Abstract
The CELF family of RNA-binding proteins regulates many steps of mRNA metabolism. Although their best characterized function is in pre-mRNA splice site choice, CELF family members are also powerful modulators of mRNA decay. In this review we focus on the different modes of regulation that CELF proteins employ to mediate mRNA decay by binding to GU-rich elements. After starting with an overview of the importance of CELF proteins during development and disease pathogenesis, we then review the mRNA networks and cellular pathways these proteins regulate and the mechanisms by which they influence mRNA decay. Finally, we discuss how CELF protein activity is modulated during development and in response to cellular signals. We conclude by highlighting the priorities for new experiments in this field.
Keywords: CELF (CUGBP, ELAV-Like Family) proteins; mRNA decay; GU-rich element; deadenylation; ELAV; myotonic dystrophy
1. Introduction
CELF (CUGBP, ELAV-Like Family) proteins have been called a plethora of names (BRUNO-like proteins, CUGBP proteins, CUGBP and ETR-3 like factors, EDEN-BP, NAPOR etc), because they were independently discovered and cloned by distinct research groups working with organisms ranging from humans to fruit flies. The inconsistent nomenclature has created much confusion; thus for the sake of simplicity we will primarily use the CELF acronym approved by the Human Genome Organization to describe this family of proteins. This acronym maintains the historical links to the prototypical and best studied member of the family, CELF1 (previously known as CUGBP1), and also highlights that the CELF proteins are related to ELAV (Embryonic Lethal Abnormal Vision) proteins (CELF2 was formerly known as ETR3, which stands for ELAV-Type RNA binding protein-3). ELAV factors form an evolutionarily conserved family of RNA-binding proteins that interact with U-rich sequences [1, 2].
The story of human CELF proteins began when CELF1/CUGBP1 was isolated as a factor that bound (CUG)8 RNA in vitro [3, 4]. As CUG-repeat expansion is the root cause of myotonic dystrophy type 1 (DM1) [5] this discovery spawned an avalanche of investigations into the role of CELF proteins in DM1 and in muscle. Many of these focused on CELF proteins as regulators of pre-mRNA splice site choice and collectively such studies demonstrated that altered CELF function is a major contributor to pathology in DM1 [6].
Just before human CELF1 was identified, the Drosophila CELF protein, Bruno, was determined to play an essential role as a translational regulator in early development [7] and a Xenopus homolog (EDEN-BP) was shown to regulate deadenylation [8]. Since these early discoveries, research in each model system has tended to focus on one aspect of CELF protein function: splicing in mammals, translation in Drosophila and mRNA deadenylation in Xenopus. These processes are often coupled, however, and there is now substantial evidence that CELF proteins are multifunctional and regulate splicing, translation and mRNA decay in mammals. Moreover, these functions of CELF1 in modulating mRNA decay and translation are undoubtedly as important in development and disease as its role in pre-mRNA splicing.
In this review, after describing the biological effects of CELF-mediated regulation, we specifically examine the impact of CELF proteins on mRNA decay, a process that profoundly influences gene expression in all cells. For most mRNAs, decay initiates with removal of the poly(A) tail (deadenylation) and this rate-limiting step is modulated by CELF proteins through interaction with deadenylase enzymes. Below, we discuss how CELF proteins recognize their target transcripts in order to initiate coordinated and rapid changes in gene expression that occur during development and in response to external stimuli. Despite an overabundance of studies implicating CELF proteins as splicing regulators, emerging studies related to their role in controlling mRNA decay indicate that this function is likely just as essential.
2. The CELF protein family
CELF proteins are found in almost all eukaryotes with many organisms encoding multiple homologs. Six members of the CELF family have been identified in humans and mice: CELF1 (CUGBP1/NAB50/BRUNOL2) and CELF2 (CUGBP2/ETR3/BRUNOL3/NAPOR) proteins are expressed widely in many different tissues. CELF3, CELF4 and CELF5 proteins are restricted to adult tissues and found almost exclusively in the nervous system [9, 10]. CELF6 is expressed in kidney, brain and testes [11].
CELF proteins bear RNA Recognition Motifs (RRMs), responsible for binding to RNA, and are characterized by the arrangement of these domains as well as their conservation (Figure 1). Each CELF protein has two N-terminal RRMs followed by a linker and a third C-terminal RRM [12–15]. This structural arrangement is shared with the ELAV proteins which also play important roles in mRNA metabolism [2]. However, beyond the arrangement of RRMs, the similarity between ELAV and CELF proteins is limited to the highly conserved residues that define the RRM motif (Figure 1). In contrast, CELF1 and CELF2 are virtually identical within their RNA-binding domains (>90% conserved) but the linker region is more divergent. At the sequence level, CELF3–6 are more closely related to each other than to CELF1 and CELF2, but homology with CELF1 is still readily detected. In addition to the three RNA-binding domains, other notable functional domains have been characterized including a C-terminal lysine/arginine-rich nuclear localization signal conserved in all CELF proteins, and a region with nuclear export activity found in the linker domain of CELF2 [16] (Figure 1). The roles and regulation of these, and other features of these proteins, are discussed below.
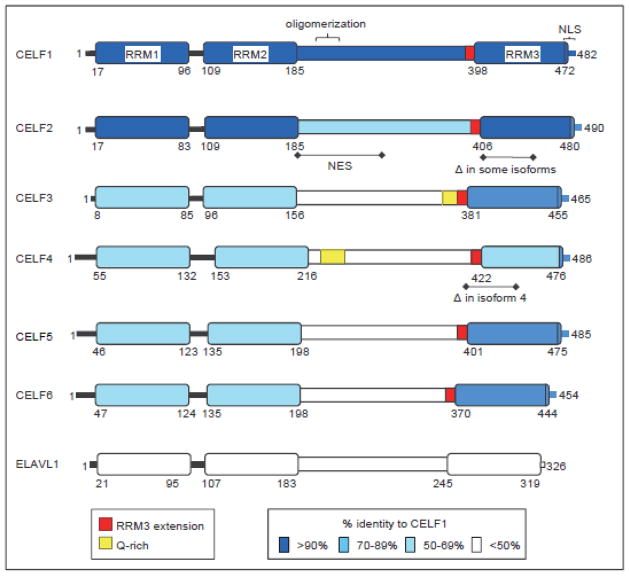
Fig 1.
CELF protein domain structure and conservation. Each human CELF protein is depicted with the level of identity to CELF1 denoted by the color of blue shading for each RR, the linker region and the C-terminal nuclear localization signal (NLS). Darker shading indicates more homology. Other motifs including Q-rich domains, oligomerization domain, the conserved N-terminal extension of RRM3 and a nuclear export signal (NES) found in CELF2 (as defined in [16]) are also shown. Regions of RRM3 affected by alternative splicing in CELF2 and CELF4 are indicated. ELAVL1 is shown for comparison, but has less than 50% identity in all domains. Protein sequences used for this alignment were CELF1 Isoform 1 (NP_006551.1), CELF2 Isoform 1 (NP_001020247.1), CELF3 Isoform 1 (NP_009116.3), CELF4 Isoform 1 (NP_064565.1), CELF5 Isoform 1 (NP_068757.2), CELF6 Isoform 2 (NP_001166155.1), ELAVL1 (NP_001410.2).
3. CELF proteins in mRNA metabolism
In this section we will examine how CELF proteins mediate their wide-ranging effects by binding to diverse mRNA targets to modulate gene expression at a post-transcriptional level. We will focus specifically on mRNA decay, but it is important to note that this is just one of many processes that CELF proteins influence. Indeed, depending on where CELF proteins associate with their targets (5′UTR, introns, 3′UTR), and what other factors they interface with, they can impact polyadenylation, splicing, editing, localization, and translation as well as mRNA turnover. Although CELF protein functions outside of mRNA decay regulation must be glossed over here in the interests of brevity, they have been reviewed elsewhere [17–20]. In addition, studies investigating other aspects of mRNA metabolism that have provided insights relevant to CELF protein roles in mRNA decay regulation will be discussed.
3.1. RNA recognition by CELF proteins
3.1.1. Sequence preferences in vitro and in vivo
An astounding number of studies have examined the binding preferences of CELF proteins and particularly CELF1 using in vitro techniques such as three-hybrid [21], Systematic Evolution of Ligands by Exponential enrichment (SELEX) [22, 23], Nuclear Magnetic Resonance (NMR) [14], Surface Plasmon Resonance (SPR) [24], Electrophoretic Mobility Shift Assay (EMSA) [23, 25], as well as RNA-Immunoprecipitation followed by microarray [26–29] or Cross-Linking Immunoprecipitation followed by sequencing [30, 31] (RIP-Chip/CLIP-Seq) in cultured cells. The findings are generally in agreement and clearly demonstrate that CELF1 and its homologs (EDEN-BP in Xenopus [29, 32], Bru [7] and Bru-3 [33] in Drosophila, Danio Rerio Brul [34]), and mammalian CELF2 [22] and CELF4 [35]) all recognize GU-rich RNA sequences with high affinity. The binding sites can take two forms; either a UG-repeat or a U-rich sequence interspersed with G nucleotides. The prominent similarity in binding preference amongst family members is perhaps not surprising given their high level of conservation. However, it is curious to note that CELF1 was first identified through an association with CUG repeat-containing RNA [4] and it has been reported to favor GC-rich sequences [36, 37]. CELF2 was also recently found to associate with the androgen receptor (AR) mRNA through the expanded CUG repeats that cause Spinal Bulbar Muscular Atrophy [38]. These observations encourage further investigation as the affinity of recombinant CELF1 protein for CUG repeats is much lower (~100 fold) in vitro [24], than for UG-rich sequences and there is no evidence for significant enrichment of CUG sequences in RIP-seq analyses which are designed to entrap natural targets of RBPs [30, 31]. At this point, re-visiting the binding preferences following post-translational modification or in the presence of other RBPs would likely be very informative.
In addition to binding GREs, CELF1 and CELF2 have also been shown to associate with AU-rich elements (AREs) in TNF [39] and COX2/PTGS2 mRNAs, respectively [40]. These interactions appear to be biologically significant as disrupting them alters stability of the target transcripts, but there is little evidence to suggest that AREs are a common recognition site for these proteins.
The sequence elements recognized by CELF proteins can differ somewhat depending on their context. CELF proteins regulate splicing in the nucleus, primarily through interaction with intronic sequences adjacent to alternatively spliced exons [22, 31, 41]. Intronic CELF binding sites closely resemble those found in 3′UTRs; both are generally GU-rich [30, 31]. In the cytoplasm, CELF proteins bind the 3′UTR to regulate mRNA decay [31] and translation [7, 40, 42, 43]. They have also been reported to associate with 5′UTR sequences to influence translation [44, 45]. To date only a few 5′UTR binding sites have been characterized. Most 5′UTR CELF1 binding sites appear to be more GC-rich [44, 46, 47], apart from that of the p27KIP1/CDKN1B mRNA, which is GU-rich and resembles the canonical CELF1 motif [45]. It seems that the CELF protein must be post-translationally modified or cooperate with other RBPs to function at this end of the transcript.
