Abstract
Evolutionary changes are determined by a complex assortment of ecological, demographic and adaptive histories. Predicting how evolution will shape the genetic structures of populations coping with current (and future) environmental challenges has principally relied on investigations through space, in lieu of time, because long-term phenotypic and molecular data are scarce. Yet, dormant propagules in sediments, soils and permafrost are convenient natural archives of population-histories from which to trace adaptive trajectories along extended time periods. DNA sequence data obtained from these natural archives, combined with pioneering methods for analyzing both ecological and population genomic time-series data, are likely to provide predictive models to forecast evolutionary responses of natural populations to environmental changes resulting from natural and anthropogenic stressors, including climate change.
Keywords: environmental genomics, climate change, resurrection ecology, adaptation, evolution, paleogenomics, network analysis
From genes to the genomes of populations
In recent years, the decreasing costs and higher accessibility of high-throughput DNA sequencing technologies have sparked a shift from genetics (the study of genes) to genomics (the wholesale study of genomes). These innovations have fueled the development of an unbiased reverse ecology or forward genetics - genome scan approach for measuring DNA variation [1,2], whereby genes with functions tied to ecologically relevant traits can be identified by contrasting patterns of neutral and adaptive genetic variation within and between populations. Neutral genetic variation provides a baseline view of both demographic (e.g., population size, migration rate) and genetic (e.g., mutation rate, recombination) processes, whereas adaptive genetic variation underlies traits that impact fitness. The growing application of this genomic approach to species with well-studied ecologies will likely identify genes and functional gene networks underlying adaptive responses in the wild (see [3], [4] and [5] for recent examples). Such studies provide an extraordinarily detailed view of the molecular basis of recurrent adaptive divergence and illustrate the importance of processes such as gene duplication in functional responses to environmental conditions. Overall, the recent application of genomics in population studies is helping to identify the types of genetic variation (variation in gene regulatory regions or gene polymorphisms) that matter most for adaptation (e.g. [6]), and to discover the sources of genetic variation (new mutations, migration, standing genetic variation) driving adaptive responses to environmental changes (e.g. [7]).
A next step is to extend these inquiries beyond straightforward descriptive measures of the spatial distribution of genetic variation to reconstruct the evolutionary processes that contributed to present-day population genomic structures in the wild. This endeavor requires observation of evolution in action to connect the genetic architecture of natural populations to the environmental context of selection, drift and migration that drive phenotypic outcomes. A mechanistic approach to the study of evolution is still comparatively rare and the temporal perspective we discuss here is likely to produce important insights, not least for aiding better predictions of adaptive responses to future environmental challenges.
A novel way of documenting the course of evolution in nature during the time-frame of a single research project is to study organisms that produce dormant life-stages (i.e. seeds, eggs, cysts, spores; [8]), which accumulate in lacustrine and marine sediments, soil or ice to build up dormant propagule banks (note: we will use the term “propagule bank” to describe all dormant seed, cyst, spore, and egg banks). These temporally stratified propagule banks can be accurately dated (Box 1) thereby aligning their local population genomic and community histories to known changes in the natural landscapes, or to environmental changes inferred from analyses of the sediments or soils [9–12]. By resuscitating past populations in the laboratory and competing isolates against their modern descendants, the function and fitness effects of genes evolving in step with the changing environment can also be experimentally inferred.
Box 1. Dating of sediment, soil, and ice cores.
A variety of methods are used for dating lacustrine and marine sediments, soils, and ice cores, ranging from visual characterization of annual laminated sediments (e.g. varved sediments in lakes containing algal pigments deposited during annual algal blooms; annual dust layers deposited in ice cores) to radiometric (i.e. radio-isotope) dating [58, 59]. For dating soil and sediment cores, traditionally researchers have used isotopes such as 210Pb (lead) and 137Cs (cesium) [60]. The dating of horizons from known historical events (e.g. 137Cs signal from the end of nuclear bomb testing in the 1960’s; historical volcanic eruptions that deposited unique “dust signatures” in ice), has been used as an aid in calibrating sediment deposition rates and as a cross-check with radiometric methods. For more accurate dating of older soil and sediment samples dating back >50,000 years before present (YBP), 14C (carbon) has been the method of choice [59]. For dating material in ice cores, in addition to visual analyses (i.e. annual dust layers), a combination of radiometric and isotopic ratios, as well as chemical signatures have been used [61]. Additional use of subfossils (e.g. pollen grains, invertebrate or plant remains, calcified structures such as shells) often augments the accuracy of the dating methods mentioned above.
Figure 1.
Examples of a (A) lacustrine (photo credit M. Wessels) and (B) ice core (http://www.globalwarmingart.com). (C) Lacustrine sediment core showing varved layers (photo credit J. Wiethold).
In this article, we discuss the use of stratified propagule banks to reconstruct the evolutionary processes that contributed to present-day population genomic structures in the wild. We posit that a parallel temporal analysis conducted on multiple populations distributed across a landscape allows for the reconstruction of the processes that drive evolutionary dynamics at regional and continental scales with unprecedented resolution. Finally, we suggest that informing predictive modeling of populations with data from biological archives is a powerful approach to forecast population adaptation to multiple environmental challenges, including climate change.
Resurrection Ecology and Paleogenomics of dormant propagule banks
Dormant propagule banks, which are increasingly used in the field of “resurrection ecology” [13], are hatched or germinated from layers of sediment, soil or ice cores to obtain representative samples of populations inhabiting environments in the recent past (from decades to centuries). The populations revived from dormant propagules are kept in the laboratory and studied for their traits, genetic polymorphisms and allelic gene frequencies, enabling a reconstruction of evolutionary dynamics in the face of environmental change [11, 14]. To date, resurrection ecology has been applied mainly in paleolimnological reconstructions of the evolutionary dynamics of aquatic invertebrates [11, 14]. Species of the genus Daphnia are among the most well studied in this respect. They are key models in evolutionary biology and the study of adaptive responses of ecological traits to the environment [15–17]. Fewer studies have examined propagule banks of terrestrial and marine invertebrates [8] or of plants [18, 19] but there is increasing potential to do so.
