Abstract
The Xin repeat-containing proteins, Xinα (Xirp1) and Xinβ (Xirp2), localize to the intercalated discs (ICDs) of mammalian hearts. Mouse Xinα (mXinα) directly interacts with β-catenin and actin filaments, potentially coupling the N-cadherin/β-catenin complexes to the underlying actin cytoskeleton and modulating ICD integrity and function. Supporting this possibility, mXinα-null hearts develop ICD structural defects and cardiomyopathy with conduction defects. However, the underlying mechanisms leading to these defects remain unclear. Here, we showed that mXinα also interacted with p120-catenin and cortactin. Different from the β-catenin binding domain, there existed multiple p120-catenin binding sites on mXinα, while only the extreme N-terminus of mXinα containing a SH3-binding motif could interact with cortactin. In mouse heart, a significant fraction of cortactin was co-localized with N-cadherin to ICDs, whereas in mXinα-null heart, this fraction of cortactin was drastically reduced. Therefore, mXinα may modulate ICD integrity and function through its interactions with catenins and cortactin. Analyses of the in vivo consequence of p120-catenin and mXinα interaction revealed that force-expressed mXinα or its fragments significantly suppressed the p120-catenin-induced branching phenotypes. It is known that p120-catenin directly regulates Rho GTPases, leading to the branching phenotype. Thus, mXinα may sequester the p120-catenin from inhibiting RhoA activity and/or from activating Rac1 activity.
Introduction
A vast majority of Xin repeat-containing proteins co-localize with N-cadherin and β-catenin to the intercalated discs (ICDs) of the heart, where they may perform important roles in maintaining the ICD structural integrity (Gustafson-Wagner et al., 2007) and initiating the ICD maturation (Wang et al., 2012a; Wang et al., 2010; Wang et al., 2012b). A pair of paralogous Xin repeat-containing genes, mXinα and mXinβ, exists in the mouse heart. Earlier statistical analyses of differential gene expression and co-expression data have identified that the human homologs of mXinα and mXinβ, called cardiomyopathy-associated 1 (CMYA1) and 3 (CMYA3), respectively, are significantly co-expressed with 13 other known cardiomyopathy-associated genes (Walker, 2001). Later, it was shown that these two gene products contain multiple Xin-repeating units, which are responsible for actin binding (Choi et al., 2007; Grosskurth et al., 2008; Lin et al., 2005; Pacholsky et al., 2004; Wang et al., 1999; Wang et al., 2010; Wang et al., 2012b). Therefore, their human orthologs are also called Xin actin-binding repeat-containing 1 and 2 (XIRP1 and XIRP2). Adult mXinα-deficient hearts exhibit ICD structural defects and develop progressive cardiomyopathy with conduction defects (Chan et al., 2011; Gustafson-Wagner et al., 2007; Lai et al., 2008; Otten et al., 2010). The mXinβ-null hearts fail to form ICDs at the cell termini, which results in severe growth retardation, diastolic dysfunction and early postnatal lethality (Wang et al., 2012a; Wang et al., 2010). The molecular mechanisms of how the loss of Xin proteins leads to these cardiac defects remain to be determined.
Studies on mXinα and XIRP1 proteins have revealed that (1) a minimum of 3 Xin repeating units is required for actin binding (Cherepanova et al., 2006; Pacholsky et al., 2004), (2) mXinα not only binds but also bundles actin filaments (Choi et al., 2007), (3) mXinα directly interacts with β-catenin (Choi et al., 2007), (4) the β-catenin-binding domain on mXinα is in the region (aa# 535-636) overlapping with the Xin repeat region (Choi et al., 2007; Grosskurth et al., 2008), (5) the N- and C-terminal regions of XIRP1 contain Mena/VASP- and filamin c-binding domains, respectively (van der Ven et al., 2006). In addition, screenings of heart cDNA library by yeast two hybrid assays have further identified several actin binding proteins as mXinα-interacting proteins, such as filamin b, tropomyosin, and gelsolin (Choi et al., 2007), suggesting that one of mXinα’s functions may be involved in tightly regulating the dynamics of the actin cytoskeleton underneath the ICD. Through its β-catenin-binding ability, mXinα may be able to further link the N-cadherin/β-catenin complex to the underlying actin cytoskeleton. This linkage may allow mXinα to perform its regulatory role of influencing the cell adhesiveness and signaling at the ICD. Although the molecular details of such linkage and regulation at the ICD are not completely understood, mXinα might function at least in analogy to the recently identified EPLIN, an actin binding protein in epithelial cells, which is capable of stabilizing apical actin bundles to form cadherin-catenin-EPLIN complexes (Abe and Takeichi, 2008). Using yeast two hybrid and co-immunoprecipitation assays, we previously found that mXinα was also associated with p120-catenin (Choi et al., 2007). This association may likely be a direct interaction, and if this should be the case, mXinα would have a great impact on the ICD structure and function. Previous studies in epithelial cells have shown that through their bindings to the cytoplasmic domains of cadherin molecules, p120-catenin and β-catenin are able to regulate cadherin stability and turnover, thereby controlling cellular adhesiveness (Gates and Peifer, 2005; Hong et al., 2010; Nelson, 2008; Provost and Rimm, 1999; Thoreson et al., 2000; Xiao et al., 2007). In addition, p120-catenin has been shown in many non-muscle cells to be able to modulate Rho GTPases activities. Through these mechanisms, p120-catenin can influence many biological processes such as cell-cell junction formation, cell shape change, and cell motility (Anastasiadis et al., 2000; Anastasiadis and Reynolds, 2001; Grosheva et al., 2001; Noren et al., 2000; Noren et al., 2001; Xiao et al., 2007). Whether these mechanisms can also operate within ICD formation and stability remains to be determined. Supporting this possibility, we have previously shown that mXinα-null hearts with ICD defects have decreased p120-catenin expression (Gustafson-Wagner et al., 2007), whereas mXinβ-null hearts which fail to form ICDs had a significant reduction in Rac1 activity (Wang et al., 2010). Toward understanding how mXinα could regulate the ICD adhesion and integrity, here we first characterized the nature of the interaction between mXinα and p120-catenin, and then examined the effects of mXinα expression on the p120-catenin-induced shape change in the context of non-muscle cells. In addition, we showed that mXinα interacted with the ICD-localized cortactin, which was drastically reduced in mXinα-null cardiomyocytes.
Materials and Methods
Animals
All animal procedures were approved and performed in accordance with institution guidelines. The mXinα-null mouse line was generated as described previously (Gustafson-Wagner et al., 2007) and has been backcrossed to and maintained in C57BL/6J strain. Age-matched wild-type and mutant mice were used in all experiments for comparisons.
Constructs of cDNA expression plasmids
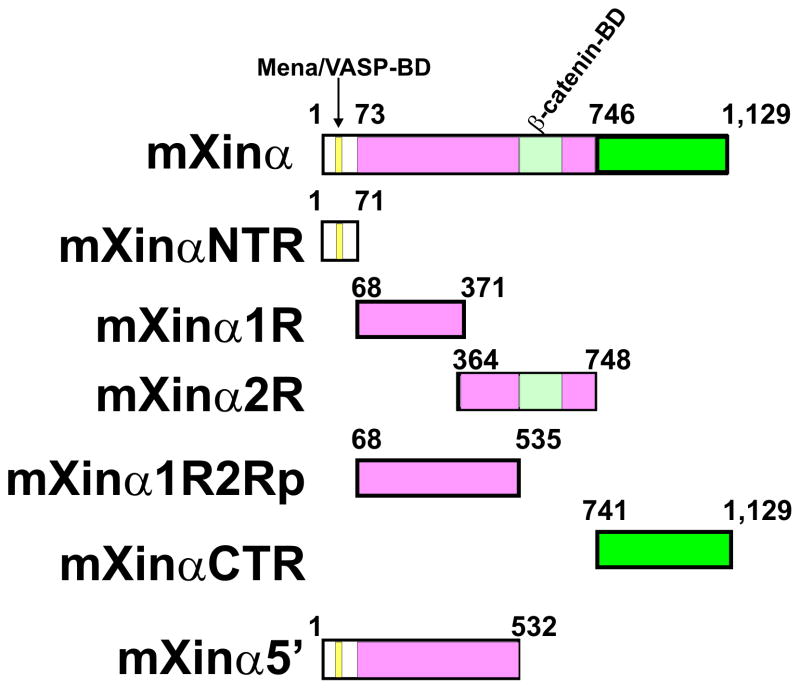
As previously described (Choi et al., 2007; Sinn et al., 2002), expression plasmid pGEX-mXinα was used to produce recombinant proteins GST-mXinα (full-length aa#1-1,129) in E. coli BL21(DE3)pLysS cells. The GST fusion proteins were affinity-purified by glutathione sepharose column (GE Healthcare, Pistcataway, NJ) according to the manufacturer’s protocols. The previously reported bait construct, pGBKT7-mXinα, for yeast two hybrid assays (Choi et al., 2007) was used to further generate various mXinα cDNA fragments (diagram shown in Fig. 1) by PCR-amplification with specific pairs of primers carrying EcoRI adapter sites. The sequences of these primer pairs are listed in Table 1. The EcoRI-digested, PCR-amplified fragments were individually subcloned into the EcoRI site of the pGEX4T-1 vector (GE Healthcare) and the pGBKT7 vector (Clontech). The resulting pGEX plasmids were transformed to BL21 (DE3)pLysS cells to generate GST tagged mXinα fragments, including GST-mXinαNTR (aa#1–71, containing Mena/VASP binding domain (Mena/VSAP-BD)), GST-mXinα1R (aa# 68-371, containing the first half of the Xin repeats), GST-mXinα2R (aa# 364-748, containing the second half of the Xin repeats and overlapped β-catenin binding domain (β-catenin-BD)), GST-mXinα1R2Rp (aa# 68-535, containing the first two third of Xin repeats and lacking of β-catenin-BD), and GST-mXinαCTR (containing aa# 741-1,129). Similarly, the resulting pGBKT7 plasmids together with pGBKT7-mXinα plasmids were further used to release SfiI/SalI-digested inserts and to subclone into the same sites of eukaryotic expression pCMV-Myc vector to generate plasmids for transient transfection experiments. Other control eukaryotic expression plasmids used in co-transfection experiments included pGFPTC22 (Lin et al., 2002) and pcDNA-vinculin-GFP (a generous gift from Dr. Wolfgang Goldmann, University of Erlangen, Germany) encoding GFP-tropomyosin and viculin-GFP, respectively, driven by similar CMV promoter.
