, Fdf for Week 2v1a | P, Fdf for Time of Blood Collectiona | P, Fdf for Week × Timea | P, Fdf for Week × Light Groupa |
|---|---|---|---|---|
| BMAL | 0.4069, 0.701,59 | 0.3483, 0.891,59 | 0.2344, 1.441,59 | 0.9140, 0.011,59 |
| CLOCK | 0.9196, 0.011,56 | 0.5150, 0.431,56 | 0.0087**, 7.401,56 | 0.0949, 2.891,56 |
| CRY1 | 0.0433*, 4.281,54 | 0.2557, 1.321,54 | 0.000**, 18.391,54 | 0.8562, 0.031,54 |
| CRY2 | 0.042*, 4.321,59 | 0.1785, 1.851,59 | 0.000**, 20.561,59 | 0.5992, 0.281,59 |
| CSNK1e | 0.122, 2.461,59 | 0.5323, 0.391,59 | 0.001**, 11.551,59 | 0.8696, 0.031,59 |
| NPAS2 | 0.3770, 0.791,59 | 0.3403, 0.921,59 | 0.000**, 18.701,59 | 0.7208, 0.131,59 |
| PER1 | 0.5345, 0.391,59 | 0.6560, 0.201,59 | 0.000**, 15.491,59 | 0.5763, 0.321,59 |
| PER2 | 0.1045, 2.721,59 | 0.0805, 3.161,59 | 0.000**, 15.461,59 | 0.6431, 0.221,59 |
| PER3 | 0.1685, 1.941,58 | 0.9133, 0.011,58 | 0.000**, 28.191,58 | 0.7187, 0.131,58 |
| TIMELESS | 0.0112*, 6.971,47 | 0.2364, 1.441,47 | 0.000**, 18.121,47 | 0.8640, 0.031,47 |
Wilk’s lambda P-value for within-subject comparisons
P < 0.05
P < 0.01
3.4 Effects of blue light on circadian gene expression
We further tested the impact of the morning “blue” light intervention on circadian gene expression by examining the interaction between light intervention group and study week for gene expression. No differences between pre- and post-pDLMO expression levels were found to be significantly different between light groups for any circadian gene (Table 1). Circadian gene expression levels for both study weeks, both sampling times and both light treatment groups are illustrated in Figure 2.
Figure 2. Circadian gene expression levels before and after sleep/wake schedule advance intervention, subdivided by “dim” light and “blue” light treatment groups.
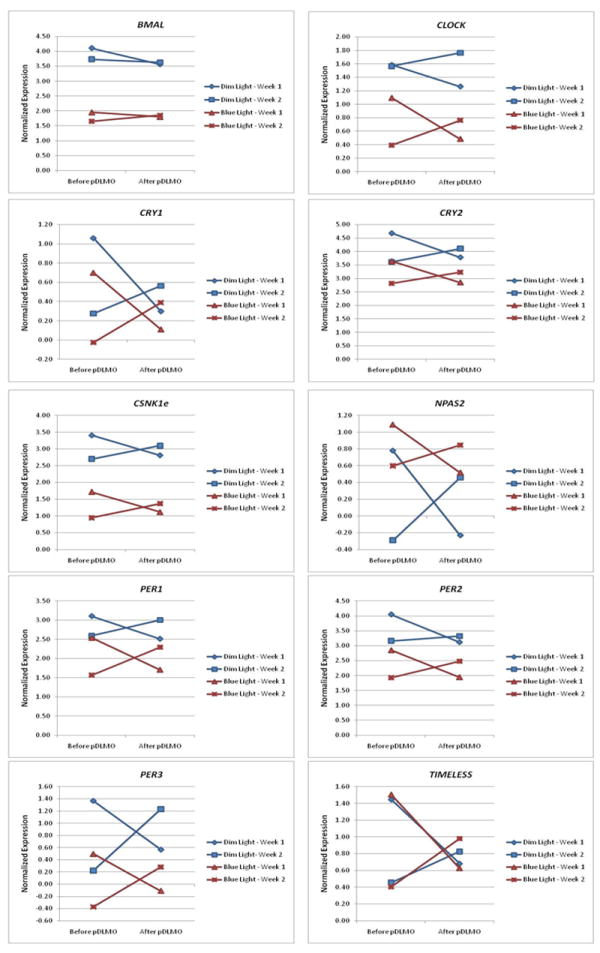
9 out of the 10 circadian genes tested exhibited changes in peripheral blood expression patterns following adherence to a fixed, advanced sleep-activity schedule, independent of the morning light exposure intervention. No differences between pre- and post-pDLMO expression levels were found to be significantly different between “blue” light and “dim” light groups for any circadian gene.
