Summary
RNA polymerase II (Pol II) transcribes hundreds of kilobases of DNA, limiting the production of mRNAs and lncRNAs. We used Global Run-on Sequencing (GRO-seq) to measure the rates of transcription by Pol II following gene activation. Elongation rates vary as much as 4-fold at different genomic loci and in response to two distinct cellular signaling pathways [i.e., 17β-estradiol (E2) and TNFα]. The rates are slowest near the promoter and increase during the first ~15 kb transcribed. Gene body elongation rates correlate with Pol II density, resulting in systematically higher rates of transcript production at genes with higher Pol II density. Pol II dynamics following short inductions indicate that E2 stimulates gene expression by increasing Pol II initiation, whereas TNFα reduces Pol II residence time at pause sites. Collectively, our results identify previously uncharacterized variation in the rate of transcription and highlight elongation as an important, variable, and regulated rate-limiting step during transcription.
Keywords: Elongation, Estrogen, Estrogen receptor (ERα), Gene regulation, GRO-seq, Nuclear factor kappa B (NF-κB), Rate, RNA Polymerase II, Signal-regulated transcription, Transcription, Tumor necrosis factor alpha (TNFα)
Introduction
The production of mRNAs by transcription is a complex and tightly regulated biological process consisting of discrete stages of RNA Polymerase II (Pol II) recruitment, initiation, elongation, and termination. During transcription elongation, Pol II tracks through gene bodies, extending an elongating, nascent RNA polymer in its wake. In humans and other metazoans, genes can be many hundreds of kilobases in size, and consequently the rate of Pol II transcription is the primary factor limiting the rate of mRNA production.
While efficient transcription elongation is required for mRNA production, it has not historically been regarded as a widely used step in gene regulation. However, studies published over the past five years have challenged this view. In particular, promoter proximal ‘pausing’ of transcriptionally engaged, early elongating Pol II ~20 to 60 nucleotides from the transcription start site (TSS) is now widely accepted as an important regulatory step (reviewed by (Fuda et al., 2009). In addition, many features of RNA processing are tightly coupled with transcription elongation dynamics. Furthermore, considerable evidence now supports a direct role for elongation rates in the regulation of alternative splicing of mRNA transcripts (Nogues et al., 2002). Pol II rates are also observed to slow near polyadenylation sites, presumably to facilitate mRNA cleavage (Core et al., 2008), and regulate alternative polyadenylation (Pinto et al., 2011). Together these observations have resulted in intense interest in understanding the kinetics and regulation of Pol II elongation.
In spite of considerable recent interest, the rate of transcription by Pol II in cells remains poorly characterized. Published estimates differ by as much as 6-fold, ranging from ~1 to 6 kilobases per minute (kb/min) (reviewed by (Ardehali and Lis, 2009). Some of this variation likely reflects bona fide biological differences. However, to date, studies that have systematically measured transcription rates on multiple genes in side-by-side experiments have not observed evidence of systematic variation that could account for the differences reported in the literature (Singh and Padgett, 2009; Wada et al., 2009). Further studies are required to sort out the potential biological or technical reasons that underlie the observed variation in Pol transcription rates, as well as the potential implications for the cellular regulatory processes noted above.
A number of different approaches for measuring Pol II elongation rates have been used. Single-molecule techniques are outstanding in their temporal and basepair-level resolution, but are labor intensive and do not scale well beyond a few genes. Alternatively, several cell-based assays have been used to determine Pol II elongation rates by monitoring the accumulation of nascent RNA or Pol II using qPCR or tiling arrays for small sets of native genes (Singh and Padgett, 2009; Wada et al., 2009), or imaging techniques for a transgene (Darzacq et al., 2007). Some of these cell-based approaches provide reasonably detailed spatial resolution across gene bodies and are modestly scalable, yet have found little variation in the transcription rate between different genes in the same cells (Singh and Padgett, 2009; Wada et al., 2009). Many unanswered questions, such as the kinetics of Pol II during co-transcriptional splicing, differences in the rate of release of paused Pol II across different genes, and variability in Pol II elongation rates across the genome, require genome-scale observations.
Global Run on and Sequencing (GRO-seq) is a high-resolution, high-sensitivity assay useful for determining the location and orientation of actively transcribing Pol II genome wide (Core et al., 2008). GRO-seq provides a clear, dynamic picture of the instantaneous levels of Pol II transcription with outstanding temporal resolution. Indeed, we have recently observed that the initial ‘wave’ of Pol II transcription can be clearly resolved as changes in Pol II move across gene bodies following activation by 17β-estradiol (E2) (Hah et al., 2011). Here, we developed an approach to measure transcription rates by following the progression of this Pol II ‘wave’ after short treatments with physiological, non-toxic inducers (ligands). Our approach allowed us to measure Pol II rates at long genes whose expression increases significantly following induction (in our case, 166 human genes). This study represents the most comprehensive and detailed picture of both gene-body and promoter-proximal transcription kinetics to date, and serves as a template for future studies measuring transcription elongation.
Results
Determining Rates of Transcription by RNA Pol II
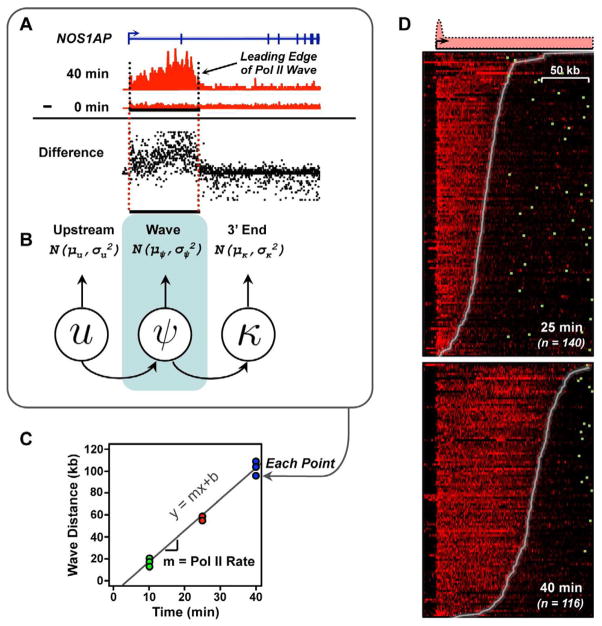
To determine the rate of transcription elongation by Pol II in mammalian cells, we used GRO-seq (Core et al., 2008; Hah et al., 2011) to monitor the global localization of Pol II following a short time course of E2 treatment (0, 10, 25, and 40 min) in MCF-7 human breast cancer cells. The “leading edge” of the Pol II, which is clearly visible in our GRO-seq data (Fig. 1A), indicates the distance Pol II has traveled following induction for a specified time and allows calculation of the rate (distance/time). To automate the identification of the leading edge of the Pol II wave, we developed a statistical inference procedure based on a three-state hidden Markov model (Fig. 1, A–C), which divides each gene into three regions: (1) upstream of the transcription start site (TSS), (2) the Pol II wave, and (3) the 3′ end of the gene downstream of the wave (Fig. 1, A and B). A key component of our pipeline is the use of “difference maps” (Fig. 1A, see “Difference”), which allow robust detection of true signals above background in a quantitative manner and dramatically reduce errors in wave identification. To control for technical errors and biological variation, we fit the wave end separately in two (25 min.) to five (0, 10, and 40 min.) independent biological replicates at each time point, and calculated the elongation rate by linear regression (Fig. 1C). Our statistical approach facilitates the systematic discovery of wave ends over the time course of induction in our experiments and is clearly illustrated in heatmaps over a large population of genes (Fig. 1D).
