Abstract
The RNA chaperone protein Hfq is required for the function of all small RNAs (sRNAs) that regulate mRNA stability or translation by limited base pairing in E. coli. While there have been numerous in vitro studies to characterize Hfq activity and the importance of specific residues, there has been only limited characterization of Hfq mutants in vivo. Here we use a set of reporters as well as co-immunoprecipitation to examine 14 Hfq mutants expressed from the E. coli chromosome. The majority of the proximal face residues, as expected, were important for the function of sRNAs. The failure of sRNAs to regulate target mRNAs in these mutants can be explained by reduced sRNA accumulation. Two of the proximal mutants, D9A and F39A, acted differently from the others in that they had mixed effects on different sRNA/mRNA pairs and, in the case of F39A, showed differential sRNA accumulation. Mutations of charged residues at the rim of Hfq interfered with positive regulation, and gave mixed effects for negative regulation. Some, but not all, sRNAs accumulated to lower levels in rim mutants, suggesting qualitative differences in how individual sRNAs are affected by Hfq. The distal face mutants were expected to disrupt binding of ARN motifs found in mRNAs. They were more defective for positive regulation than negative regulation at low mRNA expression, but the defects could be suppressed by higher levels of mRNA expression. We discuss the implications of these observations for Hfq binding to RNA and mechanisms of action.
Keywords: DsrA, ArcZ, McaS, RyhB, ChiX
Introduction
The Sm-like Hfq binds more than 30% of the known small, regulatory RNAs (sRNAs) in Escherichia coli1 and Salmonella2. The protein both stabilizes these sRNAs and is required for their base pairing with mRNA targets (reviewed in). For some targets (for example, the DsrA target rpoS and the McaS target flhD), base pairing can lead to increased translation, due to the opening of a secondary structure that blocks translation of the mRNA. However, for the majority of targets (for example, the ArcZ target flhD and the RyhB target sodB), base pairing leads to decreased expression by inhibiting ribosome binding and/or promoting mRNA degradation (reviewed in5). Some of the effects on mRNA stability may be due to the reported association of Hfq with components of the degradosome complex, including RNase E and polynucleotide phosphorylase.
Given its central role in the functions of base-pairing sRNAs, Hfq has been subject to numerous in vitro studies. Structural analysis of Hfq showed that the protein forms a hexameric ring with proximal and distal faces similar to Sm proteins. The solution of the first crystal structure of a bacterial Hfq with an 5’-AUUUUUG oligoribonucleotide revealed that the RNA bound in a circular conformation around the pore on the proximal face of the Staphylococcus aureus protein.8 Subsequent mutational studies suggested that E. coli Hfq has distinct interaction surfaces for DsrA and poly(A). Mutations of the proximal face residues and a charged amino acid on the rim led to decreased binding to the DsrA RNA in vitro; none of the proximal face mutations had significant effects on poly(A) binding. In contrast, mutations of distal face residues led to decreased binding to poly(A) but had little effect on binding to DsrA. Recent crystallographic studies of E. coli Hfq bound to an A18 oligoribonucleotide showed that the poly(A) RNA binds to the distal face.11 This structure, together with oligonucleotide binding assays, led to the proposal that the distal face of Hfq binds repeats of an ARN motif (where R is adenine or guanine and N is any nucleotide). This proposal was consistent with the earlier finding that the upstream (AAN)4 sequence motif in the rpoS mRNA contributes to tight Hfq binding and formation of stable ternary complexes between Hfq, rpoS and DsrA.12 Similar ARN sequences also were shown to be important for sRNA-dependent regulation for other sRNA/mRNA pairs. Recent studies revealed that the 3’-end terminal poly(U) tail found in Hfq-binding sRNAs2 significantly contributes to the recognition of sRNA by Hfq and is essential for the ability of sRNA to bind to the central cavity of the Hfq hexamer and regulate mRNA targets. Finally, a charged patch at the outer rim of the hexamer also has been implicated in sRNA binding.18 Together, these data suggest that the Hfq ring has at least three RNA binding surfaces: a proximal face for U-rich sRNAs, the distal face for A-rich mRNA targets, and a rim region that may provide additional binding sites.
Only a few studies have examined the effects of specific amino acid substitutions on the in vivo function of Hfq. Proximal mutants Q8A, F42A, and K56A, expressed from a plasmid, showed significant defects for Hfq-dependent activation of rpoS.9 In another study, a plasmid-expressed V43R mutant abrogated rpoS activation but both plasmid- and chromosomally-expressed V43R retained the ability to repress oppA.19
Despite the extensive in vitro studies and limited in vivo studies, many questions regarding Hfq binding to mRNAs and the mechanism by which Hfq facilitates the interaction between sRNAs and mRNAs remain. Furthermore, most of the in vitro studies have been focused on one model system, activation of rpoS by DsrA and RprA; thus it is not clear if the lessons learned with this system will extrapolate to negative regulation or even other sRNA-based positive regulation. To contribute to answering these questions and learn more about the mechanism of Hfq action in vivo, we constructed isogenic sets of strains in which 18 mutant Hfq derivatives were expressed from the chromosome. We then examined the effects of the 14 alleles expressed at significant levels on in vivo activity in a selected set of assays and on sRNA accumulation. Association of specific sRNAs and mRNAs with the Hfq mutants also was analyzed by co-immunoprecipitation (co-IP). In addition, we examined the effects of the Q8A, R16A and K31A mutations on RNA association with Hfq on a whole transcriptome scale by probing tiling arrays with total RNA and co-IP RNA isolated from these strains. The implications of these findings for the mechanism of Hfq binding to RNAs and facilitating base pairing are discussed.
Results
Most Hfq mutants are well expressed from the chromosomal locus
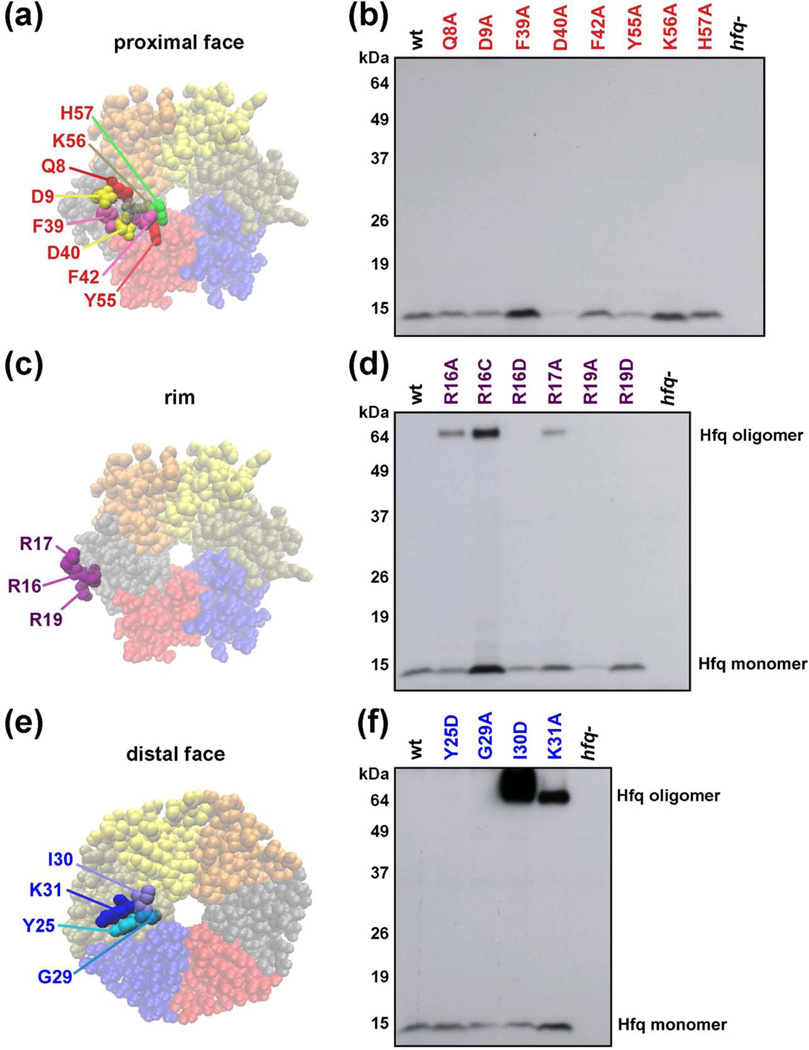
To assay the effects of Hfq mutants expressed at endogenous levels, we replaced the wild-type hfq gene with derivatives carrying alanine substitutions of the amino acids around the central pore on the proximal side of the hexamer (Q8, D9, F39, D40, F42, Y55, K56 and H57, Fig. 1a), alanine, cysteine or aspartic acid substitutions of charged amino acids on the outer rim (R16, R17 and R19, Fig. 1c) and alanine or aspartic acid substitutions of amino acids on the distal face (Y25, G29, I30 and K31, Fig. 1e). The mutations were introduced into the chromosome at the native hfq locus and moved between strains as described in Materials and Methods.
Fig. 1.
Chromosomal Hfq mutants. (a, c, e) Space-filling representation of the E. coli Hfq crystal structure (PDB 1HK9) showing the locations of amino acids mutated viewed from the proximal face (Q8, D9, F39, F42, K56 and H57), the rim (R16, R17 and R19), or distal face (Y25, G29, I30 and K31). (b, d, f) Hfq protein levels in mutant strains. Extracts were prepared from derivatives of SG30200 (PBAD-rpoS-lacZ) carrying wild-type and mutant hfq alleles (see Table S5); cells were grown in LB medium at 37 °C to early stationary phase (OD600 ~ 1.0). The levels of Hfq protein were determined by immunoblot analysis using anti-Hfq serum and ECL Western Blotting System. Ponceau S staining of the immunoblots showed that equal total protein amounts were loaded.
The levels of the different mutant proteins were then examined by Western blot analysis under denaturing conditions (Fig. 1b, d, f). The levels of the D40A, Y55A, R16D and R19A derivatives were low and were eliminated from further study. The levels of the Q8A, F42A, H57A, R16A, R17A, R19D, and Y25D proteins were similar to those of the wild-type protein, while D9A and G29A levels were slightly lower. The F39A, K56A, R16C, and K31A levels were somewhat higher and the I30D levels were significantly higher than the wild-type protein.