Fig. 1.
Schematic representation of mXinα fragments illustrating the amino acid (aa) positions relative to the full-length mXinα. In this study, both Myc and GST tags were fused in frame to the N-termini of these fragments for their expression in CHO and E. coli cells, respectively. The Xin repeat region (aa#89-742) contains 15 Xin repeating units responsible for binding and bundling actin filaments (Wang et al., 2012b). Within the Xin repeat region, a conserved β-catenin binding domain (β-catenin-BD) is located from aa#535 to aa#636 (Choi et al., 2007; Grosskurth et al., 2008). Both mXinαNTR and mXinα5′ fragments contain a previously defined Mena/VASP-binding domain (Mena/VASP-BD) (van der Ven et al., 2006), whereas only mXinα2R fragment possesses β-catenin-BD.
Table 1.
List of primer pair sequences used in PCR to generate various mXinα cDNA fragments.
| Fragment Name | Forward Primer | Reverse Primer | DNA Template |
|---|---|---|---|
| mXinαNTR (aa#1-71) | 5′gccgaattcTGCAGTCGACGAAGGATGG 3′ | 5′cccgaattcCTCCTCGAGGTTCTTCCGAA 3′ | pGBKT7-mXinα |
| mXinα1R (aa#68-371) | 5′cccgaattcAACCTCGAGGAGGCTGTGGCT 3′ | 5′cccgaattcTCCTTGGGTGGCACCTCTGC 3′ | pGBKT7-mXinα |
| mXinα2R (aa#364-748) | 5′cccgaattcGAGGCAGAGGTGCCACCCAAG 3′ | 5′cccgaattcGCTCTCAGCTGCCAGGGAGCC 3′ | pGBKT7-mXinα |
| mXinα1R2Rp (aa#68-535) | 5′cccgaattcAACCTCGAGGAGGCTGTGGCT 3′ | 5′cccgaattcGATGGTACTGGGGCTTCTACC 3′ | pGBKT7-mXinα |
| mXinαCTR (aa#741-1,129) | 5′cccgaattcATGGGCTCCCTGGCAGCTGAG 3′ | 5′cccgaattcTTGGGTGGTCAGGATCTTCTG 3′ | pGBKT7-mXinα |
Full length p120-catenin cDNA was released from pRc/RSV-p120-catenin 1A plasmid (a generous gift from Dr. Albert Reynolds, Vanderbilt University, Nashville, TN) by digestions with EcoRI (fill-in)/KpnI. The released fragment was subcloned into the BglII (fill-in)/KpnI sites of pCMV-HA vector (Clontech Lab, Inc., Mountain View, CA) to generate eukaryotic expression plasmid, pCMV-HA-p120-catenin. To construct pCMV-HA-β-catenin, full-length β-catenin cDNA from pGEX-KG-β-catenin (a generous gift from Dr. Janne Balsamo, University of Iowa) was first subcloned into BamHI/SalI sites of pBluscript SK vector. The released β-catenin cDNA by BamHI (fill-in)/XhoI digestion was then subcloned in frame into SalI (fill-in)/XhoI sites of pCMV-HA vector.
To construct a bacterial expression plasmid for His-p120-catenin recombinant protein, PCR-amplified full length p120-catenin cDNA fragment from pCMV-HA-p120-catenin was directionally cloned into pET160/GW/D-TOPO vector (Invitrogen). The primer pairs used in PCR amplification were 5′caccATGGACGACTCAGAGG 3′ & 5′GGGAACTACGTCTTCTAAATCTTC 3′ for p120-catenin. The resulting plasmids, pET160-p120-catenin were transformed into TOP10 competent cells for DNA propagation and into BL21Star(DE3) competent cells for recombinant protein induction (Invitrogen). His-tagged recombinant protein produced in E. coli was purified by Ni-NTA resin column according to the manufacturer’s protocol (Invitrogen).
All expression clones were sequenced at the Roy J. Carver Center for Comparative Genomics, Department of Biology, University of Iowa, to ensure their fidelity before use. Their expressed products were further confirmed by SDS polyacrylamide gel electrophoresis (SDS-PAGE) and Western blots.
Cell culture, DNA transfection and immunofluorescence microscopy
Chinese hamster ovary (CHO) cells were maintained in Dulbecco’ modified Eagle’s medium plus 10% fetal bovine serum in a humidified incubator at 37°C with 5% CO2. Cells grown on glass coverslips in culture dish were transiently transfected with various combinations of expression plasmids using Lipofectamine 2000 reagent (Invitrogen, Life Technologies, Grand Island, NY) as described previously (Li et al., 2004). 24 hours after transfection, cells were harvested for immunofluorescence staining and microscopy as described previously (Warren and Lin, 1993). The primary antibodies included mouse monoclonal (mAb) 15D2 anti-p120-catenin (Invitrogen) and rabbit polyclonal (pAb) anti-Myc (Abcam, Cambridge, MA). The secondary antibodies used were fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG and trimethylrhodamine isothiocyanate (TMRITC)-labeled goat anti-rabbit IgG.
The expression plasmids used in transient transfection included pCMV-HA, pCMV-HA-p120-catenin, pCMV-Myc, pCMV-Myc-mXinα, pCMV-Myc-mXinα5′, pCMV-Myc-mXinα1R, pCMV-Myc-mXinα2R, pCMV-Myc-mXinα1R2Rp, pCMV-Myc-mXinαCTR, pEGFP-C2, pEGFPTC22 (encoding GFP fused to one of tropomyosin isoform, TC22) (Lin et al., 2002) and pcDNA-vinculin-GFP (a generous gift from Dr. Wolfgang H. Goldmann, University of Erlangen, Germany). To minimize a possible competition between two plasmids for co-expression, each transfection experiment was carried out with equal molar amount of DNAs equivalent to a combination of 1.5 μg pCMV-HA and 1.5 μg pCMV-Myc.
For scoring the branching activity induced by HA-p120-catenin overexpression in combination with the expression of Myc-tag, Myc-mXinα and its fragments, vinculin-GFP, or GFP-TC22, each double transfection experiments were performed three times, and transfected cells expressed both tagged proteins were randomly identified, imaged and scored. Transfected cells with branching phenotype was morphologically defined by the presence of 3 or more dendrite-like cell extensions characterized by long (>10 μm) extensions usually having secondary and/or tertiary branches. The percentage of transfected cells with branching phenotypes was calculated from total cell number counted, and the mean±SD was used for statistical comparison. Under this assay condition, only 2.9±0.8% of CHO cells expressing neither HA-p120-catenin nor Myc-mXinα exhibited branching phenotype.
Co-immunoprecipitation (co-IP) and Western blot
The co-transfection and co-IP experiments were performed to examine the interaction between mXinα and p120-catenin. At 24 hours after co-transfection as described above, cells were harvested and total cell lysates were prepared. Subsequent pre-cleaning and immunoprecipitation with mAb anti-Myc antibody (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) were performed as described previously (Choi et al., 2007). The resulting immunoprecipitates were separated by SDS-PAGE, transferred to nitrocellulose membrane and immunoblotted with rabbit pAb anti-HA (BD Biosciences Clontech, Palo Alto, CA) or mouse mAb anti-p120-catenin for Western blot analysis as described (Choi et al., 2007), except that IRDye 800 conjugated secondary antibodies (Rockland, Gilbertsville, PA) and an Odyssey Imager (Li-Cor Biosciences, Lincoln, NE) were used for the detection of bound primary antibody.
Preparation of epitope-tagged recombinant proteins and pull-down assay
For the pull-down assay from adult mouse heart lysate, the GST-mXinα, GST-mXinαNTR, GST-mXinα1R, GST-mXinα2R and GST-mXinαCTR (Fig. 1) were individually expressed in BL21(DE3)pLysS bacterial cells and bound to glutathione-Sepharose beads (GE Healthcare). After washing with phosphate buffered saline (PBS), GST fusion protein-bound beads were used to pull-down the potential interacting proteins from total heart lysate. The final pull-down materials were solubilized in SDS gel sample buffer and analyzed by Western blot analysis with mouse mAb anti-p120-catenin (Invitrogen) or rabbit mAb anti-cortactin (Cell Signaling Technology, Danvers, MA).
To examine whether mXinα could directly interact with p120-catenin, the pull-down assay was performed using purified recombinant proteins. Purified GST-mXinα (320 nM) and GST (320nM and 0 nM) as negative controls were mixed with 90 nM His-p120-catenin each in binding buffer containing 20 mM HEPES pH7.5, 100mM KCl, 1 mM dithiothreitol, 0.1% Triton X-100, and 2.5 mM phenylmethanesulfonyl fluoride for 2 hours at 4 °C. The glutathione Sepharose beads were then added to each reaction mixture to pull-down GST-tagged fusion protein together with its interacting partners. The bound proteins were further analyzed by Western blot analysis with mouse mAb anti-p120-catenin antibody or mouse mAb anti-His (27E8) antibody (Cell Signaling Technology).