Figure 1. An approach for measuring rates of Pol II transcription using GRO-seq data.
(A and B) The Pol II wave was identified in GRO-seq data using a three state HMM applied to difference maps in each biological replicate.
(C) Pol II elongation rates were fit using linear regression.
(D) GRO-seq difference maps in the 25 and 40 min. E2 treatment conditions, versus untreated MCF-7 cells. Genes are ordered using the Pol II wave distance in each time point. Different numbers of genes reflect waves traveling past the end of the gene following 40 min. of treatment. Genes are aligned on the transcription start site; polyadenylation sites are denoted by green boxes.
To systematically evaluate the accuracy of our inference procedure, we conducted a series of validation studies, using bootstrapped, simulated data, and by comparing wave ends between individual biological replicates. First, we used bootstrapping methods to construct a large number of ‘simulated’ waves that behave in a similar manner to real data. Applying our full inference pipeline on the simulated dataset suggests that our approach can recover both wave ends (Supplemental Fig. S1, A and B) and elongation rates (Supplemental Fig. S1, C and D) with very high accuracy (R > 0.95). Using this approach, we estimate our median absolute error rate is only 0.064 kb/min. (that is, an error rate of 64 bp over the ~2000 bp traveled, on average, per minute).
This estimate of error evaluates only our wave end inference approach, and does not test biological variation. To obtain an independent ‘upper bound’ of our error, we next compared wave end (Supplemental Fig. S1, E – G) and rate measurements (Supplemental Fig. S1H) across separate, independent biological replicates. This analysis revealed a significant correlation in both wave end position (ρ = 0.83–0.92) and elongation rates (ρ = 0.74), and translates into an upper estimate of median absolute error of less than 0.312 kb/min. We note that our actual accuracy is likely to be even better than this estimate, because accuracy scales with the square root of the number of samples using regression methods.
In addition, we also conducted several additional gene-specific and dataset-wide validations. In particular, we note that visual inspection of ‘meta’-representations (Supplemental Fig. S1, I – K), and individual traces (Fig. 2A) of genes typically have a clean, clearly defined wave end position that is accurately inferred using our statistical pipeline. Finally, to rigorously evaluate the dependence of all results presented below on our methods for choosing genes and inferring elongation rates, we have repeated all analysis with two distinct sets of genes, and found that they produce qualitatively identical results (Supplemental Fig. S1L). We conclude that our approach constitutes a careful, unbiased and systematic pipeline useful for measuring rates of transcription in living cells.
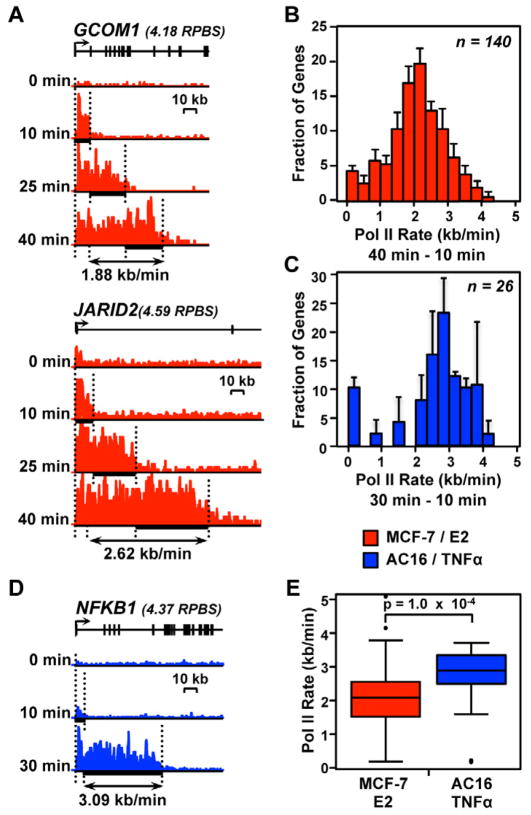
Figure 2. Pol II transcription rates vary across the genome and between different biological systems and inducers.
(A and D) Pol II density following a time course of treatment with inducer. (A) Red, two genes responding to E2 treatment in MCF-7 cells. (B) Blue, a gene responding to TNFα treatment in AC16 cells. Different time points are shown at the same Y-axis scale for each gene. Y-axes vary between genes based on their expression level, which are shown in units of reads per base scaled (RPBS) above each gene.
(B and C) Histograms of Pol II elongation rates for 140 genes in MCF-7 cells (B) or 26 genes in AC16 cells (C) for which high confidence elongation rates could be determined. Error bars represent standard error of the mean between biological replicates.
(E) Comparison of elongation rates between MCF-7 and AC16 cells. The p-value was calculated using a two-sided Wilcoxon rank sum test.
Comparing Rates of Transcription in MCF-7 and AC16 Cells
We used our approach to measure elongation rates for 140 native genes that were up-regulated by E2 in MCF-7 cells (Figs. 1D, 2A, and 2B; Supplemental Fig. S2A). Elongation rates were determined over the entire time course (10 to 40 min.) for 81 genes. We also added an additional 59 genes for which we could infer elongation rates in only the 10 to 25 or 25 to 40 min. time intervals, either because the genes are too short to provide a high-confidence wave at 40 min. (i.e., Pol II has reached the end of the gene by that time point; <120 kb, n = 28), or because they do not have a high confidence wave beginning before 10 min. of E2 treatment due to a delay in gene activation (n = 31). Interestingly, this analysis revealed that Pol II rate varies between different genomic regions. For example, we observed a rate of 1.88 kb/min. for GCOM1 and 2.62 kb/min. for JARID2 (Fig. 2A). For all 140 genes, we found that the rates ranged between 0.37 and 3.57 kb/min for the middle 90% of genes, with a median rate of 2.1 kb/ min. (Fig. 2B). This observed variation is much greater than our estimates of wave inference error (0.06–0.31 kb/min.), indicating that the observed variation is biologically relevant. Our results indicate that elongation rates are often much lower than the theoretical maximum ‘burst’ rate for Pol II (4.3 kb/min) and are consistent with reports that pausing leads to significant, local variations in average elongation rates in mammalian cells (Darzacq et al., 2007).