Additionally, we always observed a higher molecular weight band consistent with the size of a hexamer for three of the rim mutants (R16A, R16C, R17A) and two of the distal face mutants (I30D and K31A), suggesting that these mutants might form particularly stable rings recalcitrant to denaturation. The oligomerization state of the mutants was further examined using semi-native gels previously used to distinguish between Hfq monomers and oligomers20 (Supplementary Fig. S1). In these gels, the wild-type Hfq migrates primarily as an oligomer, with barely detectable levels of the monomer. The R16A, R16C, R17A, I30D and K31A, along with the other rim and distal face mutants, showed elevated levels of oligomeric forms of Hfq. The physiological implications of more stable oligomeric forms are not known, but the partial correlation with increased levels of Hfq suggests these forms might be more resistant to proteolysis, and/or be defective in negative autoregulation. In contrast, with the exceptions of the D9A and F42A mutants, the proximal face mutants were predominantly monomeric, suggesting that while these derivatives are stable enough to accumulate to wild-type levels (Fig. 1b), oligomers of the mutants are more likely to dissociate than the wild-type protein.
sRNA-dependent activation is defective in proximal, rim, and distal face mutants
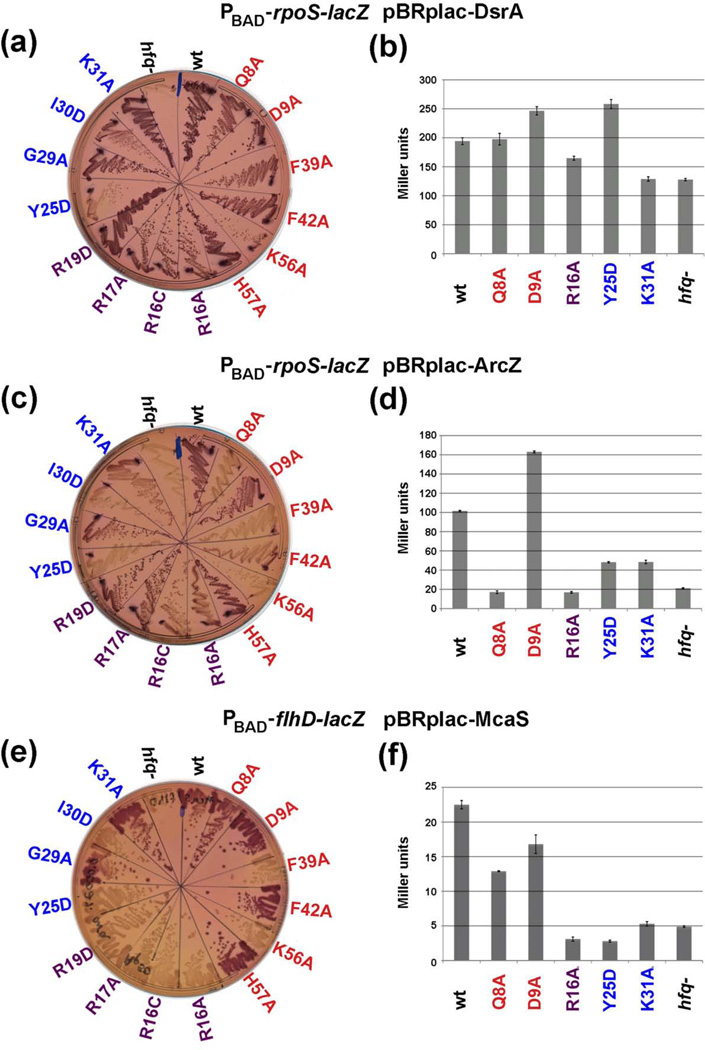
To examine the consequences of the proximal, rim and distal mutants on the ability of Hfq to facilitate sRNA-mediated activation of gene expression in vivo, we assayed the 14 hfq alleles that expressed approximately wild-type or higher levels of protein in strains carrying either PBAD-rpoS-lacZ (used for the Western blot analysis in Fig. 1) or PBAD-flhD-lacZ translational fusions. Three sRNAs, DsrA, RprA, and ArcZ, positively regulate rpoS translation; McaS positively regulates fhlD23. We examined the consequences of DsrA and ArcZ overexpression on rpoS-lacZ induction and McaS overexpression on flhD-lacZ induction by first streaking all of the strains together with the wild-type and Δhfq mutant control strains on lactose MacConkey agar plates containing ampicillin and arabinose concentrations sufficient to induce the reporter fusions to allow a significant color difference between the wild-type and Δhfq mutant strains (Fig. 2a, 2c and 2e). We also measured the levels of β-galactosidase activity for a selected set of mutants representative of each category: proximal face alleles Q8A and D9A, rim allele R16A and distal face alleles Y25D and K31A (Fig. 2b, 2d and 2f). Table 1 summarizes the results of the qualitative plate assays with the complete set of 14 Hfq mutants and Table 2 summarizes the quantitative assays with the subset of five mutant alleles; the results for all quantitative assays are in Supplementary Table S1.
Fig. 2.
Effects of hfq mutations on sRNA-dependent activation of rpoS and flhD. (a, c) Derivatives of SG30200 (PBAD-rpoS-lacZ) and (e) derivatives of NRD688 (PBAD-flhD-lacZ) carrying the indicated sRNA overexpression plasmids were grown on lactose MacConkey plates containing 50 µg/ml ampicillin and arabinose (at 0.0001% for (a) and (c) and at 0.0002% for (e)) at 37 °C for 16 h. (b, d, f) β-galactosidase activity measured in wild-type and a subset of the mutant cells shown in (a, c, e). Cells were grown in LB medium containing 100 µg/ml ampicillin, 100 µM IPTG, 0.0002% arabinose at 37 °C to early stationary phase (OD600 ~ 1.0) and assayed. Data is an average of three assays and brackets denote the standard deviation of the mean.
Table 1. Summary of in vivo assays.
The plate and growth assays shown in Fig. 2a, 2c, 2e, 3a, 3c, 3e and 3g are summarized here. Hfq alleles are given on the far left of the table. The residues investigated in the study from the three faces of Hfq are grouped in the table as proximal (red), rim (purple), and distal (blue). The top line of the table describes the assay in which the given Hfq allele was examined. Results for each Hfq allele for a given assay are reported as follows: green = an allele was functional for a given assay, yellow = an allele showed partial function, or orange = an allele was defective.
| Hfq allele | DsrA/rpoS | ArcZ/rpoS | McaS/flhD | ArcZ/flhD | ChiX/chiP | RyhB/sodB | RyhB/sdh |
|---|---|---|---|---|---|---|---|
| (activity on lac plates) |
(activity on lac plates) |
(activity on lac plates) |
(activity on lac plates) |
(activity on lac plates) |
(Northern) | (growth on succinate) |
|
| wt | Functional | Functional | Functional | Functional | Functional | Functional | Functional |
| hfq- | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional |
| Q8A | Functional | Nonfunctional | Functional | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional |
| D9A | Functional | Functional | Functional | Functional | Nonfunctional | Partial | Nonfunctional |
| F39A | Functional | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional | Partial |
| F42A | Functional | Partial | Functional | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional |
| K56A | Partial | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional | Nonfunctional |
| H57A | Functional | Functional | Functional | Functional | Functional | Functional | Partial |
| R16A | Functional | Partial | Nonfunctional | Nonfunctional | Nonfunctional | Functional | Functional |
| R16C | Functional | Partial | Nonfunctional | Nonfunctional | Nonfunctional | Functional | Functional |
| R17A | Functional | Functional | Nonfunctional | Functional | Nonfunctional | Functional | Functional |
| R19D | Functional | Functional | Nonfunctional | Functional | Nonfunctional | Functional | Functional |
| Y25D | Nonfunctional | Nonfunctional | Nonfunctional | Functional | Partial | Functional | Functional |
| G29A | Functional | Functional | Functional | Functional | Functional | Functional | Functional |
| I30D | Functional | Nonfunctional | Nonfunctional | Functional | Partial | Partial | Partial |
| K31A | Functional | Functional | Functional | Functional | Functional | Functional | Functional |
Table 2. Summary of β-galactosidase assays for cell growth in low and high arabinose.
Representative alleles from each face of Hfq were examined by β-galactosidase assays (Fig. 2b, 2d, 2f, 3b, 3d, 3f, and 3h). Hfq alleles and color-coding are as for Table 1. The levels of L-arabinose used in each assay are depicted as percentages. The percentage activity for each allele was calculated as follows: positively-regulated targets: (Miller units of hfq allele − Miller units of Δhfq)/(Miller units of wild-type hfq − Miller units of Δhfq) × 100% = percent function for Hfq allele; negatively-regulated targets: (Miller units of Δhfq − Miller units of hfq allele)/(Miller units of Δhfq − Miller units of wild-type hfq) × 100% = percent function for Hfq allele. Activities of 75% or more are colored green, activities between 50% and 74% are colored yellow, and activities <50% are colored orange. β-galactosidase values for each assay are found in Supplemental Table S1.
| Hfq allele | ArcZ/rpoS | ArcZ/rpoS | McaS/flhD | McaS/flhD | ArcZ/flhD | ArcZ/flhD | ChiX/chiP | ChiX/chiP | RyhB/sodB | RyhB/sodB | RyhB/sdhC | RyhB/sdhC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % ara | 0.0002% | 0.02% | 0.0002% | 0.02% | 0.0002% | 0.02% | 0.0002% | 0.02% | 0.0002% | 0.02% | 0.0002% | 0.02% |
| wt | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| hfq- | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% |
| Q8A | <0% | <0% | 45% | 62% | 45% | 61% | 59% | 65% | 13% | <0% | 11% | 51% |
| D9A | 176% | 85% | 68% | 87% | 64% | 94% | 75% | 80% | 81% | 76% | 81% | 70% |
| R16A | <0% | <0% | <0% | <0% | 71% | 57% | 64% | 47% | 97% | 95% | 97% | 96% |
| Y25D | 34% | 95% | <0% | 47% | 54% | 77% | 83% | 81% | 77% | 55% | 23% | 21% |
| K31A | 34% | 66% | 2% | 76% | 96% | 89% | 100% | 94% | 96% | 83% | 92% | 72% |
In agreement with previous findings that DsrA activation of rpoS-lacZ is partially independent of Hfq24, the activation of the rpoS-lacZ fusion by DsrA on plates was only partially defective in most of the mutants. The strongest difference from the wild-type strain was observed for Y25D, though the K56A and I30D mutants and possibly some of the rim alleles also showed somewhat less activation. In addition, while the strain deleted for hfq was defective on plates (Fig 2a), it was only partially defective in liquid assays (Fig. 2b). The R16A and K31A mutants also were slightly defective in liquid assays; however, the Y25D mutant was not. As discussed below, we think a reason for the lack of concordance between the plate and liquid assays may be differences in levels of the target mRNA relative to Hfq available for binding. Regardless, DsrA activation of rpoS-lacZ was largely insensitive to the effects of the hfq mutations.
