Abstract
Protein synthesis on ribosomes is carefully quality controlled to ensure the faithful transmission of genetic information from mRNA to protein. Many of these mechanisms rely on communication between distant sites on the ribosomes, and thus on the integrity of the ribosome structure. Furthermore, haploinsufficiency of ribosomal proteins, which increases the chances of forming incompletely assembled ribosomes, can predispose to cancer. Finally, release of inactive ribosomes into the translating pool will lead to their degradation together with the degradation of the bound mRNA. Together, these findings suggest that quality control mechanisms must be in place to survey nascent ribosomes and ensure their functionality. This review gives an account of these mechanisms as currently known.
Keywords: Ribosome assembly, quality control, RNA-protein complex, translation
The importance of ribosomal quality control
Actively growing cells must synthesize thousands of ribosomes every minute [1], while ensuring that they faithfully translate genetic information from mRNA to protein. The importance of producing enough functional ribosomes is illustrated by diseases linked to defects in ribosome assembly. Diamond Blackfan Anemia (DBA) and 5q-syndrome are caused by haploinsufficiency of ribosomal proteins (RPs), and characterized by low numbers of the rapidly dividing cells of the bloodline, as well as a generally diminutive phenotype [2-4]. Similarly, Shwachman Bodian syndrome (SDS) results from the inability to release the assembly factor eIF6 [2]. Interestingly, all three diseases predispose patients to cancer with 23% of patients suffering from DBA being diagnosed with cancer by the age of 46 [2, 5]. One possible scenario to explain the increased cancer incidence is the release of misassembled ribosomes into the translating pool [4, 6], underscoring the importance of mechanisms to ensure that only fully assembled and fully functional ribosomes be allowed to translate protein.
Ribosome assembly involves the coordinated transcription, modification and processing of precursor rRNAs and the binding of RPs. Assembly in vivo requires about 200 assembly factors (AFs) in yeast, or about 40 in bacteria (see [7-14] for recent reviews). Ribosomes can be reconstituted from their components under certain conditions [15-18], and many RPs of the small subunit bind co-transcriptionally [19, 20]. Thus, it appears that AFs provide a platform for the regulation of rRNA modification and processing, and for quality control of ribosome assembly steps, instead of simply facilitating the binding of RPs.
Recent work in prokaryotes and eukaryotes has provided insight into the mechanisms by which ribosomes are matured, and how their quality is controlled. Quality control can be subdivided into mechanisms to ensure that nascent subunits do not enter the translation cycle, as well as those to provide for functional checks of the nascent subunits. Excitingly, but perhaps not surprisingly, functional checks employ parts of the translational apparatus, and are thus “real-life” situations for the assembling ribosomes. Below, we will describe in detail how translation is blocked on nascent ribosomes, as well as the quality control pathways that gate release of these blocks. Finally, pathways to degrade non-functional ribosomes are discussed. Each mechanism is first described for the small and then for the large subunit, and data from eukaryotes and prokaryotes are presented. No studies on ribosome assembly have been performed in archaea, however, a number of the players are conserved and this is mentioned where applicable. Finally, most of the assembly mechanisms and factors are conserved from yeast to human. If there are known differences between yeast and humans these are explicitly described.
Blocking premature translation
Separation of assembly from translation in the nucleus
The simplest way to prevent translation by premature ribosomes is to separate ribosome maturation in the nucleolus/nucleus from translation in the cytoplasm, as is done in eukaryotes. Nevertheless, the final maturation steps for both subunits occur in the cytoplasm, where all components of the translation machinery, including tRNAs, mRNAs, translation factors and ribosomal subunits are highly abundant. Thus, blocking premature translation initiation is expected to be especially important for the cytoplasmic steps of maturation in eukaryotes, and later steps in prokaryotes and archaea, where assembly is sufficiently progressed to allow for binding of ligands, such as tRNA, mRNA etc.
Blocking RP binding
Most RPs are essential and therefore blocking their binding is expected to render the subunits incompetent for translation. Interestingly, several AFs block RP binding. Among these, binding of the AFs Enp1/Ltv1 blocks binding of S10, and binding of Pno1 blocks binding of S26, on the head and platform of the maturing small subunit [21]. While these AFs have no structural similarity to the RPs they replace, several cases of RP mimicry are known, including Mrt4, which has strong sequence homology to the stalk base protein P0, except it does not allow for stalk assembly, Rlp7, which resembles L7 and Rlp24, which resembles L24. Furthermore, Imp3 has sequence homology to S9. It is assumed that these AFs block binding of their counterpart RPs, although that has only been rigorously shown for Mrt4 [22-25].