4. Discussion
Mammalian circadian oscillators occur in both central and peripheral tissues, and the (suprachiasmatic nuclei) SCN are presumed to coordinate cyclic gene expression in the periphery by neural and/or humoral signals [40-44]. Robust daily oscillations in gene expression are seen in almost all investigated tissues [45]. Studies have shown that the peak timing of clock gene expression in peripheral tissues is delayed by up to six hours with respect to the phase of the SCN daily rhythm [46, 47], which suggests that the variation in mRNA expression of circadian genes may be useful for assessing human circadian systems [48].
In the current study, nine out of the 10 circadian genes tested exhibited changes in peripheral blood expression patterns following adherence to a fixed, advanced sleep-activity schedule for 6 consecutive days, independent of the morning light exposure intervention. Daily patterns of light and dark promote entrainment of the master clock to the 24-hour solar day which, in turn, provides timing cues to peripheral clocks. Expression changes of circadian genes detected in peripheral blood after adherence to a strict advanced sleep/wake schedule may reflect internal synchronization at the molecular level to the changed daily light/dark pattern. Interestingly, our findings indicate that post-pDLMO expression was significantly diminished relative to pre-pDLMO expression in week 1 for all circadian genes measured, while the reverse pattern was observed in week 2 for all genes with the exception of BMAL. Taken into context the 1.4 hour average phase advance in salivary DLMO at the end of week 2, this finding suggests that our observation window captured at least a segment of the falling phase of expression in week 1 and a segment of the rising phase of expression in week 2. Consistent with our observation, previous studies have found that PER genes exhibit circadian expression patterns in human blood, and that the nadirs of PER1, PER2, and PER3 expression roughly coincide with DLMO in peripheral blood mononuclear cells [27, 49, 50]. Furthermore, it has been demonstrated that DLMO in blood and saliva occur at approximately the same circadian time [51].
The observation that PER gene expression patterns may shift in a manner consistent with phase shifts in sleep/wake schedules has been previously noted [30, 52]. In contrast, the finding that CLOCK, CRY1, CRY2, CSNK1E, NPAS2, and TIMELESS may respond to sleep entrainment in a similar manner as that observed for PER has never been reported. However, detailed information on the circadian rhythmicity of these genes, including relative peak and nadir times, are lacking. To our knowledge, only one study has been conducted to examine the circadian rhythmicity of CRY1, CRY2, CLOCK, and TIMELESS in human peripheral blood, which failed to find statistically significant diurnal variation of expression of CRY1, CRY2, and TIMELESS in most of the subjects examined [28]. Of the genes we investigated, only BMAL did not exhibit a change in expression pattern following sleep/wake schedule advancement. One possible explanation is that BMAL expression may exhibit only weak circadian patterns in peripheral blood leucocytes [29] and mononuclear cells [28]. Thus, it is possible that our sampling window did not allow us to adequately capture the subtle circadian rhythmicity characteristic of BMAL expression in peripheral blood.