Paleogenetics, the study of polymorphic traits and DNA markers in biological archives, can be seen as a complimentary approach to resurrection ecology and it has produced some surprising discoveries (see Box 2 for key examples). This approach provides estimates of microevolutionary responses of populations under stress [11], changes in population- or community-level structure through time [20], and changes in neutral genetic diversity [21]. Paleogenetics has also been used to measure the rate of adaptation to environmental perturbations, such as the response to nutrient enrichment (eutrophication) and nutrient reduction (re-oligotrophication) in aquatic systems [22, 23]. Fueled by the advent of high-throughput genomics, paleogenetics has scaled up to paleogenomics -- the study of genome-wide changes in organisms from the past (e.g. [24], Box 2). Paleogenomics provides a way to reconstruct past evolutionary dynamics and to identify past demographic and adaptive processes that have contributed to genomic structure in present-day populations. By reconstructing genome-wide variation through time, we can establish if adaptive phenotypic responses result from new mutations, standing genetic variation or a combination of both mechanisms. By reconstructing population history through time, we can identify mutations underlying adaptations linked to specific environmental conditions and to further unveil the physiological basis of novel traits. Except for rare cases where investigations span decades (e.g. [25]), these questions can currently be addressed only with experimental populations of species having a short generation time (e.g. E. coli [26] and other microbes, [27]) propagated in controlled laboratory conditions. Propagule banks offer the unique opportunity to extend these findings to natural environments and to organisms with longer generation times, enabling us to forge a link between genome architecture and specific environmental challenges.
Box 2. Case studies of genetics and genomics applied to ancient (a) DNA and dormant propagule banks.
Ecological and environmental genomics: Orsini et al. [24] used genomics on resurrected Daphnia specimens. They studied spatial dynamics of Daphnia magna across a complex landscape of shallow ponds identifying adaptive responses to a suite of environmental stressors (e.g. parasites, land-use changes, fish predation), and validated these findings in time, using sediment cores with known histories for specific stressors, and in experimental evolution trials. Candidate genes linked to specific and general stress response were identified in space and validated in time and spatial evolution trials.
Evolution of microbial communities through time: Bidle et al. [62] studied community diversity in ice cores using 16S rDNA as a genetic marker. Analyses of five ice cores, spanning the last 8 million years, demonstrated an exponential decline in the average community DNA size with a half-life of 1.1 million years, with implications for the geological preservation of microbes in icy environments.
Changes in plant and invertebrate communities along an extended time axis. Willerslev et al. [10] studied the DNA composition of buried organisms recovered from the basal sections of deep ice cores, reconstructing past floral and faunal community composition. They reconstructed changes in a diverse array of conifer trees and insects within the past million years by comparing ancient biomolecules from basal ice with current floral and faunal composition. This approach offers a means for environmental reconstruction from ice-covered areas and yields insights into the ecology of communities from the distant past. The future application of genomics to microbial and other communities trapped in ice or sediment samples opens unprecedented opportunities for the study of community evolutionary dynamics in response to changing environments.
Genetic rescue using ancient specimens: Yashina et al. [63] were able to resurrect permafrost-entombed plant tissues of Silene stenophylla more than 30,000-years old, germinate and establish plants. They determined that the ancient phenotype is distinct from the modern one. Success in resurrecting centennial-scale or millennial-scale-old fertile organisms opens unprecedented opportunities to study long-term evolutionary dynamics and to use ancient specimens to restore genetic diversity in modern specimens suffering from risk of extinction.
Figure 4.
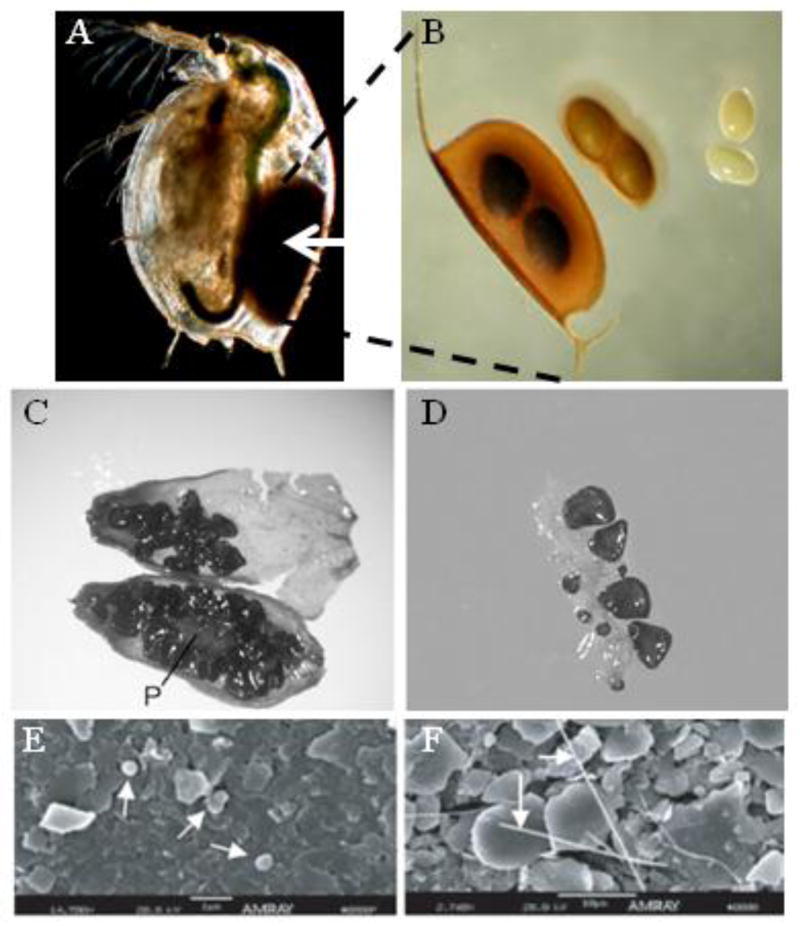
Figure 1. (A) Daphnia magna (Crustacea: Cladocera) adult female carrying an ephippium (arrow) (photo credit Joachim Mergeay) and (B) light micrograph of an ephippium with details of decapsulated eggs (photo credit Tom De Bie). (C) Silene stenophylla (Caryophyllaceae) dissected fruit from burrow buried in permafrost (> 30,000 y) showing seed and placenta (P) and (D) fragment of placenta with seeds at different developmental stages (picture modified from Yashina et al., 2012). (E-F) Scanning electron micrographs from Antarctic ice core samples illustrating DNA containing bacterial cells (arrows) (picture modified from Bidle et al., 2007).