3.1.2. Structural insights into CELF-RNA recognition
As mentioned above, CELF proteins have three RRMs and each can interact with RNA. Unfortunately, it has not yet been possible to generate structure information for an intact CELF protein but, despite this, we can glean some important insights from studies of RRM1/2 and RRM3 fragments.
NMR solution studies demonstrated that RRM1, RRM2 [14, 15] and RRM3 [13] each recognize (U)UGU(U) motifs. The tandem RRM1/2 domains together show increased affinity compared to the binding by each domain separately thus indicating binding cooperativity between the two RRMs [14, 15]. The divergent linker domain between RRM2 and RRM3 also appears to be important for RNA-binding since it was able to increase RNA-binding affinity, perhaps by conveying important conformational changes necessary for RNA-binding [21, 24, 48]. In support of this, recent studies of Drosophila Bruno RRM3 and CELF1 RRM3 show that both require an N-terminal extension in order to achieve high affinity interaction with RNA [13, 49]. Close examination of this region reveals a highly conserved seven amino acid stretch (Q(K/R)EGP(E/D)G) adjacent to RRM3 in all CELF proteins (Figure 1). This short motif is unique to CELF proteins and is not found in any other RNA-binding protein. Interestingly, RRM3 in conjunction with the N-terminal extension adopts an unusual conformation to interact with the UGU sequence [13]. This may facilitate regulation of RNA-binding affinity through post-translational modification and/or protein-protein interactions. Overall, it seems likely that all three RRMs contribute to recognition of substrate RNAs, but their relative impact and/or binding preferences may change in response to cellular signals.
3.1.3. GU-Rich Elements
While the binding preferences of CELF proteins were being uncovered, it became clear that the GU-rich elements (GREs) they recognize are essential regulators of mRNA stability in mammalian cells [18, 50]. Genome-wide analyses of mRNA decay rates allowed isolation of mRNAs that exhibited rapid turnover, and computational methods were then used to search for conserved sequence elements in their 3′UTRs [25]. In activated human T lymphocytes, transcripts that exhibited rapid mRNA decay were found to frequently contain one or more consensus “UGUUUGUUUGU” elements [25] or GU-repeats [27]. These elements were recognized by CELF1 and conferred instability onto reporter mRNAs [25, 27]. Both GU-rich sequences and GU-repeats are also enriched in unstable myoblast mRNAs [26]. It has since become clear that both types of GRE (GU-repeats and UGUUUGU sequences) recruit CELF1 to induce mRNA decay thus the GRE should perhaps be loosely defined as “UGUKUGU”.
Based on their overall U-richness, their location in the 3′UTR and their ability to modulate rates of mRNA decay, GREs bear significant similarity to the better known AU-rich element (ARE) found in the 3′UTR of many mRNAs encoding cytokines, transcription factors and growth factors. Indeed, with hindsight it seems likely that GREs may be contained within the Class III “non-AUUUA” AREs defined by Peng et al in 1996 [51] which were originally found in the JUN and FOS 3′UTRs. Nonetheless, the GRE has several distinct functional characteristics that are summarized in Figure 2.
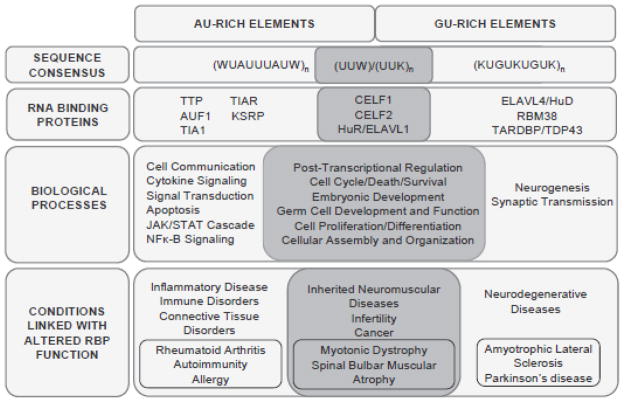
Fig. 2.
Comparison of AU-Rich and GU-Rich Elements. Functions and factors unique to each type of element are depicted with shared properties shown in the darker overlapping region. Over-represented biological processes were identified using Ingenuity Pathway Analysis (Ingenuity Systems, Inc.).
First, GREs may be less potent destabilizing elements than AREs, at least in some cell types. In Huh-7 cells, GUUUG-type GREs had a milder destabilizing effect on a reporter RNA than AREs [52]. In addition, at least in T-cells, GRE-containing mRNAs generally have longer half-lives than ARE-containing transcripts [52]. However, in C2C12 myoblasts, GREs were more closely associated with instability than AREs, suggesting that potency may vary in different cell types [26]. This could reflect tissue specific differences in expression or activity of GRE-binding factors. Second, while AREs have additive effects on mRNA decay, the number of overlapping GUUUG pentamers in the GRE does not seem to correlate with mRNA decay rate. Third, the repertoire of proteins reported to interact with GREs (CELF1, CELF2, ELAVL1, TDP43, ELAVL4) is smaller than for AREs (HuR, AUF1, TIA1, TIAR, TTP, KSRP, CELF2, CELF1 and others) but there is considerable overlap that warrants further investigation. Finally, there is a clear propensity for mRNAs encoding cytokines, apoptosis factors and members of the NF-κB cascade to contain AREs rather than GREs. Nevertheless, several functional classes of mRNAs can contain either of these two types of instability element (e.g. cell proliferation, differentiation, transcription, signal transduction). Surprisingly, very few transcripts have both types of regulatory element. Perhaps the mRNA metabolism pathways that GREs and AREs influence are mutually exclusive; the two types of transcript might be localized differently or even be expressed in different cell types.
4. Effects of CELF proteins on mRNA decay
At this point it is clear that CELF1, and likely other CELF proteins, associate with GREs in the 3′UTRs of a large number of mRNAs, but what impact does this have on mRNA stability? For CELF1, simply tethering the protein to the 3′UTR of a reporter was sufficient to destabilize the mRNA [53]. In addition, reporters bearing artificial GREs are destabilized following over-expression of CELF1 in COS-6 monkey kidney cells [31]. This is very consistent with observations made in vitro that show CELF1 association with RNA correlates with enhanced deadenylation in both HeLa [54] and Xenopus [8, 55] cell-free systems. Further investigation uncovered a direct interaction between CELF1 and the PARN deadenylase [54] which nicely explains these findings and leads to a very simple model where binding of CELF1 allows recruitment of the PARN deadenylase and results in accelerated deadenylation (Figure 3A). This phenomenon has been characterized in mammalian cell extracts [54], and also occurs in Xenopus extracts. However, the impact, if any, of PARN/CELF1 collaboration is not well-characterized in living cells. Several groups have identified mRNAs associated with CELF1 and shown that at least some of these are subject to CELF1-dependent decay, but the role of PARN in this process is not really known. PARN does impact decay rates for some mRNAs [56], but so far too few PARN substrates have been identified to allow analysis of whether they are more likely to be bound by CELF1. Identification of PARN substrates has been hindered by the fact that PARN is just one of a number of deadenylase enzymes present in mammalian cells; other deadenylases may compensate when PARN function is depleted. In addition, CELF1 might recruit multiple deadenylases, or other as yet undefined components of the mRNA decay machinery, to enhance mRNA decay rates.
Fig. 3.
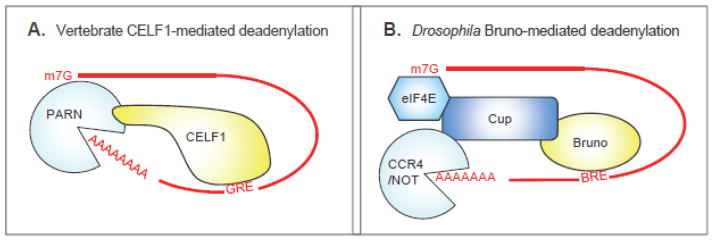
CELF proteins recruit deadenylases through direct or indirect interaction with the mRNA 5′ cap structure. (A) PARN is recruited directly to its substrates through interaction with CELF1. PARN activity is stimulated by the 5′ cap. (B) Bruno interacts indirectly with the 5′ cap through the eIF4E binding protein, Cup. Cup can recruit the CCR4/NOT deadenylase to mRNAs.
Despite strong evidence that both GREs and CELF1 act to enhance mRNA decay, it would be a huge over-simplification to state that CELF protein association always results in destabilization of substrate RNAs. On the contrary, CELF2 has clear stabilizing influences on several of its substrates, including COX2/PTGS2 and the AR mRNAs [38, 57]. This is quite surprising given that CELF1 and CELF2 are virtually identical at the amino acid level and thus would be expected to have comparable rather than opposing effects on mRNA decay. A clue as to how this might be explained comes from a recent study that showed CELF1 is required for stabilization of p21/CDKN1A mRNA during bortezomib-mediated apoptosis [58]. Under these conditions CELF1 relocalizes to stress granules (SGs) and pulls the p21 mRNA along with it, where it is protected from mRNA decay enzymes and its translation is repressed. The conditions under which CELF2-induced mRNA stabilization has been documented (e.g. γ-irradiation [59–61], curcumin treatment [62]) induce apoptosis and therefore should also be considered stressful. It seems possible that the function of CELF2 in mRNA decay has fortuitously been studied predominantly under stress conditions where CELF2 acts to stabilize mRNAs by relocating them to SGs. To our knowledge this possibility has not been directly investigated, but it is interesting to note that curcumin also induces expression of the TIA1 RNA-binding protein which is essential for SG formation and for relocalization of mRNAs to stress granules [62].
Aside from CELF1 and CELF2, the other four mammalian CELF proteins have not been extensively studied with respect to their role in mRNA degradation. However, CELF4 associates with the 3′UTRs of up to 20% of mRNAs expressed in the murine hippocampus and cortex. Abundance, localization and translation of these transcripts are impacted by CELF4 knockout indicating that CELF4 at least has the potential to influence mRNA stability [35].
4.1 Bruno, Cup and CCR4 in Drosophila
There is no clear homolog of PARN in the Drosophila genome and there’s little evidence so far that the Drosophila CELF protein, Bruno, modulates mRNA decay. Nevertheless, as mentioned previously Bruno is essential for normal oogenesis and early embryogenesis where it mediates translational repression of localized mRNAs such as Oskar [48, 63, 64]. Multiple mechanisms are involved, but one in particular has been linked with the eIF4E-binding protein Cup [65]. Cup interacts with Bruno bound to the Osk 3′UTR and presumably prevents eIF4E from recruiting eIF4G to initiate translation [66]. There is an interesting twist to this story however: Cup has the ability to recruit the CCR4 deadenylase and decapping activators to an mRNA substrate [67]. As Bruno can bind Cup, this finding strongly implies that the Drosophila Bruno protein may indirectly engage deadenylases (Figure 3B). Further investigation will be required to ascertain whether this is indeed the case, but it is intriguing that the ability of CELF proteins to induce mRNA deadenylation may be conserved from flies up to man. As PARN has the ability to interact with, and be stimulated by, the 5′ cap structure [68, 69]; the fact that Cup has connections with both the cap (through eIF4E) and deadenylase activity (through CCR4) is intriguing.