Morning “blue” light exposure alone did not differentially alter the expression of circadian genes in peripheral blood. This observation seems to stand in contrast to the findings of previous studies, which observed measurable effects of blue light on CRY1, BMAL [34], PER2 [31, 32], and PER3 [53] expression. However, of these four studies, only one examined the independent effect of morning blue light exposure, but in buccal mucosa and not peripheral blood. Viewed in the context of our own results, this distinction raises the possibility that blue light may affect circadian gene expression in a tissue-specific manner. Perhaps more revealing, however, the Daysimeter did not report differences in overall daily personal light/dark exposure patterns between the two light groups (data not shown), suggesting that the effect of morning “blue” light exposure may have been masked by the effects of light exposure events at other times of the day. Moreover, our findings are consistent with our observation that a morning “blue” light exposure, on average, did not produce a differentially larger DLMO phase advance than a morning “dim” light exposure during the advanced sleep/wake schedule. Taken in the context of previous studies which reported significant changes in both melatonin and circadian gene expression [31, 32, 34, 53], the concordance between melatonin and circadian gene responses suggests that the mechanisms that govern the synchronization of both circadian components to blue light stimulus may exist within the same biological pathway.
A potential methodological limitation is that our study was conducted primarily in a field setting, which made it impossible for us to directly monitor participants for adherence to prescribed sleep/wake schedules and light treatment protocols. However, noncompliance with the prescribed protocols appears unlikely based on the data obtained from wrist activity, Daysimeter light assessments, and laboratory voice mail recordings at bedtime and wake time that helped confirm schedule compliance. We cannot rule out the possibility that observed changes in circadian gene expression may stem from non-circadian fluctuations in gene expression, given the relatively narrow sampling window. However, the clear relationship with melatonin response, or the absence thereof, and expression change suggests that this is unlikely to be the case. In addition, as light treatments were carried out in conjunction with the sleep schedule advance intervention, it is impossible to isolate the independent effects of the schedule advance from those that may have been introduced by social cues associated with light treatment. Furthermore, in measuring clock gene expression, we assumed 100% PCR amplification efficiency for both clock genes and the HPRT1 control. Any empirical deviation from this assumption could have lead to erroneous measures of expression, although the problem is somewhat alleviated by our use of similar starting RNA concentrations.
In conclusion, our study demonstrated significant changes in expression of most of the best characterized circadian genes following adherence to a strict, advanced sleep/wake schedule in young adults, with or without morning “blue” light treatment. These results should be cautiously interpreted given the relatively narrow sampling window. A wider sampling window with more sampling times is needed to confirm whether clock gene phase angles advanced in concert with DLMO. Nevertheless, our findings suggest that synchronization of the peripheral circadian clock to even subtle changes in one’s sleep/wake schedule can result in measureable changes in the patterns of circadian gene expression.
Supplementary Material
Acknowledgments