40S assembly factors block binding of initiation factors, mRNA, tRNA and 60S subunits
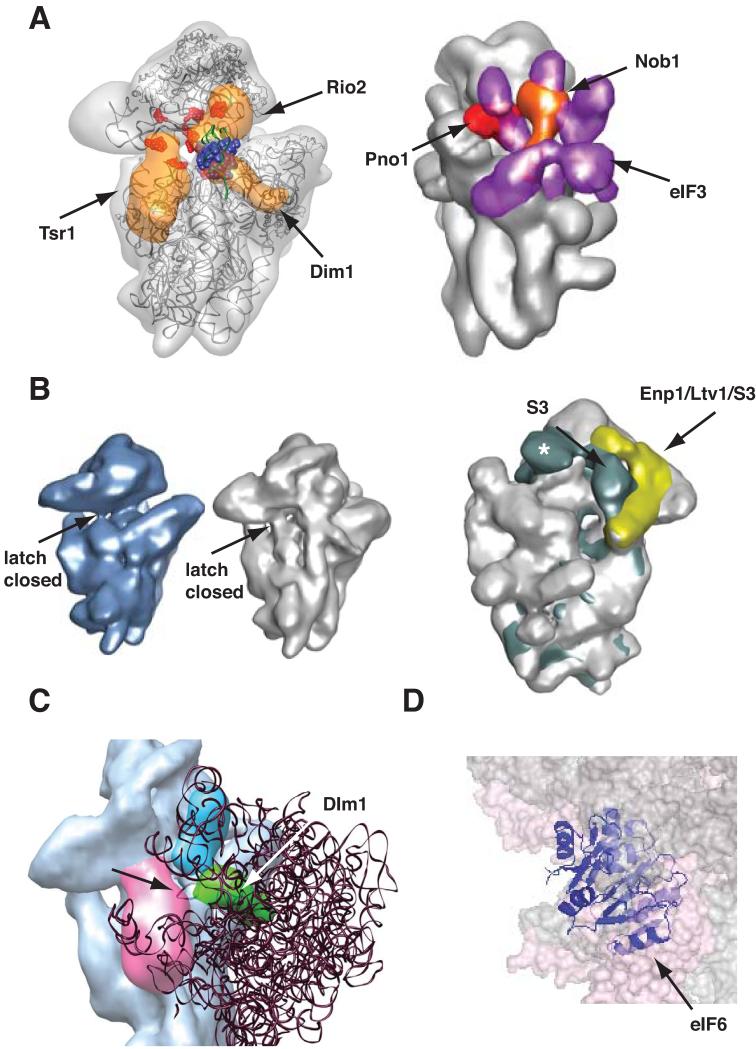
Translation initiation begins on the small subunit, which recruits translation factors, tRNAs, mRNA and finally the large subunit to begin protein production (see [26] and references therein). Thus, the small subunit (40S in eukaryotes, 30S in prokaryotes and archaea) has more ligand binding sites, and blocking these is likely more important for assembling small subunits than large subunits. Indeed, in eukaryotes and prokaryotes AFs block premature translation initiation. A recent cryo-EM structure of a cytoplasmic 40S assembly intermediate purified from yeast cells shows that all seven bound AFs cooperate to prevent every step in the translation initiation pathway [21]: AFs on the subunit interface (Tsr1, Rio2 and Dim1) and on the platform (Nob1, Pno1) block the binding of essential translation initiation factors (Figure 1A), required to render 40S subunits competent for the subsequent steps in translation initiation, including mRNA and tRNA recruitment. Furthermore, the kinase Rio2 also blocks binding of initiator tRNA (Figure 1A), as predicted based on footprinting data [27], and all three subunit interface factors together block access to the mRNA binding channel. mRNA recruitment is further blocked by an allosteric mechanism (Figure 1B): opening of the mRNA channel is stabilized by interactions on the solvent side involving S3 [28]. In pre-40S subunits the entry channel is closed, as the AFs Enp1/Ltv1 block the interaction of S3 on the solvent side. Next, subunit joining is blocked by Dim1, as predicted based on footprinting of this protein [27], and by the decoding site helix (Figure 1C), which contains many of the contact points with the large subunit. This helix is dislodged towards the large subunit and to the left (viewed from the large subunit). Thus, subunit bridges cannot form, and the decoding site helix sterically blocks subunit joining. Loss of Dim1 alone is not sufficient to release the block in 60S binding, but depletion of Tsr1, the largest factor on the subunit interface, leads to premature subunit joining [21]. Tsr1 itself does not block subunit joining (Figure 1C), but in the absence of Tsr1 the decoding site helix is not dislodged, indicating that the conformation of this helix is largely responsible for the block in subunit joining. However, because Pno1 is co-depleted with Tsr1, loss of Pno1, Tsr1, or both could be responsible for changes in the decoding site. It is also important to note that all three factors, Dim1, Pno1, and Tsr1 can be found in 80S-like ribosomes [29], suggesting that any block to 80S formation is not absolute. Perhaps the 60S subunit is accommodated in a different way in these 80S-like ribosomes. Finally, the rearrangements of the decoding site helix distort the decoding site, which is not able to provide the minor groove interactions critical for productive recognition of amino-acylated tRNA to start the elongation cycle. Thus, all seven AFs bound to cytoplasmic 40S assembly intermediates in eukaryotes contribute to block every step in the translation initiation pathway in a redundant manner, underscoring the importance of ensuring that only mature ribosomes begin translation. Importantly, four (Dim1, Nob1, Pno1 and Rio2) of the AFs are conserved in archaea, suggesting that archaea use similar mechanisms to prevent premature translation initiation on assembling subunits.
Figure 1.
Assembly Factors Block Premature Translation Initiation. Adapted from [21]. (A) Left: Tsr1, Rio2 and Dim1 on the subunit interface block binding of eIF1 (in blue spacefill, [100]), eIF1A (binding site highlighted in red, [101]), and P-site tRNA (in green, [102]). Right: Nob1 (orange) and Pno1 (red) block binding of eIF3 (purple, [103]). (B) Opening of the mRNA binding channel is destabilized. The entry latch on the mRNA binding channel (indicated with the arrow) is closed in mature 40S subunits (left, [28]) and pre-40S subunits (middle, [21]). Channel opening in mature subunits (right panel) is stabilized by an interaction between S3 and H16 (shown in dark green, [28]) on the solvent side. The AFs Enp1 and Ltv1 form a complex with S3 (in yellow, [21]), which does not allow for this interaction of S3. Asc1, present only in mature 40S subunits, is indicated with a white asterisk. (C) In 80S ribosomes the A-site finger from the 60S subunit (indicated with the arrow), overlaps with H44 and Dim1 (in green), but not Tsr1 (in magenta). (D) In 80S ribosomes eIF6 (in purple, [37]) overlaps with the 40S subunit (shown in spacefill, [104]). For simplicity the 60S subunits is not shown.
Prokaryotic assembly factors block binding of initiation factors, mRNA, tRNA and 50S subunits
The function of AFs in preventing translation initiation on small subunits is echoed in prokaryotes, although the mechanisms and AFs differ, reflecting differences in the translation initiation pathway between the two kingdoms. The AFs RbfA and KsgA are positioned to block binding of the translation initiation factors IF1 and IF3, respectively [30-32]. Similarly, the GTPase Era binds the anti-Shine-Dalgarno sequence at the 3′-end of 16S rRNA, thus likely preventing mRNA recruitment [33, 34]. Finally, the assembly factor KsgA, homologous to eukaryotic Dim1, overlaps the position of the large subunit A-site finger, thus likely blocking joining of the 50S subunit in the normal conformation ([32], see also Figure 1C).