Results
mXinα directly interacts with p120-catenin
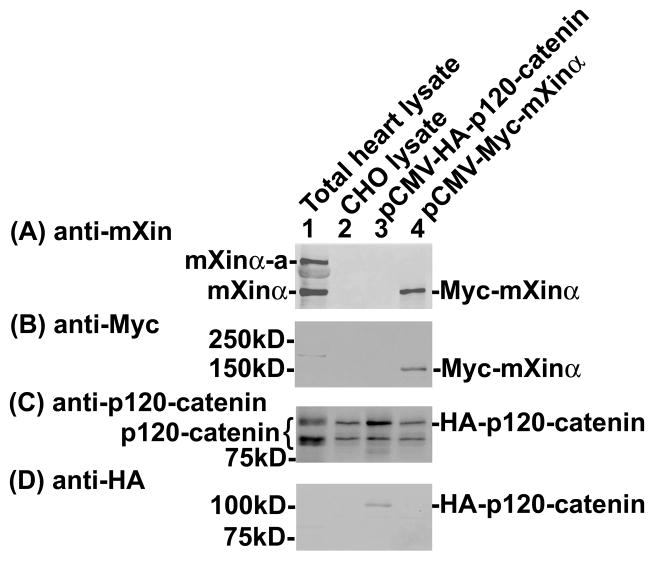
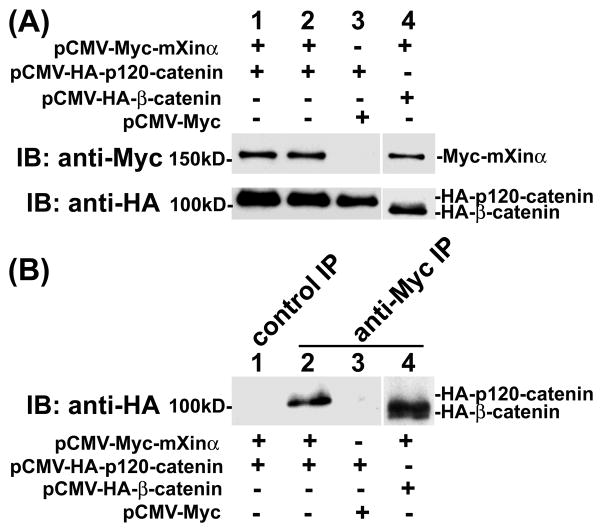
Previously, we showed by yeast two hybrid assay that p120-catenin was one of mXinα-interacting proteins (Choi et al., 2007). Co-transfection, co-IP, and pull-down assays were used in this study to demonstrate that the nature of this interaction was a direct one. As shown in Fig. 2, force-expressed Myc-tagged mXinα and HA-tagged p120-catenin (isoform 1A) had similar sizes as endogenous mXinα and p120-catenin, respectively, from mouse heart. Both force-expressed proteins were readily recognized not only by their respective anti-tag antibodies (Fig. 2B and D) but also by their specific antibodies (Fig. 2A and C). In addition to force-expressed HA-p120-catenin 1A, anti-p120-catenin antibody also recognized CHO endogenous p120-catenin isoform 1 and 3 (lane 2 in Fig. 2C). Similar two isoforms of p120-catenin were detected by this antibody in the total heart lysate (lane 1 in Fig. 2C). These results suggest that the constructed plasmids are capable of encoding epitope-tagged authentic proteins in CHO cells. Transient co-transfections of pCMV-Myc-mXinα with pCMV-HA-p120-catenin or pCMV-HA-β-catenin led to express respective tagged proteins in their cell lysates (Fig. 3A), and subsequent co-IPs from these lysates with mouse mAb anti-Myc revealed that similar to HA-β-catenin (lane 4), HA-p120-catenin was present in the immunoprecipitate for the co-expressed Myc-mXinα (lane 2), but not in the immunoprecipitate for the co-expressed Myc (lane 3 in Fig. 3B). In addition, normal mouse serum could not immunoprecipitate HA-p120-catenin from the co-transfected lysates (control IP, lane 1 in Fig. 3B). Consistent with our previous findings (Choi et al., 2007), these results supported that mXinα interacted with p120-catenin. To show the direct interaction between mXinα and p120-catenin, pull-down assays were carried out with purified recombinant GST-mXinα or GST mixed with purified His-p120-catenin (Fig. 4). Glutathione beads pre-bound with GST-mXinα (lane 1) but not with GST (lane 2 in Fig. 4A) readily pulled down the His-p120-catenin protein, whereas the beads only could not pellet any significant amount of His-p120-catenin as detected by Western blots with both anti-p120-catenin antibody and anti-His antibody (Fig. 4A). The intact GST-mXinα protein was readily detected in the reaction mixture with anti-mXin antibody (lane 1 in Fig. 4B), whereas some of its degraded fragments remained to contain GST-tag, which was detected by anti-GST antibody (Fig. 4C).
Fig. 2.
Western blot analyses of force-expressed Myc-mXinα and HA-p120-catenin proteins in CHO cells. Both force-expressed proteins as detected by anti-tag antibodies (B, D) were also recognized by their own specific antibodies (A, C). In addition, they had their expected sizes similar to that detected in the total heart extract (compare lane 1 with lane 4 in A and lane 1 with lane 3 in C).
Fig. 3.
Association of Myc-mXinα and HA-p120-catenin. CHO cells were co-transfected with plasmids encoding Myc-mXinα and HA-p120-catenin (lanes 1 & 2), with plasmids encoding Myc tag and HA-p120-catenin as a negative control (lane 3), or with plasmids encoding Myc-mXinα and HA-β-catenin as a positive control (lane 4). Equal molar amounts of each plasmids and empty vectors were used in each co-transfection to minimize the promoter competition between two different expressing plasmids. (A) Western blots of total lysates prepared from various co-transfected CHO cells with anti-tag antibodies show the input amounts of force-expressed proteins in each co-transfection. (B) Co-IP assay. Co-transfected cell lysates were immunoprecipitated with control normal mouse serum (lane 1, control IP) or with anti-Myc antibody (lanes 2~4, anti-Myc IP). The resulting immunoprecipitates were immunoblotted with anti-HA antibody to detect the associated proteins. Both HA-p120-catenin (lane 2) and HA-β-catenin (lane 4) were readily found to be associated with Myc-mXinα. Neither the control IP (lane 1) nor anti-Myc IP from lysates of cells co-transfected with pCMV-HA-p120-catenin and pCMV-Myc empty vector (lane 3) detected HA-p120-catenin.
Fig. 4.
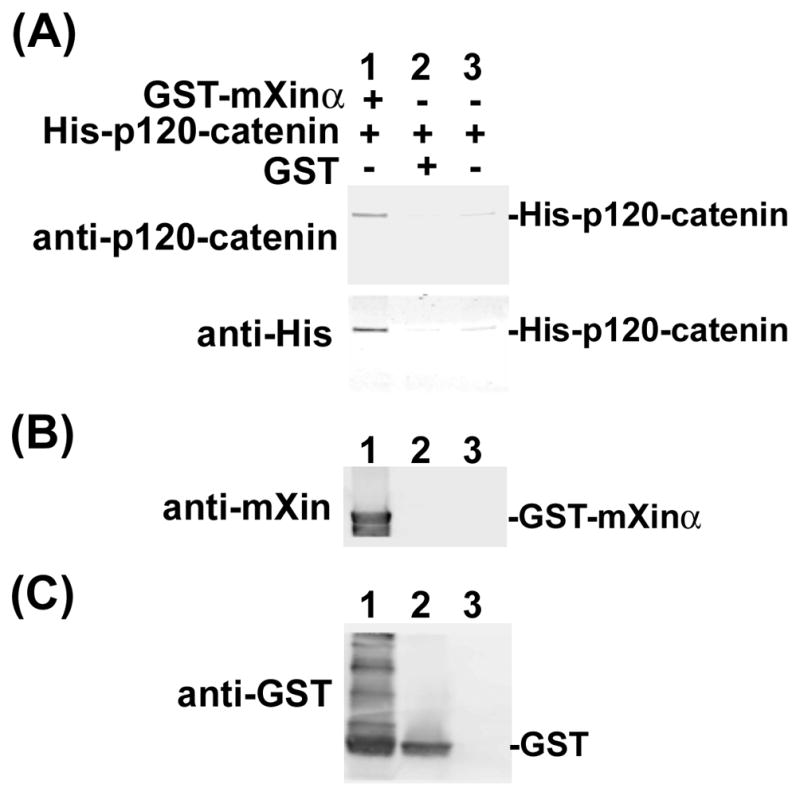
Recombinant GST-mXinα directly binds to His-p120-catenin. The pull-down assay was performed with glutathione-Sepharose beads to pull down GST-containing proteins and its interacting proteins from a mixture of purified His-p120-catenin (90 nM) and purified GST-mXinα (320 nM) (lane 1). Purified GST (320 nM) was used as a negative control (lane 2), whereas the other negative control contains no GST protein (lane 3). The pulling-down proteins were eluted from beads and analyzed by Western blot with mouse mAb anti-p120-catenin and anti-His (A), with rabbit polyclonal U1013 anti-mXinα (B) and with mouse mAb anti-GST (C).