This work was funded by NIH U01DA023822. The authors thank Andrew Baum, Jena Burgner, David Bushnell, William Coon, Danni Dunlap, Marcy D’Uva, Ellyn Ferriter, Margaret Gordon-Fogelson, Denise Maceroni, Clayton Kim, Jennifer Norton King, Nischal Nadig, Ellen Sweeney, and Celso Teixeira from Bradley Hospital for technical assistance. We also thank Andrew Bierman, Brian Donlan, Jim Dunshee, and Nicolas Meyer at the Lighting Research Center.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–53. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki T, Ishikawa A, Yoshimura T, Namikawa T, Abe H, Honma S, et al. Quantitative trait locus analysis of abnormal circadian period in CS mice. Mamm Genome. 2001;12:272–7. doi: 10.1007/s003350010280. [DOI] [PubMed] [Google Scholar]
- 3.Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang RP, Todo T, et al. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry. 1996;35:13871–7. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- 4.Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–9. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 5.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, et al. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–6. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 6.Reick M, Garcia JA, Dudley C, McKnight SL. NPAS2: an analog of clock operative in the mammalian forebrain. Science. 2001;293:506–9. doi: 10.1126/science.1060699. [DOI] [PubMed] [Google Scholar]
- 7.Unsal-Kacmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–16. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cermakian N, Whitmore D, Foulkes NS, Sassone-Corsi P. Asynchronous oscillations of two zebrafish CLOCK partners reveal differential clock control and function. Proc Natl Acad Sci U S A. 2000;97:4339–44. doi: 10.1073/pnas.97.8.4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu W, Ikeda M, Abe H, Honma S, Ebisawa T, Yamauchi T, et al. Characterization of three splice variants and genomic organization of the mouse BMAL1 gene. Biochem Biophys Res Commun. 1999;260:760–7. doi: 10.1006/bbrc.1999.0970. [DOI] [PubMed] [Google Scholar]
- 10.Johansson C, Willeit M, Smedh C, Ekholm J, Paunio T, Kieseppa T, et al. Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacol. 2003;28:734–9. doi: 10.1038/sj.npp.1300121. [DOI] [PubMed] [Google Scholar]
- 11.Katzenberg D, Young T, Finn L, Lin L, King DP, Takahashi JS, et al. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–76. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- 12.Ebisawa T, Uchiyama M, Kajimura N, Mishima K, Kamei Y, Katoh M, et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. Embo Rep. 2001;2:342–6. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Archer SN, Robilliard DL, Skene DJ, Smits M, Williams A, Arendt J, et al. A length polymorphism in the circadian clock gene Per3 is linked to delayed sleep phase syndrome and extreme diurnal preference. Sleep. 2003;26:413–5. doi: 10.1093/sleep/26.4.413. [DOI] [PubMed] [Google Scholar]
- 14.Ebisawa T, Uchiyama M, Kajimura N, Mishima K, Kamei Y, Katoh M, et al. Association of structural polymorphisms in the human period3 gene with delayed sleep phase syndrome. EMBO Rep. 2001;2:342–6. doi: 10.1093/embo-reports/kve070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freedman MS, Lucas RJ, Soni B, von Schantz M, Munoz M, David-Gray Z, et al. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science. 1999;284:502–4. doi: 10.1126/science.284.5413.502. [DOI] [PubMed] [Google Scholar]
- 16.Lucas RJ, Freedman MS, Munoz M, Garcia-Fernandez JM, Foster RG. Regulation of the mammalian pineal by non-rod, non-cone, ocular photoreceptors. Science. 1999;284:505–7. doi: 10.1126/science.284.5413.505. [DOI] [PubMed] [Google Scholar]
- 17.Brainard GC, Hanifin JP, Greeson JM, Byrne B, Glickman G, Gerner E, et al. Action spectrum for melatonin regulation in humans: evidence for a novel circadian photoreceptor. J Neurosci. 2001;21:6405–12. doi: 10.1523/JNEUROSCI.21-16-06405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thapan K, Arendt J, Skene DJ. An action spectrum for melatonin suppression: evidence for a novel non-rod, non-cone photoreceptor system in humans. J Physiol. 2001;535:261–7. doi: 10.1111/j.1469-7793.2001.t01-1-00261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright HR, Lack LC. Effect of light wavelength on suppression and phase delay of the melatonin rhythm. Chronobiol Int. 2001;18:801–8. doi: 10.1081/cbi-100107515. [DOI] [PubMed] [Google Scholar]