60S AFs block 40S joining
Large subunits (60S in eukaryotes, 50S in prokaryotes and bacteria) join the translation initiation complex late and do not interact with mRNA or tRNA on their own. Thus, to block premature translation by 60S precursors it is sufficient to block joining with 40S subunits. In eukaryotes, the conserved AFs Nmd3 and eIF6/Tif6 bind to the subunit interface of the 60S subunit (Figure 1D, [35-37]), and biochemical experiments indicate that both block the formation of 80S ribosomes [35, 36]. Similarly, Mrt4 blocks binding of the stalk protein P0 and subsequent assembly of the stalk, an important binding site for the ribosomal GTPases. Thus, it is likely that the GTPase eIF5B cannot promote joining of Mrt4-containing pre-60S ribosomes to 40S subunits. For additional discussion on the separation of premature 60S subunits from the translational machinery see [10].
Testing ligand binding in nascent ribosomes
The section above describes how AFs block ligand-binding sites to prevent premature translation initiation. However, blocking translation does not allow for separation of functional from non-functional ribosomes. Here, we describe mechanisms to distinguish functional from non-functional intermediates based on the ability to bind RPs, translation factors or AFs mimicking them.
40S export requires late-binding r-proteins from the head
As ribosomes mature they acquire ligand-binding sites, which resemble more and more those found in mature ribosomes. The simplest example of how recognition of ligand binding sites is linked to progress in the assembly cascade is the release of assembling subunits from the nucleus into the cytoplasm. S15/S18 and S5/S28 are required for nuclear export of assembling small subunits [19, 38]. These are late-binding proteins that bind as pairs to the head [19]. Thus, linking the ability to be exported to the acquisition of S15, S18, S5 and S28 ensures that early intermediates cannot escape into the cytoplasm. Furthermore, in humans Rio2 and Tsr1 are required for export [39, 40], with Rio2 a suggested export adaptor [39]. Linking the ability to be exported to the successful binding of Rio2 and Tsr1 ensures that all exported small subunits are blocked for translation initiation as Rio2 and Tsr1 are key contributor to this process as described above.
60S export requires completion of the exit tunnel
The main export adaptor for the nascent 60S subunit is Nmd3, whose binding site includes the sarcin-ricin loop (SRL), which is important for function of EF-G/eEF2 [35, 41]. Furthermore, Arx1 is a second export adaptor that functions in yeast. Arx1 has substantial sequence homology to aminopeptidases, which cleave the N-terminal methionine from nascent peptide chains [42, 43]. As a result, Arx1 binds the exit tunnel akin to aminopeptidases [44, 45], and can thus signal completion of the exit tunnel.
Furthermore, Arx1 also overlaps the binding sites for the translocon and the SRP [44, 45]. Thus, export of 60S subunits can only occur if the SRL and exit tunnel are completely assembled. Interestingly, the export factors for both subunits (Rio2 and Tsr1 and Nmd3) also function to block premature translation as described above, thus ensuring that the released subunits cannot enter the translating pool.
Removal of AFs requires completion of functional centers
Interestingly, Arx1 removal requires Jjj1 [46, 47], whose binding site also overlaps that of the SRP and the translocon [44], and which functions together with the Hsp70 Ssa, a ribosome-bound chaperone. Thus, Arx1 dissociation requires the completion of the Ssa, translocon and SRP binding sites, which signal the ability to promote co-translational protein folding and membrane insertion.
3′-end formation of 40S subunits requires completion of the GTPase site
Successful assembly of functional sites is also a prerequisite for several of the characterized rRNA maturation steps. rRNA cleavage steps are irreversible and, based on observations in other metabolic pathways, therefore likely regulated. In eukaryotes, three of the four rRNAs are co-transcribed. This pre-rRNA is matured in a series of largely ordered steps to release the mature rRNAs for the small and large subunits. In yeast, rRNA processing requires eight endonucleolytic steps and six exonucleases. Four of the endonucleases and all of the exonucleases have been identified; among them both nucleases required for formation of the 3′-end of 18S rRNA, Rcl1 and Nob1, respectively [48-53]. Rcl1 and Nob1 function successively, Rcl1 separating the precursors destined for the small and large subunits [53], before Nob1 cleaves at the 3′-end of 18S rRNA [50-52].
The GTPase Bms1 promotes delivery of Rcl1 to early pre-40S subunits [54, 55]. Bms1 has extensive sequence homology to Tsr1, a late-acting AF, which binds at the GTPase site on the small subunit [21, 56]. The sequence similarity between these two proteins, their successive binding to assembling 40S subunits, and the observation that Tsr1 binds at the GTPase site, all suggest that Bms1 occupies Tsr1’s spot in early assembly intermediates. Together, these data indicate that Bms1-dependent Rcl1 binding and the resulting rRNA cleavage require at least partial completion of the GTPase site on small subunits.
Furthermore, Nob1-dependent cleavage at the 3′-end of 18S rRNA requires prior Rcl1-dependent cleavage, which allows for a conformational change to provide Nob1 access to the cleavage site [52]. This conformational change involves formation of the decoding site helix. Interestingly, the cryo-EM structure of a later cytoplasmic 40S assembly intermediate shows that this conformational change is only partially complete, with the uppermost helical element remaining unpaired, thus allowing the decoding site helix to be “pulled out” from the body of the subunit to block 60S joining as described above [21]. Thus, formation of 80S ribosomes must somehow be linked to the formation of the decoding site. Nob1 cleavage can occur in 80S-like ribosomes, composed of pre-40S subunits and 60S subunits [29, 57], and mutations in the large subunit RP L3 affect the Nob1-dependent step (J. de la Cruz, personal communication), indicating that Nob1-dependent formation of the 3′-end of 18S rRNA must occur in 80S-like ribosomes, and is thus linked to formation of the decoding site. It is unlikely that formation of the decoding site in 80S-like ribosomes alone is sufficient to promote Nob1-dependent cleavage, as 18S rRNA formation is blocked in the absence of the ATPase Fap7, which is stalled in 80S-like complexes. Nevertheless, the absence of S14 and S1 in these assembly intermediates [29] could account for the lack of Nob1-dependent cleavage, plausible as S14 directly binds Nob1 [50], and S1 binds Pno1 [21], a regulator for Nob1 [58].