The effects of the mutations on ArcZ activation of rpoS and McaS activation of flhD were more dramatic. The pattern of the defects for these two assays was similar with some notable differences for the proximal and rim mutants. Of the proximal face mutants, D9A and H57A were functional for all the sRNA-dependent activation tested. Among the other proximal face mutants, F39A and K56A were most defective. The Q8A and F42A mutants were also defective for ArcZ activation of the rpoS-lacZ fusion but less so for McaS activation of the flhD-lacZ fusion. The four rim mutants also showed differences on MacConkey plates depending on the sRNA/mRNA pair. The ArcZ/rpoS pair was partially defective with the two R16 alleles, while the McaS/flhD pair was fully defective for all rim mutants. However, both pairs were fully defective in quantitative assays of R16A derivatives (Fig. 2d and 2f, Table 2). The results suggest that differences in binding and/or pairing at the proximal face and rim must exist between ArcZ activation of rpoS and McaS activation of flhD. Of the four distal mutants, the Y25D mutant was defective for all activation on plates and in liquid culture, and the I30D mutant showed a partial defect on plates (Fig. 2c and 2e, Tables 1 and 2). The G29A and K31A mutants were Lac+ (functional) for both sRNA/mRNA pairs in the plate assays, but the K31A mutant did show a defect in liquid assays. Together these results indicate that residues on all RNA binding surfaces are important for sRNA-mediated gene activation but that there are differences between sRNA/mRNA pairs.
In a control experiment, expression of the rpoS-lacZ fusion was also measured in wild-type and mutant backgrounds in the absence of sRNA-overexpression plasmids (Supplementary Fig. S2). Under our growth conditions, endogenous DsrA, ArcZ, and to a lesser extent RprA are likely to contribute to expression of the fusion.21 On plates, these cells were less permissive than those overexpressing DsrA (Fig. 2a) or ArcZ (Fig. 2c). The wild-type strain and the D9A, H57A and R19D mutants were still Lac+ (Supplementary Fig. S2a), but the F42A, R16C, G29A and K31A mutants were pale red and all other mutants showed even less expression, consistent with decreased activation. In liquid assays, the Q8A and R16A mutants were more defective than the D9A and K31A mutants (Supplementary Fig. S2b) in concordance with the plate assays. In contrast, the Y25D mutant, which was clearly defective on plates, gave wild-type or greater activation in the liquid assays for as yet unknown reasons.
sRNA-dependent repression is defective in proximal and rim mutants
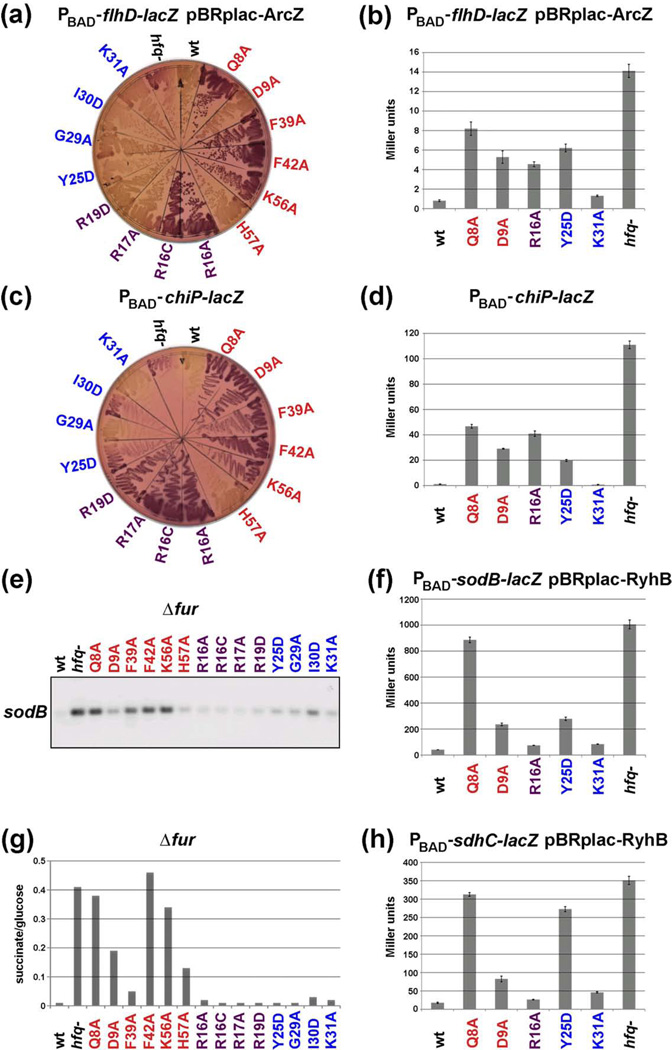
The hfq alleles were also tested in strains appropriate for the evaluation of negative regulation by three different sRNAs. flhD-lacZ is negatively-regulated by ArcZ25, in addition to being activated by McaS23. The effects of the hfq alleles on this repression were determined both on lactose MacConkey agar plates (Fig. 3a) and in β-galactosidase activity assays for representative mutants (Fig. 3b). Chromosomal levels of ChiX are sufficient for strong repression of chiP, allowing plate (Fig. 3c) and liquid (Fig. 3d) assays of a chiP-lacZ fusion in the absence of a plasmid expressing the sRNA. The mutant alleles also were introduced into a strain lacking the Fur repressor, in which the levels of the RyhB RNA are constitutively high. Two RyhB targets were investigated in the fur hfq mutants. RyhB represses sodB28, and this was followed by Northern blot analysis in the fur mutant host (Fig. 3e) and by assays of a PBAD-sodB-lacZ fusion in a subset of hfq mutants expressing RyhB from a plasmid in a fur+ host (Fig. 3f). In an hfq+ background, fur mutants are unable to grow on succinate minimal medium due to the constitutive RyhB repression of sdhCDAB expression28; growth is restored by a deletion of hfq. Thus, the hfq alleles were tested for their ability to grow on succinate minimal media (Fig. 3g). In addition, we assayed a PBAD-sdhC-lacZ fusion in a subset of mutants expressing RyhB from a plasmid in a fur+ background (Fig. 3h). Summaries of all of these results and comparisons to the tests of positive regulation are given in Tables 1, 2 and S1.
Fig. 3.
Effects of hfq mutations on sRNA-dependent repression of flhD, chiP, sodB and sdhC. (a) Derivatives of NRD688 (PBAD-flhD-lacZ) carrying a plasmid expressing McaS were grown on a lactose MacConkey plate containing 50 µg/ml ampicillin and 0.001% arabinose for 24 h. (c) Derivatives of DJS2677 (PBAD-chiP-lacZ) were grown on lactose MacConkey plates with 0.0005% arabinose for 11 h at 37 °C. (e) Derivatives of DJS2546 (fur::kan) were grown in MOPS EZ rich defined medium with 0.4% glycerol to late exponential phase at 37 °C, and RNA was extracted and sodB RNA levels were analyzed as described in Materials and Methods. (g) The same strains as for (e) were grown in minimal succinate medium at 37° C overnight and growth determined by OD600 normalized to growth in minimal glucose medium. (b, d, f, h) β-galactosidase activity measured in wild-type and a subset of hfq mutants shown in (a, c, e, g). Strains in (a, c), derivatives of DJS2676 (PBAD-sodB-lacZ) overexpressing RyhB, and derivatives of DJS2729 (PBAD-sdhC-lacZ) overexpressing RyhB were grown in LB medium containing 100 µg/ml ampicillin, 100 µM IPTG, 0.0002% arabinose at 37 °C to early stationary phase (OD600 ~ 1.0) and assayed for β-galactosidase activity. Data is an average of three assays and brackets denote the standard deviation of the mean.
As for positive regulation, the proximal mutants, with the exception of H57A and D9A, were completely or partially defective for most of these assays, with Q8A, F42A and K56A showing the strongest effects. These defects were clear on plates. In liquid assays, partial activity was found for ArcZ repression of flhD and ChiX repression of chiP in the Q8A mutant hosts but not for RyhB repression of sodB or sdhC. As mentioned above, the D9A mutant was functional for positive regulation. The mutant was also functional for ArcZ repression of the flhD-lacZ fusion on plates and partially functional for RyhB repression of sodB, but was more defective for ChiX repression of the chiP-lacZ fusion and RyhB repression of succinate growth. Note that the D9A allele was identified in a genetic selection for failure to regulate sdh by RyhB and was also defective for sRNA OmrA repression of cirA.29 D9A was not fully defective for RyhB repression of the sdhC-lacZ fusion (Fig. 3h); we do not currently have an explanation for this difference from the succinate growth results. F39A also was notable in that, although it was defective for negative regulation of flhD-lacZ and chiP-lacZ on plates, and sodB in culture, it was reasonably effective for regulation of growth on succinate (Fig. 3g). These results suggest qualitative differences between Hfq action in different cases of negative regulation, with D9 and F39 defining sites that distinguish between different sRNA/mRNA regulatory pairs. H57A was partially functional for both positive and negative regulation of all targets tested, and thus this site is either redundant with other sites or not essential.
For positive regulation, with the exception of DsrA, the rim mutants were Lac− (McaS/flhD) or less Lac+ (ArcZ/rpoS) than wild-type on plates and completely defective in liquid assays (Tables 1 and 2). For the tests of negative regulation, defects of the rim mutants were not as striking. On plates, the R16 rim alleles were defective for both ArcZ repression of flhD-lacZ and ChiX repression of chiP-lacZ, but the R16A mutant was only partially defective for repression for both of these targets in liquid assays. In addition, the R16 alleles showed almost wild-type function for RyhB repression of sodB and sdh (Fig. 3 and Tables 1 and 2). Similarly, the R17A and R19A alleles were functional in all plate assays except ChiX repression of chiPlacZ. Based on our findings, the rim interactions have stronger consequences for positive regulation, although our results suggest that these sites also make some contribution to efficient negative regulation.
The distal mutants Y25D and I30D were quite defective for positive regulation. In contrast, these alleles were relatively active for repression on plates and Y25D was partially active in most quantitative assays of negative regulation (Fig. 3 and Tables 1 and 2). As for D9A, Y25D differed between the succinate growth assay (Fig. 3g) and RyhB repression of the sdhC-lacZ fusion (Fig. 3h) but in the opposite direction (functional for succinate growth, non-functional for repression of sdhC-lacZ). Overall, the distal side residues were more important for positive then negative regulation.
We note that none of the hfq alleles tested in the experiments described above gave phenotypes worse than the deletion (Tables 1 and S1) and most were less defective. This observation indicates that, not surprisingly, there is redundancy in the contacts made between specific Hfq residues and the RNAs.