4.2 Coordination of gene expression by CELF proteins
CELF1 binds to a wide variety of mRNAs, but of what value is this to the cell? It is hard to estimate the global importance of CELF binding, but its main purpose is to allow coordinate control of mRNAs encoding factors required for an appropriate cellular response. This coordination is evident in a number of studies and occurs at multiple levels from splicing to translation, to mRNA decay. First, during T-cell activation a large number of GRE-containing mRNAs exhibit increased stability and abundance, many of them encoding factors important for cell proliferation, apoptosis and activation. Second, in Xenopus oocytes, CELF1 works with PARN to coordinate the deadenylation of GRE-containing mRNAs immediately following fertilization. The substrate mRNAs are predominantly maternally deposited transcripts within the oocyte that must be translationally silenced after fertilization and eventually degraded. The process is initiated by calcium-dependent dephosphorylation of CELF1, but the effect of this event on CELF1 interactions with RNA or other proteins remains to be deciphered. Third, there is also evidence to suggest that CELF1 coordinates important changes in gene expression during muscle development, although the targets are not well defined. In C2C12 cells, CELF1 is associated with a number of mRNAs that encode factors important for myogenesis [26] and as such is likely to influence the dramatic changes that occur in the abundance of some of these transcripts as cells differentiate. In addition, down-regulation of CELF1 has been linked with the global switch from embryonic to adult splicing patterns that occur in neonatal murine hearts [70]. This down-regulation would also be expected to influence stability of CELF-associated transcripts. Given the close ties between CELF1 and myotonic dystrophy, it will be imperative to fully characterize changes in stability of CELF associated transcripts that occur during myogenesis and in DM patient cells.
The wide-ranging impact of CELF1 on networks of mRNAs is a property that is almost certainly shared with other CELF proteins. CELF4, for example, binds to over 2000 mRNAs in mouse brain tissue, many of which encode factors involved in the regulation of synaptic function. As such, it may be intimately involved in coordinating changes in protein synthesis and/or mRNA decay in response to synaptic stimuli [35].
4.3 Regulation of CELF activity
CELF proteins do not serve mere housekeeping functions; they are essential to many of the changes in gene expression that occur during development and in response to extracellular stimuli. As such, CELF proteins exhibit regulation at multiple levels including abundance, localization, and RNA-binding affinity.
4.3.1. Regulation of CELF protein abundance
During embryonic and early post-natal development gene expression must be extensively reprogrammed as cells differentiate into specific tissues. CELF proteins undoubtedly play a role in this reprogramming by influencing splice site choice and presumably mRNA decay rates and translation as well. Changes in the abundance of CELF proteins that favor certain splice events have been well documented. For example, expression of CELF1 and CELF2 proteins decreases dramatically in the murine heart after birth and this correlates with a global change from embryonic to adult splicing patterns [70]. Abundance of both CELF1 and CELF2 also changes during myogenesis in C2C12 cells with comparable effects on splice site choice [71].
In heart, the developmental changes in CELF protein abundance have been attributed to miRNA-mediated regulation as there is little change in the amount of the CELF mRNAs. The two miR-23 family miRNAs (miR-23a, miR-23b) have a reciprocal expression pattern to CELF1 and CELF2 proteins; miRNA abundance increases as the heart becomes more mature. Moreover, inhibition of miR-23a function resulted in elevated expression of CELF proteins in vivo [72]. CELF2 is also targeted by miR-144/miR-451 in a pathway that influences cardiomyocyte survival [73].
CELF protein abundance is modulated during certain cellular responses. Polyamine depletion increases abundance of CELF1 protein in intestinal epithelial cells by down-regulating miR-503, a miRNA that targets the ORF of CELF1 to repress translation [74]. CELF2 expression is induced in colon cancer cells in response to ionizing radiation but in this case regulation is likely to be at the level of transcription or mRNA decay as the abundance of CELF2 mRNA is increased [60, 61]. Curiously, this response can be prevented by pre-treatment with lipopolysaccharide which completely blocks induction of CELF2 expression [60]. Finally, CELF1 expression in pre-adipocytes increases with age. This increase also occurs at the mRNA level and may be linked with the increased abundance of TNF in aging fat tissue as treatment of pre-adipocytes with either TNF or LPS induces CELF1 expression [75]. A great deal more work is needed to uncover the mechanisms that regulate CELF expression at the transcriptional and post-transcriptional levels.
4.3.2 Post-translational modifications of CELF proteins
Post-translational modifications are known to modulate the activity of a wide range of RNA-binding proteins and CELF proteins are no exception. CELF1 and CELF2 have quite extensive Ser/Thr-rich segments within their linker domain that could represent phosphorylation sites (Figure 4). Phosphorylation of CELF1 has been detected at Ser28 (Akt kinase) and Ser302 (Cyclin D/cdk4/6) and in each case these modifications increase the affinity of the protein for certain mRNA substrates [76] and affect the ability of CELF1 to interact with translation factors [77]. CELF1 also becomes phosphorylated during T-cell activation. In this case the site of phosphorylation is not known, but it results in lower affinity of CELF1 for GRE-containing target transcripts [28]. As a result, many of these transcripts are more stable following activation. CELF2 RNA-binding can also be modulated through phosphorylation; when rat smooth muscle cells are treated with PDGF, C-SRC tyrosine kinase is activated and phosphorylates CELF2 on Tyr49 [78]. This modification enhances association of CELF2 with COX2/PTGS2 mRNA and results in mRNA stabilization.
Fig. 4.
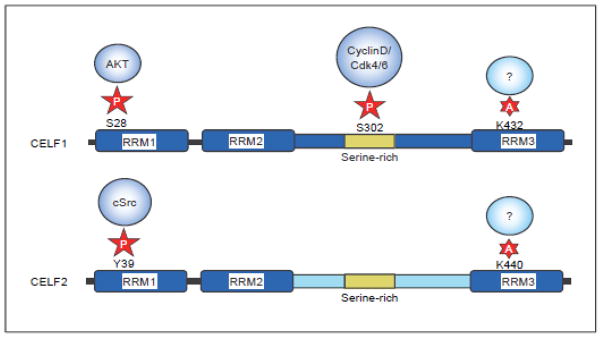
Post-translational modifications of CELF1 and CELF2 proteins. Experimentally verified phosphorylation and acetylation sites and the likely enzymes responsible are shown. In addition, serine-rich regions within the linker domain are depicted as they represent possible sites for phosphorylation by kinases such as PKC.
Hyperphosphorylation of CELF1 has been reported as a major contributor to pathogenesis in myotonic dystrophy [79, 80]. Although the specific site(s) are again not known, phosphorylation requires Protein Kinase C (PKC) and increases stability of CELF1 protein leading to its over-expression [80]. PKC inhibitors can reverse the symptoms exhibited by mouse models of DM1 presumably by limiting CELF1 phosphorylation [81]. It is not known whether the RNA-binding preferences of CELF1 are affected by hyperphosphorylation, but splicing patterns are consistent with increased CELF1 activity in DM1 patient cells and mouse models of the disease [82]. This is expected given the increased accumulation of the CELF1 protein in the nucleus. The effects on cytoplasmic functions, such as mRNA decay, await further characterization.
In Xenopus oocytes, in order for CELF1/EDEN-BP to activate deadenylation of its maternal mRNAs following fertilization it must undergo calcium-dependent dephosphorylation [83]. It is not known whether activation involves enhanced RNA-binding or improved CELF1 interaction with the deadenylase. Again, characterization of the precise sites of phosphorylation is needed to shed light on this phenomenon.
Finally, acetylation of Lys436 has been reported for CELF1 (as well as the analogous residue in CELF2) [84, 85]. The role of this particular modification has been explored only superficially for cytoplasmic proteins. However, as this lysine lies within RRM3 it seems very possible that it could impact RNA-binding affinity or protein-protein interactions. Interestingly, similar modifications were recently reported for the cytoplasmic poly(A)-binding protein where they potentially influence many aspects of mRNA metabolism including deadenylation and translation [86].
One of the major priorities must be to identify sites of modification and characterize their impact on CELF protein function. Many functions, from RNA-binding preferences and protein-protein interactions, to localization and decay activity, may be extensively impacted. Dramatic changes in substrate preference following modification might explain why unmodified recombinant CELF1 prefers GU-rich sequences while some studies report affinity for CUG- and GC-rich sequences in cell extracts. Such information could lead to exciting new avenues for therapeutics for DM1 patients.
4.3.3 Localization and Relocalization of CELF Proteins
In order for an RNA-binding protein to influence decay of its substrate mRNAs, the two must be able to come together and also to interact with effector factors such as the mRNA decay enzymes. Thus one of the main ways that activity can be regulated is through restricting the RBP to certain subcellular compartments. Like many RBPs, in most cells CELF1 is found in the nucleus where it modulates splicing and in the cytoplasm where it influences translation and mRNA decay. However, the distribution of CELF1 can vary in disease or in response to stress and this almost certainly impacts its function.
Various environmental stresses, such as heat shock and viral infection, converge on the translation initiation factor eIF2α [87]. Phosphorylation of eIF2α inhibits translation and induces the aggregation of mRNAs and RNA-binding proteins into cytoplasmic domains known as stress granules (SGs) [88]. From there, mRNAs are stored and either eventually returned to the cytoplasm for translation, or targeted for mRNA decay [89]. The TIA1 and TIAR RNA-binding proteins are essential components of SGs and their C-terminal glutamine/asparagine- rich domains are vital for SG formation [90]. During stress, cytoplasmic CELF1 relocalizes to SGs, likely through a direct interaction with TIA1 [91]. As an example, the relocalization of CELF1 during heat shock is shown in Figure 5. Importantly, although CELF1 is not required for SG formation, it is required for relocalization of the p21/CDKN1A mRNA to SGs, and also for stabilization of p21 mRNA during stress [58]. It seems likely that other CELF1 mRNA targets experience this type of regulation too. Importantly, as SGs can be induced under cellular conditions that mimic DM1, and they have been observed in DM1 patient myoblasts [77], relocalization of CELF1 to SGs may result in changes in mRNA translation and decay that could play a role in DM pathogenesis. In support of this, CELF1-mediated translation is impacted in DM1 myoblasts [92]. Other CELF proteins may also be relocalized during stress, but this has not been investigated to date.
Fig. 5.