5′-end formation of 5.8S rRNA requires r-proteins near the exit tunnel
Early rRNA maturation steps for the 60S subunit require a group of proteins, referred to as the A3 cluster proteins for the precursor that accumulates in their absence [59-66]. Depletion of any of the A3 cluster proteins results in the failure to recruit the exonuclease Rrp17, required for formation of the 5′-end of 5.8S rRNA, as well as four neighboring RPs, L17, L26, L35 and L37, which bind adjacent to the 5′-end of 5.8S rRNA, and form part of the exit tunnel [67]. Because the A3 cluster proteins bind distant from the Rrp17 site at the 5′-end of 5.8S rRNA [68], it is tempting to speculate that failure to recruit Rrp17 in the absence of the A3 cluster proteins is an indirect effect resulting from the failure to recruit the four r-proteins. Additionally, failure to recruit these RPs also leads to Rat1-dependent degradation of 60S precursors, while normally Rat1 stops at the 5′-end of 5.8S rRNA, presumably due to the presence of these four RPs [67]. These data indicate that the decision between maturation or degradation at the 5′-end of 5.8S rRNA depends on assembly of the exit tunnel.
Using ligands to mimic translation
In the examples above AFs structurally mimic RPs or translation factors to couple successful formation of ligand binding sites on assembling ribosomes to subsequent maturation steps. However, these only approximate the binding function, without providing for quality control of functionality. The next section describes mechanisms in place to test the functionality of nascent ribosomal subunits.
Parts of the translation machinery are used to “test-drive” 40S ribosomes
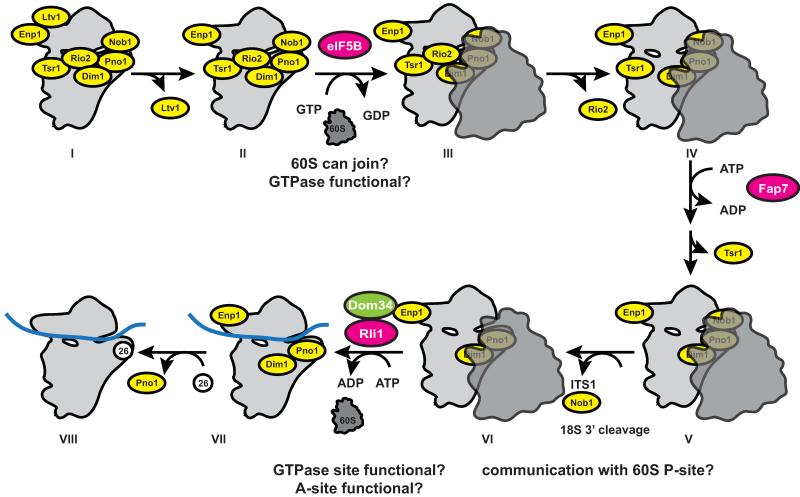
AFs that prevent premature translation initiation are stably bound to subunit precursors, providing an opportunity to regulate their release, which eventually leads to translation initiation. Excitingly, release of AFs from the subunit interface of nascent 40S subunits and subsequent translation initiation require a translation-like cycle, during which translation factors must function (Figure 2, [29]). The starting point for this cycle is the stable 40S assembly intermediate containing all seven stably bound AFs shown in Figure 1A-C. After Ltv1 dissociation the translation factor eIF5B promotes joining of 60S subunits with the nascent 40S subunits in a GTP-dependent manner [29, 57]. However, because mRNA and initiator tRNA recruitment are blocked as described above, the resulting 80S-like ribosomes do not contain mRNA or initiator tRNA and thus do not produce proteins [29]. The assembly factors Tsr1 and Rio2 on the subunit interface dissociate after or with formation of 80S-like ribosomes. However, mRNA and initiator tRNA recruitment are blocked by the bound 60S subunit, which thus prevents translation initiation until the 80S-like ribosomes are dissociated by the termination factors Rli1 and Dom34, which also dissociate 80S ribosomes after translation is complete [69-72]. Thus, this cycle has the ability to probe key functional sites on assembling 40S subunits, including the ability to bind and position large subunits, the ability to bind and activate the translation initiation factor eIF5B, the ability to bind Dom34 in the decoding site [73, 74], and the ability to bind the termination factor Rli1 and activate its ATPase activity. Furthermore, it has been suggested that Dom34, a homolog of the release factor RF1 is delivered to ribosomes by the GTPase Hbs1, a close homolog of release factor RF3 [75]. Thus, release factor function is also inspected by homology. Importantly, the key players in this translation-like cycle (eIF5B, Fap7, Rli1 and Dom34) are conserved from archaea to humans, suggesting the conservation of this cycle, and demonstrating its importance. Interestingly, this cycle also has the ability to test nascent 60S subunits against key translation factors. It remains to be tested if this cycle also quality controls nascent 60S subunits.
Figure 2.
A translation-like cycle during 40S ribosome maturation. Adapted from [29]. The cycle starts with the dissociation of Ltv1 from the intermediate visualized by cryo-EM (I). The translation factor eIF5B, a GTPase, then promotes joining of 60S subunits, analogous to its function during translation, to give an 80S-like complex, lacking both mRNA and tRNA (III). Rio2 dissociates with or soon after 60S joining to give a stable 80S-like complex (IV), which accumulates in the absence of the ATPase Fap7. Dissociation of Tsr1 (V) allows for Nob1-dependent rRNA maturation (VI) and access of Dom34 and Rli1 to separate 80S-like ribosomes (VII). Exchange of S26 for Pno1 appears to be the final step in maturation. The small and large ribosomal subunits are shown in light and dark grey, respectively. Stably bound assembly factors are shown in yellow. ATP or GTP-hydrolyzing transiently bound factors are shown in magenta, other transiently bound factors are shown in green and mRNA is shown as a blue band.