Understanding processes and mechanisms of adaptation requires “Time”
Spatial analyses, commonly used in studies of the genetic diversity among populations provide useful insights into the processes contributing to present-day population-genetic structure [28, 29]. However, these analyses are often limited to a single snapshot in time, limiting our ability to infer the processes that may have lead to the population genetic diversity and structure we observe in present-day populations. The inference of past processes via contemporary patterns of genetic diversity may be confounded by phylogeographic signals, complicating the identification of the causes of adaptive and demographic changes. By contrast, monitoring genetic diversity and adaptation through time, and comparing changes in genetic diversity with expectations of neutral and adaptive evolutionary models, circumvents these limitations and allows a shift from measuring patterns, to determining processes of evolution. This temporal analysis of genomes is key to establishing the interaction between genome architecture and the environment that drives phenotypic evolution in natural populations. Tracking genome-wide changes in allele frequencies through time allows us to identify the type and rate of mutations underlying adaptive responses to specific selective regimes. This temporal analysis conducted in parallel on several populations distributed across a landscape allows to reconstruct the processes that drive evolutionary dynamics at regional and continental scales (Figure 1). Temporally stratified propagule banks are a powerful resource to reconstruct evolutionary dynamics at different spatial scales. They are also useful for studying fluctuations in selective forces (i.e., environmental variation) among years (e.g. [30]). As high-throughput sequencing technologies continue to improve, and the range of application to low-copy and degraded DNA increase [31], the study of evolutionary dynamics using paleogenomics becomes more accessible for a growing number of species producing dormant propagules along extended time axes (≫500 years, Box 2). In particular, third generation sequencing technologies offer unprecedented opportunities for the sequencing of degraded and ancient (a)DNA, often recovered from stratified propagule banks. At this early stage, a number of technological challenges still exist for investigating dormant propagule banks (Box 3). However, technical developments in our laboratories and those of others are minimizing these challenges and maximizing the future impacts of paleogenomic studies.
Figure 1.
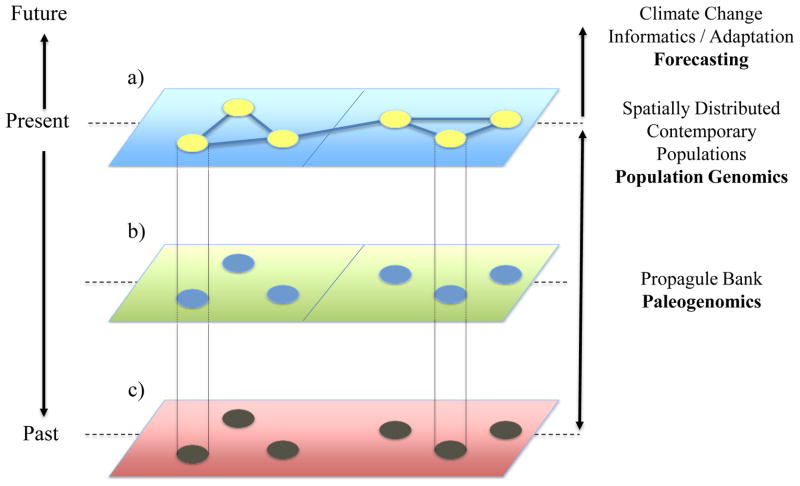
Genomics of propagule banks in space and time
Dormant propagules sampled from sediment, soil or ice cores can be used to reconstruct evolutionary processes, consisting of demographic and adaptive changes, over extended time axes. The reconstruction of evolutionary processes conducted in parallel through time in several populations distributed at regional or continental scales informs our understanding of adaptive responses of natural populations to environmental changes. Spatially-structured contemporary populations (a) are amenable to population genomic and landscape genomic analysis. Dormant propagules from the recent past (b) can be resurrected and compared directly to contemporary populations using the full range of available DNA- and RNA-based genomic techniques as well as using trait-based approaches. Ancient (a)DNA from the far past (c) provides an archive of genetic material that can be compared to the one of contemporary populations and indexed to reconstruct past environmental conditions and historical selective regimes. The joint consideration of past and present genetic structure in a time-series provides critical parameterization for future forecasting.
Box 3. Challenges of using dormant propagule banks.
Germination or hatching of dormant propagules
If aged propagules cannot be induced to germinate or hatch, there is no possibility for direct experimentation and the application of reverse genetics. Thus genome-based studies will remain descriptive in some systems.