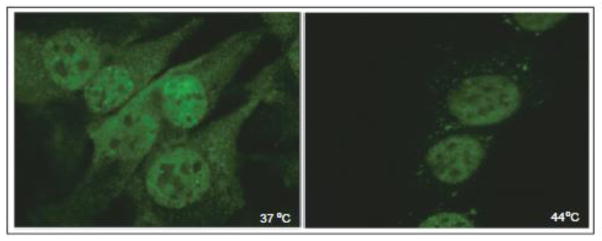
CELF1 relocalizes to cytoplasmic granules upon heat shock in mouse myoblasts. C2C12 cells were incubated at 37°C (left) or 44 °C for 2 hours prior to fixing and staining for CELF1 using 3B1 monoclonal antibody (Santa Cruz Biotechnology Inc.) and Cy2 conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch). These images are intended to illustrate CELF1 relocalization as reported by others following heat shock in HeLa cells [91].
CELF proteins have also been found in other types of RNA granules. In particular, CELF4 is present in large neuronal RNA granules of as yet unknown function [35], and the Drosophila Bruno protein participates in aggregating mRNAs into silencing particles that are required for spatially restricted expression of its target transcripts [93]. It seems likely that sequestration of mRNAs into such aggregates would restrict access of mRNA decay enzymes and alter mRNA decay rates.
While CELF1 redistribution can be a natural and normal function of this protein, there are occasions where CELF1 mislocalization may be pathogenic. In cancer cells, CELF1 can be found in a specialized region of the cell termed the perinucleolar compartment (PNC) [91] where it is associated with the RNA component of RNAse MRP (mitochondrial RNA-processing) and PTBP (polypyrimidine tract binding protein) [94]. If CELF1 is sequestered within the PNC, its ability to perform its normal cellular functions, including cytoplasmic mRNA decay, would obviously be impaired. The function of the PNC remains obscure, but as its presence correlates closely with metastasis and poor prognosis, the localization of CELF1 to this region will be a high priority for further investigation. Sequestration of CELF1 may also contribute to pathogenesis in oculopharyngeal muscular dystrophy (OPMD), as it accumulates in the nuclear inclusions that form in this disease [95].
Finally, although CELF1 and 2 usually are distributed in both the nucleus and cytoplasm, under certain conditions cytoplasmic abundance may be elevated. This is the case for CELF2 following γ-irradiation, where the majority of the protein becomes cytoplasmic [59]. This change correlates with increased expression of CELF2 splice isoforms that localize to the cytoplasm [96]. Increased cytoplasmic accumulation is a common response for many RBPs (reviewed in [97, 98] and almost certainly influences the decay of mRNAs targeted by these proteins.
4.3.4 Alternative processing events generate different CELF protein isoforms
Although one might imagine that six different genes encoding CELF proteins would be sufficient, there is evidence for multiple isoforms of each of these six proteins. Most of the CELF genes are encode multiple variant transcripts through use of different promoters and/or alternative mRNA processing (according to the UCSC genome browser, http://genome.ucsc.edu/). Different transcripts can encode different protein isoforms or allow for tissue-specific or developmental regulation of expression. CELF2, for example, has three promoters that generate three different isoforms each of which has a unique N-terminal sequence and 5′UTR [96]. The encoded proteins are expressed differently with the shortest isoform being preferentially induced in response to ionizing radiation and more highly expressed in colon cancer cells [96, 99]. Notably, the N-terminally extended isoforms are more cytoplasmic and thus may impact decay of mRNA targets. It remains to be seen whether the altered localization might be achieved through a nuclear export signal encoded only in the extended isoforms.
NCBI RefSeq Gene records show that CELF2 and CELF5 also have alternative 3′UTRs and C termini generated by alternative splicing which could lead to differences in regulation and protein function as well [100]. Finally, both CELF2 and CELF4 have an alternatively spliced exon that encodes the N terminal of the third RRM (exon 14 for CELF2 [101], exon 9 for CELF4). For CELF2, skipping of this exon occurs to different extents in different tissues and the loss of the N-terminal part of RRM3 prevents this domain from binding RNA. Moreover, the CELF2 isoform lacking exon 14 has altered function with regards to its ability to modulate splice site choice [101]. This may result either from altered RNA-binding affinity or from effects on protein-protein interactions. Further studies are needed to assess the global influence of different CELF protein isoforms in various tissues and at different developmental stages.
4.3.5 Hetero- and homo-oligomerization of CELF proteins
The human and Xenopus CELF1 proteins and Drosophila Bru-3 bind to RNA as dimers [32, 33] and may require GU-rich sequences of sufficient length to allow dimer formation. Surrounding U-rich sequences may be necessary for assembly of CELF proteins on RNA by allowing optimal secondary structure to facilitate the formation of RNA-protein complexes. Interestingly, at least for Xenopus CELF1 (EDEN-BP), the ability to interact with other CELF1 molecules is essential for RNA-binding and for induction of deadenylation. The N-terminal region of the linker domain is required for this interaction [102].
In addition to being required for RNA-binding, CELF-CELF interactions can facilitate mRNP formation in other ways. Oligomerization of the Drosophila CELF protein, Bruno, can lead to formation of large silencing particles that prevent access of ribosomes [93]. These large RNPs perhaps facilitate translational silencing and transport of mRNAs. It is not clear whether this type of function is conserved among CELF proteins in higher eukaryotes.
The fact that CELF1 can dimerize with itself gives rise to the possibility that heterodimers between different CELF proteins also exist. Such heterodimers might have different binding specificities or functions. In support of this idea, a dominant negative CELF4 (DNCELF4Δ) protein lacking all of RRM1 and part of RRM2 is able to interfere with the splicing activity of wild-type CELF2 and CELF4 in cultured cells [103]. Similar effects on splicing are seen when this mutant is expressed in either heart or skeletal muscle. DNCELF4Δ expression in the heart causes dilated cardiomyopathy and cardiac hypertrophy [104] while expression in skeletal muscle has milder effects on muscle organization and fiber size [105]. As the DNCELF4Δ mutant does not bind RNA [103], the effect is most likely mediated through direct interactions between the mutant CELF4 protein and other CELF proteins. One implication of this is that sequestration or inactivation of one CELF protein could impact the function of other CELF proteins by influencing the availability of heterodimers. To date, this is a virtually unexplored area of CELF protein function, especially with respect to effects on mRNA decay.
4.4 CELF competition/collaboration with other RBPs
Each mRNP is a complex and ephemeral entity that can be rapidly remodeled in response to changing conditions. As such it is important to consider the influence of other RNA-binding proteins (and RNAs) on CELF function. Such factors can hinder, or help RNA binding either through steric mechanisms or by altering the sub-cellular location of the transcript to make it more or less accessible to CELF.
4.4.1 MBNL1
Muscleblind (MBNL) proteins have an extremely intimate and wide-ranging relationship with CELF proteins. They are zinc finger-type RBPs that, like CELF proteins, have been strongly implicated as one of the main cellular proteins impacted in myotonic dystrophy [106, 107]. Although CELF and MBNL proteins have distinct binding preferences (MBNL1 prefers YCGY [108]) they associate with many of the same mRNAs to regulate splicing. Evidence to date indicates that these factors oppose each other with CELF binding favoring one splice pattern and MBNL encouraging the opposite [70]. Recent studies have shown that MBNL1 is also associated with a large number of mRNA 3′UTRs in C2C12 cells and many substrates are shared with CELF1 [31]. MBNL1 binding to the 3′UTR has multiple effects on mRNA metabolism as both mRNA stability and subcellular localization can be altered [31, 109]. Interestingly, both MBNL1 and CELF1 appear to favor destabilization of their substrates, but more work is required to determine whether they have additive effects when both associate with the same substrate mRNAs. It also is not known whether MBNL1 affects deadenylation rates.
4.4.2 HuR and other RBPs that recognize GRE-like sequences
One of the most effective ways to block binding of CELF proteins is through competition by another RBP that recognizes the same sequence elements. Many RBPs have similar binding preferences including several of those that recognize AREs (e.g. TIA1, AUF1/HNRNPD, HuR/ELAVL1, HuD/ELAVL4) (Figure 2). Of note, HuR and RBM38 (RNPC1) both favor U-rich sequences that are deficient in cytosine residues, which have obvious overlap with the UGUUUGU type CELF binding sites. Moreover, CELF1, HuR and RBM38 are all associated with CDKN1A/p21 mRNA [26, 110] and HuR and RBM38 are essential for myogenesis [111, 112], a process in which CELF1 has been clearly implicated [113]. In fact, there is significant overlap between the sets of transcripts bound by HuR and CELF1 [26]. In addition, CELF2 has been shown to compete with HuR for an AU-rich binding site on the COX2 mRNA leading to translational silencing [114].
The TDP43/TARDBP RNA binding protein is also worth a mention as it binds avidly to long UG repeats in introns to regulate splicing, and also to UG-rich regions in 3′UTRs to enhance mRNA decay, much as CELF proteins do [115]. As an example, both CELF2 and TDP43 can bind to a UG-rich region in intron 8 of the CFTR1 transcript to promote exon skipping [116]. More analysis is required to determine whether CELF proteins and TDP43 actually share RNA substrates, but if they do, CELF protein function may be altered in diseases where TDP43 is mutated, such as in Amyotrophic Lateral Sclerosis.
4.5. Interplay with miRNAs
It is likely that CELF proteins also influence miRNA function as has been seen for HuR [117], RBM38 [118] and several other RBPs. This could be achieved through direct competition for a shared binding site, or through remodeling of the mRNA structure to favor (or impede) miRNA association nearby. In support of this, a recent analysis determined that UUUGUUU motifs, which bear an uncanny resemblance to CELF binding sites, are enriched adjacent to miRNA binding sites and their presence tends to augment miRNA activity [119]. Such cooperation between miRNAs and CELF proteins could potentiate the ability of these factors to induce rapid mRNA decay and translational repression.
Any miRNA that contain a UGUKUGU seed sequence could in theory bind mRNA and occlude its association with CELF proteins. Although this has not been investigated, miR-495 is worth mentioning as it has perfect seed alignment to the GUUUGUU motif. This miRNA is differentially expressed in cancer cells [120, 121] and also in several inherited muscular disorders, including DM [122]. We note that several validated targets of miR-495, including REDD1 [26, 120], are also associated with CELF1 by co- immunoprecipitation [26].
In addition, CELF proteins could potentially indirectly influence mRNA decay rates by controlling miRNA processing. Other RNA-binding proteins, such as TDP43 [123], and KSRP [124], have been shown to associate with specific miRNAs to modulate their processing efficiency and thereby influence their activity.
Interplay between CELF proteins and miRNAs is essentially unstudied to date, but should perhaps be a high priority, given recent observations that miRNA expression and/or processing are affected in myotonic dystrophy [125–127].