In a related finding, it was shown that prokaryotic 16S and 23S precursors can enter polyribosomes [76-78], indicating that maturation can occur after translation initiation. Subsequent work indicates that 23S rRNA processing requires conditions that allow for protein synthesis. Lastly, in dictyostelium it has been suggested that 18S rRNA maturation requires active translation [79].
Mimicking translocation to release eIF6 from nascent 60S subunits
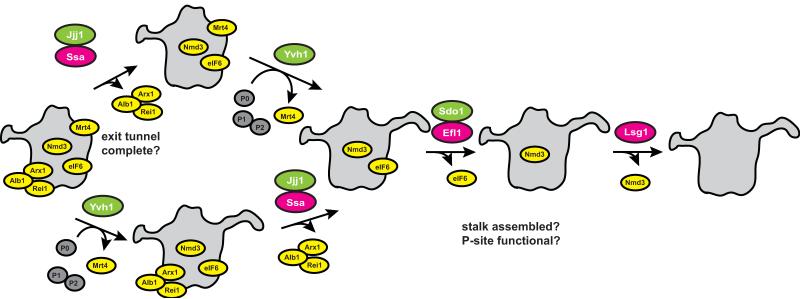
Dissociation of eIF6 is promoted by the GTPase Efl1 [80, 81], aided AF Sdo1 [82], the protein mutated in SDS patients, and requires stalk assembly ([83], Figure 3). Furthermore, Efl1 has strong homology to the translation factor eEF2, which promotes translocation of tRNA from the A/P to the P/P site, and the shape of Sdo resembles that of tRNA [82, 84, 85]. These findings prompted the suggestion that Efl1/Sdo1 function analogously to eEF2/tRNA, “translocating” and releasing Sdo1 and eIF6. This proposal is consistent with the eIF6 binding site adjacent to the GTPase binding site, and explains the requirement for the stalk, which plays a role in GTPase stimulation [83]. Furthermore, mutations in a loop of L10, located in the P-site of the large subunit, disrupt Efl1-dependent release of eIF6 [86]. This linkage of two proteins separated by ~90 Å, is explained by Efl1-binding, and suggests that Efl1 binds the P-site in a manner similar to that observed with eEF2. Thus, Efl1-dependent release of eIF6 during maturation appears to mimic the function of eEF2-mediated translocation of tRNAs during translation, and therefore provides an opportunity to inspect the integrity of the P-site and the stalk base on the large subunit [86]. Interestingly, eEF2 functions in the context of 80S ribosomes. Perhaps the release of eIF6 by Efl1 also requires 40S joining, or, alternatively, the mechanism of translocation differs. Of note, archaea have homologs for Sdo1 and eIF6 [87], suggesting conservation of this pathway.
Figure 3.
Pathway for Cytoplasmic 60S maturation. Adapted from [83]. Release of the Arx1/Alb1 complex from the exit tunnel requires the Hsp70 ATPase Ssa and its co-chaperone Jjj1, and has the ability to check the binding site for Ssa on the exit tunnel. Exchange of Mtr4 with P0 and subsequent stalk assembly requires Yvh1. Both events can occur independently of each other but are both required for the Efl1/Sdo1-dependent release of eIF6/Tif6. Efl1 function tests the P-site [86] as well as the stalk [83]. The final step in 60S maturation is the Lsg1-dependent release of Nmd3.
Discard pathways
Quality control also requires mechanisms to provide for degradation of intermediates identified as non-functional [88]. Non-functional mature ribosomes are identified as they are stalled on mRNAs [89]. Because defective ribosomes stalled on an intact mRNA are indistinguishable from intact ribosomes stalled on a defective mRNA, degradation is initiated in both cases by Hbs1-mediated binding of Dom34 to the ribosomal A-site (see [75] and references therein). In principle this mechanism could be used to eliminate misassembled ribosomes based on their inability to translate. However, because disassembly of the stalled mRNA*ribosome complex involves endonucleolytic degradation of the mRNA, this is not desired to occur on a large scale, thus necessitating quality control and degradation before translation starts.
The recent work on the translation-like cycle also suggests several intermediates that are sensitive to degradation, as well as others resistant to it. In particular, depletion of Rli1 leads to degradation of 80S-like ribosomes [29]. The only difference between the intermediate that accumulates in the absence of Rli1 (Rli1-intermediate), which is exceptionally labile, and the intermediate that accumulates in the absence of Fap7 (Fap7-intermediate), which is exceptionally stable, is the presence of Tsr1 and the absence of S14 and S1 in the Fap7-intermediate. Furthermore, in the Rli1-intermediate 18S rRNA is already matured [29]. It is possible that Tsr1 in the Fap7-intermediate blocks access of Hbs1 to the 80S-like ribosome to deliver Dom34. Dom34 could initiate Rli1-dependent maturation, but also degradation, albeit by mechanisms poorly understood. Alternatively, or additionally, the presence of the precursor RNA sequence might protect the subunit, perhaps by providing a 3′-end that is largely double-stranded and thus less sensitive to exonucleolytic degradation.
Furthermore, the translation initiation factor eIF5B is required for 40S ribosome maturation, but its deletion has only a relatively small (3-fold) effect on accumulation of the 18S precursor [29, 57, 90]. This is in contrast to about 30-fold increases observed in the absence of Fap7 or Nob1 [29, 91]. This difference is likely partially a reflection of the fact that Fap7 or Nob1 are essential proteins, while eIF5B is not [92]. Nevertheless, eIF5B deletion results in a profound slow growth phenotype, suggesting that the modest accumulation of 20S rRNA could result from degradation of the assembly intermediate that accumulates in the absence of eIF5B. The difference between the stable intermediate purified for cryo-electronmicroscopy and the unstable intermediate that accumulates without eIF5B is likely the presence of Ltv1 and perhaps Rio2. How these proteins affect the stability of the assembly intermediate remains to be tested.