Low quantity/low quality of ancient (a)DNA
Ancient (a)DNA is often limited in amount and quality, because aged propagules might be degraded and yield limited amounts of DNA material. Low quality and/or limited starting material has impeded the application of high-throughput technologies [64] in a paleogenetic setting. Requirements on starting material are several orders of magnitude lower for third generation sequencing technologies [65], offering promise for the sequencing of degraded DNA contained in ancient propagule banks. The high DNA sequencing error rate of this technology, ranging from 5 to 10%, will almost certainly be reduced as these technologies are refined and can be partially alleviated by increasing the fold coverage per run (number of reads per nucleotide position) with a finished read accuracy of 99% [66]. Bioinformatics tools able to account for sequencing errors are rapidly being generated, improving the quality of the data produced by second-generation sequencing [67]. Similar improvements are to be expected for third-generation sequencing platforms.
Sources of contamination for ancient (a)DNA
If aged propagules are hatched or germinated, they do not pose concerns for DNA/RNA analysis. Conversely, if propagules cannot be hatched or germinated, measures to avoid contamination should be adopted. One of the most common sources of contaminations comes from drilling fluids during sampling (e.g. ice coring). While this kind of contamination cannot be avoided, it can be controlled by spiking the drilling fluid or the surface of the core with recognizable microorganisms, used as trace control to measure the penetration of contaminants in the core. The second source of contamination derives from handling cores in the laboratory. To reduce contamination at this stage, the outer surface of the cores should be removed in a laminar flow hood in a cold room to avoid the formation of a water film on the surface of ice cores and degradation of DNA/RNA in all cases. The third source of contamination is the DNA laboratory. Contamination with modern DNA can happen if aDNA samples are processed in the same environment as modern samples. An additional source of contamination is bacterial DNA that can be present in reagents and solutions. For a complete overview of the precautions that should be adopted when working with aDNA samples we direct the reader to several comprehensive reviews on the topic (e.g. [68–70].
From patterns to processes
The analysis of time-series data combined with environmental data increases the power to identify selective forces shaping allele frequencies. Time-series analysis for identifying natural selective processes pre-dates the “omics” era, when changes in allele frequencies were followed at target genes [32]. However, only recently have analytical methods become available to study long-term data series [33]. Recent analytical tools to infer selection and demographic processes from time-series data of allele frequencies [34] have been validated using data from artificial selection experiments on yeast [35]. The approach suggested by Illingworth and co-workers [34] compares changes in allele frequencies over time with predictions of different evolutionary scenarios based on population genetic theory and calculates likelihoods of fit for each scenario. The highest likelihood is used to identify the most probable dynamics of the system under study. Applying this approach to time-series data obtained from dormant propagules will enable the reconstruction of evolutionary dynamics over extended time axes in response to known selection pressures and to disentangle adaptive from neutral processes with unprecedented resolution [34, 36]. Initially developed for clonal lineages, this analytic approach can be extended to sexually-reproducing populations by considering selection and recombination simultaneously and can be used to discover complex multi-locus interactions underlying fitness traits [34].
The study of gene frequency changes through time can be complemented by a quantitative genetics analysis of traits and experimental evolution trials using populations resurrected from different time-periods of dormant propagule banks. Experimental evolution studies are a key tool for evolutionary genomics in non-model taxa, for which reverse genetic techniques are not possible. Resurrection ecology, even if it provides experimental validation of evolutionary processes only over a portion of the time-frame, is a powerful complementary approach to paleogenomics and enables a deeper understanding of the processes steering evolutionary dynamics in response to well-documented environmental changes.
The integrated analysis of complex landscapes: network analysis
The identification of evolutionary processes in natural landscapes requires an understanding of the interactions between genome architecture and the environment that drives phenotypic evolution. Understanding these complex interactions requires analysis of multiple levels of biological organization, ranging from molecules to environmental and phenotypic variation. In recent years, network analysis has emerged as a powerful tool across numerous disciplines of science to analyze complex interactions related to public health, social (reviewed in [37, 38]), and medical science [39, 40]. In the field of biology, network approaches have found wide application to diverse topics including the study of metabolic and biochemical pathways [41], the evolution of proteins [42] and interactions among community members [43]. Because of its versatility, this tool can be also applied in a paleogenomic setting to identify patterns of co-variation in genetic and phenotypic variation in space and along the time axis in response to known environmental changes. For example, variation in metabolic and chemical pathways linked to known environmental changes can be linked to patterns of variation in genotypic trait values identified in natural and experimental populations. Patterns identified with network analysis can be then used to develop informed models to forecast adaptive responses of natural populations to environmental changes (see “Future avenues of research”).
Eco-evolutionary dynamics
An increasing number of observations shows that eco-evolutionary feedbacks can have substantial consequences for population persistence [44, 45], trophic interactions [46, 47], community assembly [48] and changes in ecosystem characteristics [49]. An outstanding issue is to understand over what time-frame (years, decades, centuries) ecological dynamics drive evolutionary changes and eco-evolutionary feedbacks. It is critical to establish if, and to what extent, population-driven evolutionary processes can influence population dynamics, community composition and ecosystem functioning on relatively short time scales. An analytical approach that will help fill this gap is the method developed by Ellner et al. [45] that identifies the relative contribution of environment and evolutionary change to changes in population, community or ecosystem properties. The applicability of this method has been shown in an elegant predator-prey microcosm experiment [50]. Applying this approach to layered dormant propagule banks offers unique opportunities to infer the role of changes in gene frequencies through time to explain dynamics in population densities and species composition. Combining paleogenomics with resurrection ecology will be a powerful approach in documenting eco-evolutionary feedbacks. Not only can genotypic or phenotypic trait values of hatched, or germinated, individuals be linked to a specific environment, but transplant experiments can also be conducted, by replacing individuals from “evolved” subpopulations with individuals from “ancestral” subpopulations in experimental trials [12]. This type of experiment allows direct quantification of the impact of evolutionary changes on ecological interactions and viceversa, and therefore represents an important validation step for the study of eco-evolutionary dynamics.