Multiple p120-catenin-binding sites exist in mXinα
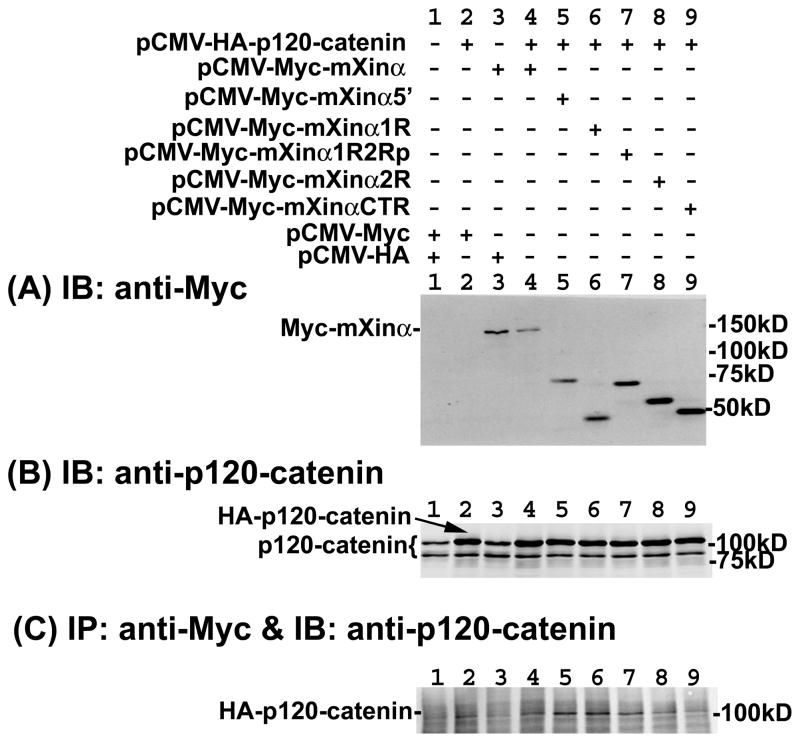
To map the p120-catenin-binding sites on mXinα, we first generated both eukaryotic and prokaryotic expressing plasmids encoding mXinα and its various fragments (Fig. 1) fused to Myc or GST tag. We then performed (i) co-IP from co-transfected CHO cells to detect the association of co-expressed HA-p120-catenin (Fig. 5) and (ii) pull-down experiments to detect proteins from total heart lysates that bind to GST fusion protein-containing beads (Fig. 6). Force-expressed Myc-mXinα and its various fragments in CHO cells all migrated to their respective sizes as detected by anti-Myc Western blot (lanes 3~9 in Fig. 5A). CHO cells co-transfected with empty vectors expressed two endogenous p120-catenin isoforms, one of which co-migrated with the force-expressed HA-p120-catenin (lane 2 and 4~9 in Fig. 5B). The immunoprecipitates with anti-Myc antibody co-pelleted down HA-p120-catenin band from co-transfected cell lysates with full-length mXinα (lane4) and all of its fragments (lanes 5~9 in Fig. 5C), but not significantly from co-transfected cell lysates with both empty vectors (lane 1) or with either one of empty vectors (lanes 2~3 in Fig. 5C). These results suggest that mXinα may have multiple binding sites for p120-catenin. This multiple binding site possibility was further supported by the pull down assays with glutathione beads pre-bound with various GST-mXinα fragments (Fig. 6A). Beads containing all mXinα fragments, including mXinαNTR, mXinα1R, mXinα2R and mXinαCTR were capable of pulling down the p120-catenin from total mouse heart lysates (lanes 1~4), whereas beads only did not pull down p120-catenin (lane 5 in Fig. 6A). Although this pull-down experiment was not carried out by equal molar amounts of mXinα fragment pre-bound beads, the results were consistent with the presence of multiple p120-catenin-binding sites on mXinα. Additionally, mXinα fragments seemed to preferentially associate with p120-catenin isoform 1, as evident from differences in the relative ratio of isoform 1 and 3 detected in the pull-down lanes (lanes 1~4) versus endogenous heart lysate lane (lane 6 in Fig. 6A). Therefore, multiple p120-catenin-binding sites exist in different regions of mXinα.
Fig. 5.
Force-expressed mXinα and its various fragments are able to immunoprecipitate HA-p120-catenin from co-transfected cell lysates. (A, B) Western blots of cell lysates with anti-Myc (A) and anti-p120-catenin (B) to show the input amounts of force-expressed fusion proteins. CHO cells did not express mXinα (lanes 1–2 of A), but expressed both p120-catenin isoform 1 and 3 (lane 1 of B). Force-expressed HA-p120-catenin migrated to the similar position as that of endogenous p120-catenin isoform 1 (indicated by arrow in lanes 2 of B). (C) Western blot of the anti-Myc immunoprecipitates. The anti-Myc immunoprecipitates from lysates of cells transfected with a mixture of empty vectors (lane 1) or with mixtures of one of plasmids and one of empty vectors (lanes 2 and 3) did not contain HA-p120-catenin. On the other hand, all of the immunoprecipitates from co-transfection of HA-p120-catenin with Myc-tagged full-length mXinα (lane 4) and its various fragments (lanes 5~9) co-pelleted a significant amount of HA-p120-catenin, suggesting the presence of multiple p120-catenin interacting sites on mXinα.
Fig. 6.
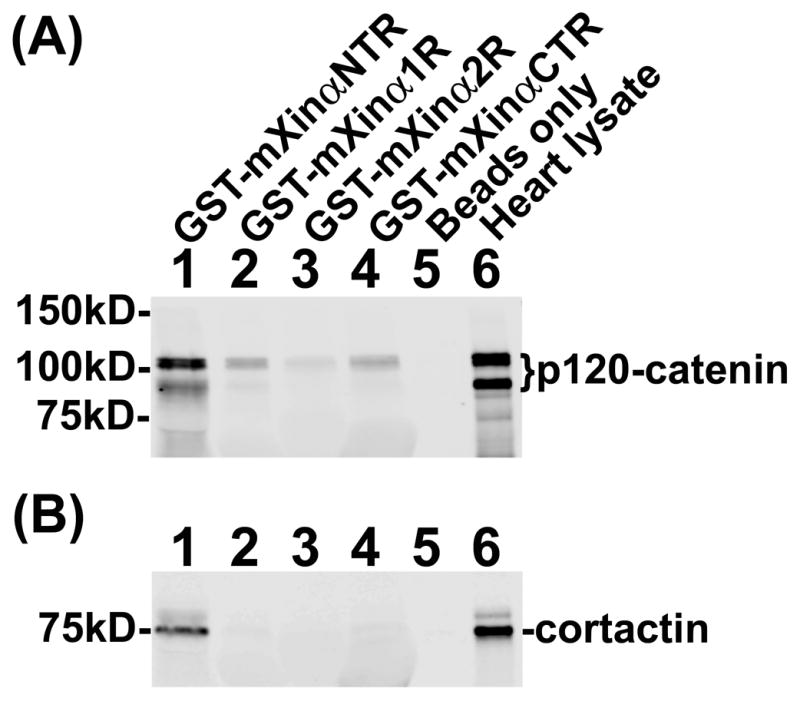
Pull-down assays from heart lysates. Glutathione Sepharose beads pre-bound with various GST-mXinα fragments were used to pull down associated proteins from total heart lysates. After washing the beads, the bound proteins were eluted from beads by gel sample buffer and analyzed on Western blot with anti-p120-catenin (A) and with anti-cortactin (B). The heart lysates contained p120-catenin isoform 1 and 3 (lane 6 in A), which were clearly associated with the N-terminal region of mXinα (GST-mXinαNTR, lane 1). The p120-catenin isoform 1 could be pulled down by mXinα fragments, including the first half of Xin repeats (mXinα1R), the second half of Xin repeats (mXinα2R) and the C-terminal region of mXinα (mXinαCTR) (lanes 2, 3 and 4, respectively). These results also suggest multiple p120-catenin-interacting sites on mXinα. This nature appears to be specific to p120-catenin, because only beads pre-bound with GST-mXinαNTR could pull down cortactin, another mXinα-interacting protein, from total heart lysates (lane 1 in B). For a negative control, glutathione Sepharose beads only pulled down neither p120-catenin nor cortactin (beads only, lane 5 in A and B).
The N-terminal region of mXinα interacts with cortactin
The Xin repeat-containing proteins were initially recognized as proline-rich proteins, which contained consensus Src homology 3 (SH3)-binding motifs (Wang et al., 1999). Scansite (http://scansite3.mit.edu/#home) motif serach have predicted many SH3-binding motifs on mXinα, which would potentially interact with SH3 domains on various adaptor/scaffold proteins, including cortactin. We have then subcloned some of these SH3 domains in bacterial expression vector to produce His-tagged fusion proteins. Initial pull down screen confirmed that beads coated with His-cortactin SH3 was able to pull-down GST-mXinα (data not shown). Furthermore, it is recently reported that cortactin associates with Kv1.5 channel and is required for N-cadherin-mediated enhancement of Kv1.5 channel activity in cardiomyocytes (Cheng et al., 2011). Therefore, the same pulled-down materials described above (Fig. 6A) were further used in Western blot with anti-cortactin antibody to test the interaction between mXinα and cortactin. The results shown in Fig. 6B confirmed that only GST-mXinαNTR containing a predicted SH3-binding motif (aa#30-44) could pull down cortactin from the heart lysates.
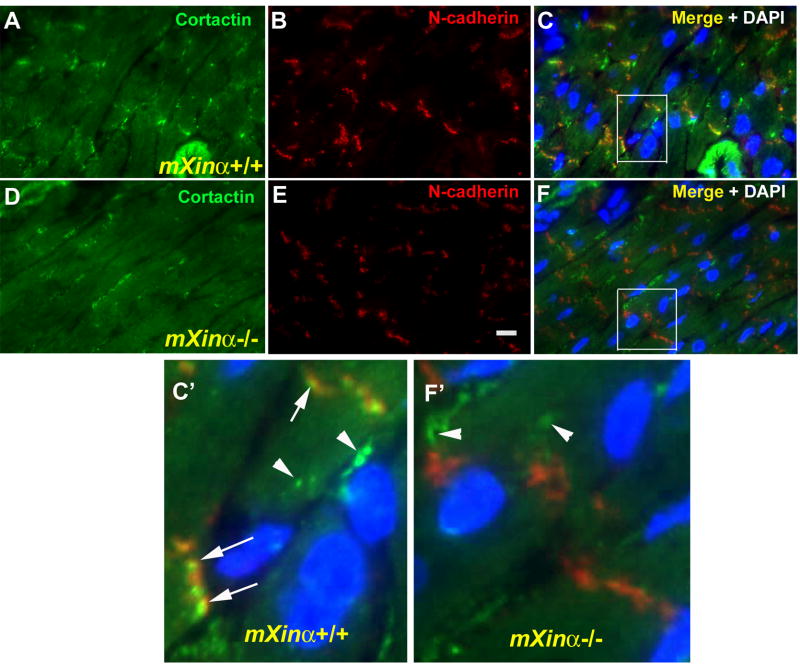
A population of cortactin colocalizes with N-cadherin to ICDs of the heart and this ICD-localized coratctin was greatly reduced in mXinα-null cardiomyocytes
To investigate whether cortactin was pulled down by GST-mXinαNTR from cardiomyocytes or from non-muscle cells such as cardiac fibroblasts, smooth muscle cells or endothelial cells, double-label immunofluorescence microscopy with anti-cortactin and anti-N-cadherin was performed on heart frozen sections prepared from wild-type and mXinα-null littermates. As shown in Fig. 7A–C and C′, a significant amount of cortactin (indicated by arrows in Fig. 7C′) was localized with N-cadherin to the ICDs of wild-type cardiomyocytes, consistent with the previous report that in isolated cardiomyocytes, cortactin has been localized to the ICD and the cortex near the cell surface (Cheng et al., 2011). Studies in cardiac-specific N-cadherin conditional knockout mice further revealed that cortactin is required for N-cadherin regulation of Kv1.5 channel function (Cheng et al., 2011), although the molecular mechanism for this requirement remained unclear. In mXinα-null cardiomyocytes, the ICD-localized fraction of cortactin was largely diminished, whereas the diffused and cortical-puncta staining of cortactin remained unchanged (Fig. 7D~F and F′). Together, our results suggest that mXinα interacting with the ICD-localized cortactin may provide an account for the requirement of cortactin in N-cadherin-mediated Kv1.5 channel function.