The best-characterized pathway responsible for degradation of ribosome assembly intermediates is the nuclear pathway mediated by the TRAMP complex [93-95]. This complex, composed of the polyA polymerase Trf4/5, the RNA binding protein Air1/2, and the helicase Mtr4, marks nuclear RNAs with short polyA tails, and then targets them for degradation by the exosome. Recent data indicate that the Air proteins have some sequence specificity, although they can complement for each other [96]. Similarly, Trf4 and Trf5 proteins also have specificity, but can complement for each other [97]. Nevertheless, this limited specificity likely only segregates different clients for these complexes, and is unlikely to explain how misassembled ribosomes are distinguished from correctly assembled ribosomes. Although not directly shown, it is likely that TRAMP-mediated polyadenylation and decay is kinetically controlled [98, 99], such that RNAs that linger longer are more likely to be recognized and degraded. In this context, the mechanisms described above to link progress in assembly and export to successful completion of important sites becomes important, as this will decrease the time spent in states vulnerable to degradation.
Concluding remarks
Mechanisms to provide for quality control of assembling ribosomes can be subdivided into three categories: (i) those ensuring that later assembly events require successful construction of key structural features recognized by AFs that mimic translation factors or RPs; (ii) those ensuring that translation initiation on premature ribosomes is effectively blocked sterically and/or allosterically; (iii) functional tests of the nascent ribosomal subunits, which employ the translational apparatus, or mimic elements of the translational cycle. Failure to succeed in (i) or (iii) predisposes nascent subunits to degradation, while mechanism (ii) could in principle also used to safely store assembly intermediates, perhaps under stress conditions.
Future work will be required to better understand how release of AFs blocking translation initiation is coupled to functional tests, as well as the mechanisms leading to decay, or even why certain intermediates are more vulnerable than others. Furthermore, conspicuously absent in all quality control pathways is the testing of the real function of ribosomes, to produce protein in a codon-dependent manner, although it has been reported in dictyostelium that translation is required for rRNA maturation [79]. Thus, it is tempting to speculate that there is at least one more layer in the quality control that remains to be discovered. Finally, no progress has been made to test if the absence of quality control pathways actually leads to larger populations of malfunctioning ribosomes, and if these can affect the proteome in a manner that explains the increased cancer incidence observed in DBA and 5q-syndrome. It will be exciting to see future progress in these areas, which will go hand-in-hand with new insights into maturation.
Highlights.
Assembly factors can mimic binding of ribosome ligands to promote maturation
Assembly factors block premature translation initiation
Release of assembly factors requires translation-like cycles to test function
Table 1. Assembly factors and their roles in quality control.
| AF name | Archaeal homolog? |
Function | |
|---|---|---|---|
| 40S assembly factors | Diml | yes | Blocks 60S joining, eIFl |
| Enpl+Ltvl | Block Sl0 binding and mRNA channel opening |
||
| Nobl | yes | Blocks eIF3 binding, nuclease | |
| Pnol | yes | Blocks eIF3 binding, S26 binding, Nobl co-factor |
|
| Rio2 | yes | Blocks initiator tRNA, eIFlA | |
| Tsrl | Blocks eIFlA | ||
| eIF5B | yes | Checks GTPase site integrity | |
| Rlil | yes | Checks GTPase site integrity | |
| Dom34 | yes | Checks decoding site | |
| 60S assembly factors | Arxl | Export factor, Blocks exit tunnel factors | |
| Nmd3 | yes | Export factor, Blocks subunit joining | |
| eIF6/Tif6 | yes | Blocks subunit joining | |
| Efll+Sdol | no/yes | Test P-site integrity, remove eIF6 | |
| Ssal | yes | Hsp70 chaperone to remove Arxl, tests chaperone site |
|
| Mrt4 | Blocks premature P0 binding | ||
| Rpl7 | Blocks premature L7 binding | ||
| Rpl24 | Blocks premature L24 binding |
Acknowledgements
I would like to thank A. Johnson and all members of my lab for fruitful discussion, J. de la Cruz for communicating unpublished results, and S. Khoshnevis and B. Strunk for help with Figure 1. Work on ribosome assembly in our lab is supported by NIH grant R01GM086451 and NSF grant MCB0845156.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 2.Burwick N, et al. Non-Diamond Blackfan anemia disorders of ribosome function: Shwachman Diamond syndrome and 5q-syndrome. Semin Hematol. 2011;48:136–143. doi: 10.1053/j.seminhematol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis SR, Lipton JM. Diamond Blackfan anemia: a disorder of red blood cell development. Curr Top Dev Biol. 2008;82:217–241. doi: 10.1016/S0070-2153(07)00008-7. [DOI] [PubMed] [Google Scholar]
- 4.Stumpf CR, Ruggero D. The cancerous translation apparatus. Curr Opin Genet Dev. 2011;21:474–483. doi: 10.1016/j.gde.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlachos A, et al. Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood. 2012;119:3815–3819. doi: 10.1182/blood-2011-08-375972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amsterdam A, et al. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henras AK, et al. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strunk BS, Karbstein K. Powering through ribosome assembly. Rna. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phipps KR, et al. The small subunit processome in ribosome biogenesis-progress and prospects. Wiley Interdiscip Rev RNA. 2011;2:1–21. doi: 10.1002/wrna.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci. 2010;35:260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karbstein K. Inside the 40S ribosome assembly machinery. Curr Opin Chem Biol. 2011;15:657–663. doi: 10.1016/j.cbpa.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shajani Z, et al. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 13.Connolly K, Culver G. Deconstructing ribosome construction. Trends Biochem Sci. 2009;34:256–263. doi: 10.1016/j.tibs.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol. Mol. Biol. Rev. 2007;71:477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traub P, Nomura M. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl. Acad. Sci. U. S. A. 1968;59:777–784. doi: 10.1073/pnas.59.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Culver GM, Noller HF. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol. 2000;318:446–460. doi: 10.1016/s0076-6879(00)18069-3. [DOI] [PubMed] [Google Scholar]