Future avenues of research
Processes and mechanisms of adaptation in the wild
Species producing propagule banks offer the unique opportunity to follow evolution in time by comparing changes in genetic diversity at numerous points in time, as opposed to classic studies of genetic diversity in museum specimens when only one or a few time points have been compared [51] (but see [52]). In addition, the analysis of propagule banks exposed to known selective pressures [11, 14, 22, 24] offers unique opportunities to link phenotypes to the underlying genotype and to specific environments. This linkage is made possible by leveraging historical reservoirs of genotypes to monitor changes through time within natural populations with known histories. With the recent advances of third-generation sequencing and the most recent statistical tools to analyze time-series data discussed in this opinion paper, it will be possible to disentangle demographic and selective processes shaping genetic diversity and to understand mechanisms of adaptation in the wild. A parallel temporal analysis conducted on multiple populations distributed across a landscape allows for the reconstruction of the processes that drive evolutionary dynamics at regional and continental scales.
Predicting future adaptive responses
Through the study of evolutionary dynamics over extended time axes in dormant propagule banks it is possible to identify environmental processes that drive phenotypic evolution in natural settings. Combining these results with predictive models of future change in environmental conditions (e.g. climate change, land use change), we can predict adaptive responses of natural populations to these future environmental challenges. Thus far, predictions regarding the response of extant biodiversity to environmental challenges have been limited to climate change and have been generated primarily with climate envelope modeling strategies using species distributional data [53], projecting species range shifts (e.g WorldClim database) while ignoring evolutionary dynamics. However, the lack of information on ecological and evolutionary processes that allow species to persist in the landscape strongly affects the accuracy of predictive models to forecast the long-term consequences of climate and other anthropogenic changes [54]. An additional level of complexity that limits the predictive ability of such models is the lack of information on the interspecific interactions within communities [55]. A first attempt to unify population modeling with both ecological responses as well as evolutionary processes is the “box-in-a-box” modeling approach that couples population models to phenological change [54]. This approach unifies population modeling with both ecological responses to climate change as well as evolutionary processes, using a mechanistic embedded correlative approach, where the link from genes to population is established using a periodic matrix population model. This approach can be readily expanded beyond climate change to other environmental changes. A next step is informing this predictive model with data from biological archives to forecast population adaptation to multiple environmental challenges.
Metagenomics
Combining paleogenomics and resurrection ecology of layered propagule banks will allow reconstruction of eco-evolutionary dynamics of interacting species (e.g. [12]), given that a number of species in the community produces resting stages. Dynamics of evolving metacommunities can be reconstructed, documenting how adaptation at the genome level in single species interacts with community dynamics in response to environmental changes [55], particularly if keystone species are incorporated into these studies. As technology advances and genomic resources are developed for a wider range of species, one can imagine applying a metagenomics approach to all species within a guild to analyze emergent dynamics [56].
Paleogenomics combined with resurrection ecology applied at the community level can provide unique opportunities to study how local genetic changes might impact community responses to environmental changes, including climate change. Advances in bioinformatics will, in the near future, allow direct tackling of questions in eco-evolutionary dynamics using a metagenomics approach.
Concluding remarks
The parallel analysis of neutral and adaptive variation of dormant propagule banks on multiple populations distributed in the landscape, in some cases combined with resurrection ecology, allows research to go beyond descriptive measures of patterns of genetic variation and instead to reconstruct evolutionary processes that lead to present-day population genomic structure in natural populations (Figure 2). A genomic analysis will simultaneously describe the patterns of adaptive variation of specific traits and reveal the underlying genetic mechanisms, providing an assessment of the repeatability of evolutionary dynamics. The identification of evolutionary processes is key to forecasting future adaptation to environmental changes and to designing conservation plans to prevent loss of biodiversity, which has an effect on ecosystem(s) persistence comparable to the one induced by anthropogenic changes [57].
Figure 2.
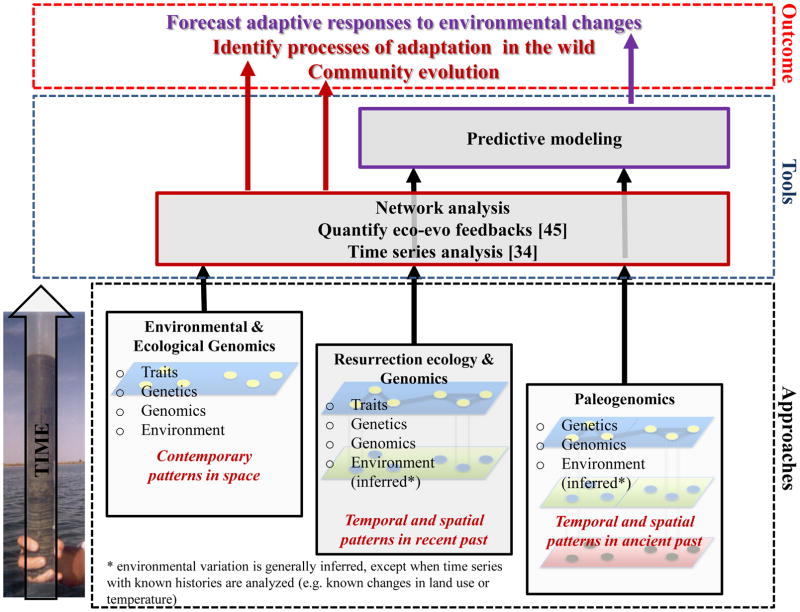
Showcasing paleogenomics
Environmental and ecological genomic approaches are used to measure patterns of phenotypic, genotypic and environmental variation in spatially distributed populations at a single point in time (present-day populations). These patterns can be correlated using network analysis and eco-evolutionary feedbacks can be unraveled for a single point in time. When resurrection ecology is combined with the analysis of genomes, patterns of phenotypic, genotypic and environmental variation are reconstructed in the recent past (decades to centuries) at different time points. Paleogenomics applied directly to propagules allows to establish a link between the genome and environmental variation along extended time axes (≫ 500 years). Patterns of variation in phenome, genome and environment are correlated with network analysis for the recent and ancient past and eco-evolutionary feedbacks measured along the time axis to reconstruct species and community dynamics in the face of environmental changes. The comparison of changes in allele frequencies over time (analysis of time series in recent and ancient past) with predictions of different evolutionary scenarios identifies the prevailing dynamics of the system under study. Combining the study of evolutionary dynamics with predictive models is used to forecast adaptive responses of natural populations to future environmental changes. By combining “Space” and “Time” (approaches), we can identify processes of adaptation in the wild (tools), forecast adaptive responses of natural populations to future environmental changes, and study the evolutionary dynamics of communities (outcome).