- 20.Lockley SW, Brainard GC, Czeisler CA. High sensitivity of the human circadian melatonin rhythm to resetting by short wavelength light. J Clin Endocrinol Metab. 2003;88:4502–5. doi: 10.1210/jc.2003-030570. [DOI] [PubMed] [Google Scholar]
- 21.Cajochen C, Munch M, Kobialka S, Krauchi K, Steiner R, Oelhafen P, et al. High sensitivity of human melatonin, alertness, thermoregulation, and heart rate to short wavelength light. J Clin Endocrinol Metab. 2005;90:1311–6. doi: 10.1210/jc.2004-0957. [DOI] [PubMed] [Google Scholar]
- 22.Wright HR, Lack LC, Kennaway DJ. Differential effects of light wavelength in phase advancing the melatonin rhythm. J Pineal Res. 2004;36:140–4. doi: 10.1046/j.1600-079x.2003.00108.x. [DOI] [PubMed] [Google Scholar]
- 23.Revell VL, Arendt J, Terman M, Skene DJ. Short-wavelength sensitivity of the human circadian system to phase-advancing light. J Biol Rhythms. 2005;20:270–2. doi: 10.1177/0748730405275655. [DOI] [PubMed] [Google Scholar]
- 24.Smith MR, Revell VL, Eastman CI. Phase advancing the human circadian clock with blue-enriched polychromatic light. Sleep Med. 2009;10:287–94. doi: 10.1016/j.sleep.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warman VL, Dijk DJ, Warman GR, Arendt J, Skene DJ. Phase advancing human circadian rhythms with short wavelength light. Neurosci Lett. 2003;342:37–40. doi: 10.1016/s0304-3940(03)00223-4. [DOI] [PubMed] [Google Scholar]
- 26.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–64. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 27.Boivin DB, James FO, Wu A, Cho-Park PF, Xiong H, Sun ZS. Circadian clock genes oscillate in human peripheral blood mononuclear cells. Blood. 2003;102:4143–5. doi: 10.1182/blood-2003-03-0779. [DOI] [PubMed] [Google Scholar]
- 28.Kusanagi H, Hida A, Satoh K, Echizenya M, Shimizu T, Pendergast JS, et al. Expression profiles of 10 circadian clock genes in human peripheral blood mononuclear cells. Neurosci Res. 2008;61:136–42. doi: 10.1016/j.neures.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 29.Fukuya H, Emoto N, Nonaka H, Yagita K, Okamura H, Yokoyama M. Circadian expression of clock genes in human peripheral leukocytes. Biochem Biophys Res Commun. 2007;354:924–8. doi: 10.1016/j.bbrc.2007.01.063. [DOI] [PubMed] [Google Scholar]
- 30.James FO, Cermakian N, Boivin DB. Circadian rhythms of melatonin, cortisol, and clock gene expression during simulated night shift work. Sleep. 2007;30:1427–36. doi: 10.1093/sleep/30.11.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cajochen C, Jud C, Munch M, Kobialka S, Wirz-Justice A, Albrecht U. Evening exposure to blue light stimulates the expression of the clock gene PER2 in humans. Eur J Neurosci. 2006;23:1082–6. doi: 10.1111/j.1460-9568.2006.04613.x. [DOI] [PubMed] [Google Scholar]
- 32.Jud C, Chappuis S, Revell VL, Sletten TL, Saaltink DJ, Cajochen C, et al. Age-dependent alterations in human PER2 levels after early morning blue light exposure. Chronobiol Int. 2009;26:1462–9. doi: 10.3109/07420520903385564. [DOI] [PubMed] [Google Scholar]
- 33.Ackermann K, Sletten TL, Revell VL, Archer SN, Skene DJ. Blue-light phase shifts PER3 gene expression in human leukocytes. Chronobiol Int. 2009;26:769–79. doi: 10.1080/07420520902929045. [DOI] [PubMed] [Google Scholar]
- 34.Chen A, Du L, Xu Y, Chen L, Wu Y. The effect of blue light exposure on the expression of circadian genes: bmal1 and cryptochrome 1 in peripheral blood mononuclear cells of jaundiced neonates. Pediatr Res. 2005;58:1180–4. doi: 10.1203/01.pdr.0000183663.98446.05. [DOI] [PubMed] [Google Scholar]