Fig. 7.
A population of cortactin is localized near N-cadherin at the ICDs of wild-type cardiomyocytes, and this ICD-localized cortacin is drastically reduced in mXinα-null cardiomyocytes. Double-label immunofluorescence with mouse mAb anti-N-cadherin (in red) and rabbit mAb anti-cortactin (in green) was performed on 4-μm frozen sections of hearts from postnatal day 19.5 wild-type (A–C) and mXinα-null (D–F) mice. (C) and (F) show the merged images of (A, B) and (D, E), respectively. The boxes regions in (C) and (F) were 5 times enlarged by Adobe Photoshop and shown in (C′) and (F′), respectively. In both wild-type and mXina-null cardiomyocytes, most of cortactin were found in diffuse localization and in puncta near the cortex (indicated by arrowheads in C′ and F′). In contrast, ICD-localized cortactin (indicated by arrows in C′) was only detected in the wild-type cardiomyocytes. Bar in (E) equals to 10 μm for A~F images.
mXinα suppresses the p120-catenin-induced branching phenotype in CHO cells
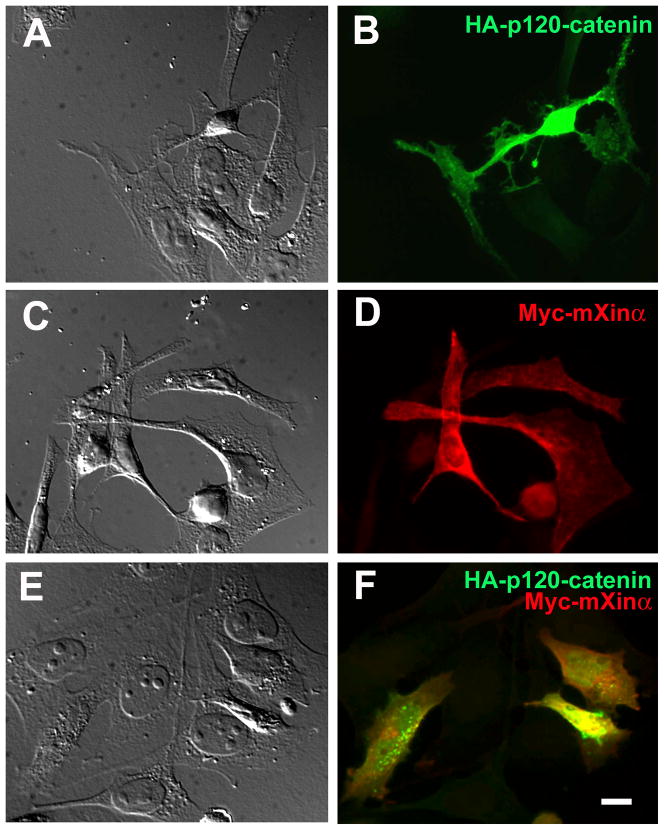
It has been shown that overexpression of p120-catenin in many cell lines induces a striking dendrite-like or branching phenotype (Anastasiadis et al., 2000; Grosheva et al., 2001; Noren et al., 2000; Reynolds et al., 1996). We used this branching phenotype as an assay to further study the in vivo consequence of the direct interaction between mXinα and p120-catenin. We found that co-expressed mXinα could significantly suppress the p120-catenin-induced branching phenotype in CHO cells (Fig. 8 and 9). Similar to previous reports and review in (Anastasiadis and Reynolds, 2001), p120-catenin overexpression in CHO cells caused an extensive branching phenotype characterized by extreme arborization of cellular processes (Fig. 8A and B). This branching phenotype was not significantly observed in the transfected cells overexpressing only mXinα (Fig. 8C and D). In contrast, co-expression of mXinα and p120-catenin in CHO cells appeared to suppress the p120-catenin-induced arborizing processes (Fig. 8E and F).
Fig. 8.
Force-expressed Myc-mXinα suppresses the p120-catenin-induced branching phenotype. CHO cells were co-transfected with a mixture of pCMV-HA-p120-catenin and pCMV-Myc vector (A, B), a mixture of pCMV-HA vector and pCMV-Myc-mXinα (C, D), or a mixture of pCMV-HA-p120-catenin and pCMV-Myc-mXinα (E, F). 24 hours after transfection, the cells were indirectly labeled with anti-p120-catenin (in green) (A, B), or anti-Myc (in red) (C, D), or both antibodies (E, F). Phase-contrast micrographs were shown in A, C and E, and their corresponding immunofluorescent micrographs were shown in B, D and F. Bar = 10 μm. Transient expression of HA-p120-catenin in CHO cells resulted in extensive branching in a large population of transfected cells (B). This branching phenotype was not frequently observed in the transfected cells with Myc-mXinα overexpression (D). Furthermore, the co-transfection with both plasmids encoding HA-p120-catenin and Myc-mXinα resulted in significantly suppressing the p120-catenin-induced branching phenotype (F).
Fig. 9.
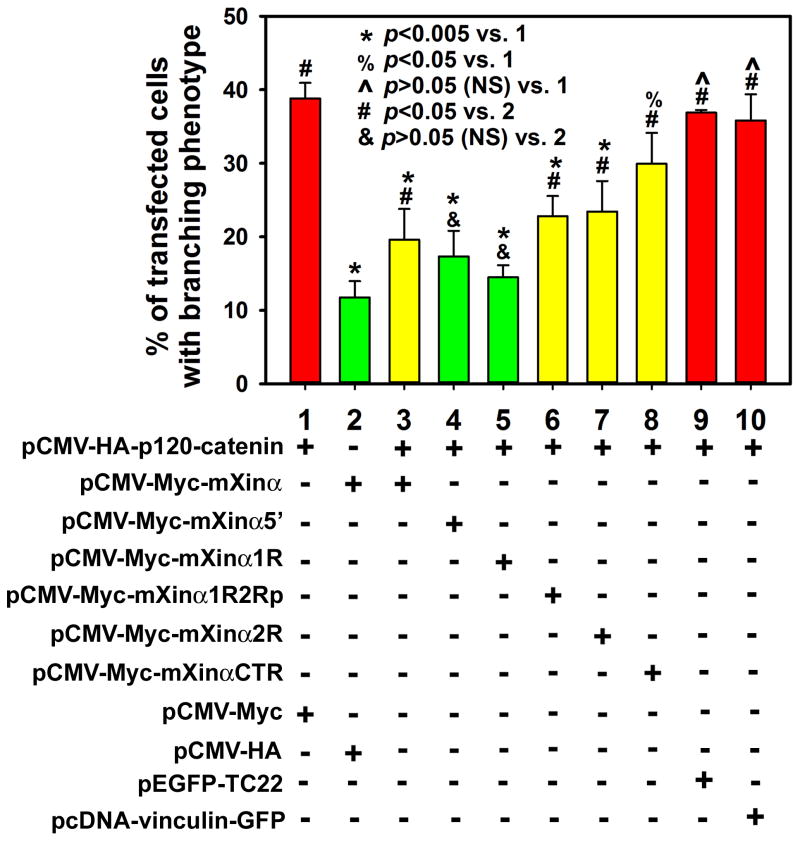
The p120-catenin-induced branching phenotype can be suppressed by co-expressed Myc-full-length mXinα and its various fragments. Transfected cells from each co-transfection were scored for their ability to generate extensive branching phenotype, using criteria as described under Materials and Methods. Data were plotted as % of transfected cells with branching phenotype and the means±SD of three separate experiments performed in each co-transfection case were compared using t test. When compared to HA-p120-catenin expression control (co-transfection #1), the expression of mXinα and all of its fragments significantly suppressed the p120-induced branching phenotype (*, p<0.05 and %, p<0.05). When compared to Myc-mXinα expression control (co-transfection #2), the expression of only mXinα5′ (co-transfection #4) and mXinα1R (co-transfection #5) could reduce the p120-induced branching phenotype to the same level of mXinα control (&, p>0.05, NS, not significant), suggesting a full rescue. On the other hand, the co-expression of full-length mXinα, mXinα1R2Rp, mXinα2R or mXinaCTR only partially suppressed the branching phenotype, because the percentages of cells with branching phenotypes in these co-transfections remained significantly higher than the control co-transfection #2 (#, p<0.05). Co-expression of GFP-TC22 (a non-muscle tropomyosin) (co-transfection #9) or vinculin-GFP (a focal adhesion component) (co-transfection #10) in the co-transfected cells did not suppress the p120-catenin induced braching phenotype (^, p>0.05, NS).