Acknowledgments
This work is part of the STRESSFLEA project of the European Science Foundation EUROCORES Programme EuroEEFG. LO and LDM acknowledge Centre of Excellence funding by the KU Leuven Research Fund (PF/2010/007) and FWO projects G.0614.11 and G. 0468.10. LJW gratefully acknowledges funding from the U.S. National Science Foundation (NSF award #0924289) during the manuscript preparation stage of this project. KS acknowledges financial support by the Biodiversity and Climate Research Centre Frankfurt am Main (BiKF; ‘LOEWE–Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz’ of Hesse’s Ministry of Higher Education, Research and the Arts). MEP gratefully acknowledges funding from the U.S. National Science Foundation (NSF – award #0922251) and U.S. National Institutes of Health (NIH – award #GM078274) during the manuscript preparation stage of this project. We thank Paul Craze and three anonymous reviewers for constructive comments on earlier versions of the manuscript.
Glossary
- Reverse ecology
the use of genomics to study ecological responses in a given organism with no a priori assumptions. The term is used as an analogy to reverse genetics in which the function of a given gene is studied by comparing the phenotypic effects of specific gene sequences
- Forward genetics
term used here in association with reverse ecology. The forward genetics approach studies the genetics of a given organism without any a priori knowledge of its adaptive responses to environmental changes. It seeks signatures of selection at the genome-wide scale in opposition to a candidate gene approach – a method that assesses the functional link between a gene variant and a phenotype
- Genome scans (or genome-wide linkage scans)
a genome-wide multi-locus screening of genetic variation. Genome scan studies identify genomic regions under selection, either directly or more often through linkage, through the screening of multiple loci. Genomic regions under selection are the ones that depart from neutral expectations in a multi-locus distribution of Fst
- Resurrection ecology
hatching of dormant propagules of a still living (or potentially extirpated) species from a different time than the present, to study the traits and responses to the environment of populations that lived in the past
- Genome/genetic architecture
genetic architecture refers to the underlying genetic basis of phenotypic traits and genotypic trait values (defined below). The study of genetic architecture is the study of the functional link between genotypes and phenotypes
- Third generation or single-molecule sequencing
direct DNA, cDNA and RNA sequencing. This technology, in full development, does not include amplification (DNA, cDNA) and reverse transcriptase steps (RNA)
- Ancient (a)DNA
DNA isolated from ancient specimens. Examples of aDNA include DNA recovered from archaeological and historical skeletal material, mummified tissues, archival collections of non-frozen medical specimens, preserved plant remains from ice, soil, sediment and permafrost cores
- DNA sequencing error rate
reading error generated by sequencing platforms often associated with low quality of starting DNA or RNA. There are three types of errors that can be introduced by automated sequencing, deletion, insertion and mismatches. The error rate occurring during the sequencing process differs between platforms, but it is commonly larger in Next Generation Sequencing and Third Generation Sequencing technologies as compared to Sanger sequencing
- Reverse genetics
through genetic analyses, the function of genes is investigated by studying organisms where gene function is altered. In classical forward genetic screening, individuals are treated with mutagens to induce DNA lesions and mutants with a phenotype of interest are sought. After a mutant is found, the gene mutated is identified through standard molecular techniques
- Eco-evolutionary feedbacks
reciprocal interactions between ecological and evolutionary processes. Eco-evolutionary feedbacks require that micro-evolution of a given population (driven by environmental change) impacts ecological responses at the population, community or ecosystem level
- Genotypic trait value
average value of the phenotype of a specific trait for a genotype in a particular environment
- Phenotypic trait value
result of the combined effect of environmental influences (including the maternal environment) and the genotypic trait value
- Climate envelope model
Climate envelope models use current distributions of species to construct a projected set of climatic conditions that suit a given set of species. This ‘envelope’ can then be used to visualize where species could live under predictions of future climate change
- Complex network
a graph (network) with non-trivial topological features—features that do not occur in simple networks such as lattices or random graphs
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luikart G, et al. The power and promise of population genomics: from genotyping to genome typing. Nat Revi Genet. 2003;4:981–994. doi: 10.1038/nrg1226. [DOI] [PubMed] [Google Scholar]
- 2.Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- 3.Jones FC, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colbourne JK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331:555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner TL, et al. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet. 2010;42:260–263. doi: 10.1038/ng.515. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Maury L, et al. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet. 2008;9:583–593. doi: 10.1038/nrg2398. [DOI] [PubMed] [Google Scholar]