Amounts and/or ratios between sRNAs, mRNAs, and Hfq matter for mutant phenotypes
It has been observed previously, both in vivo and in vitro, that while Hfq is needed for optimal sRNA regulation, the Hfq requirement can be bypassed, particularly when the sRNA is present at high levels24. Thus, it was not surprising that multicopy DsrA could bypass most of the hfq mutants (Fig. 2a and 2b, Table S1). We similarly observed that ChiX expressed from a plasmid could bypass most of the hfq mutants with respect to chiP-lacZ repression (data not shown). These observations support the role of Hfq as chaperone rather than an essential component, consistent with the work of others30.
We also noted that the penetrance of some Hfq alleles for other small RNA/mRNA target pairs varied with different levels of mRNA expression. Most of the sRNA/mRNA reporter pairs described above were constructed such that the reporter was under the control of the arabinose-inducible PBAD promoter in single copy in the chromosome. Each sRNA (with the exception of ChiX) was expressed from a multicopy plasmid under the control of a Plac promoter inducible by either IPTG or lactose. Thus, both on MacConkey lactose plates and in liquid assays, the sRNA should be made at high (fully induced) levels. However, the levels of the mRNA target will vary depending on the extent of induction of the PBAD promoter. On the MacConkey plates in Fig. 2 and 3, we added just enough arabinose to optimize differences between hfq+ and hfq− cells; generally this was a very low level of arabinose. The liquid assays in Figs. 2 and 3 were similarly carried out at a low level of arabinose (0.0002%). In parallel experiments, these assays were repeated at a higher arabinose level (0.02%). A comparison of these results (Tables 2 and S1) provided further insight into how Hfq is acting.
On plates and in assays at low arabinose levels, the Y25D and K31A distal site mutants were generally defective for positive regulation [32% and 33% for ArcZ/rpoS and <0% and 3% for McaS/flhD]. Both mutants were significantly less defective at high arabinose levels (87% and 67% for ArcZ/rpoS, and 50% and 77% for McaS/flhD). This observation that higher mRNA levels can lead to suppression of the Y25D and K31A alleles is consistent with the model that the distal site is required for mRNA binding. The negatively-regulated targets flhD, chiP, and sodB were well-regulated by the distal site mutants at either low or high arabinose levels while sdhC was poorly regulated by Y25D at both arabinose levels (Table 2). Overall, effective distal site binding seems to be particularly important for positive regulation. The observation that defects can be overcome by higher levels of mRNA implies that the distal site mutants tested reduce but do not fully lose distal site binding. Other alleles (Q8A, D9A and R16A) do not show the same pattern of changes with different mRNA levels, consistent with them playing roles at steps other than the initial binding of the mRNA target.
Proximal face mutants have decreased sRNA levels and rim mutants differentially affect sRNA levels
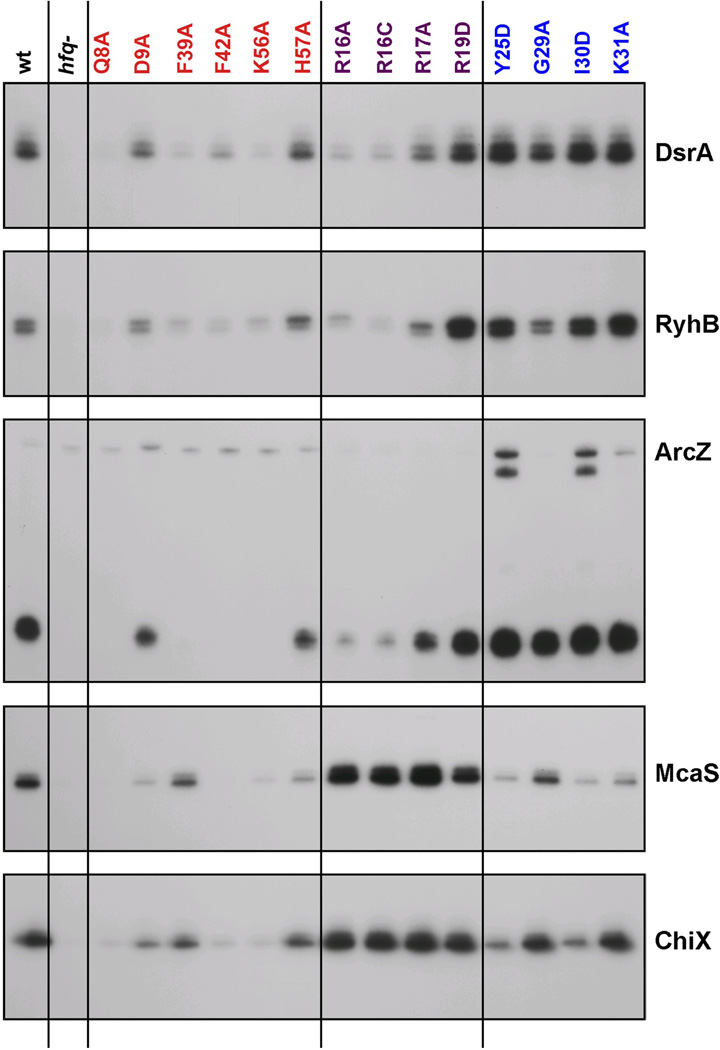
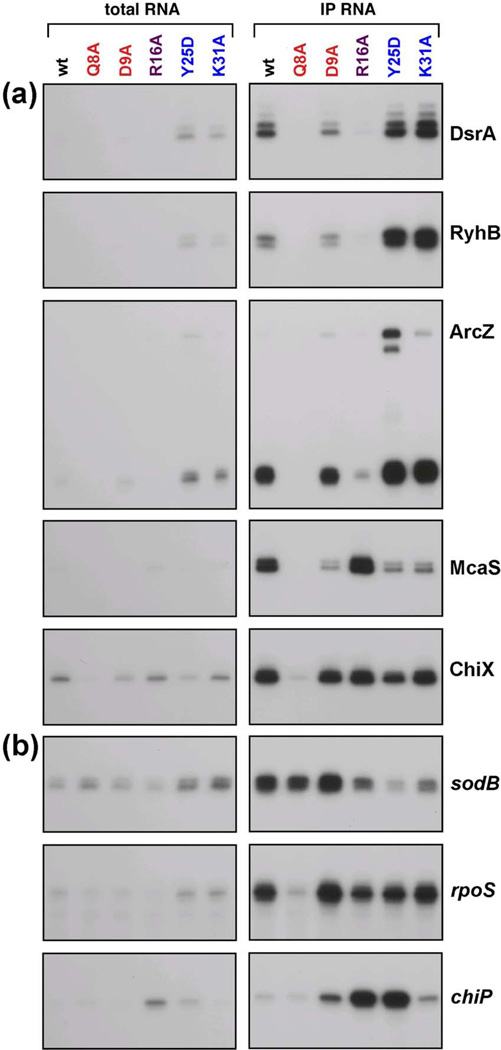
The stability of most base-pairing sRNAs is dependent upon Hfq. In addition, even after binding Hfq, sRNAs can be degraded as they are used.31 We thus examined the levels of the sRNAs assayed above as a reflection of their ability to bind to and be protected by Hfq. Using the same set of strains used to measure Hfq levels in Fig. 1, we assayed the endogenous levels of five sRNAs in cells grown in rich medium by primer extension or Northern blot analysis (Fig. 4). As expected, the sRNAs were barely detectable in the hfq deletion strain.
Fig. 4.
Effects of hfq mutations on sRNA levels under specific growth conditions. Extracts were prepared from wild-type and mutant derivatives of SG30200 (PBAD-rpoS-lacZ) grown in LB medium at 37 °C to early stationary phase (OD600 ~ 1.0). ArcZ levels were analyzed by Northern blots while the levels of all other sRNAs were analyzed by primer extension. In both cases, 5 µg of total RNA and primers specific to the indicated RNAs were used.
Consistent with previous models that sRNAs bind to the proximal face of Hfq, the levels of all the sRNAs were reduced in the Q8A, F42A and K56A mutant backgrounds, the proximal face alleles with the strongest defects in the functional assays. In contrast, the H57A mutant had wild-type levels of sRNAs, consistent with this allele not disrupting sRNA function. sRNA levels were only partially reduced in the D9A background, despite somewhat decreased Hfq levels (Fig. 1b) and some defects in activity (Table 1), again distinguishing this allele from other proximal site mutants. The F39A mutation also was unique in that it led to decreased levels of DsrA, RyhB and processed ArcZ but had lesser effects on McaS or ChiX levels.
The rim mutants similarly distinguished between the sRNAs. The levels of DsrA, RyhB and processed ArcZ were significantly reduced, particularly for R16A and R16C (Fig. 4); these same two rim alleles were inactive in the activity tests (Figs. 2 and 3, Table 1), while R17A and R19D had both somewhat higher activities and higher sRNA levels. Thus, for this group of sRNAs, higher sRNA levels correlated with higher activity. For the second group, which included McaS and ChiX, the sRNA levels were high (similar to wild-type or higher) even though these rim alleles were quite inactive in the activity assays (Tables 1 and 2). Therefore, for this group, loss of activity cannot be due simply to instability of the sRNA.
Distal site mutants generally had wild-type levels of all the sRNAs, whether or not the sRNAs were functional for the tested targets (compare Fig. 4 to Table 1). One striking observation was that while ArcZ processing to the predominant 56-nucleotide species was similar in all distal mutants, the levels of the full length sRNA and a slightly shorter form, truncated at the 5’ end, were higher in the Y25D and I30D mutants. Possibly, processing was slower, was occurring by a somewhat different pathway, or the unprocessed forms were stabilized in these distal site mutants. Y25D and I30D mutants also had somewhat lower McaS and ChiX levels. Overall, these results indicate that residues on the proximal face of the Hfq hexamer are most important for sRNA stability, and most likely binding, but also show that sRNAs differ in their association with Hfq or in the consequences of this interaction.
The experiments reported here were all carried out with derivatives of MG1655. An earlier set of experiments was conducted with a subset of the hfq alleles in a MC4100 background (Fig. S3). The results are generally in agreement, with low levels of essentially all the sRNAs for proximal mutants except H57A, high levels for distal site alleles G29A and K31A, and mixed results for R16A, the only rim allele in this earlier experiment. For the R16A allele, McaS and ChiX levels again were high compared to DsrA, RyhB, Spot42 and GlmZ, while ArcZ, GcvB, SgrS and GadY were present at intermediate levels. Consistent with previous observations that GcvB stability is less dependent on Hfq32, GcvB was easily detectable in the hfq deletion strain, while some other sRNAs were not. Overall, the similarity between the results in MC4100, a relA mutant, and MG1655, a relA+ strain, suggest that the behavior of the hfq alleles is not strain specific and is not dependent on RelA, which has been shown to affect Hfq behavior33. The one difference between the two strains is that somewhat more full-length than fully-processed ArcZ was detected in the MC4100 background.