Our measurements in MCF-7 cells reflect a variation of more than two-fold in the Pol II elongation rate for different genomic loci, yet most were on the lower end of the reported range (~1 to 6 kb/min) (Ardehali and Lis, 2009). We postulated that faster rates are possible in different cell types or in response to different cellular signaling pathways, consistent with reports that rapidly acting stimuli can elicit faster rates than we measure here (Wada et al., 2009). To test this hypothesis, we stimulated AC16 cardiomyocytes with TNFα (0, 10, and 30 min), a rapidly acting cytokine that elicits acute responses, and determined Pol II elongation rates by GRO-seq as described above. We observed a median rate of 2.80 kb/min. for 26 genes (Fig. 2C; e.g., NFKB1 was transcribed at a rate of 3.09 kb/min, Fig. 2D and Supplemental Fig. S2B), almost 50% faster than the median rate in MCF-7 cells (Fig. 2E; p = 1.0 × 10−4; two-sided Wilcoxon rank sum test). Moreover, for two genes in common between the MCF-7 and AC16 systems, KIAA1199 and SPSB1, the rate of Pol II transcription was ~25 to 40% faster in AC16 cells compared to MCF-7 cells (Supplemental Fig. S2C), suggesting that DNA sequence is not the primary factor underlying the observed differences in elongation rate. These results demonstrate that not only can Pol II elongation rates vary between different genes, but they can also vary at the same gene in different cell types or in response to different signaling contexts.
Comparing Pol II Elongation Rates During Early and Late Transcription
We tested whether gene body elongation rates vary between different regions within the same gene. To address this question, we compared elongation rates during the 10 to 25 and 25 to 40 min. intervals in MCF-7 cells. We observed a systematic increase in rates during the second (25 to 40 min.) interval (Fig. 3A; p = 2.8 × 10−12, two-sided Wilcoxon rank sum test). The increase was observed to have a median magnitude of 1.05 kb/ min (range 0.01 and 1.9 kb/ min for the middle 75% of genes; see Supplemental Fig. S3A). There are two potential hypotheses to explain this result, including: (1) a universal increase in Pol II rate at all genes as Pol II elongates further into gene bodies or (2) the effects of sustained activation on elongation rates at E2-responsive genes later in the time course.
Figure 3. Pol II accelerates into gene bodies.
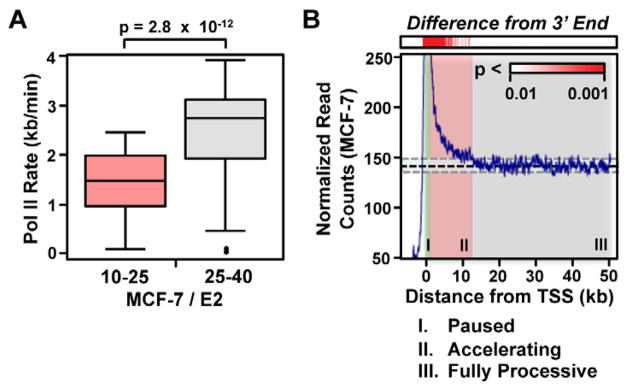
(A) Boxplots comparing elongation rates in the 10 to 25 and 25 to 40 min. windows. A two-sided Wilcoxon rank sum test was used to test significance.
(B) Metagene analysis showing steady-state Pol II density in resting MCF-7 cells at genes longer than 50 kb (n = 3,384). The red scale (top) shows the p-value of each window relative to a standardized distribution in the region between 30 to 50 kb downstream of the transcription start site.
If the observed increases in transcription rate at later time points are caused by the sustained effects of E2 treatment, we should observe uniform transcription rates in the untreated conditions. Therefore, we determined whether a similar increase in rate is observed prior to treatment with inducer (i.e., E2 or TNFα). To this end, we carefully examined the distribution of GRO-seq read density along the body of longer genes (>50 kb) using GRO-seq data sets from untreated cells. We found that read density dropped slowly from its maximum value at the pause peak over ~10 to 15 kb at the 5′ end of genes in both MCF-7 and AC16 data sets (Fig. 3B, Supplemental Fig. S3B). This 15 kb window overlaps the region in which the Pol II wave is currently undergoing transcription at ~75% of genes analyzed in the 10 to 25 min. interval, suggesting that the same biological process explains both results. Therefore, the simplest explanation for these observations is an elevated read density due to an increased residence time for Pol II in this region (i.e. slower transcription rates, on average, at the 5′ end of genes), although we cannot eliminate the possibility that premature termination contributes to this profile as well. Moreover, the average rates of transcription do not differ systematically for genes showing maximal induction at 10 min. versus 40 min. (Supplemental Fig. S3C), further suggesting that our results do not reflect effects of sustained activation. Together, these results strongly suggest that the average Pol II transcription rates increase systematically during transcription in the first ~10 to 15 kb of the gene body for most genes in human cells.
Mechanistic Basis for Variation in Pol II Elongation Rates
The variation in Pol II elongation rates between different genes could have several mechanistic explanations. First, structural differences in chromatin could influence elongation rates. For example, preexisting nucleosome occupancy (Deal et al., 2010; Dion et al., 2007), post-translational modifications (Zhou et al., 2011), or polymerase density (Epshtein and Nudler, 2003; Saeki and Svejstrup, 2009) could lead to gene-specific differences in elongation rate. All of these features have been shown to correlate with gene expression and, thus, we asked whether elongation rates correlate with gene expression levels. Indeed, we found a striking correlation between elongation rate and the gene body density of Pol II (Fig. 4A; ρ = 0.46; p = 4.5 × 10−10), as well as statistically significant correlations with several histone marks related to gene activation (Roh et al., 2006), such as promoter H3K4me3 and gene body H3K36me3 (Supplemental Fig. S4A). While we computed gene expression using the untreated condition to avoid correlations driven by technical factors, similar results were also obtained using gene expression levels from treated cells. Subsampling experiments indicate that the former observation does not reflect a density-related bias in our inference of wave position (Supplemental Fig. S4, B – D). The observed correlation is highly significant in both cell systems separately (MCF-7: ρ = 0.30, AC16: ρ = 0.61; p < 1.2 × 10−3) and is extremely strong during the first 25 to 30 min. of treatment with inducer (combined MCF-7 and AC16: ρ = 0.60; p = 6.3 × 10−9), suggesting a possible relationship to the ‘acceleration’ phase of gene transcription.
Figure 4. Elongation rates correlate with Pol II density.