- 7.Khan AI, et al. Negative Epistasis Between Beneficial Mutations in an Evolving Bacterial Population. Science. 2011;332:1193–1196. doi: 10.1126/science.1203801. [DOI] [PubMed] [Google Scholar]
- 8.Evans ME, Dennehy JJ. Germ banking: bet-hedging and variable release from egg and seed dormancy. Q Rev Biol. 2005;80:431–451. doi: 10.1086/498282. [DOI] [PubMed] [Google Scholar]
- 9.Parducci L, et al. Glacial survival of boreal trees in northern Scandinavia. Science. 2012;335:1083–1086. doi: 10.1126/science.1216043. [DOI] [PubMed] [Google Scholar]
- 10.Willerslev E, et al. Ancient biomolecules from deep ice cores reveal a forested southern Greenland. Science. 2007;317:111–114. doi: 10.1126/science.1141758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cousyn C, et al. Rapid, local adaptation of zooplankton behavior to changes in predation pressure in the absence of neutral genetic changes. PNAS. 2001;98:6256–6260. doi: 10.1073/pnas.111606798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decaestecker E, et al. Host-parasite Red Queen dynamics archived in pond sediment. Nature. 2007;450:870–874. doi: 10.1038/nature06291. [DOI] [PubMed] [Google Scholar]
- 13.Kerfoot WC, et al. A new approach to historical reconstruction: Combining descriptive and experimental paleolimnology. Limnol Oceanog. 1999;44:1232–1247. [Google Scholar]
- 14.Hairston JNG, et al. Rapid evolution revealed by dormant eggs. Nature. 1999;401:446. [Google Scholar]
- 15.Jeyasingh PD, et al. How do consumers deal with stoichiometric constraints? Lessons from functional genomics using Daphnia pulex. Mol Ecol. 2011;20:2341–2352. doi: 10.1111/j.1365-294X.2011.05102.x. [DOI] [PubMed] [Google Scholar]
- 16.Latta LC, et al. The evolution of salinity tolerance in Daphnia: a functional genomics approach. Ecol Lett. 2012;15:794–802. doi: 10.1111/j.1461-0248.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 17.Scoville AG, Pfrender ME. Phenotypic plasticity facilitates recurrent rapid adaptation to introduced predators. PNAS. 2010;107:4260–4263. doi: 10.1073/pnas.0912748107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalisz S. Experimental-Determination of Seed Bank Age Structure in the Winter Annual Collinsia-Verna. Ecology. 1991;72:575–585. [Google Scholar]
- 19.Cozzolino S, et al. Genetic variation in time and space: the use of herbarium specimens to reconstruct patterns of genetic variation in the endangered orchid Anacamptis palustris. Conserv Genet. 2007;8:629–639. [Google Scholar]
- 20.Mergeay J, et al. Invasion of an asexual American water flea clone throughout Africa and rapid displacement of a native sibling species. Proc R Soc B. 2006;273:2839–2844. doi: 10.1098/rspb.2006.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Härnström K, et al. Hundred years of genetic structure in a sediment revived diatom population. PNAS. 2011;108:4252–4257. doi: 10.1073/pnas.1013528108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weider LJ, et al. Long-term genetic shifts in a microcrustacean egg bank associated with anthropogenic changes in the Lake Constance ecosystem. Proc R Soc Lond B. 1997;264:1613–1618. [Google Scholar]
- 23.Brede N, et al. The impact of human-made ecological changes on the genetic architecture of Daphnia species. PNAS. 2009;106:4758–4763. doi: 10.1073/pnas.0807187106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orsini L, et al. Genomic signature of natural and anthropogenic stress in wild populations of the waterflea Daphnia magna: validation in space, time and experimental evolution. Mol Ecol. 2012;21:2160–2175. doi: 10.1111/j.1365-294X.2011.05429.x. [DOI] [PubMed] [Google Scholar]
- 25.Grant PR, Grant BR. Unpredictable evolution in a 30- year study of Darwin’s finches. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- 26.Blount ZD, et al. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 28.Lowry DB. Landscape evolutionary genomics. Biol Lett. 2010 doi: 10.1098/rsbl.2009.0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storfer A, et al. Putting the “landscape” in landscape genetics. Heredity. 2007;98:128–142. doi: 10.1038/sj.hdy.6800917. [DOI] [PubMed] [Google Scholar]
- 30.Charmantier A, et al. Adaptive phenotypic plasticity in response to climate change in a wild bird population. Science. 2008;320:800–803. doi: 10.1126/science.1157174. [DOI] [PubMed] [Google Scholar]
- 31.Millar CD, et al. New developments in ancient genomics. Trends Ecol Evol. 2008;23:386–393. doi: 10.1016/j.tree.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Mueller LD, et al. Natural selection vs. random drift: evidence from temporal variation in allele frequencies in nature. Genetics. 1985;111:517–554. doi: 10.1093/genetics/111.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bollback JP, et al. Estimation of 2Nes from temporal allele frequency data. Genetics. 2008;179:497–502. doi: 10.1534/genetics.107.085019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Illingworth CJ, et al. Quantifying selection acting on a complex trait using allele frequency time series data. Mol Biol Evol. 2012;29:1187–1197. doi: 10.1093/molbev/msr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parts L, et al. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res. 2011;21:1131–1138. doi: 10.1101/gr.116731.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woods RJ, et al. Second-order selection for evolvability in a large Escherichia coli population. Science. 2011;331:1433–1436. doi: 10.1126/science.1198914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borgatti SP, et al. Network analysis in the social sciences. Science. 2009;323:892–895. doi: 10.1126/science.1165821. [DOI] [PubMed] [Google Scholar]