- 17.Nierhaus KH, Dohme F. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1974;71:4713–4717. doi: 10.1073/pnas.71.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mangiarotti G, Chiaberge S. Reconstitution of functional eukaryotic ribosomes from Dictyostelium discoideum ribosomal proteins and RNA. J Biol Chem. 1997;272:19682–19687. doi: 10.1074/jbc.272.32.19682. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira-Cerca S, et al. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 20.Ferreira-Cerca S, et al. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 2007;28:446–457. doi: 10.1016/j.molcel.2007.09.029. [DOI] [PubMed] [Google Scholar]
- 21.Strunk BS, et al. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 2011;333:1449–1453. doi: 10.1126/science.1208245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo KY, et al. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J Cell Biol. 2009;186:849–862. doi: 10.1083/jcb.200904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kemmler S, et al. Yvh1 is required for a late maturation step in the 60S biogenesis pathway. J Cell Biol. 2009;186:863–880. doi: 10.1083/jcb.200904111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez-Mateos M, et al. The amino terminal domain from Mrt4 protein can functionally replace the RNA binding domain of the ribosomal P0 protein. Nucleic Acids Res. 2009;37:3514–3521. doi: 10.1093/nar/gkp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Mateos M, et al. Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae. Nucleic Acids Res. 2009;37:7519–7532. doi: 10.1093/nar/gkp806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinnebusch AG, Lorsch JR. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harbor perspectives in biology. 2012:4. doi: 10.1101/cshperspect.a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Granneman S, et al. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. Embo J. 2010;29:2026–2036. doi: 10.1038/emboj.2010.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Passmore LA, et al. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Strunk BS, Novak MN, Young CL, Karbstein K. A Translation-Like Cycle is a Quality Control Checkpoint for Maturing 40S Ribosome Subunits. Cell. 2012 doi: 10.1016/j.cell.2012.04.044. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta PP, et al. Structural aspects of RbfA action during small ribosomal subunit assembly. Mol Cell. 2007;28:434–445. doi: 10.1016/j.molcel.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Z, et al. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat. Struct. Mol. Biol. 2008;15:534–536. doi: 10.1038/nsmb.1408. [DOI] [PubMed] [Google Scholar]
- 32.Boehringer D, et al. Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J Biol Chem. 2012;287:10453–10459. doi: 10.1074/jbc.M111.318121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu C, et al. The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3′ end of 16S rRNA. Proc Natl Acad Sci U S A. 2011;108:10156–10161. doi: 10.1073/pnas.1017679108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu C, et al. Structure of ERA in complex with the 3′ end of 16S rRNA: implications for ribosome biogenesis. Proc Natl Acad Sci U S A. 2009;106:14843–14848. doi: 10.1073/pnas.0904032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sengupta J, et al. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol. 2010;189:1079–1086. doi: 10.1083/jcb.201001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gartmann M, et al. Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J Biol Chem. 2010;285:14848–14851. doi: 10.1074/jbc.C109.096057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klinge S, et al. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334:941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 38.Leger-Silvestre I, et al. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. Embo J. 2004;23:2336–2347. doi: 10.1038/sj.emboj.7600252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zemp I, et al. Distinct cytoplasmic maturation steps of 40S ribosomal subunit precursors require hRio2. J Cell Biol. 2009;185:1167–1180. doi: 10.1083/jcb.200904048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carron C, et al. Analysis of two human pre-ribosomal factors, bystin and hTsr1, highlights differences in evolution of ribosome biogenesis between yeast and mammals. Nucleic Acids Res. 2011;39:280–291. doi: 10.1093/nar/gkq734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi X, et al. Functional role of the sarcin-ricin loop of the 23S rRNA in the elongation cycle of protein synthesis. J Mol Biol. 2012;419:125–138. doi: 10.1016/j.jmb.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hung NJ, et al. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol Biol Cell. 2008;19:735–744. doi: 10.1091/mbc.E07-09-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bradatsch B, et al. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol Cell. 2007;27:767–779. doi: 10.1016/j.molcel.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 44.Greber BJ, et al. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol. 2012;19:1228–1233. doi: 10.1038/nsmb.2425. [DOI] [PubMed] [Google Scholar]
- 45.Bradatsch B, et al. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol. 2012;19:1234–1241. doi: 10.1038/nsmb.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demoinet E, et al. The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae. Rna. 2007;13:1570–1581. doi: 10.1261/rna.585007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer AE, et al. The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60 S subunit factor Arx1. J Biol Chem. 285:961–968. doi: 10.1074/jbc.M109.038349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lygerou Z, et al. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 49.Elela SA, et al. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 50.Lamanna AC, Karbstein K. Nob1 binds the single-stranded cleavage site D at the 3′-end of 18S rRNA with its PIN domain. Proc Natl Acad Sci U S A. 2009;106:14259–14264. doi: 10.1073/pnas.0905403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pertschy B, et al. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem. 2009;284:35079–35091. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lamanna AC, Karbstein K. A Conformational Change Regulates Pre-18S Cleavage. J. Mol. Biol. 2011;405:3–17. doi: 10.1016/j.jmb.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horn DM, et al. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J Biol Chem. 2011;286:34082–34087. doi: 10.1074/jbc.M111.268649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karbstein K, Doudna JA. GTP-dependent formation of a ribonucleoprotein subcomplex required for ribosome biogenesis. J. Mol. Biol. 2006;356:432–443. doi: 10.1016/j.jmb.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 55.Karbstein K, et al. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol. Cell. 2005;20:633–643. doi: 10.1016/j.molcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 56.Gelperin D, et al. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. Rna. 2001;7:1268–1283. doi: 10.1017/s1355838201013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lebaron S, et al. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol. 2012;19:744–753. doi: 10.1038/nsmb.