Using extensive branching phenotype as criteria, we scored the percentage of transfected cells with branching phenotype from each co-transfection of pCMV-HA-p120-catenin with various plasmids encoding full-length Myc-mXinα or its fragments. The means ± SD of three separate experiments for each co-transfection were shown in Fig. 9. Overexpression of HA-p120-catenin with Myc control resulted in a large fraction (38.8±2.1%) of transfected cells with extensive branching (column 1 in Fig. 9), whereas only 11.8±2.2% of the transfected cells overexpressing Myc-mXinα and HA control had branching phenotypes (column 2 in Fig. 9). Co-expression of Myc-mXinα with HA-p120-catenin significantly reduced the fraction of transfected cells with branching phenotype to 19.6±4.2% (comparison between columns 1 and 3, p=0.002, t test). However, this percentage although reduced was still significantly higher than 11.8% mXinα control (comparison between columns 2 and 3, p=0.042), suggesting that under the co-transfection condition used here, full-length mXinα only partially suppressed the p120-catenin-induced branching phenotype. Interestingly, the co-expression of all of different mXinα fragments resulted in significantly suppressing the p120-catenin-induced branching phenotype (columns 4~8 in Fig. 9, p<0.05), consistent with the idea that mXinα contained multiple p120-catenin-binding sites as suggested from the above co-IP and pull-down results. In contrast, co-expression of vinculin-GFP (a focal adhesion component) or GFP-TC22 (one of non-muscle tropomyosin isoforms) in the co-transfected cells did not suppress the p120-catenin-induced branching phenotype (35.8±3.6% or 36.9±0.4%, respectively). Although all fragments tested showed suppression activity, only co-expression of mXinα5′ (aa#1-532) or mXinα1R (aa#68-371) could fully suppress the p120-catenin-induced branching phenotype to the level (17.3±3.5% or 14.5±1.6%, respectively) not significantly different from the mXinα alone control (11.8±2.2%). It is worthwhile to note that the most effective regions on mXinα in suppressing the p120-catenin-induced branching phenotype located outside the previously defined β-catenin binding domain (Choi et al., 2007). It is known that overexpression of p120-catenin not only induces branching morphology but also disrupts stress fibers and focal adhesions (Anastasiadis et al., 2000; Grosheva et al., 2001; Noren et al., 2000). Whether mXinα or its fragments can also rescue the disrupted stress fibers and/or focal adhesions remains to be determined.
Discussion
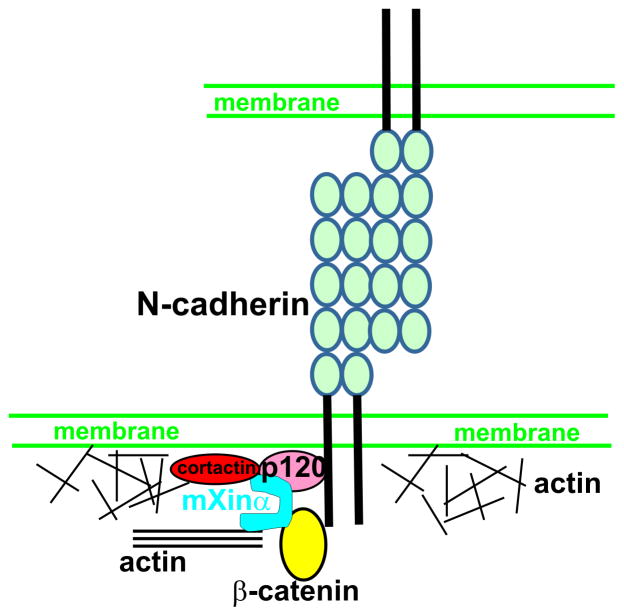
In this study, we have shown that mXinα directly interacts with p120-catenin. Results from pull-down and co-transfection/co-IP with different mXinα fragments suggest multiple p120-catenin binding sites on mXinα protein. This is further supported by the evidence that different mXinα fragments are able to suppress the p120-catenin-induced branching phenotype in co-transfected CHO cells. It is known that cells with arborized morphology induced by p120-catenin overexpression are accompanied by a loss of actin stress fibers and a reduction in the number and size of focal adhesions (Anastasiadis et al., 2000; Grosheva et al., 2001; Noren et al., 2000). To test whether overexpression of either tropomyosin (stabilizing stress fibers) or vinculin (enhancing focal adhesions) could have any effect on the p120-catenin-induced branching, we performed co-transfection of pCMV-HA-p120-catenin with either pEGFPTC22 or pcDNA-vinculin-GFP plasmid. The results revealed that overexpression of neither protein could rescue the p120-catenin-induced branching phenotype, suggesting that the p120-catenin overexpression acts upstream of stabilization of stress fibers and focal adhesions. In contrast, force-expression of mXinα or its various fragments all significantly suppressed the p120-catenin-induced branching phenotype. The most effective fragments for this morphological rescue are mXinα5′ (aa#1-532) and mXinα1R (aa# 68-371, the first half of the Xin repeats of mXinα, which do not contain the previously identified β-catenin-BD (Choi et al., 2007). Interestingly, both mXinα2R and mXinα1R2Rp fragments have similar effectiveness in suppressing the p120-catenin-induced branching phenotype (Fig. 9) and pulling down p120-catenin in co-IP (Fig. 5), despite that only mXinα2R contains β-catenin-BD. These results suggest that the binding of endogenous β-catenin to force-expressed mXinα may have very little effect on the further interaction between mXinα and p120-catenin. Coincidently, two most effective fragments in suppressing the p120-catenin-induced branching phenotype (mXinα5′ and mXinα1R) also have a strong ability to co-IP the force-expressed HA-p120catenin when co-expressed (lanes 5 & 6 in Fig. 5C). The observed suppression of the p120-catenin-induced branching phenotype by mXinα or its various fragments is very similar to that by the co-expressed cadherin with a functional p120-catenin binding domain (Grosheva et al., 2001; Noren et al., 2000). The results from the present study have further suggested that the N-terminal SH3-binding motif of mXinα is capable of interacting with ICD-localized cortactin. Loss of mXinα in mXinα-null cardiomyocytes results in drastically reducing the ICD-localized cortactin, which may in part account for the development of the ICD structural defects in adult mXinα knockout heart. It is previously reported that p120-catenin in cooperation with cortactin regulates lamellipodial dynamics and cell adhesion (Boguslavsky et al., 2007). Based on these results and others (Boguslavsky et al., 2007; Choi et al., 2007; Pacholsky et al., 2004; van der Ven et al., 2006), we propose a model (Fig. 10) that mXinα via direct interactions with both β-catenin and p120-catenin links the N-cadherin/catenin complex to the underlying actin cytoskeleton and via its complex with cortactin and p120-catenin modulates the dynamics of cortical actin network. Thus, mXinα can play important roles in maintaining the integrity and function of the adherens junctions at ICDs of the heart.
Fig. 10.
A schematic model suggesting that mXinα plays important roles in maintaining the integrity and function of the adherens junctions at ICD of the heart. mXinα via its directly interactions with β-catenin, p120-catenin and actin filaments can link the N-cadherin/catenin complex to actin cytoskeleton, and regulate the stability and function of the ICD of the heart. To carry out these roles, mXinα control the dynamics of cortical actin cytoskeleton underneath the ICD membrane through its ability to interact with cortactin and p120-catenin.
In the cardiac muscle, both long isoform 1 and short isoform 3 of p120-catenin have been shown to be expressed and localized to the adherens junctions of the ICDs (Montonen et al., 2001). Unlike β-catenin, the p120-catenin is relatively stable in the cytosol. However, in normal epithelial cells, the majority (>90%) of p120-catenin is found to be associated with cadherins at the adherens junctions (Thoreson et al., 2000). Although the roles of p120-catenin in cardiomyocytes remain largely unknown, several lines of evidence from studies in other cell types and cancer cells indicate two major functions for p120-catenin. First, depending on the cell context (e.g., normal cells, cancer cells, or cadherin-deficient cells), the binding of p120-catenin to cadherin can exert a positive (Navarro et al., 1998; Thoreson et al., 2000; Yap et al., 1998) or negative (Aono et al., 1999; Ohkubo and Ozawa, 1999; Ozawa and Kemler, 1998) effect on the cadherin clustering and the strength of adhesion. Furthermore, p120-catenin regulates cell surface cadherin levels by controlling their turnover and trafficking (Davis et al., 2003; Ireton et al., 2002; Xiao et al., 2003; Xiao et al., 2007). Second, p120-catenin directly interacts with RhoA leading to decrease in RhoA activity and with RhoGEF effectors activating Rac1 and Cdc42 activities (Anastasiadis et al., 2000; Anastasiadis and Reynolds, 2001; Grosheva et al., 2001; Noren et al., 2000). These small GTPases can further modulate the dynamics of actin cytoskeleton, leading to changes in cell shape (branching phenotype), motility and adhesion (Anastasiadis, 2007; Anastasiadis and Reynolds, 2001). In this study, we show that cardiac muscle-restricted mXinα protein is capable of binding to p120-catenin and rescuing the p120-catenin-induced branching phenotype. The existence of common functions of p120-catenin in diverse cell types as shown by these studies indicate that p120-catenin may play the same roles, stabilizing N-cadherin and modulating Rho GTPases, in cardiomyocytes. Furthermore, our results suggest that mXinα together with cortactin is able to modulate the p120-catenin activity for shape change and adhesion at the ICDs. Supporting this possibility, mXinα-deficient hearts express significantly decreased amounts of N-cadherin and p120-catenin, resulting in weak adhesion and progressive ICD structural defects and cardiomyopathy (Gustafson-Wagner et al., 2007). The most effective mXinα fragment (mXinα1R) in rescuing the p120-catenin-induced branching phenotype contains the first half of the Xin repeats, which is highly conserved among all Xin proteins including mXinβ (Grosskurth et al., 2008). Therefore, mXinβ that has been shown to be essential for the initiation of ICD formation (Wang et al., 2012a) is likely able to interact and modulate p120-caetinin activity and Rho GTPases in the hearts. A significant decrease in the active Rac1 level has been detected in the mXinβ-null hearts (Wang et al., 2010) further support this mXinβ’s role. The most of p120-catenin interacting sites appear to be different from the previously defined and highly conserved β-catenin binding domain (Choi et al., 2007). Potentially simultaneous binding of mXinα and mXinβ to both β-catenin and p120-catenin may cooperatively modulate the integrity and function of the ICD, although the underlying molecular mechanisms are not completely understood. During postnatal development, plasma membrane polarization in cardiomyocytes leads to the formation of the locally specialized architecture of ICDs. mXinβ through its ability to modulate p120-catenin and Rho GTPase activity as well as to potentially translate from ICD-localized messages (Wang et al., 2012b) could play an initiating role in the polarization of N-cadherin, mXinα, and other known ICD components to the cell termini (Wang et al., 2012a). How local domains in the plasma membrane are formed and how they control local ICD architecture in conjunction with the cytoskeleton remain a mystery.