- 35.Sharkey KM, Carskadon MA, Figueiro MG, Zhu Y, Rea MS. Effects of an advanced sleep schedule and morning short wavelength light exposure on circadian phase in young adults with late sleep schedules. Sleep Med. 2011;12:685–92. doi: 10.1016/j.sleep.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rea MS, Figueiro MG, Bullough JD, Bierman A. A model of phototransduction by the human circadian system. Brain Res Brain Res Rev. 2005;50:213–28. doi: 10.1016/j.brainresrev.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Rea MS, Figueiro MG, Bierman A, Bullough JD. Circadian light. J Circadian Rhythms. 2010;8:2. doi: 10.1186/1740-3391-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bierman A, Klein T, Rea MS. The Daysimeter: a device for measuring optical radiation as a stimulus for the human circadian system. Measurement Science and Technology. 2005;16:2292. [Google Scholar]
- 39.Burgess HJ, Eastman CI. The dim light melatonin onset following fixed and free sleep schedules. J Sleep Res. 2005;14:229–37. doi: 10.1111/j.1365-2869.2005.00470.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–7. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 41.Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, et al. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–10. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- 42.Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, et al. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–5. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 43.Le Minh N, Damiola F, Tronche F, Schutz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. Embo J. 2001;20:7128–36. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McNamara P, Seo SP, Rudic RD, Sehgal A, Chakravarti D, FitzGerald GA. Regulation of CLOCK and MOP4 by nuclear hormone receptors in the vasculature: a humoral mechanism to reset a peripheral clock. Cell. 2001;105:877–89. doi: 10.1016/s0092-8674(01)00401-9. [DOI] [PubMed] [Google Scholar]
- 45.Schibler U, Ripperger J, Brown SA. Peripheral circadian oscillators in mammals: time and food. J Biol Rhythms. 2003;18:250–60. doi: 10.1177/0748730403018003007. [DOI] [PubMed] [Google Scholar]
- 46.Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, et al. Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J Biol Chem. 1998;273:27039–42. doi: 10.1074/jbc.273.42.27039. [DOI] [PubMed] [Google Scholar]
- 47.Zylka MJ, Shearman LP, Weaver DR, Reppert SM. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20:1103–10. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 48.Takata M, Burioka N, Ohdo S, Takane H, Terazono H, Miyata M, et al. Daily expression of mRNAs for the mammalian Clock genes Per2 and clock in mouse suprachiasmatic nuclei and liver and human peripheral blood mononuclear cells. Jpn J Pharmacol. 2002;90:263–9. doi: 10.1254/jjp.90.263. [DOI] [PubMed] [Google Scholar]
- 49.Kusanagi H, Hida A, Satoh K, Echizenya M, Shimizu T, Pendergast JS, et al. Expression profiles of 10 circadian clock genes in human peripheral blood mononuclear cells. Neuroscience Research. 2008;61:136–42. doi: 10.1016/j.neures.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 50.Fukuya H, Emoto N, Nonaka H, Yagita K, Okamura H, Yokoyama M. Circadian expression of clock genes in human peripheral leukocytes. Biochem Bioph Res Co. 2007;354:924–8. doi: 10.1016/j.bbrc.2007.01.063. [DOI] [PubMed] [Google Scholar]
- 51.Voultsios A, Kennaway DJ, Dawson D. Salivary melatonin as a circadian phase marker: Validation and comparison to plasma melatonin. J Biol Rhythm. 1997;12:457–66. doi: 10.1177/074873049701200507. [DOI] [PubMed] [Google Scholar]
- 52.Archer SN, Viola AU, Kyriakopoulou V, von Schantz M, Dijk DJ. Inter-individual differences in habitual sleep timing and entrained phase of endogenous circadian rhythms of BMAL1, PER2 and PER3 mRNA in human leukocytes. Sleep. 2008;31:608–17. doi: 10.1093/sleep/31.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ackermann K, Sletten TL, Revell VL, Archer SN, Skene DJ. Blue-Light Phase Shifts PER3 Gene Expression in Human Leukocytes. Chronobiology International. 2009;26:769–79. doi: 10.1080/07420520902929045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