5. CELF proteins in development and disease
Mammalian CELF proteins are expressed throughout development and have been linked with a variety of human conditions including cancer and neuromuscular disease. Many of the developmental roles of CELF proteins are conserved in lower organisms, thus models ranging from Drosophila to Danio rerio have been informative with regards to the biological significance of this family of proteins.
5.1. CELF1 in muscle and myotonic dystrophy
Much of the attention focused on CELF proteins today was fueled by the finding that CELF1 is overexpressed in DM1, an inherited neuromuscular disease affecting 1 in 8000 adults ([5], reviewed in [6, 128]). Recent studies have determined that CELF1 overexpression in mice is sufficient to reproduce many of the symptoms of DM1 such as dilated cardiomyopathy and muscle wasting [113, 129, 130]. CELF1 is also overexpressed and/or mislocalized in other neuromuscular disorders, supporting the hypothesis that it plays a special role in muscle tissue [95, 104, 131]. Deletion of CELF1 in mice is neonatally lethal in a pure genetic background [132] and results in greatly reduced viability in mixed genetic backgrounds. Those mice that survive have significantly retarded growth, and compromised fertility in both sexes [133]. Assessment of possible muscle-specific defects in these knockout mice awaits further investigation.
Abnormal CELF1 expression in DM1 has been closely linked with the aberrant splicing patterns observed in patient tissues [129, 130, 134–136]. Indeed, the myotonia seen in patients appears to be entirely due to mis-splicing of the CLCN1 mRNA which encodes a chloride channel [137]. In addition to myotonia, DM1 patients exhibit a large number of additional signs including muscle wasting, cardiac abnormalities, cataracts and cognitive impairment. Given the wide range of other activities CELF proteins possess, it is likely that some aspects of DM1 pathogenesis are independent of mis-splicing. For example, aberrant CELF1 function reduces translation of p21/CDKN1A in DM1 patient cells perhaps explaining impaired differentiation of these cells [92]. There is also compelling evidence that in DM1 mouse models a significant number of dysregulated genes exhibit altered mRNA abundance rather than changes in splicing [138]. Such effects are potentially due to altered transcription and/or mRNA decay. Further work will be required to separate the impact of various CELF1 functions on genes whose expression is altered in this disease.
5.2. CELF proteins in neurodegenerative and neurological disease
Myotonic dystrophy also impacts tissues other than muscle, particularly the central nervous system (reviewed in [139–141]). The majority of DM1 patients suffer from excessive fatigue, mental deficits and executive dysfunction while the most severely affected congenital DM1 patients exhibit severe mental retardation. Consistent with these symptoms, white matter lesions and neurofibrillary tangles have been reported in the brains of patients [142]. At least some of these problems are likely to result from direct effects on splicing of the tau mRNA [143]. Tau is a protein that has been linked with many neurodegenerative diseases [144] thus it is possible that altered splicing of Tau may contribute to neurocognitive deficits associated with DM1. A wide range of other splicing changes have also been detected in DM1 patients [6, 128]. Although there is no direct evidence implicating changes in mRNA decay as a cause of brain pathology in DM1, neuronal cells derived from patients have defects in synaptogenesis and neurite outgrowth that are linked with reduced expression of SLITRK2 and SLITRK4 genes [145]. Such down-regulation could well be induced by changes in mRNA decay.
CELF proteins have also been linked with other neurodegenerative diseases; CELF1 overexpression suppresses the neurodegenerative phenotype in a Drosophila model of Fragile-X-associated Tremor/Ataxia Syndrome, FXTAS [47], while CELF2 (BRUNOL3) is up-regulated in Spinal Muscular Atrophy (SMA) [146]. The role that CELF proteins play in these disorders remains unclear but it is interesting to note that CELF1 can associate with the CGG-repeat RNA that causes FXTAS through binding to hnRNPA1/B2 [47]. This interaction might sequester CELF1 from its normal cytoplasmic functions. It would be interesting to determine whether CELF1-mediated mRNA decay is affected in FXTAS patient cells.
Finally, advances in high-resolution genome sequencing recently led to the discovery that a neurodevelopmental disorder associated with deletions of 18q12.2 is almost certainly caused by haploinsufficiency of CELF4 [147]. The resulting phenotype includes borderline IQ, behavioral problems, seizures, myopia and obesity. The phenotype is remarkably similar to that of CELF4−/− mice which display a complex seizure disorder [10, 148]. Deletion of CELF4 at seven weeks of age induced convulsive seizures, while earlier loss of the gene resulted in absence-like non-convulsive seizures. These results are consistent with the expression pattern of CELF4 which is restricted to neural tissues in adults, primarily in excitatory neurons [10]. Such phenotypes are particularly interesting in light of the fact that CELF4 has a glutamine-rich domain similar to those found in the prion protein PrP and the RNA-binding protein TDP43 [149], both of which have also been linked with neurological conditions [150–152].
5.3. CELF proteins in apoptosis and cancer
Although CELF proteins clearly play a special role in muscle and neuronal tissues, an increasing number of studies are linking these RNA-binding proteins with more fundamental cellular functions and particularly with cell proliferation, apoptosis and cancer. Moreover, many of these connections involve regulation of mRNA decay and/or translation.
CELF protein over-expression has been reported in several cancers including esophageal cancer [153] and glioma [154]. Many of the mRNAs targeted by CELF proteins seem to encode proteins with roles in apoptosis including TP63, AKT, MYD88, SIAH1, UBE2C, RUNX1, SERPINE1 etc. [27], suggesting that CELF1 is involved in coordinating the apoptotic response. Apoptosis is a cellular mechanism that often goes awry in cancer cells and CELF1 overexpression could therefore be involved in the aberrant apoptotic response in cancer. In esophageal cancer cells, the survivin transcript, which encodes an inhibitor of apoptosis, is stabilized through association with CELF1 [153]. CELF1 has also been implicated in modulating the stability of the mRNA encoding the apoptosis regulator p21/CDKN1A in cancer cells [58]. In these two studies [58, 153], CELF1 over-expression led to undesirable resistance to the chemotherapeutics camptothecin or bortemozib, which kill cancer cells by inducing apoptosis. CELF1 knockdown was shown to have the opposite effect; in HeLa cells, reduced expression of CELF1 resulted in increased sensitivity to apoptosis induced by TNF and cycloheximide [27]. Although further work is needed, together, these studies suggest that CELF1 binds to numerous transcripts involved in apoptosis and its overexpression can inhibit this process in cancer cells.
CELF2 is also associated with apoptosis; it is induced in response to apoptotic stimuli such as gamma irradiation and curcumin [59, 62]. One well-characterized target of CELF2, the COX2/PTGS1 mRNA, encodes a protein with strong anti-apoptotic activity that facilitates survival following genotoxic stress. CELF2 functions to reduce expression of COX2 following irradiation by simultaneously stabilizing the COX2 mRNA and inhibiting its translation [40, 59]. Similar effects on the MCL1 transcript, which also encodes an anti-apoptotic protein, have been reported in colon cancer cell lines [99] suggesting that CELF2 may coordinate down-regulation of multiple anti-apoptotic factors during stress.
The compelling links between CELF1 and cancer prompted recent investigations of cancer incidence in DM1 patients as aberrant CELF1 function has been established in this group. These studies revealed an increased overall risk of cancer in DM1 patients, especially thyroid cancer, choroidal melanoma and pilomatricoma [155–158]. In addition, indirect links between the abnormal function of CELF1 and liver [159], breast [59, 160] and blood [161] cancer pathogenesis have been described.
In conclusion, although studies of the influence of CELF1 and CELF2 proteins in cancer are in their infancy, it is already quite clear that they are important regulators of apoptosis making them viable targets for anti-cancer therapeutics.
5.4. CELF proteins and immune cell function
In primary human T-cells, CELF1 regulates gene expression observed following T cell receptor-mediated activation. T-cell receptor stimulation induces stimulus-dependent changes in the decay rates of hundreds of mRNA transcripts containing GU-rich elements (GREs) in their 3′UTRs [25, 162]. Identification of the cytoplasmic binding targets of CELF1 before and after T-cell activation led to the observation that CELF1 dissociates from GRE-containing transcripts following immune activation and associates with a different array of targets [28]. This change correlated with the transient up-regulation of GRE-containing mRNAs, many of which encoded proteins necessary for the transition from a quiescent state to a state of cellular immune activation such as JUN, JUNB, PP1CC, ETS2, ELF4, TGIF2, TNFRSF1B etc. [163]. This data supports a model whereby CELF1 functions in resting T-cells to down-regulate a network of transcripts involved in activation and proliferation. Subsequent activation-induced phosphorylation of CELF1 results in stabilization and accumulation of these transcripts within activated immune cells [28]. Thus it appears that CELF1 plays an essential role in facilitating the immune cell response to external stimuli and coordinates regulation of a gene network involved in cellular activation.
5.5. CELF proteins in fertility
Numerous RNA-binding proteins (RBPs) are important for fertility [164]. This is not surprising because during the process of meiosis, transcription ceases, leaving only post-transcriptional mechanisms to regulate gene expression. As mentioned above, CELF1−/− mice have reduced fertility, and in males this coincides with spermatogenesis defects [132, 133]. CELF1 and CELF3 (BRUNOL1) are both highly expressed in testes, and CELF3−/− mice have reduced sperm count and motility but are otherwise normal and fertile [165]. The true influence of CELF proteins in gametogenesis may be obscured by redundancy in their functions. There is still much to be learned as there is clear evidence that these proteins are involved in germ cell development in lower organisms (see below).
6. CELF proteins in non-mammalian model organisms
CELF proteins are found throughout the animal kingdom as well as in plants, and much has been learned from studying their expression and mutant phenotypes in various models from fruit flies to fish. The critical role of CELF1 during development has been illustrated by knockdown and over-expression experiments. In Xenopus, neutralizing or knocking down expression of CELF1/EDEN-BP caused developmental defects, such as the loss of somitic segmentation [166] primarily due to overexpression of the Notch pathway protein, XSu(H) [167]. In this case, CELF1 directly associated with the 3′UTR of XSu(H) and modulated deadenylation and subsequent decay of this transcript and likely other transcripts involved in segmentation. In zebrafish, CELF1 overexpression profoundly affected somite symmetry and left-right patterning predominantly through destabilization of the Dmrt2a transcription factor mRNA [168]. The experiments in Xenopus and zebrafish strongly imply that CELF1 is a major coordinator of post-transcriptional events during vertebrate somite segmentation. Moreover, as somites are the precursors of muscle, this indicates that CELF1 may play important roles throughout muscle development. In support of this, genetic deletion of CELF1/ETR-1 in Caenorhabditis elegans caused embryonic lethality with a phenotype similar to that exhibited by mutants affecting muscle formation [169]. This confirms that CELF1 is an important factor in muscle development in diverse species and also suggests that targets of CELF1-mediated mRNA decay may be regulated during somitogenesis.