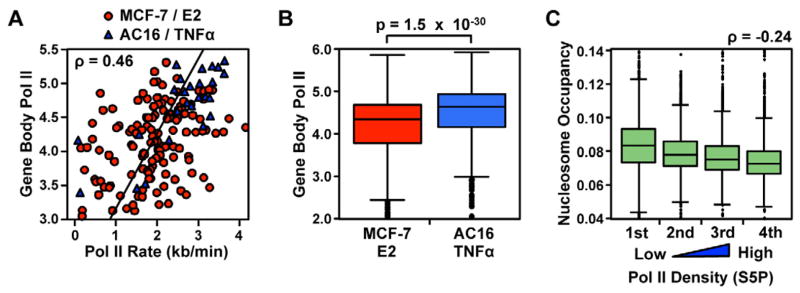
(A) Scatter plot showing Pol II rates as a function of gene expression levels, in RPBS. Red points show 140 genes in MCF-7 cells and blue points show 26 genes in AC16 cells.
(B) Expression levels of E2 target genes in MCF-7 cells, compared with TNFα target genes in AC16 cells. Significance was tested using a two-sided Wilcoxon rank sum test.
(C) Nucleosome occupancy in human T-cells, as determined by MNase digestion, presented in quartiles of gene expression level.
Pol II density also largely explains the differences observed between rates in the MCF-7/E2 and AC16/TNFα systems, with slower rates and less density (i.e., less transcription) at E2 target genes compared to TNFα target genes. In this regard, note that TNFα signaling targets a subset of genes with higher basal levels of Pol II density (p = 1.5 × 10−30; Fig. 4C), corresponding with faster rates of transcription than are observed in MCF-7 cells. We propose that this systemic difference in the initial transcriptional activity of target genes reflects differences in the mechanistic basis by which E2 and TNFα activate transcription (explored in detail below).
Chromatin represents one of the most important obstacles to transcription elongation in the gene body (Izban and Luse, 1992). Prior studies in yeast (Dion et al., 2007) and Drosophila (Deal et al., 2010) have shown that gene body nucleosome densities correlate negatively with gene expression levels. This suggests that the nucleosome barrier may be systematically lower at genes with higher expression levels, explaining the faster rates of transcription that we observe here. We examined the strength of this negative correlation in higher eukaryotes using existing nucleosome occupancy data from human T-cells (Schones et al., 2008). We observed a weak inverse correlation between nucleosome occupancy and the density of Pol II (CD4+ T-cells; ρ = −0.24; p < 0.01; Fig. 4C), consistent with chromatin occupancy contributing to variation in the Pol II transcription rate. However, the correlation is surprisingly weak and is characterized by significant overlap in nucleosome densities between genes in different quartiles of expression (compare whiskers in Fig. 4C). This suggests that other factors, such as the histone modifications noted above or chromatin-independent factors, may contribute to variation in transcription rate as well.
We also asked whether elongation rates are affected by co-transcriptional splicing. We found no significant difference in the median elongation rate for Pol II transcribing through intron-exon junctions during the first 40 minutes, compared to Pol II that does not (Supplemental Fig. S5A). A careful analysis of how our statistical power is influenced by pause duration suggests that we can reliably detect pauses at exons that are as short as 90 seconds (Supplemental Fig. S5B). These findings suggest that Pol II continues to transcribe during splicing, but leaves open the possibility that short transcriptional pauses facilitate 3′ splice site recognition and splicing complex assembly.
Consequences of Rate Variation: Effects on mRNA Production
Next, we considered whether variations in transcription rate will have downstream consequences on the rate and dynamics of mRNA production. Intuitively, the observation that elongation rates are faster at genes with higher Pol II density suggests that mRNA production will be systematically faster as well, more than can be accounted for by variations in Pol II density alone. To explore this consequence systematically, we derived a model that relates Pol II density to mRNA levels. We assumed that changes in mRNA over time are equal to the rate of mRNA production by transcription, minus the rate of mRNA decay, which can be expressed as:
Here, [R] represents the level of mRNA, [G] is the density of Pol II in the gene body, v is the degradation rate of mRNA, and r is the elongation rate. For mathematical simplicity, we focused on modeling steady-state conditions, during which the change in mRNA (d[R]/dt) is assumed to be 0.
To determine how the observed rate variation affects the relationship, we substituted r for the empirical relationship between elongation rate and Pol II density (see Experimental Procedures for details). This yields a model that describes mRNA levels in relation to Pol II levels, which accounts for the observed, systematic variations in elongation rate.
To evaluate how accurately our model accounts for ongoing processes occurring in living MCF-7 cells, we compared steady-state levels of mRNA before induction with Pol II gene body density. We found a nonlinear relationship between Pol II density and mRNA levels (Fig. 5A, red line denotes the best fit by LOESS), consistent with the intuition that variation in elongation rate systematically affects the rate of mRNA production. Indeed, comparison of this empirical relationship to our model of mRNA production (green line), derived above, indicates a strong match between our model and the empirical data. In addition, this analysis also suggests that the observed relationship between gene expression level and elongation rate occurs under steady-state conditions, prior to gene activation (i.e. used to measure mRNA and Pol II densities) because such a strong fit is unlikely to occur unless a very similar relationship holds.
Figure 5. Variation in Pol II elongation rate impacts mRNA production.
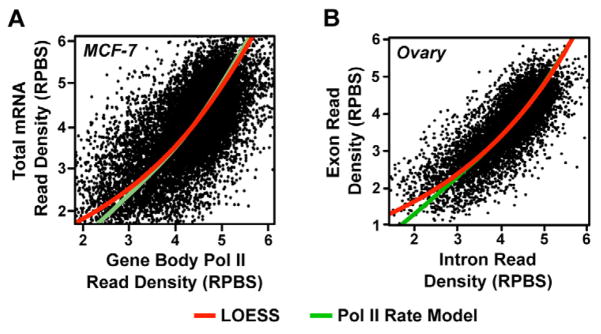
(A and B) Comparison of primary transcription using GRO-seq data (A) or intron reads from non-polyA selected mRNA-seq data (B) to mRNA levels in the indicated cell type. Red lines reflect the best fit using LOESS local regression. Green lines reflect the expected relationship, accounting for Pol II elongation rate variation.
We also sought to address whether technical differences between GRO-seq and mRNA-seq (e.g., sensitivity and dynamic range) could be responsible for the observed relationship and shape of the curve. To address this, we analyzed existing RNA-seq data sets generated from ribosomal RNA-depleted total RNA. We assumed that read density mapping in introns is proportional to the density of primary transcription on the gene (Supplemental Fig. S5C). Reads in exons primarily reflect stable, steady-state mRNA, with some component of primary transcription that can be removed by subtracting intronic read densities. Under these assumptions, these mRNA-seq data contain measurements of both primary transcription and steady-state mRNA in the same cell population, taken using a single extraction procedure. These RNA-seq data reveal a qualitatively similar relationship to what we observed for our GRO-seq data (Fig. 5, B and Supplemental Fig. S5D), which fits very well with our model accounting for systematic variation between elongation rate and Pol II density.
Finally, an alternative formulation of the relationship between mRNA and Pol II, which makes no assumptions about the functional form of the relationship (Mayer et al., 2010), is also consistent with the hypothesis that systematic variation in elongation rate affects mRNA levels (Supplemental Fig. S5E). We conclude that variation in elongation rate has substantial consequences for the rate of mRNA production at different genes and exerts a measurable effect on mRNA expression levels.