- 38.Lazer D, et al. Social science. Computational social science Science. 2009;323:721–723. doi: 10.1126/science.1167742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Lopez B, et al. Social network analysis. Review of general concepts and use in preventive veterinary medicine. Transbound Emerg Dis. 2009;56:109–120. doi: 10.1111/j.1865-1682.2009.01073.x. [DOI] [PubMed] [Google Scholar]
- 41.Jeong H, et al. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 42.Kim PM, et al. Relating three-dimensional structures to protein networks provides evolutionary insights. Science. 2006;314:1938–1941. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 43.Bascompte J. Disentangling the Web of Life. Science. 2009;325:416–419. doi: 10.1126/science.1170749. [DOI] [PubMed] [Google Scholar]
- 44.Hanski I, Mononen T. Eco-evolutionary dynamics of dispersal in spatially heterogeneous environments. Ecol Lett. 2011;14:1025–1034. doi: 10.1111/j.1461-0248.2011.01671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellner SP, et al. Does rapid evolution matter? Measuring the rate of contemporary evolution and its impacts on ecological dynamics. Ecol Lett. 2011;14:603–614. doi: 10.1111/j.1461-0248.2011.01616.x. [DOI] [PubMed] [Google Scholar]
- 46.Hairston NG, et al. Rapid evolution and the convergence of ecological and evolutionary time. Ecol Lett. 2005;8:1114–1127. [Google Scholar]
- 47.Yoshida T, et al. Rapid evolution drives ecological dynamics in a predator-prey system. Nature. 2003;424:303–306. doi: 10.1038/nature01767. [DOI] [PubMed] [Google Scholar]
- 48.Urban M, et al. The evolutionary ecology of metacommunities. Trends Ecol Evol. 2008;23:311–317. doi: 10.1016/j.tree.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 49.Harmon LJ, et al. Evolutionary diversification in stickleback affects ecosystem functioning. Nature. 2009;458:1167–1170. doi: 10.1038/nature07974. [DOI] [PubMed] [Google Scholar]
- 50.Becks L, et al. The functional genomics of an eco-evolutionary feedback loop: linking gene expression, trait evolution, and community dynamics. Ecol Lett. 2012;15:492–501. doi: 10.1111/j.1461-0248.2012.01763.x. [DOI] [PubMed] [Google Scholar]
- 51.Wandeler P, et al. Back to the future: museum specimens in population genetics. Trends Ecol Evol. 2007;22:634–642. doi: 10.1016/j.tree.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 52.Glenn TC, et al. Effects of a population bottleneck on Whooping Crane mitochondrial DNA variation. Conserv Biol. 1999;13:1097–1107. [Google Scholar]
- 53.Thomas CD, et al. Extinction risk from climate change. Nature. 2004;427:145–148. doi: 10.1038/nature02121. [DOI] [PubMed] [Google Scholar]
- 54.Jenouvrier S, Visser ME. Climate change, phenological shifts, eco-evolutionary responses and population viability: toward a unifying predictive approach. Int J Biometeorol. 2011;55:905–919. doi: 10.1007/s00484-011-0458-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Urban MC, et al. A crucial step toward realism: responses to climate change from an evolving metacommunity perspective. Evol Appl. 2012;5:154–167. doi: 10.1111/j.1752-4571.2011.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 57.Hooper DU, et al. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature. 2012;486:105–108. doi: 10.1038/nature11118. [DOI] [PubMed] [Google Scholar]
- 58.Appleby P. Chronostratigraphic techniques in recent sediments. In: Last W, Smol JP, editors. Tracking Environmental Change Using Lake Sediments. Basin Analysis, Coring, and Chronological Techniques. Kluwer Academic Publishers; 2001. pp. 171–201. [Google Scholar]
- 59.Walker M. Quaternary Dating Methods. John Wiley & Sons; 2005. [Google Scholar]
- 60.Engstrom DR, et al. Historical changes in sediment and phosphorus loading to the upper Mississippi River: mass-balance reconstructions from the sediments of Lake Pepin. J Paleolimnol. 2009;4:563–588. [Google Scholar]
- 61.Legrand M, Meyewsky P. Glaciochemistry of polar ice cores: A review. Reviews in Geophysics. 1997;35:219–243. [Google Scholar]
- 62.Bidle KD, et al. Fossil genes and microbes in the oldest ice on earth. PNAS. 2007;104:13455–13460. doi: 10.1073/pnas.0702196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yashina S, et al. Regeneration of whole fertile plants from 30,000-y-old fruit tissue buried in Siberian permafrost. PNAS. 2012;109:4008–4013. doi: 10.1073/pnas.1118386109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knapp M, Hofreiter M. Next Generation Sequencing of Ancient DNA: Requirements, Strategies and Perspectives. Genes. 2010;1:227–243. doi: 10.3390/genes1020227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pushkarev D, et al. Single-molecule sequencing of an individual human genome. Nat Biotechnol. 2009;27:847–850. doi: 10.1038/nbt.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schadt EE, et al. A window into third-generation sequencing. Hum Mol Genet. 2010;19:R227–R240. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 67.Hubisz MJ, et al. Error and error mitigation in low-coverage genome assemblies. PLoS One. 2011;6:e17034. doi: 10.1371/journal.pone.0017034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hebsgaard MB, et al. Geologically ancient DNA: fact or artefact? Trends Microbiol. 2005;13:212–220. doi: 10.1016/j.tim.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 69.Willerslev E, Cooper A. Review Paper. Ancient DNA. Proc R Soc Lond B. 2007;272:3–16. doi: 10.1098/rspb.2004.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Willerslev E, et al. Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol Evol. 2004;19:141–147. doi: 10.1016/j.tree.2003.11.010. [DOI] [PubMed] [Google Scholar]