Another C. elegans CELF protein, UNC-75, is most closely related to CELF3 and CELF4 and like them is expressed primarily in the nervous system [170]. Interestingly, Unc-75 mutants have a phenotype suggestive of defects in synaptic transmission reminiscent of that seen in CELF4 knockout mice. This supports a conserved function for CELF4 in regulating mRNA metabolism in excitatory neurons.
Drosophila has provided a particularly fruitful and perhaps undervalued model for the study of CELF protein function. The Drosophila Bruno protein encoded by the arrest (aret) gene is essential for gametogenesis and mutations in aret result in a complete failure of oogenesis as well as male sterility [171]. Weak aret alleles allow production of viable larvae, but they exhibit complex defects in segmentation patterning. Bruno over-expression causes defects in both antero-posterior and dorso-ventral patterning – a phenotype that is perhaps reminiscent of the patterning defects seen in zebrafish over-expressing CELF1 [168]. Two other Drosophila CELF proteins, Bru-2 and Bru-3, remain virtually uncharacterized.
Taken together the plethora of studies applying knockdown, knockout and over-expression approaches, support that CELF proteins have conserved roles in gametogenesis, and in somitic segmentation as well as special functions in muscle and nerves. Some of these roles may well be redundant as there are multiple CELF genes present in most organisms studied to date. As a result, knockout phenotypes may underestimate the real impact of these proteins in development and disease.
7.0 Conclusions and Perspectives
In conclusion, CELF proteins are powerful regulators of mRNA metabolism in many biological pathways from apoptosis to myogenesis, but it seems that we have only glimpsed the tip of the iceberg so far. In particular, in mammalian cells most studies have focused on how CELF proteins modulate splice site choice in muscle cells while their roles in other cell types and mechanisms have been understudied. With the advent of more powerful approaches to assess global mRNA abundance [172] and mRNA decay rates [173], we anticipate that the true impact of these proteins on gene expression in cancer, infertility, neuromuscular disease and other conditions will be better appreciated in the future. We expect that studies in model organisms will shed new light on the importance of CELF proteins in early embryonic development and especially in germ cells.
Simply identifying the mRNAs targeted for CELF-mediated decay will not be sufficient to provide much insight into how these proteins function. We need to identify the enzymes, RBPs and miRNAs that CELF proteins interface with and determine how these interactions are altered in diseased tissues. We especially need to work towards characterizing the post-translational modifications that CELF proteins experience, and isolating the enzymes responsible for their addition and removal. Such enzymes may represent invaluable targets for therapeutics. In addition, it will be important to assess how each post-translational modification influences RNA-binding affinity and possibly interactions with other proteins and enzymes. This is particularly important in light of the continuing debate as to whether and how CELF1 recognizes CUG repeats as well as GREs.
While CELF1 and CELF2 have been quite extensively investigated, the other CELF proteins are understudied, especially with regards to possible roles in mRNA decay. Studies in neural cell lines and tissues might be particularly informative for CELF3 and CELF4. The CELF proteins that have more restricted expression patterns could play important roles in many neurological and neuromuscular conditions, including DM1, but this simply has not been investigated. Building of biological networks and pinning down specific interactions would be a tremendous asset to understanding how these RBPs are involved in pathogenesis.
The links between CELF proteins and the stress response are also in need of more focused experimentation. As we discussed, CELF1 relocalizes to SGs and interacts with the TIA1 RNA-binding protein that initiates assembly of these cytoplasmic bodies. It would be interesting to know how this affects stability of CELF1 target transcripts on a global scale, and also whether CELF1 facilitates decay or translation as the stress is resolved [174]. Moreover, as there is evidence for formation of cytoplasmic granules during differentiation of both T-cells [175] and muscle cells [176], similar effects may be occurring during stress responses and normal developmental changes.
Highlights.
CELF proteins bind to GU-rich elements in mRNAs and induce cytoplasmic mRNA decay as well as splicing and translation.
CELF proteins coordinate post-transcriptional events for extensive networks of genes in response to cellular conditions
Changes in expression, function and localization of CELF proteins occur during development, cellular activation and disease.
Post-translational modification influences CELF protein activity.
Acknowledgments
C.J.W. is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Number AR059247. P.R.B. and I.A.V-S. are supported by NIH grant AI072068. A.M.D. is supported by a postdoctoral fellowship from the Myotonic Dystrophy Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Myotonic Dystrophy Foundation. We thank Dr. Ying Zhang (University of Minnesota Supercomputing Institute) and Dr. Eric Ross (Colorado State University) for helpful discussions regarding protein structures and prion domains.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Antic D, Keene JD. Embryonic lethal abnormal visual RNA-binding proteins involved in growth, differentiation, and posttranscriptional gene expression. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srikantan S, Gorospe M. HuR function in disease. Front Biosci. 2012;17:189–205. doi: 10.2741/3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Timchenko LT, Timchenko NA, Caskey CT, Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum Mol Genet. 1996;5:115–121. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- 4.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- 6.Sicot G, Gourdon G, Gomes-Pereira M. Myotonic dystrophy, when simple repeats reveal complex pathogenic entities: new findings and future challenges. Hum Mol Genet. 2011;20:R116–123. doi: 10.1093/hmg/ddr343. [DOI] [PubMed] [Google Scholar]
- 7.Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 8.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagnon JL, Mahaffey CL, Sun W, Yang Y, Chao HT, Frankel WN. Etiology of a genetically complex seizure disorder in Celf4 mutant mice. Genes Brain Behav. 2011;10:765–777. doi: 10.1111/j.1601-183X.2011.00717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ladd AN, Nguyen NH, Malhotra K, Cooper TA. CELF6, a member of the CELF family of RNA-binding proteins, regulates muscle-specific splicing enhancer-dependent alternative splicing. J Biol Chem. 2004;279:17756–17764. doi: 10.1074/jbc.M310687200. [DOI] [PubMed] [Google Scholar]
- 12.Han J, Cooper TA. Identification of CELF splicing activation and repression domains in vivo. Nucleic Acids Res. 2005;33:2769–2780. doi: 10.1093/nar/gki561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, Sugano S, Guntert P, Muto Y, Yokoyama S. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37:5151–5166. doi: 10.1093/nar/gkp546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards J, Malaurie E, Kondrashov A, Long J, de Moor CH, Searle MS, Emsley J. Sequence determinants for the tandem recognition of UGU and CUG rich RNA elements by the two N--terminal RRMs of CELF1. Nucleic Acids Res. 2011;39:8638–8650. doi: 10.1093/nar/gkr510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teplova M, Song J, Gaw HY, Teplov A, Patel DJ. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18:1364–1377. doi: 10.1016/j.str.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladd AN, Cooper TA. Multiple domains control the subcellular localization and activity of ETR-3, a regulator of nuclear and cytoplasmic RNA processing events. J Cell Sci. 2004;117:3519–3529. doi: 10.1242/jcs.01194. [DOI] [PubMed] [Google Scholar]
- 17.Gallo JM, Spickett C. The role of CELF proteins in neurological disorders. RNA Biol. 2010;7:474–479. doi: 10.4161/rna.7.4.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol. 2008;5:201–207. doi: 10.4161/rna.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dasgupta T, Ladd AN. The importance of CELF control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip Rev RNA. 2012;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladd AN. CUG-BP, Elav-like family (CELF)-mediated alternative splicing regulation in the brain during health and disease. Mol Cell Neurosci. 2012 doi: 10.1016/j.mcn.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi N, Sasagawa N, Suzuki K, Ishiura S. The CUG-binding protein binds specifically to UG dinucleotide repeats in a yeast three-hybrid system. Biochem Biophys Res Commun. 2000;277:518–523. doi: 10.1006/bbrc.2000.3694. [DOI] [PubMed] [Google Scholar]
- 22.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori D, Sasagawa N, Kino Y, Ishiura S. Quantitative analysis of CUG-BP1 binding to RNA repeats. J Biochem. 2008;143:377–383. doi: 10.1093/jb/mvm230. [DOI] [PubMed] [Google Scholar]
- 25.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, Bohjanen PR. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One. 2010;5:e11201. doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rattenbacher B, Beisang D, Wiesner DL, Jeschke JC, von Hohenberg M, St Louis-Vlasova IA, Bohjanen PR. Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol Cell Biol. 2010;30:3970–3980. doi: 10.1128/MCB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beisang D, Rattenbacher B, Vlasova-St Louis IA, Bohjanen PR. Regulation of CUG-binding protein 1 (CUGBP1) binding to target transcripts upon T cell activation. J Biol Chem. 2012;287:950–960. doi: 10.1074/jbc.M111.291658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graindorge A, Le Tonqueze O, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis. Nucleic Acids Res. 2008;36:1861–1870. doi: 10.1093/nar/gkn031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda A, Andersen HS, Doktor TK, Okamoto T, Ito M, Andresen BS, Ohno K. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Sci Rep. 2012;2:209. doi: 10.1038/srep00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonnet-Corven S, Audic Y, Omilli F, Osborne HB. An analysis of the sequence requirements of EDEN-BP for specific RNA binding. Nucleic Acids Res. 2002;30:4667–4674. doi: 10.1093/nar/gkf586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delaunay J, Le Mee G, Ezzeddine N, Labesse G, Terzian C, Capri M, Ait-Ahmed O. The Drosophila Bruno paralogue Bru-3 specifically binds the EDEN translational repression element. Nucleic Acids Res. 2004;32:3070–3082. doi: 10.1093/nar/gkh627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki H, Jin Y, Otani H, Yasuda K, Inoue K. Regulation of alternative splicing of alpha-actinin transcript by Bruno-like proteins. Genes Cells. 2002;7:133–141. doi: 10.1046/j.1356-9597.2001.00506.x. [DOI] [PubMed] [Google Scholar]