The Effects of E2 on Early Transcription Dynamics are Explained by Pol II Pausing and the Recruitment of NELF
Gene expression is regulated at multiple rate-limiting steps early during transcription, including the opening of chromatin, Pol II recruitment, transcription initiation, and the residence time for Pol II at the 5′ end of genes (known as promoter-proximal pausing (Fuda et al., 2009)). We sought to understand which rate-limiting step is targeted by the transcription factors transducing the E2 and TNFα signals [estrogen receptor alpha (ERα) and NF-κB, respectively] by analyzing the distance traveled by the Pol II wave following short (10 min.) treatments using the statistical methods described above for determining Pol II wave ends (Supplemental Fig. S6, A and B). Specifically, we were interested in asking whether ERα and NF-κB regulate gene expression by improving the efficiency of transcription through promoter-proximal pause sites, or rate-limiting steps earlier in transcription (e.g. promoter accessibility, preinitiation complex formation, or initiation).
We reasoned that three factors may influence the distance Pol II travels following short treatments with inducer, including: (1) a delay between the addition of the inducer to the culture medium and the activation of each gene, (2) the time Pol II spends paused near the promoter, and (3) the rate of transcription during productive elongation (Supplemental Fig. S6C). We assume that newly initiated Pol II will encounter an additional rate-limiting step (i.e. pausing) before beginning productive transcription. Notably, under this assumption the residence time of Pol II in the pause site will affect wave distance if the transcription factor activates gene expression at a step prior to Pol II pausing. Therefore, to determine whether ERα or NF-κB regulate gene expression by improving efficiency of transcription through pause sites or an earlier step, we evaluated whether the occurrence and degree of pausing influences the distance traveled following 10 min. of treatment with inducer.
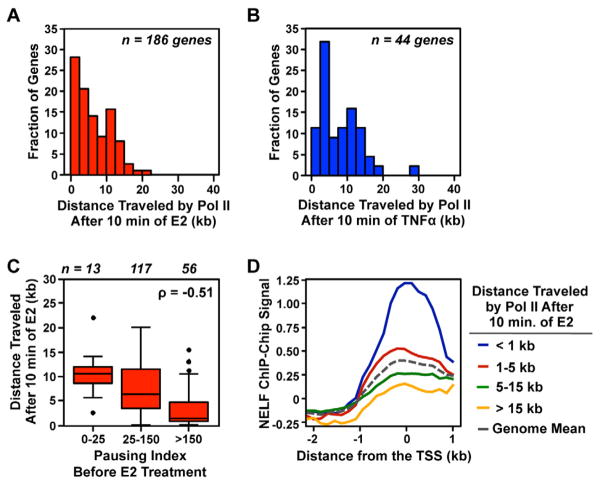
To address this possibility, we measured the distance Pol II travels following 10 min. of treatment. Our focus on the 10 min. time point allowed us to use a shorter minimum gene length (50 kb) than outlined above and thus increased the number of genes available for analysis (MCF-7, n = 186; AC16, n = 44). We found that at the majority of genes, Pol II travels between ~0 and 20 kb into the gene body following a 10 min. treatment with E2 or TNFα (Fig. 6, A and B). This represents a much shorter distance than expected based on the observed elongation rates in the gene body, consistent with the effects of either a systematic delay, or Pol II pausing, acting to reduce transcribed distances in both systems.
Figure 6. Pausing influences Pol II wave distance following a 10 min. treatment with E2, but not TNFα.
(A and B) Histograms show the distance travelled by Pol II following a 10 min. treatment with E2 (A) or TNFα (B).
(C) The pausing index of untreated MCF-7 cells compared to the distance traveled by Pol II following a 10 min. treatment with E2.
(D) Localization of NELF near the transcription start site of genes moving the distance indicated following 10 min. of E2.
The pausing index is a measurement of residence time for Pol II in a paused state, and directly reflects the time that Pol II spends paused at each gene. Therefore, if these slower rates are caused by Pol II becoming transiently “stuck” in a paused state, we expected to see a negative correlation between the pausing index and Pol II distance traveled following 10 min. of treatment with inducer. Indeed, we found a highly significant negative correlation between the distance traveled by the Pol II wave following E2 treatment and the pausing index, wherein genes with a high pausing index travel a significantly shorter distance compared to genes with a lower pausing index (ρ = −0.51; p = 7.0 × 10−14; Fig. 6C). Conversely, only a weak negative correlation was observed following TNFα treatment (ρ = −0.28; p = 0.06).
These correlations are potentially influenced by elongation rates after Pol II enters fully productive transcription. Although we are not able to measure elongation rates during the first 10 min. due to confounding factors (e.g. differences in the stringency of gene-length filters, slower transcription rates at the 5′ ends of genes, etc.), we can directly measure a correlated quantity: gene expression level. To determine whether the negative correlation (reported above; Fig. 6C) is caused by a delay at the pause site, we used a semi-partial correlation approach to statistically remove the effects of gene expression level from wave distance following 10 min. of ligand treatment. This analysis showed that the effects of pausing index on distance at 10 min. cannot be explained be gene expression level at E2-regulated genes (ρ = −0.44; p = 6.6 × 10−10; Supplemental Fig. S6D). Conversely, in the AC16 system, where pausing index was observed to have only a relatively weak negative correlation with distance following 10 min., this relationship is almost entirely explained by the effects of expression level (ρ = −0.11; p = 0.48; Supplemental Fig. S6D). This strongly suggests that the weak negative correlation between pausing index and Pol II distance travelled at TNFα regulated genes is an indirect effects of confounding variables (most likely elongation rate during active transcription). Together, these results suggest that NF-κB works by releasing paused Pol II at TNFα target genes, whereas ERα stimulates transcription at a prior rate-limiting step.
If the shorter distances traveled by Pol II following 10 min. of E2 treatment are a direct result of pausing, we also expected to find an enrichment of pausing-related factors at the promoter of these genes. To determine if this is so, we monitored the localization of subunit A of the negative elongation factor (NELF) complex, which is partially responsible for establishing a stable pause (Yamaguchi et al., 1999). We used available ChIP-Chip data (Kininis et al., 2009) to evaluate the relationship between NELF deposition and the distance traveled following 10 min. of E2. Indeed, we found that genes for which Pol II traveled very short distances (e.g. <1 kb) following 10 min. of E2 had an accumulation of NELF at their promoter relative to the genome mean, whereas genes for which Pol II traveled longer distances (e.g. >15 kb) showed a depletion of NELF (Fig. 6D). This result strongly suggests that a NELF-dependent decrease in Pol II distance following 10 min. of E2 slows the initial E2-dependent response, as newly added Pol IIs are delayed from entering the gene-body.