In the present study, we have also identified a cortactin-binding site at the N-terminal fragment of mXinα. Cortactin, a known actin-binding protein, is capable of interacting with many other actin-associated proteins, such as Arp2/3 complex, caldesmon, and influencing the organization of the cortical actin network. The ability of cortactin to alter the cortical actin network renders this protein a critical modulator for many cellular processes such as adhesion, migration, endocytosis and tumor invasion (Ammer and Weed, 2008). Furthermore, it has been shown that the N-terminus of cortactin interacts and cooperates with p120-catenin to regulate lamellipodial dynamics and cell adhesion (Boguslavsky et al., 2007). The motifscan program predicts a direct interaction between mXinα and the SH3 domain on the extreme C-terminus of cortactin. The cortactin-SH3-binding motif is predicted to be located at the N-terminal aa#30-44 (PEGLPPPPPKETFSK) of mXinα. The results from our pull-down assay (Fig. 6B), showing that cortactin binds only to GST-mXinαNTR beads, confirmed this prediction. This interaction is further supported by the localization of a population of cortactin with N-cadherin to the ICDs and by the reduction of this population of cortactin in mXinα-null cardiomyocytes (Fig. 7). The cortactin-SH3-binding motif sequence is homologous to that of human Xinα (hXinα or XIRP1), which is partially overlapped with the previously defined Mena/VASP-binding domain (Mena/VASP-BD) (aa #19–36, AEDLPLPPPPALEDLPLP) from hXinα/XIRP1 protein (van der Ven et al., 2006). The Mena/VASP protein family at the lamellipodia and filopodia can spatially regulate actin polymerization (Eigenthaler et al., 2003; Scott et al., 2006). While the Mena/VASP-BD is only present in the placental mammal Xinα proteins, the sequences #34–44 of the cortactin-SH3-binding motif are highly conserved among all vertebrate Xinα proteins except lamprey Xin protein (Grosskurth et al., 2008). However, both Mena/VASP-BD and cortactin-SH3-binding motif sequences are not found in Xinβ and may represent the novel and likely derived functional roles of mammalian Xinα. This is consistent with the notion that mXinβ function cannot fully compensate for the loss of mXinα in the adult heart, because mXinα-deficient hearts even with the up-regulation of mXinβ develop ICD structure defects and cardiomyopathy (Gustafson-Wagner et al., 2007). Another binding region interacting with actin-crosslinking protein, filamin c (muscle-specific isoform), has been previously reported on the large variant of hXinα/XIRP1 protein (van der Ven et al., 2006). The conserved filamin c-binding region is also found in the C-terminus of mXinα-a but not in mXinα or mXinβ (Grosskurth et al., 2008).
Using yeast two hybrid screening, we have previously found that mXinα can interact with many other actin-binding proteins such as filamin b (crosslinking actin filaments), tropomyosin (stabilizing actin filaments), gelsolin (severing actin filaments and binding to plus end of filament fragments), and vinculin (anchoring actin filaments to the membrane at cell-cell and cell matrix junctions) (Choi et al., 2007), although their binding regions are awaiting to be mapped. Furthermore, both mXinα and mXinβ with their conserved Xin repeats are able to bind and bundle actin filaments (Choi et al., 2007; Pacholsky et al., 2004). Taken together, the ability of mXinα to interact with these actin regulatory proteins at the cortical actin network near the membrane suggests an important role for mXinα in modulating the strength and integrity of ICDs as well as in transmitting signals through the N-cadherin/catenin complex. Supporting this role, it has been reported that conditional deletion of non-muscle myosin IIB in the heart results in progressive hypertrophic cardiomyopathy with disrupted ICDs (Ma and Adelstein, 2012; Ma et al., 2010), very much similar to mXinα knockout mouse hearts (Gustafson-Wagner et al., 2007). Non-muscle myosin II acts on bundled/organized actin filaments near the cortex, providing tension for the ICD strength and signaling. Coincidently, loss of non-muscle myosin IIB in the heart results in marked reduction of mXinα, suggesting that non-muscle myosin IIB in cooperation with mXinα regulates the integrity and function of ICDs. Non-muscle myosin IIC also localizes to the ICDs. Complete loss of non-muscle myosin IIC in the hearts of non-muscle myosin IIB hypomorphic mice accelerates the development of cardiomyopathy and impairs the localization of N-cadherin and β-catenin in the ICDs (Ma and Adelstein, 2012). These results suggest that both non-muscle myosin IIB and IIC are required for maintaining the ICD integrity. The molecular mechanisms for the cooperation between mXinα and non-muscle II in regulating the cortical actin network near the ICDs remain to be determined.
Conclusions
mXinα directly interacts with p120-catenin and suppresses the p120-catenin-induced branching phenotype, likely via the alterations in Rho GTPase activities. Multiple p120-catenin-binding sites distinctive from the previously identified β-catenin-binding domain exist in mXinα. Furthermore, the N-terminal region of mXinα containing a consensus SH3-binding motif can pull down cortactin from total heart lysates. A population of cortactin is localized with N-cadherin to the ICDs of wild-type cardiomyocytes, and this fraction of cortactin is drastically reduced in the ICDs of mXinα-null cardiomyocytes. The findings from this and previous studies indicate that mXinα is able to link the adherens junction to the underlying actin cytoskeleton and to modulate the cortical actin organization for maintaining the integrity and function of the ICD in the heart.
Highlights.
mXinα interacts with p120-catenin and suppresses p120-catenin-induced branching.
Multiple p120-catenin-binding sites present on mXinα.
N-terminus of mXinα containing a consensus SH3-binding motif binds to cortactin.
mXinα links and modulates the adherens junctions to the underlying actin network.
Acknowledgments
This work was supported by US National Institutes of Health grant HL107383 to JJCL and partly by grant (#NSC 100-2320-B-039-025) support to Dr. Te-Ling Lu from the National Science Council, Taiwan, ROC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe K, Takeichi M. EPLIN mediates linkage of the cadherin catenin complex to F-actin and stabilizes the circumferential actin belt. Proc Natl Acad Sci U S A. 2008;105:13–19. doi: 10.1073/pnas.0710504105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammer AG, Weed SA. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell Motil Cytoskeleton. 2008;65:687–707. doi: 10.1002/cm.20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiadis PZ. p120-ctn: A nexus for contextual signaling via Rho GTPases. Biochim Biophys Acta. 2007;1773:34–46. doi: 10.1016/j.bbamcr.2006.08.040. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ, Moon SY, Thoreson MA, Mariner DJ, Crawford HC, Zheng Y, Reynolds AB. Inhibition of RhoA by p120 catenin. Nat Cell Biol. 2000;2:637–644. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ, Reynolds AB. Regulation of Rho GTPases by p120-catenin. Curr Opin Cell Biol. 2001;13:604–610. doi: 10.1016/s0955-0674(00)00258-1. [DOI] [PubMed] [Google Scholar]
- Aono S, Nakagawa S, Reynolds AB, Takeichi M. p120(ctn) acts as an inhibitory regulator of cadherin function in colon carcinoma cells. J Cell Biol. 1999;145:551–562. doi: 10.1083/jcb.145.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguslavsky S, Grosheva I, Landau E, Shtutman M, Cohen M, Arnold K, Feinstein E, Geiger B, Bershadsky A. p120 catenin regulates lamellipodial dynamics and cell adhesion in cooperation with cortactin. Proc Natl Acad Sci U S A. 2007;104:10882–10887. doi: 10.1073/pnas.0702731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FC, Cheng CP, Wu KH, Chen YC, Hsu CH, Gustafson-Wagner EA, Lin JL, Wang Q, Lin JJ, Lin CI. Intercalated disc-associated protein, mXin-alpha, influences surface expression of ITO currents in ventricular myocytes. Front Biosci (Elite Ed) 2011;3:1425–1442. doi: 10.2741/e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Yung A, Covarrubias M, Radice GL. Cortactin is required for N-cadherin regulation of Kv1.5 channel function. J Biol Chem. 2011;286:20478–20489. doi: 10.1074/jbc.M111.218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanova O, Orlova A, Galkin VE, van der Ven PF, Furst DO, Jin JP, Egelman EH. Xin-repeats and nebulin-like repeats bind to F-actin in a similar manner. J Mol Biol. 2006;356:714–723. doi: 10.1016/j.jmb.2005.11.082. [DOI] [PubMed] [Google Scholar]
- Choi S, Gustafson-Wagner EA, Wang Q, Harlan SM, Sinn HW, Lin JL, Lin JJ. The intercalated disc protein, mXinα, is capable of interacting with β-catenin and bundling actin filaments. J Biol Chem. 2007;282:36024–36036. doi: 10.1074/jbc.M707639200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenthaler M, Engelhardt S, Schinke B, Kobsar A, Schmitteckert E, Gambaryan S, Engelhardt CM, Krenn V, Eliava M, Jarchau T, Lohse MJ, Walter U, Hein L. Disruption of cardiac Ena-VASP protein localization in intercalated disks causes dilated cardiomyopathy. Am J Physiol Heart Circ Physiol. 2003;285:H2471–2481. doi: 10.1152/ajpheart.00362.2003. [DOI] [PubMed] [Google Scholar]