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woolls HA, et al. The Roles of Dim2 in Ribosome Assembly. J. Biol. Chem. 2011;286:2578–2586. doi: 10.1074/jbc.M110.191494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunbar DA, et al. A nucleolar protein related to ribosomal protein L7 is required for an early step in large ribosomal subunit biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13027–13032. doi: 10.1073/pnas.97.24.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pestov DG, et al. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 2001;29:3621–3630. doi: 10.1093/nar/29.17.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adams CC, et al. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. Rna. 2002;8:150–165. doi: 10.1017/s1355838202010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gadal O, et al. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J. Cell Biol. 2002;157:941–951. doi: 10.1083/jcb.200111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oeffinger M, et al. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. Rna. 2002;8:626–636. doi: 10.1017/s1355838202020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oeffinger M, Tollervey D. Yeast Nop15p is an RNA-binding protein required for pre-rRNA processing and cytokinesis. Embo J. 2003;22:6573–6583. doi: 10.1093/emboj/cdg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Horsey EW, et al. Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. Rna. 2004;10:813–827. doi: 10.1261/rna.5255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miles TD, et al. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sahasranaman A, et al. Assembly of Saccharomyces cerevisiae 60S ribosomal subunits: role of factors required for 27S pre-rRNA processing. Embo J. 2011;30:4020–4032. doi: 10.1038/emboj.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Granneman S, et al. A cluster of ribosome synthesis factors regulate pre-rRNA folding and 5.8S rRNA maturation by the Rat1 exonuclease. Embo J. 2011;30:4006–4019. doi: 10.1038/emboj.2011.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khoshnevis S, et al. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep. 2010;11:214–219. doi: 10.1038/embor.2009.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shoemaker CJ, et al. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pisareva VP, et al. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. Embo J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Becker T, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Becker T, et al. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- 75.Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ceccarelli A, et al. Immature 50 S subunits in Escherichia coli polyribosomes. FEBS Lett. 1978;93:348–350. doi: 10.1016/0014-5793(78)81137-5. [DOI] [PubMed] [Google Scholar]
- 77.Turco E, et al. Ribosome biosynthesis in Escherichia coli. Concerning the limiting step. Biochemistry. 1974;13:4752–4757. doi: 10.1021/bi00720a011. [DOI] [PubMed] [Google Scholar]
- 78.Mangiarotti G, et al. Precursor 16S RNA in active 30S ribosomes. Nature. 1974;247:147–148. doi: 10.1038/247147a0. [DOI] [PubMed] [Google Scholar]
- 79.Mangiarotti G, et al. rRNA maturation as a “quality” control step in ribosomal subunit assembly in Dictyostelium discoideum. J Biol Chem. 1997;272:27818–27822. doi: 10.1074/jbc.272.44.27818. [DOI] [PubMed] [Google Scholar]
- 80.Becam AM, et al. Ria1p (Ynl163c), a protein similar to elongation factors 2, is involved in the biogenesis of the 60S subunit of the ribosome in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2001;266:454–462. doi: 10.1007/s004380100548. [DOI] [PubMed] [Google Scholar]
- 81.Senger B, et al. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 82.Menne TF, et al. The Shwachman-Bodian-Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet. 2007;39:486–495. doi: 10.1038/ng1994. [DOI] [PubMed] [Google Scholar]
- 83.Lo KY, et al. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Savchenko A, et al. The Shwachman-Bodian-Diamond syndrome protein family is involved in RNA metabolism. J Biol Chem. 2005;280:19213–19220. doi: 10.1074/jbc.M414421200. [DOI] [PubMed] [Google Scholar]
- 85.Shammas C, et al. Structural and mutational analysis of the SBDS protein family. Insight into the leukemia-associated Shwachman-Diamond Syndrome. J Biol Chem. 2005;280:19221–19229. doi: 10.1074/jbc.M414656200. [DOI] [PubMed] [Google Scholar]
- 86.Bussiere C, et al. Integrity of the P-site is probed during maturation of the 60S ribosomal subunit. J Cell Biol. 2012;197:747–759. doi: 10.1083/jcb.201112131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Blombach F, et al. Assembling the archaeal ribosome: roles for translation-factor-related GTPases. Biochem Soc Trans. 2011;39:45–50. doi: 10.1042/BST0390045. [DOI] [PubMed] [Google Scholar]
- 88.Lafontaine DL. A ‘garbage can’ for ribosomes: how eukaryotes degrade their ribosomes. Trends Biochem Sci. 2010;35:267–277. doi: 10.1016/j.tibs.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 89.Cole SE, et al. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell. 2009;34:440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li Z, et al. Rational extension of the ribosome biogenesis pathway using network-guided genetics. PLoS Biol. 2009;7:e1000213. doi: 10.1371/journal.pbio.1000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Granneman S, et al. The putative NTPase Fap7 mediates cytoplasmic 20S pre-rRNA processing through a direct interaction with Rps14. Mol. Cell Biol. 2005;25:10352–10364. doi: 10.1128/MCB.25.23.10352-10364.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 93.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 94.Vanacova S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wyers F, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 96.Schmidt K, et al. Air proteins control differential TRAMP substrate specificity for nuclear RNA surveillance. Rna. 2012;18:1934–1945. doi: 10.1261/rna.033431.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.San Paolo S, et al. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet. 2009;5:e1000555. doi: 10.1371/journal.pgen.1000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Karbstein K. Ribosome Assembly, Eukaryotic. In: Begley T, editor. Wiley Encyclopedia of Chemical Biology. John Wiley & Sons; 2009. pp. 222–230. [Google Scholar]
- 99.Houseley J, et al. RNA-quality control by the exosome. Nature reviews. Molecular cell biology. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 100.Rabl J, Leibundgut M, Ataide S, Haag A, Ban N. Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 101.Yu Y, et al. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Voorhees RM, et al. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol. 2009;16:528–533. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Siridechadilok B, et al. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 104.Ben-Shem A, et al. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]