Inefficient Transcription by Newly Recruited Pol II Through Promoter-Proximal Pause Sites at E2, but not TNFα, Target Genes
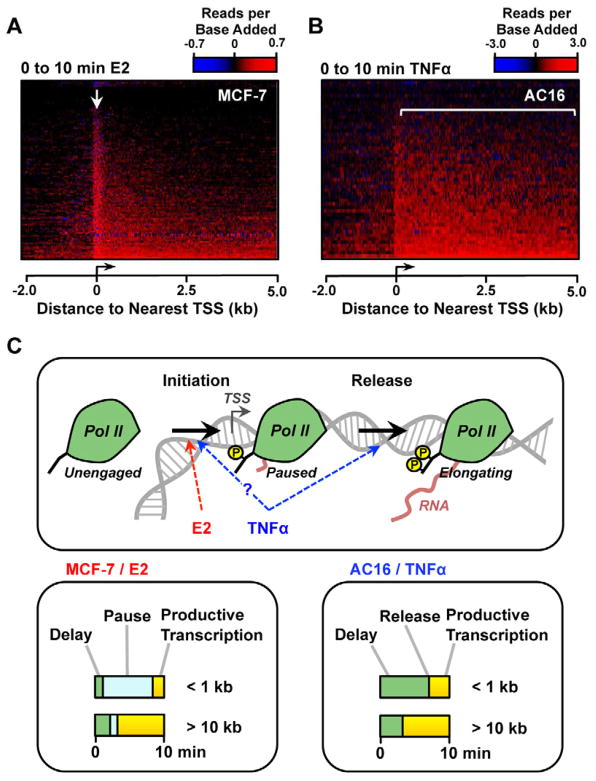
In the analysis described above, we demonstrated that for a typical induced gene, Pol II recruited to the promoter in response to E2, but not TNFα, transcribes inefficiently through the promoter proximal pause site before entering the gene body. This conclusion was based on results from a set of longer genes, for which we could identify a Pol II wave end following 10 min. of treatment with inducer. We next sought to demonstrate that these observations extrapolate to the remaining target genes which were not captured in this initial analysis. To this end, we systematically evaluated how newly recruited Pol II is distributed on all genes up-regulated by E2 and TNFα. In this analysis, we assume that Pol II recruited at each gene is proportional to the normalized read density added between 0 and 10 min. of treatment. Heatmaps of newly recruited Pol II show a strong tendency for Pol II to localize near the promoter at E2 responsive genes (Fig. 7A; see arrow), consistent with Pol II having a long residence time at the pause site following E2 treatment. Conversely, Pol II recruited following TNFα enters almost immediately into the gene body (Fig. 7B; see bracket). Comparing the relative gene activation times between treatment systems, by converting residuals from a linear model of Pol II distance to units of minutes, does not reveal any systematic differences that could provide an alternative explanation for these observations (Supplemental Fig. S7A). Finally, we noted that TNFα targets tend to have universally higher pausing indices than E2 targets (Supplemental Fig. S7B), consistent with the idea that TNFα requires paused Pol II to activate transcription.
Figure 7. Tracking the location of Pol II added following treatment with inducer suggests distinct modes for E2- and TNFα-dependent gene activation.
(A and B) Heatmaps show the amount and location of Pol II recruited during the first 10 min. of treatment with either E2 (A, n =1,712) or TNFα (B, n =732).
(C) Model depicting two distinct rate-limiting steps early in the transcription process (initiation and release from pause). At E2 target genes, newly recruited Pol II is associated with high residence times at the pause site (bottom, left), supporting a model in which E2 stimulates transcription by increasing the rate of initiation, or earlier rate-limiting steps. Pol II recruited by TNFα is not associated with a transient pause (bottom, right), suggesting that TNFα increases transcription efficiency through the pause site.
Together, these results are consistent with our observation (noted above) that Pol II recruited to E2, but not TNFα, target gene promoters transcribe inefficiently through the pause site. This finding is consistent with a model in which NF-κB activates transcription, at least partially, by improving Pol II transcription efficiency through the pause site, whereas ERα activates transcription at a prior rate-limiting step (Fig. 7C).
Discussion
We have used robust biological systems in conjunction with state-of-the art genomic and computational approaches to address fundamental questions about the transcriptional activation of signal-regulated genes, in particular the rate of transcription by Pol II under different conditions. By combining GRO-seq and a robust statistical inference approach that tracks the leading edge of the Pol II wave following gene activation by short treatments with inducers, we have made high throughput measurements of Pol II transcription rates for over 160 genes across the human genome. This represents more than order of magnitude more genes than have been assessed for transcription rates side-by-side in a single experiment (Ardehali and Lis, 2009). In addition, our strategy for measuring rates of transcription will make genome wide measurements (i.e. thousands of genes) a possibility simply by simply substituting E2 or TNFα with a small molecule (e.g. flavopiridol) that affects transcription globally (Singh and Padgett, 2009).
Variation Between Cell Types and Treatment Systems
We observed more than 2-fold difference between transcription rates in the MCF-7/ E2 and AC16/ TNFα system. There are several possible explanations for this observation, including differences intrinsic to cell types or treatment systems. Once corrected for systematic differences in gene expression level, however, elongation rates in the two cell systems are remarkably similar (Fig. 4A). This strongly suggests that differences in elongation rate between the two systems primarily reflect differences in the expression level of primary response genes targeted by each transcription factor. As noted below, we suggest that these intrinsic differences in target genes activated by E2 and TNFα are caused by different mechanisms by which these signaling pathways activate transcription.
Potential Mechanisms Underlying the Variation in Pol II Elongation Rate
A number of potential mechanisms may underlie the variation in Pol II elongation rates that we observed across the genome, although future work is needed to determine the extent to which each contributes. Although prior literature suggests that nucleosome occupancy is a likely candidate to explain the variation (Deal et al., 2010; Dion et al., 2007), we do not observe sufficient variation in occupancy to explain our observations (Fig. 4C). This suggests that nucleosomes are likely to be an obstacle to Pol II transcription regardless of gene expression level. Therefore, while nucleosome occupancy may play some role in determining the rates that we have measured in our assays, we think that other factors are likely to contribute as well, perhaps in a more significant way.
First, certain post-translational histone modifications may reduce the influence of nucleosomes as a barrier to Pol II elongation. This may occur by changing the free energy of histone-DNA interactions (Manohar et al., 2009) or through the recruitment of transcriptional regulatory factors to nucleosomes which may assist with elongation (Smith and Shilatifard, 2010). In this regard, we have observed a positive correlation between Pol II elongation rate and H3K36me3, a histone mark associated with transcribed regions of genes (Barski et al., 2007) (Supplemental Fig. S4A). Second, our results are also consistent with prior reports that multiple RNA polymerases transcribing the same template sequence can interact by ‘pushing’ a paused or backtracked polymerase forward into productive elongation (Epshtein and Nudler, 2003; Saeki and Svejstrup, 2009). Studies of single cells have revealed that Pol II is activated in ‘bursts’ (Raj et al., 2006), resulting in uneven densities along gene bodies, which could explain direct Pol II interactions even in the absence of full Pol II occupancies. Finally, the recruitment dynamics of elongation factors, which help Pol II surmount systematic barriers to transcription such as nucleosomes (Belotserkovskaya et al., 2003), may be heavily influenced by the size or concentration of the locally available pool of factors. Prior reports indicate the amount of elongation factors in yeast systematically correlate with the density of Pol II (Mayer et al., 2010). Higher Pol II levels may act to establish a larger local pool of elongation factors, which, in turn, may facilitate more efficient transcription at more highly expressed genes.