- 35.Wagnon JL, Briese M, Sun W, Mahaffey CL, Curk T, Rot G, Ule J, Frankel WN. CELF4 Regulates Translation and Local Abundance of a Vast Set of mRNAs, Including Genes Associated with Regulation of Synaptic Function. PLoS Genet. 2012;8:e1003067. doi: 10.1371/journal.pgen.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bae EJ, Kim SG. Enhanced CCAAT/enhancer-binding protein beta-liver-enriched inhibitory protein production by Oltipraz, which accompanies CUG repeat-binding protein-1 (CUGBP1) RNA-binding protein activation, leads to inhibition of preadipocyte differentiation. Mol Pharmacol. 2005;68:660–669. doi: 10.1124/mol.105.012997. [DOI] [PubMed] [Google Scholar]
- 37.Timchenko NA, Welm AL, Lu X, Timchenko LT. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyazaki Y, Adachi H, Katsuno M, Minamiyama M, Jiang YM, Huang Z, Doi H, Matsumoto S, Kondo N, Iida M, Tohnai G, Tanaka F, Muramatsu S, Sobue G. Viral delivery of miR-196a ameliorates the SBMA phenotype via the silencing of CELF2. Nat Med. 2012;18:1136–1141. doi: 10.1038/nm.2791. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for Myotonic Dystrophy. J Biol Chem. 2008 doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 41.Dembowski JA, Grabowski PJ. The CUGBP2 splicing factor regulates an ensemble of branchpoints from perimeter binding sites with implications for autoregulation. PLoS Genet. 2009;5:e1000595. doi: 10.1371/journal.pgen.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fox JT, Stover PJ. Mechanism of the internal ribosome entry site-mediated translation of serine hydroxymethyltransferase 1. J Biol Chem. 2009;284:31085–31096. doi: 10.1074/jbc.M109.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horb LD, Horb ME. BrunoL1 regulates endoderm proliferation through translational enhancement of cyclin A2 mRNA. Dev Biol. 2010;345:156–169. doi: 10.1016/j.ydbio.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 2004;23:406–417. doi: 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng Y, Miskimins WK. CUG-binding protein represses translation of p27Kip1 mRNA through its internal ribosomal entry site. RNA Biol. 2011;8:365–371. doi: 10.4161/rna.8.3.14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Timchenko NA, Wang GL, Timchenko LT. RNA CUG-binding protein 1 increases translation of 20-kDa isoform of CCAAT/enhancer-binding protein beta by interacting with the alpha and beta subunits of eukaryotic initiation translation factor 2. J Biol Chem. 2005;280:20549–20557. doi: 10.1074/jbc.M409563200. [DOI] [PubMed] [Google Scholar]
- 47.Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, Nelson DL, Botas J. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snee M, Benz D, Jen J, Macdonald PM. Two distinct domains of Bruno bind specifically to the oskar mRNA. RNA Biol. 2008;5:1–9. [PubMed] [Google Scholar]
- 49.Lyon AM, Reveal BS, Macdonald PM, Hoffman DW. Bruno protein contains an expanded RNA recognition motif. Biochemistry (Mosc) 2009;48:12202–12212. doi: 10.1021/bi900624j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim HH, Gorospe M. GU-rich RNA: expanding CUGBP1 function, broadening mRNA turnover. Mol Cell. 2008;29:151–152. doi: 10.1016/j.molcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng SS, Chen CY, Shyu AB. Functional characterization of a non-AUUUA AU-rich element from the c-jun proto-oncogene mRNA: evidence for a novel class of AU-rich elements. Mol Cell Biol. 1996;16:1490–1499. doi: 10.1128/mcb.16.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halees AS, Hitti E, Al-Saif M, Mahmoud L, Vlasova-St Louis IA, Beisang DJ, Bohjanen PR, Khabar K. Global assessment of GU-rich regulatory content and function in the human transcriptome. RNA Biol. 2011;8:681–691. doi: 10.4161/rna.8.4.16283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 54.Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paillard L, Legagneux V, Maniey D, Osborne HB. c-Jun ARE targets mRNA deadenylation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway. J Biol Chem. 2002;277:3232–3235. doi: 10.1074/jbc.M109362200. [DOI] [PubMed] [Google Scholar]
- 56.Lee JE, Lee JY, Trembly J, Wilusz J, Tian B, Wilusz CJ. The PARN Deadenylase Targets a Discrete Set of mRNAs for Decay and Regulates Cell Motility in Mouse Myoblasts. PLoS Genet. 2012;8:e1002901. doi: 10.1371/journal.pgen.1002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker MR, Brown SL, Riehl TE, Stenson WF, Stappenbeck TS. Growth factor regulation of prostaglandin-endoperoxide synthase 2 (Ptgs2) expression in colonic mesenchymal stem cells. J Biol Chem. 2010;285:5026–5039. doi: 10.1074/jbc.M109.032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gareau C, Fournier MJ, Filion C, Coudert L, Martel D, Labelle Y, Mazroui R. p21(WAF1/CIP1) upregulation through the stress granule-associated protein CUGBP1 confers resistance to bortezomib-mediated apoptosis. PLoS One. 2011;6:e20254. doi: 10.1371/journal.pone.0020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukhopadhyay D, Jung J, Murmu N, Houchen CW, Dieckgraefe BK, Anant S. CUGBP2 plays a critical role in apoptosis of breast cancer cells in response to genotoxic injury. Ann N Y Acad Sci. 2003;1010:504–509. doi: 10.1196/annals.1299.093. [DOI] [PubMed] [Google Scholar]
- 60.Murmu N, Jung J, Mukhopadhyay D, Houchen CW, Riehl TE, Stenson WF, Morrison AR, Arumugam T, Dieckgraefe BK, Anant S. Dynamic antagonism between RNA-binding protein CUGBP2 and cyclooxygenase-2-mediated prostaglandin E2 in radiation damage. Proc Natl Acad Sci U S A. 2004;101:13873–13878. doi: 10.1073/pnas.0406066101. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Natarajan G, Ramalingam S, Ramachandran I, May R, Queimado L, Houchen CW, Anant S. CUGBP2 downregulation by prostaglandin E2 protects colon cancer cells from radiation-induced mitotic catastrophe. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1235–1244. doi: 10.1152/ajpgi.00037.2008. [DOI] [PubMed] [Google Scholar]
- 62.Subramaniam D, Ramalingam S, Linehan DC, Dieckgraefe BK, Postier RG, Houchen CW, Jensen RA, Anant S. RNA binding protein CUGBP2/CELF2 mediates curcumin-induced mitotic catastrophe of pancreatic cancer cells. PLoS One. 2011;6:e16958. doi: 10.1371/journal.pone.0016958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Snee MJ, Harrison D, Yan N, Macdonald PM. A late phase of Oskar accumulation is crucial for posterior patterning of the Drosophila embryo, and is blocked by ectopic expression of Bruno. Differentiation. 2007;75:246–255. doi: 10.1111/j.1432-0436.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- 64.Reveal B, Garcia C, Ellington A, Macdonald PM. Multiple RNA binding domains of Bruno confer recognition of diverse binding sites for translational repression. RNA Biol. 2011;8:1047–1060. doi: 10.4161/rna.8.6.17542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol. 2003;163:1197–1204. doi: 10.1083/jcb.200309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]
- 67.Igreja C, Izaurralde E. CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes Dev. 2011;25:1955–1967. doi: 10.1101/gad.17136311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao M, Wilusz CJ, Peltz SW, Wilusz J. A novel mRNA-decapping activity in HeLa cytoplasmic extracts is regulated by AU-rich elements. EMBO J. 2001;20:1134–1143. doi: 10.1093/emboj/20.5.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dehlin E, Wormington M, Korner CG, Wahle E. Cap-dependent deadenylation of mRNA. EMBO J. 2000;19:1079–1086. doi: 10.1093/emboj/19.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bland CS, Wang ET, Vu A, David MP, Castle JC, Johnson JM, Burge CB, Cooper TA. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res. 2010;38:7651–7664. doi: 10.1093/nar/gkq614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24:653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang X, Wang X, Zhu H, Zhu C, Wang Y, Pu WT, Jegga AG, Fan GC. Synergistic effects of the GATA-4-mediated miR-144/451 cluster in protection against simulated ischemia/reperfusion-induced cardiomyocyte death. J Mol Cell Cardiol. 2010;49:841–850. doi: 10.1016/j.yjmcc.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cui YH, Xiao L, Rao JN, Zou T, Liu L, Chen Y, Turner DJ, Gorospe M, Wang JY. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol Biol Cell. 2012;23:151–162. doi: 10.1091/mbc.E11-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Karagiannides I, Thomou T, Tchkonia T, Pirtskhalava T, Kypreos KE, Cartwright A, Dalagiorgou G, Lash TL, Farmer SR, Timchenko NA, Kirkland JL. Increased CUG triplet repeat-binding protein-1 predisposes to impaired adipogenesis with aging. J Biol Chem. 2006;281:23025–23033. doi: 10.1074/jbc.M513187200. [DOI] [PubMed] [Google Scholar]
- 76.Salisbury E, Sakai K, Schoser B, Huichalaf C, Schneider-Gold C, Nguyen H, Wang GL, Albrecht JH, Timchenko LT. Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP1. Exp Cell Res. 2008;314:2266–2278. doi: 10.1016/j.yexcr.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huichalaf C, Sakai K, Jin B, Jones K, Wang GL, Schoser B, Schneider-Gold C, Sarkar P, Pereira-Smith OM, Timchenko N, Timchenko L. Expansion of CUG RNA repeats causes stress and inhibition of translation in myotonic dystrophy 1 (DM1) cells. FASEB J. 2010;24:3706–3719. doi: 10.1096/fj.09-151159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu K, Kitchen CM, Shu HK, Murphy TJ. Platelet-derived growth factor-induced stabilization of cyclooxygenase 2 mRNA in rat smooth muscle cells requires the c-Src family of protein-tyrosine kinases. J Biol Chem. 2007;282:32699–32709. doi: 10.1074/jbc.M705272200. [DOI] [PubMed] [Google Scholar]
- 79.Roberts R, Timchenko NA, Miller JW, Reddy S, Caskey CT, Swanson MS, Timchenko LT. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc Natl Acad Sci U S A. 1997;94:13221–13226. doi: 10.1073/pnas.94.24.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang GS, Kuyumcu-Martinez MN, Sarma S, Mathur N, Wehrens XH, Cooper TA. PKC inhibition ameliorates the cardiac phenotype in a mouse model of myotonic dystrophy type 1. J Clin Invest. 2009;119:3797–3806. doi: 10.1172/JCI37976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Orengo JP, Chambon P, Metzger D, Mosier DR, Snipes GJ, Cooper TA. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Detivaud L, Pascreau G, Karaiskou A, Osborne HB, Kubiak JZ. Regulation of EDEN-dependent deadenylation of Aurora A/Eg2-derived mRNA via phosphorylation and dephosphorylation in Xenopus laevis egg extracts. J Cell Sci. 2003;116:2697–2705. doi: 10.1242/jcs.00477. [DOI] [PubMed] [Google Scholar]
- 84.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 85.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 2011;39:D253–260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brook M, McCracken L, Reddington JP, Lu ZL, Morrice NA, Gray NK. The multifunctional poly(A)-binding protein (PABP) 1 is subject to extensive dynamic post-translational modification, which molecular modelling suggests plays an important role in coordinating its activities. Biochem J. 2012;441:803–812. doi: 10.1042/BJ20111474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Linero FN, Thomas MG, Boccaccio GL, Scolaro LA. Junin virus infection impairs stress-granule formation in Vero cells treated with arsenite via inhibition of eIF2alpha phosphorylation. J Gen Virol. 2011;92:2889–2899. doi: 10.1099/vir.0.033407-0. [DOI] [PubMed] [Google Scholar]