Co-immunoprecipitation of specific sRNAs with Hfq parallels sRNA levels
An assumption in the previous section was that lower sRNA levels reflect poor association with Hfq, resulting in instability of the sRNA. sRNA association with Hfq was directly tested by comparing the total levels of the DsrA, RyhB, ArcZ, McaS and ChiX sRNAs with the levels in the RNA fraction that co-IPs with Hfq in extracts from the MG1655 wild-type, Q8A, D9A, R16A, Y25D and K31A mutant strains (Fig. 5a and Table S2). The levels of DsrA, RyhB, Spot 42, GlmZ and SgrS that co-IP with Hfq were also analyzed in extracts from the MC4100 wild-type, Q8A, R16A and K31A mutant strains (Fig. S4a). In addition, tiling arrays were used to analyze the total and co-IP fraction in extracts from the MC4100 wild-type and mutant strain. In these latter experiments, RNA samples were hybridized directly to tiled oligonucleotide arrays with coverage of the entire E. coli genome, and hybridization was detected with antibodies specific for RNA:DNA hybrids. Data from the arrays were background-adjusted and normalized, and signals from sets of seven consecutive probes were averaged for the total RNA and co-IP samples for each sRNA annotated in RefSeq35 (Table S3). Quantitation of the assays of specific sRNAs in the MG1655 background (Fig. 5a and Table S2) and the MC4100 tiling array results (Table S3) are in good agreement, and the changes in total sRNA levels agree with observations in Fig. 4 and Fig. S3.
Fig. 5.
Effects of hfq mutations on Hfq association with select (a) sRNAs and (b) mRNAs. Extracts were prepared from wild-type and hfq mutant derivatives of SG30200 (PBAD-rpoS-lacZ) cells grown in LB medium at 37 °C to early stationary phase (OD600 ~ 1.0). co-IP was carried out with anti-Hfq antiserum. ArcZ levels were analyzed by Northern blots while the levels of all other RNAs were analyzed by primer extension. In both cases, 5 µg of total RNA or 0.5 µg of co-IP RNA and primers specific to the indicated RNAs were used.
A primary conclusion from these experiments is that the sRNA levels that co-IP with Hfq correlate reasonably well with the total levels of the sRNAs, for all of the mutants (compare IP/total for wt and mutants, Table S2 and Table S3c). This implies that each of these Hfq mutants, even if not protecting the sRNA from degradation, is still able to bind sufficiently well to immunoprecipitate these sRNAs.
Consistent with the important role of the proximal face in binding and protecting sRNAs from degradation and the low levels of all sRNAs in most proximal site mutants in Fig. 4, the proximal face Q8A mutant led to the biggest decrease in the levels of the sRNAs, both in the total and in the co-IP sample (Fig. 5a). It is clear, however, that Q8A still does interact stably enough with many of the remaining sRNAs to result in co-IP (Table S2, Table S3c). We cannot distinguish between several possibilities to explain this. Possibly, the sRNA is less stably bound to this mutant, is degraded when it is not bound, but can still co-IP during the transient periods when it is bound. Alternatively, some sRNA may escape degradation because it is bound in an alternative or indirect way to Q8A protein. The proximal face D9A allele was only examined in MG1655 (Fig. 5a). Although the levels of the D9A protein were somewhat reduced (Fig. 1), the levels of four of the five sRNAs tested in both the total and co-IP samples were close to wild-type levels (Fig. 5a, Table S2).
An examination of the total and co-IP levels of sRNAs in the rim R16A mutant suggests that the sRNAs fall into different categories, as was also noted for Fig. 4. For DsrA, RyhB, and ArcZ as well as Spot 42 and GlmZ, the total levels were reduced in R16A compared to wild-type, whether measured from gels or from the tiling arrays (Fig. 5a, Table S2 and Table S3), while the level was close to the wild-type levels for McaS and ChiX as well as MgrR and MicC. This again suggests that residues on the rim, and R16 in particular, impact the levels of some, but not all sRNAs.
It is worth mentioning that there was not a good correlation between the levels of expression and co-IP of the chromosomally encoded, endogenous sRNAs in the rim mutants in Fig. 4 and Fig. 5 and levels of activity in Fig. 2 and 3, Tables 1 and 2, measured under conditions of sRNA overproduction. Most strikingly, chromosomal McaS levels are high in the rim mutants, but overexpressed McaS is non-functional for regulation of flhD in these mutants. The lack of correlation suggests that the major effects of the rim mutants on sRNA-mediated regulation are likely to be independent of sRNA levels.
For the distal Y25D and K31A mutants, the total and co-IP sRNA levels were higher than wild-type levels for DsrA, RyhB, and ArcZ and somewhat lower than wild-type levels for McaS and ChiX in the MG1655 background (Table S2). While the Y25D allele was not tested in the MC4100 background, the results for K31A were consistent with the results from the distal mutants in MG1655; DsrA, RyhB and ArcZ as well as Spot 42 and GlmZ levels were increased compared to wild-type cells in the K31A mutant (Fig. S4a and Table S3). These observations suggest that the K31A mutant may increase the overall stability of the sRNA or bind to a larger proportion of the newly made sRNA than wild-type Hfq. In contrast, McaS and ChiX as well as MgrR and MicC, whose levels were elevated in the R16A background, were somewhat lower in the K31A background. Intriguingly, a few sRNAs such as RybB showed still a third pattern and were present at higher levels in both the R16A and K31A mutant compared to wild-type levels (Table S3c). Overall these results indicate that Hfq association with different sRNAs cannot be considered to be equivalent.
mRNAs differentially co-IP with proximal and distal face mutants
The same co-IP samples probed for sRNA levels were also assayed for the levels of the sodB, rpoS and chiP mRNAs in the MG1655 background (Fig. 5b) and for the sodB, rpoS, ptsG, maeA, glmS and shiA mRNAs in the MC4100 background (Fig. S4b). Unlike what was observed for the sRNAs, where the levels of sRNA measured after co-IP paralleled the sRNA levels, some hfq alleles specifically impacted the association of some mRNAs with Hfq. For example, while total sodB mRNA levels were not decreased (and in fact were modestly increased) in the distal face Y25D and K31A alleles, the co-IP levels were clearly lower (Fig. 5b and Table S2). A similar pattern of reduced co-IP was observed for maeA (Fig. S4b). In contrast, the rpoS mRNA was present at wild-type or higher levels in both the total and co-IP samples for the distal face mutants (Fig. 5b, Table S2 and Fig. S4b).
An examination of the known targets of sRNAs for their behavior in the tiling microarrays reinforced the conclusion that individual mRNAs show distinct patterns both in terms of association with wild-type Hfq and with respect to the consequences of the different mutant alleles. The signals for total and co-IP RNA samples from two independent sets of tiling arrays are presented for 47 known sRNA target genes in Table S4c. For each of these known sRNA target mRNAs, seven or more consecutive probes gave signals among the upper quartile of expression for all mRNAs; for many of the known sRNA targets, two sets of probes gave signals above the upper quartile expression threshold (see Materials and Methods for details). The enrichment seen upon co-IP with wild-type Hfq ranged from more than 10-fold to less than 1-fold. This range partially reflects the abundance of a particular mRNA (with more abundant mRNAs generally showing less enrichment), suggesting that only a fraction of some mRNAs may bind to Hfq or that the mRNA that does bind is depleted by degradation.
For the proximal Q8A mutant, total mRNA levels for many targets were similar to wild-type levels and there was reasonable agreement between the co-IP levels in the Q8A and wild-type backgrounds (Q8A total/wt total, Table S4c). Two exceptions with higher levels in the Q8A mutant compared to wild-type cells were ompX and eptB. These same mRNAs were present in relatively low levels in the Q8A co-IP samples compared to the Q8A total sample. Possibly the differences in the levels of the two mRNAs is a consequence of significant regulation in the presence of wild-type Hfq under our growth conditions. The sRNAs regulating these targets, CyaR and MgrR, respectively, are abundant under the conditions of the experiment so the mRNAs might be expected to accumulate in the Q8A background where regulation would be expected to be defective.
The R16A rim mutant had relatively modest effects on total levels of mRNA for all but ompX, for which levels were significantly increased. However, there were differences between mRNAs in terms of their efficiency of co-IP with the R16A allele (R16A co-IP/R16 total, Table S4c). Many had similar co-IP levels between the wild-type and R16A mutant, but some (for instance ptsG, oppA, and cycA) showed higher co-IP levels for R16A than for wild-type, while others (for instance eptB and fur) show much lower co-IP levels for the R16A allele.
The mRNA levels in the distal K31A mutant generally were similar to those in wild-type cells, consistent with the general retention of function in these mutants. The efficiency of co-IP varied more between experiments for K31A than for the other alleles. Nonetheless, it was again evident that different mRNAs showed very different behavior. Thus, significant enrichment in the K31A mutant relative to wild-type was observed for cycA, sthA, glmS, folX, shiA, and hns, while other mRNAs, including sodB, ompC, and cirA, gave K31A co-IP/K31A total ratios of 1 or less. Overall, the global co-IP analysis with selected Hfq mutants revealed that there is significant variation between how Hfq binds to different mRNAs.
Discussion
Hfq has been the subject of significant interest since it was found to act as an essential chaperone for the function of regulatory sRNAs in many bacteria (reviewed in3). Our in vivo analysis of multiple Hfq mutants confirms many of the expectations from in vitro studies but reveals significant differences in how different Hfq mutants affect regulation for specific sRNA and mRNA pairs as well as differences in the association of mutant Hfq with individual sRNAs and mRNAs.
Before we consider our findings for each Hfq region, it is worth noting some complexities inherent in interpreting our in vivo data. For example, the sRNAs amounts reflect levels of synthesis as well as rates of turnover. Thus for a few sRNAs, higher levels in hfq mutants might reflect effects on the expression of the transcription regulators that control the synthesis of the sRNA. For instance, the σE response, responsible for expression of RybB and MicA, is activated in an hfq deletion strain. Given this regulation, we suggest that RybB and MicA show only a limited decrease in the Q8A mutant (Table S3c, Q8A total/wt total) because, while their turnover may be accelerated, their synthesis likely is also increased. In addition, since co-IP with Hfq is a reflection of multiple factors including binding to and dissociation from Hfq, higher co-IP could be due to longer occupancy rather than stronger binding. Related to both of these points, since we are assaying the mutant effects in the context of other RNAs and there is known competition between sRNAs, altered levels of one sRNA due to changes in levels or binding might impact the levels and Hfq association of other RNAs. In addition, Hfq has been shown to bind to DNA40, and changes in this property could indirectly impact the outcome of the assays described here. Nevertheless, some general conclusions can be made about the effects of the Hfq alleles examined in this study.