Each of these potential hypotheses is a plausible explanation for our observations. However, because these features are so tightly linked to Pol II levels, it is not possible to determine how each parameter contributes to variation in transcription rates. New assays, combined with a broader understanding of transcription elongation, will be required to directly evaluate the contribution of each of these explanations to variations in Pol II rates on a genome-wide-scale.
Consequences of Pol II Elongation Rate Variation for mRNA Production
Regardless of the underlying mechanism, our results show that the observed variation between gene expression and Pol II rate has important consequences for mRNA production (Fig. 5). Specifically, the positive correlation between elongation rate and Pol II densities implies that mRNA will be produced more rapidly at highly expressed genes than can be explained by Pol II density alone. A similar relationship between Pol II density and mRNA levels has recently been reported in yeast (Mayer et al., 2010), suggesting that a variable Pol II elongation rate holds widely across a broad spectrum of eukaryotic species. Our observations suggest that elongation rate and Pol II density work systematically to produce a broader range of mRNA levels than could be produced based solely on Pol II densities. This observation has important implications for understanding how transcription by Pol II impacts gene regulation.
Selective Effects of Signaling Pathways on the Dynamics of the Early Transcription Process
Observing the distance traveled by Pol II following short treatments with inducer reveals the rate-limiting step by which E2/ERα and TNFα/NF-κB signaling pathways activate gene expression (Figs. 6 and 7). Because pausing is a rate-limiting step at the majority of human genes (Core et al., 2008), newly initiated Pol II will spend a variable amount of time ‘held’ in a paused state near the transcription start site. We observed a pause-related increase in residence time for Pol II recruited by E2/ERα, but no evidence of such a slowdown for Pol II recruited by TNFα/NF-κB (Figs. 6 and 7). Indeed, this increase in Pol II residence time was associated with increased density of the negative elongation factor NELF (Fig. 6D). We would also expect these genes to be associated with a slight depletion of positive regulatory factors such as CDK9.
Taken together, our results strongly support a model in which different transcription factors mediate their effects at distinct rate-limiting steps in the transcription process. Specifically, we propose a model in which TNFα-activated NF-κB stimulates transcription by reducing the residence time of Pol II at the pause site, and E2-activated ERα regulates transcription at a prior rate-limiting step (Fig. 7C). Our observations on NF-κB target genes are in general agreement with previous work in this system (Barboric et al., 2001; Hargreaves et al., 2009). Although previous studies have suggested that many E2-regulated genes are associated with “pre-loaded” Pol II prior to stimulation (Carroll et al., 2006; Kininis et al., 2007; Kininis et al., 2009; Lupien et al., 2009), the literature has been divided on whether E2/ERα stimulates release from pause (Kininis et al., 2009; Lupien et al., 2009) or Pol II recruitment and reinitiation (Kraus and Kadonaga, 1998; Welboren et al., 2009). Our direct observations of the kinetics of changes in Pol II levels following short treatments with E2 (Fig. 7A) support a model in which ERα acts to stimulate Pol II recruitment, initiation, or perhaps reinitiation.
This model potentially explains other observations on the specific properties of each signaling system. In particular, we noted above that TNFα/NF-κB tends to target a subset of genes with higher expression levels before TNFα treatment compared to E2/ERα (Fig. 4, A and B). This observation is consistent with a requirement that Pol II must be present at TNFα target genes at relatively high levels before NF-κB has a substantial effect on gene expression. In addition, we note that genes regulated by E2 tend to have systematically lower pausing indices than those regulated by TNFα (p = 1.5 × 10−6; two-sided Wilcoxon rank sum test), with both differing subtly from the genome-wide mean (Supplemental Fig. S7B). This may indicate that NF-κB requires paused Pol II to activate transcription, whereas ERα does not.
We are not able to determine whether TNFα/NF-κB acts exclusively to accelerate Pol II transcription through the pause site, or whether it acts on earlier rate-limiting steps as well. Indeed, a model in which TNFα accelerates both Pol II recruitment and transcription though the pause site seems to be supported by the large increases in Pol II observed at both the pause site and in the gene body (Fig. 7B). Together, these results demonstrate the power of comparing Pol II dynamics across the genome as a tool for exploring the mechanistic basis for transcriptional regulation.
Implications for Understanding the Function of Paused Pol II
Our results provide insights for understanding the function of promoter-proximally paused Pol II in signal-regulated transcription. Original hypotheses posited that genes with paused Pol II are “poised” for rapid activation (Lis, 1998). However, we (Hah et al., 2011) and others (Gilchrist et al., 2012; Lin et al., 2011) have recently challenged this view, showing that genes with paused Pol II may actually induce more slowly. This observation is illustrated clearly in our detailed genomic analyses in MCF-7 cells, which show that Pol II progresses the shortest distance following induction on E2-regulated genes with the highest pausing indices (Fig. 6C). Furthermore, comparisons of the MCF-7/E2 and AC16/TNFα systems show that TNFα target genes induce more slowly than E2 target genes (Supplemental Fig. S7A) in spite of having higher Pol II pausing indices (Supplemental Fig. S7B). These observations add strong support for the idea that pausing does not necessarily imply rapid gene activation.
What other functions might Pol II pausing serve? A recent report has suggested that it may act to synchronize gene activation events, allowing for gene induction in a narrow temporal window (Boettiger and Levine, 2009). The heatmap in Fig. 7B showing a nearly synchronous addition of Pol II to gene bodies across a set of TNFα target genes provides a visual representation of this effect. Quantitatively, we note that in spite of slower induction profiles, TNFα target gene activation occurs over a shorter temporal window (~5 min.; see “Residual Distance” −2.5 to +2.5; Supplemental Fig. S7A) than E2 target gene activation (~10 min.; −5 to +5; Supplemental Fig. S7A). Whether this is a general feature of all transcription factors that decrease residence time at the pause site, or whether it reflects distinct properties of the signaling and activation pathways transducing the E2 and TNFα signals, is currently unclear.
Collectively, our results demonstrate unexpected variability in the rates of Pol II transcription elongation in the gene body. By comparing Pol II densities with mRNA levels, we find that elongation rates play a significant role in establishing variations in gene expression level. Finally, our approach identifies the rate-limiting step targeted by transcription factors to activate gene expression. Our approach will serve as a template for future studies seeking to understand transcription on a genomic scale.