- Gates J, Peifer M. Can 1000 reviews be wrong? Actin, alpha-Catenin, and adherens junctions. Cell. 2005;123:769–772. doi: 10.1016/j.cell.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Grosheva I, Shtutman M, Elbaum M, Bershadsky AD. p120 catenin affects cell motility via modulation of activity of Rho-family GTPases: a link between cell-cell contact formation and regulation of cell locomotion. J Cell Sci. 2001;114:695–707. doi: 10.1242/jcs.114.4.695. [DOI] [PubMed] [Google Scholar]
- Grosskurth SE, Bhattacharya D, Wang Q, Lin JJ. Emergence of Xin demarcates a key innovation in heart evolution. PLoS One. 2008;3:e2857. doi: 10.1371/journal.pone.0002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson-Wagner EA, Sinn HW, Chen YL, Wang DZ, Reiter RS, Lin JL, Yang B, Williamson RA, Chen J, Lin CI, Lin JJ. Loss of mXinα, an intercalated disk protein, results in cardiac hypertrophy and cardiomyopathy with conduction defects. Am J Physiol Heart Circ Physiol. 2007;293:H2680–2692. doi: 10.1152/ajpheart.00806.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Troyanovsky RB, Troyanovsky SM. Spontaneous assembly and active disassembly balance adherens junction homeostasis. Proc Natl Acad Sci U S A. 2010;107:3528–3533. doi: 10.1073/pnas.0911027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton RC, Davis MA, van Hengel J, Mariner DJ, Barnes K, Thoreson MA, Anastasiadis PZ, Matrisian L, Bundy LM, Sealy L, Gilbert B, van Roy F, Reynolds AB. A novel role for p120 catenin in E-cadherin function. J Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YJ, Huang EY, Yeh HI, Chen YL, Lin JJ, Lin CI. On the mechanisms of arrhythmias in the myocardium of mXinalpha-deficient murine left atrial-pulmonary veins. Life Sci. 2008;83:272–283. doi: 10.1016/j.lfs.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lin JL, Reiter RS, Daniels K, Soll DR, Lin JJ. Caldesmon mutant defective in Ca(2+)-calmodulin binding interferes with assembly of stress fibers and affects cell morphology, growth and motility. J Cell Sci. 2004;117:3593–3604. doi: 10.1242/jcs.01216. [DOI] [PubMed] [Google Scholar]
- Lin JJC, Gustafson-Wagner EA, Sinn HW, Choi S, Jaacks SM, Wang DZ, Evans S, Lin JLC. Structure, expression, and function of a novel intercalated disc protein, Xin. J Med Sci. 2005;25:215–222. doi: 10.1901/jaba.2005.25-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JL, Geng X, Bhattacharya SD, Yu JR, Reiter RS, Sastri B, Glazier KD, Mirza ZK, Wang KK, Amenta PS, Das KM, Lin JJ. Isolation and sequencing of a novel tropomyosin isoform preferentially associated with colon cancer. Gastroenterology. 2002;123:152–162. doi: 10.1053/gast.2002.34154. [DOI] [PubMed] [Google Scholar]
- Ma X, Adelstein RS. In vivo studies on nonmuscle myosin II expression and function in heart development. Front Biosci. 2012;17:545–555. doi: 10.2741/3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Jana SS, Conti MA, Kawamoto S, Claycomb WC, Adelstein RS. Ablation of nonmuscle myosin II-B and II-C reveals a role for nonmuscle myosin II in cardiac myocyte karyokinesis. Mol Biol Cell. 2010;21:3952–3962. doi: 10.1091/mbc.E10-04-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montonen O, Aho M, Uitto J, Aho S. Tissue distribution and cell type-specific expression of p120ctn isoforms. J Histochem Cytochem. 2001;49:1487–1496. doi: 10.1177/002215540104901202. [DOI] [PubMed] [Google Scholar]
- Navarro P, Ruco L, Dejana E. Differential localization of VE- and N-cadherins in human endothelial cells: VE-cadherin competes with N-cadherin for junctional localization. J Cell Biol. 1998;140:1475–1484. doi: 10.1083/jcb.140.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ. Regulation of cell-cell adhesion by the cadherin-catenin complex. Biochem Soc Trans. 2008;36:149–155. doi: 10.1042/BST0360149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Liu BP, Burridge K, Kreft B. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J Cell Biol. 2000;150:567–580. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Niessen CM, Gumbiner BM, Burridge K. Cadherin engagement regulates Rho family GTPases. J Biol Chem. 2001;276:33305–33308. doi: 10.1074/jbc.C100306200. [DOI] [PubMed] [Google Scholar]
- Ohkubo T, Ozawa M. p120(ctn) binds to the membrane-proximal region of the E-cadherin cytoplasmic domain and is involved in modulation of adhesion activity. J Biol Chem. 1999;274:21409–21415. doi: 10.1074/jbc.274.30.21409. [DOI] [PubMed] [Google Scholar]
- Otten J, van der Ven PF, Vakeel P, Eulitz S, Kirfel G, Brandau O, Boesl M, Schrickel JW, Linhart M, Hayess K, Naya FJ, Milting H, Meyer R, Furst DO. Complete loss of murine Xin results in a mild cardiac phenotype with altered distribution of intercalated discs. Cardiovasc Res. 2010;85:739–750. doi: 10.1093/cvr/cvp345. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Kemler R. The membrane-proximal region of the E-cadherin cytoplasmic domain prevents dimerization and negatively regulates adhesion activity. J Cell Biol. 1998;142:1605–1613. doi: 10.1083/jcb.142.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacholsky D, Vakeel P, Himmel M, Lowe T, Stradal T, Rottner K, Furst DO, van der Ven PFM. Xin repeats define a novel actin-binding motif. J Cell Sci. 2004;117:5257–5268. doi: 10.1242/jcs.01406. [DOI] [PubMed] [Google Scholar]
- Provost E, Rimm DL. Controversies at the cytoplasmic face of the cadherin-based adhesion complex. Curr Opin Cell Biol. 1999;11:567–572. doi: 10.1016/s0955-0674(99)00015-0. [DOI] [PubMed] [Google Scholar]
- Reynolds AB, Daniel JM, Mo YY, Wu J, Zhang Z. The novel catenin p120cas binds classical cadherins and induces an unusual morphological phenotype in NIH3T3 fibroblasts. Exp Cell Res. 1996;225:328–337. doi: 10.1006/excr.1996.0183. [DOI] [PubMed] [Google Scholar]
- Scott JA, Shewan AM, den Elzen NR, Loureiro JJ, Gertler FB, Yap AS. Ena/VASP proteins can regulate distinct modes of actin organization at cadherin-adhesive contacts. Mol Biol Cell. 2006;17:1085–1095. doi: 10.1091/mbc.E05-07-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinn HW, Balsamo J, Lilien J, Lin JJ. Localization of the novel Xin protein to the adherens junction complex in cardiac and skeletal muscle during development. Dev Dyn. 2002;225:1–13. doi: 10.1002/dvdy.10131. [DOI] [PubMed] [Google Scholar]
- Thoreson MA, Anastasiadis PZ, Daniel JM, Ireton RC, Wheelock MJ, Johnson KR, Hummingbird DK, Reynolds AB. Selective uncoupling of p120(ctn) from E-cadherin disrupts strong adhesion. J Cell Biol. 2000;148:189–202. doi: 10.1083/jcb.148.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ven PF, Ehler E, Vakeel P, Eulitz S, Schenk JA, Milting H, Micheel B, Furst DO. Unusual splicing events result in distinct Xin isoforms that associate differentially with filamin c and Mena/VASP. Exp Cell Res. 2006;312:2154–2167. doi: 10.1016/j.yexcr.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Walker MG. Pharmaceutical target identification by gene expression analysis. Mini Rev Med Chem. 2001;1:197–205. doi: 10.2174/1389557013407034. [DOI] [PubMed] [Google Scholar]
- Wang DZ, Reiter RS, Lin JL, Wang Q, Williams HS, Krob SL, Schultheiss TM, Evans S, Lin JJ. Requirement of a novel gene, Xin, in cardiac morphogenesis. Development. 1999;126:1281–1294. doi: 10.1242/dev.126.6.1281. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lin JL-C, Chan SY, Lin JJ-C. The Xin-repeat-containing protein, mXinβ, initiates the maturation of the intercalated discs during postnatal heart development. Dev Biol. 2012a doi: 10.1016/j.ydbio.2012.12.007. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lin JL, Reinking BE, Feng HZ, Chan FC, Lin CI, Jin JP, Gustafson-Wagner EA, Scholz TD, Yang B, Lin JJ. Essential roles of an intercalated disc protein, mXinβ, in postnatal heart growth and survival. Circ Res. 2010;16:1468–1478. doi: 10.1161/CIRCRESAHA.109.212787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Lin JL, Wu KH, Wang DZ, Reiter RS, Sinn HW, Lin CI, Lin CJ. Xin proteins and intercalated disc maturation, signaling and diseases. Front Biosci. 2012b;17:2566–2593. doi: 10.2741/4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren KS, Lin JJ. Forced expression and assembly of rat cardiac troponin T isoforms in cultured muscle and nonmuscle cells. J Muscle Res Cell Motil. 1993;14:619–632. doi: 10.1007/BF00141559. [DOI] [PubMed] [Google Scholar]
- Xiao K, Allison DF, Buckley KM, Kottke MD, Vincent PA, Faundez V, Kowalczyk AP. Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J Cell Biol. 2003;163:535–545. doi: 10.1083/jcb.200306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, Oas RG, Chiasson CM, Kowalczyk AP. Role of p120-catenin in cadherin trafficking. Biochim Biophys Acta. 2007;1773:8–16. doi: 10.1016/j.bbamcr.2006.07.005. [DOI] [PubMed] [Google Scholar]
- Yap AS, Niessen CM, Gumbiner BM. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]