Proximal face of Hfq is necessary for sRNA function and stability
sRNA function was decreased or absent in most proximal face mutants (Figs. 2 and 3, Tables 1 and 2). In addition, sRNA levels were significantly decreased in the majority of these mutants. The simplest conclusion from these results is that the proximal face residues are necessary for sRNA binding to Hfq, allowing for sRNA function. However, our results do not allow us to distinguish between initial interactions with Hfq and subsequent steps that stabilize the interactions.
These observations are very consistent with the in vitro data showing sRNA interactions with the proximal face. Crystal structures first demonstrated the binding of U-rich oligonucleotides to the proximal face of the hexamer of S. aureus Hfq.8 A recent pair of studies implicated the 3’ polyU terminator sequence in Hfq-stimulated function16 and binding to the proximal domain of Salmonella Hfq17. This last paper identified Q8, F42, and K56 as involved in the binding of uracil, consistent with our finding of significant defects in the activity of these mutants. However, H57A also was implicated in binding specifically to the 3’ of the RNA, but only had a mild phenotype in our tests. Therefore, 3’ end binding may not be essential for sRNA function; possibly there is secondary binding elsewhere on Hfq, or there might be a more limited requirement for highly stable binding under the sRNA overproduction conditions of most of our assays. In another study, eight different Hfq-binding sRNAs showed reduced binding to the proximal face K56A mutant39, and all were competed by polyU but not polyA, consistent with the conclusion that the K56 residue contributes to 3’ polyU binding. An additional contribution to decreased sRNA binding could be reduced hexamer stability as observed for Q8A, Y55A, and K56A in vitro20. Consistent with this, we observed lower levels of Hfq accumulation in the Y55A mutant (Fig. 1A), and lower oligomer levels for Q8A and K56A (Fig. S1).
Given the lower sRNA levels in proximal mutants and the in vitro data on decreased sRNA binding, we expected a significant decrease in the sRNA fraction that co-IPs with Q8A. However, the efficiency of co-IP with Q8A, defined as the ratio of co-IP/total, was comparable to wild-type Hfq for most sRNAs (Tables S2 and S3c). A subset of sRNAs do show a lower efficiency of co-IP in the Q8A mutant (see Q8A co-IP/Q8A total in Table S3c). These sRNAs may more easily be excluded from binding to the mutant Hfq, due to competition by other RNAs or a more stringent requirement for the Q8 residue for binding. Very low levels in the Q8A strain interfere with a robust calculation of co-IP efficiency for still other sRNAs.
Our results regarding target mRNA levels and binding to the Q8A mutant were consistent with expectations but also provided new information on in vivo trafficking of mRNAs. Positively regulated targets would be expected to show lower levels of mRNAs whenever regulation is perturbed and negatively-regulated targets would be expected to show higher levels, assuming growth is under conditions that lead to expression of the sRNA. This pattern was seen (Table S2). The most strongly up-regulated mRNAs in the Q8A mutant were ompX and eptB, both negatively-regulated by sRNAs that are well-expressed in these samples (Table S4c, Q8A total/wt total). The co-IP of many mRNAs with Q8A was generally parallel to that seen with wild-type Hfq, suggesting that most mRNAs can bind to Hfq in the absence of sRNAs. In support of this suggestion, co-IP with Q8A was seen for mRNA targets of both relatively abundant sRNAs (GcvB target cycA) and poorly expressed sRNAs (FnrS target folX) (Table S4c).
Interestingly, mutations in two residues, D9 and F39, were distinct from other proximal face alleles in that they showed mixed effects on different sRNA/mRNA pairs. The only published studies on D9A report strong binding to DsrA9, consistent with our results (Table S2). However, sRNA binding does not necessarily reflect activity for this allele, since RyhB is efficiently bound (Table S2), but only partially regulates sodB and is unable to regulate the succinate operon (Figure 3g, Tables 1, 2). In other studies, the D9A mutant also was found to be defective for OmrB repression of cirA29. The effects of F39A also varied between sRNA/mRNA pairs. ArcZ and RyhB levels were both low in the F39A mutant, but ArcZ was defective for repression of flhD while RyhB was effective for repression of sdhC. More significantly, McaS levels and ChiX levels were only slightly lower in the F39A mutant, but both activation of flhD by McaS and repression of chiP by ChiX were defective in the F39A mutant (Table 1). We suggest that both D9 and F39 define functions in addition to sRNA binding, but these functions cannot be defined from the current in vivo experiments. Further work, including the study of these mutant proteins in vitro, will be necessary to define the roles of these residues in Hfq-mediated regulation.
Rim residues participate in the function of some sRNAs
In our studies, the rim alleles R16A and R16C, and, to a lesser extent, R17A and R19D, were defective for positive regulation, particularly in liquid assays, and showed partial defects for negative regulation. In addition, the R16A mutant showed differences in the effects on the levels of specific sRNAs (Figs. 4 and 5, Table S2). While the accumulation of many sRNAs was modestly decreased, the levels of ChiX and McaS, as well as MgrR and MicC were relatively high (Tables S2 and S3). The basis for this difference is not yet clear, but strongly suggests differences in how sRNAs bind and/or how stably they bind, even if all are dependent upon proximal face interactions.
mRNAs levels were mostly unchanged in amounts in the R16A allele, although ompX levels were significantly increased, suggesting that R16A is likely defective for negative regulation of ompX by CyaR and possibly by MicA. Interestingly, however, there were dramatic differences in the efficiency of mRNA co-IP with R16A that may reflect different roles for the rim alleles for individual mRNAs. For instance, the total level of the MgrR target eptB mRNA was modestly increased relative to wild-type cells, but co-IP with this mutant was poor (Table S4). On the other hand, hns, repressed by DsrA, also was modestly increased in total RNA levels in the R16A mutant but co-IP with the mutant was very efficient. Interestingly, even though positive regulation of flhD and rpoS was defective in the R16A mutant, co-IP of these mRNAs with R16A, as well as the positively-regulated shiA mRNA, was efficient. These results support different modes of interaction or action on individual mRNAs, some disrupted by mutations of rim residues while others not.
Distal face residues affect mRNA binding and mutants show a dose-specific loss of function
Crystal structures of Hfq bound to a polyA sequence demonstrate binding at the distal face, with recognition of ARN motifs by residues including Y25, G29, I30, and K31.11 In our tests, the Y25D and I30D alleles had the strongest phenotypes, followed by K31A, while the G29A mutant was generally functional. It is likely that there is redundancy in these sites as we see a stronger phenotype for a G29A K31A double mutant (unpublished results).
Binding of ARN motifs is critical for Hfq to promote annealing of activating sRNAs to the rpoS leader in vitro and activation of rpoS mRNA in vivo. Consistent with this finding and the role of the distal face in binding ARN motifs, we found that the Y25D and I30D mutants were defective for activation of rpoS by ArcZ on plates (Table 1, Fig. 2), and Y25D was defective in liquid assays, under conditions of low mRNA expression (Table 2). An examination of the array data also provides support for a role for the distal face K31 residue for GlmZ regulation of glmS, one other positively regulated target. GlmZ is well expressed under the conditions of the array (Table S3), and the levels of glmS mRNA were decreased below wild-type levels for K31A, consistent with a loss of positive regulation.
If the distal face binds mRNAs, and therefore helps promote annealing to sRNAs, one might expect that higher levels of mRNAs could bypass the defect in these mutants. Indeed, we see clear suppression of positive regulation, but less so for negative regulation, when the promoter expressing the mRNA target was activated by higher levels of arabinose (Table 2). Therefore, for these assays, it would appear that the distal face mutants are more critical for positive regulation than for negative regulation. Whether this reflects a qualitatively different mode of mRNA binding or a need for prolonged binding/annealing for positive regulation remains to be determined.
As expected if distal face residues bind mRNAs, but are not central to sRNA binding, sRNA levels were not decreased in the distal face mutants (Fig. 4 and Table S3). In fact, the distal site mutants had increased levels of some sRNAs. This may reflect the lack of pairing partners on Hfq for these sRNAs. When sRNA half-life is determined under conditions where mRNAs continue to be made (turning off the sRNA promoter by washing out an inducer rather than stopping all transcription with an antibiotic), the sRNAs are generally unstable.31 This has been interpreted to suggest that sRNA turnover is coupled to mRNA pairing. If so, Hfq alleles that fail to bind mRNAs might lead to high levels of sRNA accumulation.
The levels of at least two of the sRNAs, McaS and ChiX, did not increase in the K31A mutant. ChiX is known to be used catalytically rather than being degraded at each use.27 Possibly catalytic use will be found for other sRNAs with low ratios of K31A total/wt total. It is also conceivable that distal site residues are directly involved in binding of some sRNAs. We note that the behavior of sRNAs in K31A and R16A appears to be reciprocal; McaS and ChiX are present at higher levels in R16A and lower levels in K31A while DsrA, RyhB and ArcZ are present at higher levels in K31A and lower levels in R16A. It seems possible that there are two alternative modes for sRNA binding, one dependent on K31 and the other dependent on R16. Whether this is binding of the sRNA itself, binding of a target mRNA, or some other factor is not yet clear.
The results of mRNA co-IP for the distal site alleles did not necessarily reflect the findings for regulation. However, the assays for regulation were carried out with excess sRNA expressed from plasmids, while the mRNA and sRNA measurements were conducted in cells expressing only endogenous levels of the sRNAs (Fig. 5, Table S2, Table S4). Three mRNAs were tested in Fig. 5b. While there were distinctions between the mRNAs, these differences did not segregate with positive or negative behavior. Examining the set of mRNAs in Table S4, the general trend was that strong enrichment by co-IP in the K31A mutant correlated with strong enrichment by wild-type Hfq; mRNAs that were poorly enriched by co-IP with Hfq in wild-type cells were also very low for K31A (for instance gltA). We propose that further characterization of the distal face mutants will help to clarify the steps in Hfq-mediated base pairing once initial binding has taken place.
In vivo approaches raise some caveats about in vitro approaches to studying Hfq
As mentioned above, there have been many in vitro studies of Hfq and the ability of at least some sRNAs to bind to mutant versions of Hfq. A comparison of those studies to the in vivo studies described here supports the basic in vitro conclusions of multiple binding surfaces with different specificities. Our results also highlight some issues that should help guide future in vitro experiments. In general, only a limited set of sRNAs and target mRNAs have been studied in vitro, with the majority of studies focusing on DsrA and rpoS. As our results show, not all sRNAs are equivalent in their behavior, and even for a given sRNA, different targets may show different sensitivities to Hfq mutants (compare RyhB regulation of sodB and sdh, Table 1). The basis for this difference is not currently understood and will certainly benefit from in depth in vitro comparisons.