Experimental Procedures
Additional details on the experimental procedures can be found in the Supplemental Materials.
Cell culture and GRO-seq library preparation
MCF-7 and AC16 cells were cultured and nuclei prepared as previously described, with modifications (Hah et al., 2011). Cells were treated for the indicated time with either 100 nM 17β-estradiol (MCF-7) or 25 ng/ml TNFα (AC16). For MCF-7 cells, we used a GRO-seq library preparation protocol based circular ligation for adapter addition (Wang et al., 2011) to improve the efficiency of library preparation, reduce sequence bias, and allow barcoding. For AC16 cells, we used the original GRO-seq protocol (Core et al., 2008; Hah et al., 2011), which involves a direct ligation of sequencing adapters to nascent run-on RNAs.
Identifying the Pol II wave
GRO-seq data were processed and mapped as described (Hah et al., 2011), with some additional modifications necessary for efficient adapter removal in circular ligation based libraries. The Pol II wave was identified in all long (>120 kb for MCF-7; >75 kb for AC16), up-regulated genes (>2 fold; FDR q < 0.05). The start and end of the Pol II wave were identified using a three state hidden Markov model (HMM), with states that correspond to the upstream region, the Pol II wave, and the 3′ end of the gene. Differences in read counts between the control (0 min.) and later time points were used as input to the HMM. The Pol II rate was defined as the slope of the line that describes the distance traveled by the Pol II wave as a function of the treatment time, and was calculated using linear regression.
Quantifying Pol II density and mRNA levels
Pol II density was quantified in units of “reads per mappable base scaled” (RPBS), defined as the number of reads in the gene body (+1 kb to the gene end), divided by the number of unique 44-mers, and scaled to a common library and gene size. The expected relationship between mRNA levels and Pol II density was derived and fit on a log scale using non-linear least-squares. Pol II density was compared to mRNA levels in MCF7 cells taken from GEO (GSM325487). Finally, polyA neutral mRNA-seq data from spleen and ovary was collected from a recent study (Castle et al., 2010), and used to compare intron and exon read density.
Pol II pausing analysis
Gene with paused Pol II were identified, and pausing indices were estimated, directly from the GRO-seq data using the untreated condition, as described (Core et al., 2008). NELF ChIP-chip data (Kininis et al., 2009) were obtained from GEO (GSM327127). Heat maps focused on up-regulated genes were generated using R. Heat maps show the difference in read counts in 10 bp windows at the 5′ end of the appropriate subset of regulated genes (−2 to +5 kb relative to the TSS). Rows represent the average over 20 genes. Genes were ordered by the number of reads added to the gene (promoter or gene body) following 10 min. of treatment. For the scatterplots, we defined the pause peak as the 50 bp region of maximal Pol II density prior to treatment, and the gene body as the region between +1 and +15 kb.
Supplementary Material
Highlights.
Pol II rates vary by 4-fold between genes, cell types, and induction conditions.
Rates correlate strongly with gene expression, but weakly with nucleosome occupancy.
Rate variation has important consequences for mRNA and lncRNA production.
E2 signaling stimulates Pol II initiation; TNFα signaling reduces Pol II pause time.
Acknowledgments
We thank Iris Jonkers, Hojoong Kwak, and Josh Waterfall for helpful insights and suggestions, and Shrikanth Gadad and Bryan Gibson for critical comments on this manuscript. This work was supported by an NIH training award (T32HD052471) and a postdoctoral fellowship from the PhRMA Foundation to C.G.D., a predoctoral fellowship from the DOD Breast Cancer Research Program (BC093731) to X.L., a grant from the NIH/NIGMS (GM25232) to J.T.L., and a grant from the NIH/NIDDK (DK058110) to W.L.K.
Footnotes
Accession Numbers
The GRO-seq data sets used in this study are deposited in the NCBI Gene Expression Omnibus (GEO) database and can be accessed with accession numbers GSE27463, GSE41323, and GSE41324.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ardehali MB, Lis JT. Tracking rates of transcription and splicing in vivo. Nat Struct Mol Biol. 2009;16:1123–1124. doi: 10.1038/nsmb1109-1123. [DOI] [PubMed] [Google Scholar]
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Castle JC, Armour CD, Löwer M, Haynor D, Biery M, Bouzek H, Chen R, Jackson S, Johnson JM, Rohl CA, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA-neutral amplification. PLoS One. 2010;5:e11779. doi: 10.1371/journal.pone.0011779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Epshtein V, Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature. 2009;461:186–192. doi: 10.1038/nature08449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Fromm G, dos Santos G, Pham LN, McDaniel IE, Burkholder A, Fargo DC, Adelman K. Regulating the regulators: the pervasive effects of Pol II pausing on stimulus-responsive gene networks. Genes Dev. 2012;26:933–944. doi: 10.1101/gad.187781.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core L, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izban MG, Luse DS. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol. 2007;27:5090–5104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Isaacs GD, Core LJ, Hah N, Kraus WL. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol Cell Biol. 2009;29:1123–1133. doi: 10.1128/MCB.00841-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Kadonaga JT. p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC) Genes Dev. 2011;25:1486–1498. doi: 10.1101/gad.2059211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Krum SA, Rhodes DR, Liu XS, Brown M. Coactivator function defines the active estrogen receptor alpha cistrome. Mol Cell Biol. 2009;29:3413–3423. doi: 10.1128/MCB.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar M, Mooney AM, North JA, Nakkula RJ, Picking JW, Edon A, Fishel R, Poirier MG, Ottesen JJ. Acetylation of histone H3 at the nucleosome dyad alters DNA-histone binding. J Biol Chem. 2009;284:23312–23321. doi: 10.1074/jbc.M109.003202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- Nogues G, Kadener S, Cramer P, Bentley D, Kornblihtt AR. Transcriptional activators differ in their abilities to control alternative splicing. J Biol Chem. 2002;277:43110–43114. doi: 10.1074/jbc.M208418200. [DOI] [PubMed] [Google Scholar]
- Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, Wyrzykowska PS, Coelho PA, Carmo AM, Sunkel CE, Proudfoot NJ, et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki H, Svejstrup JQ. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol Cell. 2009;35:191–205. doi: 10.1016/j.molcel.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohta Y, Xu M, Tsutsumi S, Minami T, Inoue K, Komura D, Kitakami J, Oshida N, Papantonis A, et al. A wave of nascent transcription on activated human genes. Proc Natl Acad Sci U S A. 2009;106:18357–18361. doi: 10.1073/pnas.0902573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Zhou HL, Hinman MN, Barron VA, Geng C, Zhou G, Luo G, Siegel RE, Lou H. Hu proteins regulate alternative splicing by inducing localized histone hyperacetylation in an RNA-dependent manner. Proc Natl Acad Sci U S A. 2011;108:E627–635. doi: 10.1073/pnas.1103344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.