In addition, as we have learned more about how sRNAs and mRNAs are recognized by Hfq, it has become clear that not all in vitro studies were carried out under the best conditions. We now know that an essential site on the rpoS leader for Hfq-dependent activation is located far upstream of the DsrA/rpoS pairing region ; this site was not present in the rpoS substrates used in initial studies, making earlier results with mutant Hfqs somewhat difficult to interpret. Similarly, recent studies have implicated the polyU tail on sRNAs as important for regulation in vivo16, and this same 3’ end has been shown to participate in binding to Hfq in vitro17. However, many in vitro substrates, made by run-off transcription, were synthesized without this polyU tail.9 Thus, some of the earlier conclusions about what Hfq mutants affect sRNA binding should be revisited.
In vivo approaches such as ours avoid the complications of possibly missing components, truncated mRNA and sRNA substrates. The genetic systems also allow for multiple sRNA/mRNA regulatory pairs to easily be screened, uncovering intriguing variations in the ways in which Hfq stimulates pairing. Our results provide questions as well as models that are eminently testable in vitro and in vivo.
Can array signatures be used to classify RNAs?
The tiling array experiments, coupled with the analysis above, should allow us to extrapolate conclusions from the RNAs analyzed in Figs. 4 and 5 to other RNAs we can detect in our array. Thus we considered whether we could define a general signature for Hfq-binding sRNAs based on the data in Table S3. First, not surprisingly, the best predictor for an Hfq-binding sRNA is the level in the co-IP sample compared to the total RNA sample. This number was >2 for all the 25 known Hfq-binding sRNAs. Of the other 35 sRNAs in Tables S3a and S3b not classified as Hfq-binding sRNAs, only six sRNAs (IsrC, SibC, PsrO/SraG, RyfA, RyjA, IstR, PsrD/SraB) had ratios >2. Therefore, this criterion should be considered necessary but not sufficient to define an Hfq-binding sRNA. Second, the decreased amount of RNA in both the total and co-IP samples in a Q8A mutant, compared to wild-type samples, also was a reasonable predictor. For instance, if one considers the samples shown in Tables S3a and S3b, of the 40 entries with a Q8 total/wt total ratio of 0.5 or less, 34 are known Hfq-binding sRNAs, and the six that are not known Hfq-binding sRNAs did not fit the first criteria (wt IP/wt total <2). However, not every Hfq-binding sRNA (MicF, MicC, MicA, RyeB/SdsR) gave this decrease in Q8A. Third, the ratio of K31A IP/K31A total was >2 for all known Hfq-binding sRNAs (Table S3c). For the few other sRNAs with a ratio >2 for K31A IP/K31A total, the majority failed the first criterion (wt IP/wt total <2). We note, however, that these predictors do not eliminate all other RNAs, since many mRNAs have similar patterns of enrichment upon co-IP with different Hfq mutant alleles. Using these criteria, we should, however, be able to identify likely candidates for additional Hfq-dependent sRNAs.
Different sets of sRNAs and mRNAs showed significantly different patterns with respect to the effects of the Hfq mutant alleles. Given the limited numbers of characterized sRNA/mRNA pairs, it is not clear whether the sets define differences in mechanism of action. Nevertheless, the dataset is a resource that can be mined for additional Hfq-binding sRNAs as well as possible mRNA targets that might be regulated similarly.
Materials and Methods
Bacterial strains and plasmids
The bacterial strains used in this study are listed in Supplementary Table S5. All E. coli K-12 strains used in this study are derivatives of strains MG1655 or MC4100. All plasmids and olignonucleotides (obtained from Integrated DNA Technologies) are listed in Supplementary Tables S6 and S7, respectively.
The hfq mutations were first introduced into hfq carried on either pUC-hfq (where the hfq gene was amplified from MC4100 chromosomal DNA by PCR using primers AZ#654 and AZ#680, digested with EcoRI and HindIII and cloned into the corresponding sites in pUC18) or pNRD41429. For both plasmids the mutations were generated by site-directed mutagenesis using the specific primers listed in Table S7 and the QuikChange Lightning site-directed mutagenesis kit (Stratagene). All mutations were confirmed by sequencing.
The hfq mutations were introduced into the chromosome using λ-Red recombination. This was simplified by first replacing the wild-type hfq gene with either an Δhfq::cat-sacB cassette [generated by PCR amplification using NC397 chromosomal DNA and primers AZ#997 and AZ#999 carrying sequencing flanking hfq] or a Δhfq::trpAterminator-kan-PBAD-ccdB cassette [generated by amplifying the kan-PBAD-ccdB cassette from strain CR201 (Ranquet et al., submitted, deposit patent number: FR 11/60169, 08/11/2011, UJF/BGene) using primers HFQTRPATERMKANF and HFQKANCCDBR]. To construct the mutant strains, PCR fragments carrying the desired hfq mutations were generated using primers AZ#1000 or AZ1000TRUNC and AZ#1001 (carrying sequences homologous to hfq 40 bp upstream and downstream of the gene), and then electroporated into the appropriate strain at 30 °C to replace the Δhfq::cat-sacB or Δhfq::trpAterminator-kan-PBAD-ccdB cassette. Recombinants were selected by growth on sucrose or arabinose, respectively, at 37 °C (to select against the mini-λ). The resulting mutants were verified by PCR and sequencing.
To assay the effect of various Hfq alleles on different sRNA:mRNA partners, it was desirable to have a simple method for moving all of the mutations into various assay strains using phage P1 transduction46. To this end, the Δhfq::cat-sacB allele (from AZ234) was transduced by P1vir into a strain carrying a ΔpurA::kan allele (ECK4173), selecting for both chloramphenicol and kanamycin resistance to generate DJS2286, in which the hfq and purA mutations are tightly linked (within 4 kb). The linked hfq and purA mutations subsequently were transduced into the desired backgrounds and used as the recipient parental strain for P1 transduction of the desired hfq mutant alleles by selecting for growth on glucose minimal plates (PurA+) and screening for sensitivity to chloramphenicol. Isogenic purA+ Δhfq::cat-sacB derivatives were also constructed for each assay strain.
Strains carrying lacZ fusions were constructed in PM120547. Some of these fusion strains were described previously (see Table S5). For those fusions constructed for this paper, primers were designed to give a PCR product with 40 nt of homology to the PBAD promoter followed by the specific gene starting at the transcription start site (+1) through codon 9, followed by 40 nt of homology to lacZ. The PCR products were introduced into the PM1205 chromosome using λ-Red recombination. Successful recombinants were obtained by selection for growth in the presence of sucrose, and the resulting fusions were verified by PCR and sequencing. To examine the effect of hfq mutants in a strain expressing high constitutive levels of RyhB, a fur::kan mutation was transduced from MG1054 into derivatives of KM331 carrying the hfq alleles of interest, selecting for kanamycin resistance.
Plasmids expressing the sRNA of interest were described previously as noted in Table S6. The plasmids were introduced into the appropriate strain background by TSS transformation48 and plated on LB plates containing ampicillin.
Bacterial growth conditions
Bacterial strains were grown at 37 °C in Luria-Bertani (LB) rich medium, Lennox broth, MacConkey lactose, MOPS EZ rich defined medium (Teknova) with 0.4% glycerol, or M63 minimal medium with 0.001% vitamin B1 and 0.2% glucose or succinate. Ampicillin (50–100 µg/ml), chloramphenicol (25 µg/ml), kanamycin (30–50 µg/ml), or tetracycline (10 µg/ml) was added where appropriate. Isopropyl-β-D-thiogalactopyranoside (IPTG) was added at a final concentration of 0.0001%, and L-arabinose was added at indicated final concentrations.
Immunoblot analysis
Wild-type and hfq mutant derivatives of SG30200 (PM1205 lacI’::PBAD-rpoS-lacZ purA::kan Δhfq::cat-sacB) were grown in LB medium at 37 °C to early stationary phase (OD600 ~ 1.0). To examine Hfq under denaturing conditions, cell pellets were suspended in Laemmli buffer (100 ml/OD600 cells) and heated at 90 °C for 5 min. The proteins (5 µl lysate) were separated on 10–20% Tris-Glycine gels (Invitrogen). To examine Hfq under semi-native conditions, cell pellets were suspended in Laemmli buffer containing 0.5% SDS and no reducing agent (100 ml/OD600 cells) in the presence of an equal volume of glass beads (Sigma). The proteins (3 µl lysate) were separated on a 15% semi-native polyacrylamide gel20. For both types of gels, the proteins were transferred to nitrocellulose membranes by electroblotting. All blots were probed with a 1:10,000 dilution of anti-Hfq antiserum, and signal was detected by using the ECL Western Blotting System (GE Healthcare).
β-galactosidase assays
β-galactosidase activity was assayed using o-nitrophenyl-β-D-galactopyranoside (ONPG) as a substrate as described previously46.
RNA isolation
Total RNA was extracted by the hot phenol method described previously49 or using the TRIzol Reagent (Invitrogen) followed by isopropanol precipitation.
Northern blot analysis
Northern blot analysis of sodB mRNA levels (Fig. 3) was performed by fractionating 10 µg of RNA from each sample on a 1.2% agarose gel that was prerun for 5 min at 12 V/cm in a 1X morpholinepropanesulfonic acid (MOPS) buffer and subsequently run at 5 V/cm for 2 h. The RNA was transferred to a Zeta-Probe GT membrane (Bio-Rad) by capillary action overnight, and cross-linked to the membrane by UV irradiation. The membrane was probed with SodB probe (Table S7) in ULTRAhyb solution (Ambion) at 42 °C overnight. The blot was developed using the Brightstar Biodetect kit (Ambion) as per the manufacturer’s instructions.
Northern blot analysis of ArcZ expression (Fig. 4 and 5) was performed by fractionating 5 µg of total RNA or 0.5 µg of co-IP RNA from each sample on 8% polyacrylamide urea gels containing 8 M urea in 1X TBE buffer at 70 watts for 70 min. The fractionated RNA was transferred by electroblotting to a Zeta-Probe GT membrane (Bio-Rad) at 20 V for 16 h in 0.5X TBE. The RNA was cross-linked to the membrane by UV irradiation, and the membrane was probed with the 5’-32P-end labeled primer specific to ArcZ (Table S7) in ULTRAhyb solution (Ambion) at 42 °C overnight.
Primer extension analysis
The levels of sRNA and mRNA targets were analyzed by primer extension analysis using primers (listed in Supplemental Table S7) specific to the indicated genes as described50. RNA samples (5 µg total RNA or 0.5 µg co-IP RNA) were incubated with 2 pmol of 5’-32P-end labeled primer at 80 °C and then slow-cooled to 42 °C. After the addition of dNTPs (1 mM each) and AMV reverse transcriptase (10 U, Life Sciences Inc.), the reactions were incubated in a 10 µl reaction volume at 42 °C for 1 h. The reactions were terminated by adding 10 µl Stop/Loading Buffer and cDNA products then were fractionated on 8% polyacrylamide urea gels containing 8 M urea in 1X TBE buffer at 70 watts for 70 min.
Co-IP assay
RNAs that co-IP with Hfq were isolated as described previously1 with the following modifications. Cell extracts were prepared from derivatives of SG30200 cells grown to early stationary phase (OD600 ~ 1.0, Fig. 5) or MC4100 cells grown to exponential phase (OD600 = 0.4, Fig. S4) in LB medium at 37 °C, and co-IPs were carried out using 20 µl of anti-Hfq antiserum51, 24 µg of protein-A-Sepharose (Amersham Biosciences) and 200 µl of cell extract per co-IP reaction. Co-IP RNA was isolated from protein-A-Sepharose beads by extraction with phenol:chloroform:isoamyl alcohol (50:50:1), followed by ethanol precipitation.
Affymetrix tiling array assays
MC4100, GSO615 (hfq-Q8A), GSO621 (hfq-R16A), and GSO627 (hfq-K31A) cells were grown in LB medium at 37 °C to exponential phase (OD600 = 0.4), and total RNA or co-IP RNA samples were prepared as described above. Microarray analysis was carried out by hybridizing RNA directly to the Affymetrix E. coli Tiling Arrays covering the entire E. coli genome and bacteriophage λ genome (Affymetrix, Ecoli_Tab520346F, P/N 520346). The RNA samples (2 µg for co-IP RNA and 20 µg for total RNA) were added to 1X hybridization buffer (100 mM MES, pH 6.6, 1 M NaCl, 20 mM EDTA, 0.01% Tween 20) supplemented with 0.1 mg/ml herring sperm DNA, 0.5 mg/ml BSA and 50 pM of control biotin-labeled oligonucleotide B2 in 200 µl total volume, heated to 95 °C for 5 min and then incubated at 45 °C for 5 min before being placed in the microarray cartridge. Hybridization was carried out at 45 °C for 16 h on a rotary mixer at 60 rpm. Following hybridization, the sample solution was removed and the array was washed in the Affymetrix Fluidics station as recommended in the technical manual. Hybridization was detected using the RNA:DNA mouse monoclonal antibody34. The antibody was diluted to 0.02 mg/ml in 1X staining buffer (100 mM MES, pH 6.6, 1 M NaCl, 0.05% Tween 20) with 2 mg/ml BSA in 600 µl total volume, loaded on the array and incubated at 25 °C for 60 min. After 10 wash cycles in NSWB, the array was incubated with 0.035 mg/ml biotin-labeled rabbit anti-mouse IgG (Invitrogen) and 0.4 mg/ml rabbit IgG (Sigma) in 1X staining buffer with 2 mg/ml BSA in 600 ml total volume at 25 °C for 60 min. After another 10 wash cycles in NSWB, the arrays were incubated with 0.01 mg/ml streptavidin-phycoerythrin (Invitrogen) in 1X staining buffer with 2 mg/ml BSA in 600 ml total volume at 25 °C for 60 min. After a third set of 10 wash cycles in NSWB, the arrays were scanned in an Affymetrix laser scanner (at 570 nm with a resolution of 3 µm) and data (in .CEL file) were collected using Affymetrix GeneChip Operations Software (GCOS 1.4).
Analysis of tiling array data
The Affymetrix E. coli Tiling Arrays contain 25-mer probes staggered by 8 nucleotides for both strands of the genome. As a first step in data analysis, perfect match (PM) probe signals for each array experiment were background adjusted following the approach of Irizarry et al.52. In summary, for a given array experiment, optical noise and non-specific binding were modeled by a common mean background level estimated from the mode of the log distribution of mismatch (MM) probe signals, and this experiment specific background level was subtracted from each PM probe signal in the experiment. Following background adjustment, data from different array experiments were normalized using quantile normalization53.
For each RefSeq35-annotated sRNA, the set of seven consecutive probes assaying the sRNA with highest background-adjusted, normalized signal from the wild-type co-IP sample was identified. The average background-adjusted, normalized signal for this maximal set of seven consecutive probes was identified for each sRNA for all total RNA and co-IP samples (Table S3).
For each RefSeq35-annotated mRNA, the set of seven consecutive probes assaying the mRNA around the translation start site, from 100 nucleotides upstream to 50 nucleotides downstream of the start of translation, with highest background-adjusted, normalized signal from the wild-type total RNA and wild-type co-IP samples were identified. For each of the wild-type total RNA sample and wild-type co-IP sample, the maximal set of seven consecutive probes for each mRNA was used to identify an upper quartile threshold such that 25% of mRNAs had expression in the sample above the threshold and 75% did not. For 47 known sRNA target mRNAs, sets of seven consecutive probes assaying the mRNA around the translation start site with signal greater than or equal to the upper quartile threshold in both wild-type total RNA and wild-type co-IP samples were identified. The average background-adjusted, normalized signal for each set of seven consecutive probes meeting these two criteria was identified for each of the 47 known sRNA target mRNAs for all total RNA and co-IP samples (Table S4).
Supplementary Material
In vivo analysis of 14 chromosomally-expressed hfq mutants reveals differential consequences of specific amino acid substitutions.
Phenotypes confirm a critical role for the proximal face of Hfq in sRNA binding.
The rim of the Hfq hexamer is important for positive regulation by sRNAs.
Individual sRNA:mRNA pairs show different sensitivities to hfq mutants.
The results suggest unexpected complexity in how Hfq promotes sRNA-based regulation.
Acknowledgements
We thank Nadim Majdalani and Nicholas De Lay for providing strains and technical advicẽ We also thank Joyce Fu and Daniel Fu for assistance in constructing some mutants and Sarah Woodson and colleagues for helpful discussions. We thank Taylor Updegrove, Sarah Woodson, Kumaran Ramamurthi and Nadim Majdalani for their comments on the manuscript. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Abbreviations used
- sRNA
small RNA
- co-IP
co-immunoprecipitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Data
Supplementary data associated with this article can be found in separate files.
References
- 1.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 2.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000163. e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 5.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 7.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X, Wartell RM. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–4887. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- 11.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beisel CL, Updegrove TB, Janson BJ, Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31:1961–1974. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salim NN, Faner MA, Philip JA, Feig AL. Requirement of upstream Hfq-binding (ARN)x elements in glmS and the Hfq C-terminal region for GlmS upregulation by sRNAs GlmZ and GlmY. Nucleic Acids Res. 2012;40:8021–8032. doi: 10.1093/nar/gks392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013028. e13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc. Natl. Acad. Sci. USA. 2011;108:13059–13064. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauer E, Weichenrieder O. Structural basis for RNA 3'-end recognition by Hfq. Proc. Natl. Acad. Sci. USA. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. USA. 2012;109:9396–9401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziolkowska K, Derreumaux P, Folichon M, Pellegrini O, Régnier P, Boni IV, Hajnsdorf E. Hfq variant with altered RNA binding functions. Nucleic Acids Res. 2006;34:709–720. doi: 10.1093/nar/gkj464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panja S, Woodson SA. Hexamer to monomer equilibrium of E. coli Hfq in solution and its impact on RNA annealing. J. Mol. Biol. 2012;417:406–412. doi: 10.1016/j.jmb.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 23.Thomason MK, Fontaine F, De Lay N, Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soper TJ, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Lay N, Gottesman S. A complex network of small noncoding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012;86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Overgaard M, Johansen J, Møller-Jensen J, P V-H. Switching off small RNA regulation with trap-mRNA. Mol. Microbiol. 2009;73:790–800. doi: 10.1111/j.1365-2958.2009.06807.x. [DOI] [PubMed] [Google Scholar]
- 28.Massé E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Lay N, Gottesman S. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA. 2011;17:1172–1189. doi: 10.1261/rna.2531211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maki K, Uno K, Morita T, Aiba H. RNA, but not protein partners, is directly responsible for translational silencing by a bacterial Hfq-binding small RNA. Proc. Natl. Acad. Sci. USA. 2008;105:10332–10337. doi: 10.1073/pnas.0803106105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Argaman L, Elgrably-Weiss M, Hershko T, Vogel J, Altuvia S. RelA protein stimulates the activity of RyhB small RNA by acting on RNA-binding protein Hfq. Proc. Natl. Acad. Sci. USA. 2012;109:4621–4626. doi: 10.1073/pnas.1113113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu Z, Zhang A, Storz G, Gottesman S, Leppla SH. An antibody-based microarray assay for small RNA detection. Nucleic Acids Res. 2006;34:e52. doi: 10.1093/nar/gkl142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson KM, Rhodius VA, Gottesman S. SigmaE regulates and is regulated by a small RNA in Escherichia coli. J. Bacteriol. 2007;189:4243–4256. doi: 10.1128/JB.00020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Figueroa-Bossi N, Lemire S, Maloriol D, Balbontín R, Casadesús J, Bossi L. Loss of Hfq activates the sigmaE-dependent envelope stress response in Salmonella enterica. Mol. Microbiol. 2006;62:838–852. doi: 10.1111/j.1365-2958.2006.05413.x. [DOI] [PubMed] [Google Scholar]
- 38.Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. Proc. Natl. Acad. Sci. USA. 2011;108:1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olejniczak M. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry. 2011;50:4427–4440. doi: 10.1021/bi102043f. [DOI] [PubMed] [Google Scholar]
- 40.Updegrove TB, Correia JJ, Galletto R, Bujalowski W, Wartell RM. E. coli DNA associated with isolated Hfq interacts with Hfq's distal surface and C-terminal domain. Biochim. Biophys. Acta. 2010;1799:588–596. doi: 10.1016/j.bbagrm.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Updegrove TB, Wartell RM. The influence of Escherichia coli Hfq mutations on RNA binding and sRNA•mRNA duplex formation in rpoS riboregulation. Biochim. Biophys. Acta. 2011;1809:532–540. doi: 10.1016/j.bbagrm.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Updegrove TB, Wilf N, Sun X, Wartell RM. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5' rpoS mRNA leader region. Biochemistry. 2008;47:11184–11195. doi: 10.1021/bi800479p. [DOI] [PubMed] [Google Scholar]
- 43.Lease RA WS. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Court DL, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, Sawitzke J, Sharan SK. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–69. doi: 10.1016/s0378-1119(03)00728-5. [DOI] [PubMed] [Google Scholar]
- 46.Miller JH. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Plainview, NY: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 47.Mandin P, Gottesman S. A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol. Microbiol. 2009;72:551–565. doi: 10.1111/j.1365-2958.2009.06665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung CT, Miller RH. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988;16:3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Massé E, Vanderpool CK, Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005:20. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 52.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 53.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.