- 88.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Balagopal V, Parker R. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr Opin Cell Biol. 2009;21:403–408. doi: 10.1016/j.ceb.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fujimura K, Kano F, Murata M. Dual localization of the RNA binding protein CUGBP-1 to stress granule and perinucleolar compartment. Exp Cell Res. 2008;314:543–553. doi: 10.1016/j.yexcr.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 92.Timchenko NA, Iakova P, Cai ZJ, Smith JR, Timchenko LT. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol Cell Biol. 2001;21:6927–6938. doi: 10.1128/MCB.21.20.6927-6938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 94.Pollock C, Daily K, Nguyen VT, Wang C, Lewandowska MA, Bensaude O, Huang S. Characterization of MRP RNA-protein interactions within the perinucleolar compartment. Mol Biol Cell. 2011;22:858–867. doi: 10.1091/mbc.E10-09-0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Corbeil-Girard LP, Klein AF, Sasseville AM, Lavoie H, Dicaire MJ, Saint-Denis A, Page M, Duranceau A, Codere F, Bouchard JP, Karpati G, Rouleau GA, Massie B, Langelier Y, Brais B. PABPN1 overexpression leads to upregulation of genes encoding nuclear proteins that are sequestered in oculopharyngeal muscular dystrophy nuclear inclusions. Neurobiol Dis. 2005;18:551–567. doi: 10.1016/j.nbd.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 96.Ramalingam S, Natarajan G, Schafer C, Subramaniam D, May R, Ramachandran I, Queimado L, Houchen CW, Anant S. Novel intestinal splice variants of RNA-binding protein CUGBP2: isoform-specific effects on mitotic catastrophe. Am J Physiol Gastrointest Liver Physiol. 2008;294:G971–981. doi: 10.1152/ajpgi.00540.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cammas A, Lewis SM, Vagner S, Holcik M. Post-transcriptional control of gene expression through subcellular relocalization of mRNA binding proteins. Biochem Pharmacol. 2008;76:1395–1403. doi: 10.1016/j.bcp.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 99.Subramaniam D, Natarajan G, Ramalingam S, Ramachandran I, May R, Queimado L, Houchen CW, Anant S. Translation inhibition during cell cycle arrest and apoptosis: Mcl-1 is a novel target for RNA binding protein CUGBP2. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1025–1032. doi: 10.1152/ajpgi.00602.2007. [DOI] [PubMed] [Google Scholar]
- 100.Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Suzuki H, Takeuchi M, Sugiyama A, Alam AK, Vu LT, Sekiyama Y, Dam HC, Ohki SY, Tsukahara T. Alternative splicing produces structural and functional changes in CUGBP2. BMC Biochem. 2012;13:6. doi: 10.1186/1471-2091-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cosson B, Gautier-Courteille C, Maniey D, Ait-Ahmed O, Lesimple M, Osborne HB, Paillard L. Oligomerization of EDEN-BP is required for specific mRNA deadenylation and binding. Biol Cell. 2006;98:653–665. doi: 10.1042/BC20060054. [DOI] [PubMed] [Google Scholar]
- 103.Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 104.Ladd AN, Taffet G, Hartley C, Kearney DL, Cooper TA. Cardiac tissue-specific repression of CELF activity disrupts alternative splicing and causes cardiomyopathy. Mol Cell Biol. 2005;25:6267–6278. doi: 10.1128/MCB.25.14.6267-6278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Berger DS, Moyer M, Kliment GM, van Lunteren E, Ladd AN. Expression of a dominant negative CELF protein in vivo leads to altered muscle organization, fiber size, and subtype. PLoS One. 2011;6:e19274. doi: 10.1371/journal.pone.0019274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pascual M, Vicente M, Monferrer L, Artero R. The Muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- 108.Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum Mol Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 109.Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, Lecuyer E, Burge CB. Transcriptome-wide Regulation of Pre-mRNA Splicing and mRNA Localization by Muscleblind Proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cho SJ, Zhang J, Chen X. RNPC1 modulates the RNA-binding activity of, and cooperates with, HuR to regulate p21 mRNA stability. Nucleic Acids Res. 2010;38:2256–2267. doi: 10.1093/nar/gkp1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.van der Giessen K, Di-Marco S, Clair E, Gallouzi IE. RNAi-mediated HuR depletion leads to the inhibition of muscle cell differentiation. J Biol Chem. 2003;278:47119–47128. doi: 10.1074/jbc.M308889200. [DOI] [PubMed] [Google Scholar]
- 112.Miyamoto S, Hidaka K, Jin D, Morisaki T. RNA-binding proteins Rbm38 and Rbm24 regulate myogenic differentiation via p21-dependent and -independent regulatory pathways. Genes Cells. 2009;14:1241–1252. doi: 10.1111/j.1365-2443.2009.01347.x. [DOI] [PubMed] [Google Scholar]
- 113.Timchenko NA, Patel R, Iakova P, Cai ZJ, Quan L, Timchenko LT. Overexpression of CUG triplet repeat-binding protein, CUGBP1, in mice inhibits myogenesis. J Biol Chem. 2004;279:13129–13139. doi: 10.1074/jbc.M312923200. [DOI] [PubMed] [Google Scholar]
- 114.Sureban SM, Murmu N, Rodriguez P, May R, Maheshwari R, Dieckgraefe BK, Houchen CW, Anant S. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology. 2007;132:1055–1065. doi: 10.1053/j.gastro.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 115.Xiao S, Sanelli T, Dib S, Sheps D, Findlater J, Bilbao J, Keith J, Zinman L, Rogaeva E, Robertson J. RNA targets of TDP-43 identified by UV-CLIP are deregulated in ALS. Mol Cell Neurosci. 2011;47:167–180. doi: 10.1016/j.mcn.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 116.Dujardin G, Buratti E, Charlet-Berguerand N, Martins de Araujo M, Mbopda A, Le Jossic-Corcos C, Pagani F, Ferec C, Corcos L. CELF proteins regulate CFTR pre-mRNA splicing: essential role of the divergent domain of ETR-3. Nucleic Acids Res. 2010;38:7273–7285. doi: 10.1093/nar/gkq573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Srikantan S, Tominaga K, Gorospe M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr Protein Pept Sci. 2012;13:372–379. doi: 10.2174/138920312801619394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Leveille N, Elkon R, Davalos V, Manoharan V, Hollingworth D, Oude Vrielink J, le Sage C, Melo CA, Horlings HM, Wesseling J, Ule J, Esteller M, Ramos A, Agami R. Selective inhibition of microRNA accessibility by RBM38 is required for p53 activity. Nat Commun. 2011;2:513. doi: 10.1038/ncomms1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jacobsen A, Wen J, Marks DS, Krogh A. Signatures of RNA binding proteins globally coupled to effective microRNA target sites. Genome Res. 2010;20:1010–1019. doi: 10.1101/gr.103259.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY, Lee WH. miR-495 is upregulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via downregulation of E-cadherin and REDD1. Oncogene. 2011;30:2463–2474. doi: 10.1038/onc.2010.618. [DOI] [PubMed] [Google Scholar]
- 121.Li Z, Cao Y, Jie Z, Liu Y, Li Y, Li J, Zhu G, Liu Z, Tu Y, Peng G, Lee DW, Park SS. miR-495 and miR-551a inhibit the migration and invasion of human gastric cancer cells by directly interacting with PRL-3. Cancer Lett. 2012;323:41–47. doi: 10.1016/j.canlet.2012.03.029. [DOI] [PubMed] [Google Scholar]
- 122.Eisenberg I, Eran A, Nishino I, Moggio M, Lamperti C, Amato AA, Lidov HG, Kang PB, North KN, Mitrani-Rosenbaum S, Flanigan KM, Neely LA, Whitney D, Beggs AH, Kohane IS, Kunkel LM. Distinctive patterns of microRNA expression in primary muscular disorders. Proc Natl Acad Sci U S A. 2007;104:17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci U S A. 2012;109:3347–3352. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Perbellini R, Greco S, Sarra-Ferraris G, Cardani R, Capogrossi MC, Meola G, Martelli F. Dysregulation and cellular mislocalization of specific miRNAs in myotonic dystrophy type 1. Neuromuscul Disord. 2011;21:81–88. doi: 10.1016/j.nmd.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 126.Gambardella S, Rinaldi F, Lepore SM, Viola A, Loro E, Angelini C, Vergani L, Novelli G, Botta A. Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J Transl Med. 2010;8:48. doi: 10.1186/1479-5876-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D, Wahbi K, Day JW, Fujimura H, Takahashi MP, Auboeuf D, Dreumont N, Furling D, Charlet-Berguerand N. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
- 128.Schoser B, Timchenko L. Myotonic dystrophies 1 and 2: complex diseases with complex mechanisms. Curr Genomics. 2010;11:77–90. doi: 10.2174/138920210790886844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ward AJ, Rimer M, Killian JM, Dowling JJ, Cooper TA. CUGBP1 overexpression in mouse skeletal muscle reproduces features of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:3614–3622. doi: 10.1093/hmg/ddq277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Koshelev M, Sarma S, Price RE, Wehrens XH, Cooper TA. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum Mol Genet. 2010;19:1066–1075. doi: 10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu Z, Wang AM, Robins DM, Lieberman AP. Altered RNA splicing contributes to skeletal muscle pathology in Kennedy disease knock-in mice. Dis Model Mech. 2009;2:500–507. doi: 10.1242/dmm.003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cibois M, Boulanger G, Audic Y, Paillard L, Gautier-Courteille C. Inactivation of the Celf1 Gene that Encodes an RNA-Binding Protein Delays the First Wave of Spermatogenesis in Mice. PLoS One. 2012;7:e46337. doi: 10.1371/journal.pone.0046337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kress C, Gautier-Courteille C, Osborne HB, Babinet C, Paillard L. Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol Cell Biol. 2007;27:1146–1157. doi: 10.1128/MCB.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 135.Wang GS, Kearney DL, De Biasi M, Taffet G, Cooper TA. Elevation of RNA-binding protein CUGBP1 is an early event in an inducible heart-specific mouse model of myotonic dystrophy. J Clin Invest. 2007;117:2802–2811. doi: 10.1172/JCI32308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Orengo JP, Ward AJ, Cooper TA. Alternative splicing dysregulation secondary to skeletal muscle regeneration. Ann Neurol. 2011;69:681–690. doi: 10.1002/ana.22278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Osborne RJ, Lin X, Welle S, Sobczak K, O’Rourke JR, Swanson MS, Thornton CA. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 140.Meola G. Myotonic dystrophies as a brain disorder. Neurol Sci. 2010;31:863–864. doi: 10.1007/s10072-010-0414-2. [DOI] [PubMed] [Google Scholar]
- 141.Romeo V. Myotonic Dystrophy Type 1 or Steinert’s disease. Adv Exp Med Biol. 2012;724:239–257. doi: 10.1007/978-1-4614-0653-2_18. [DOI] [PubMed] [Google Scholar]
- 142.Di Costanzo A, Santoro L, de Cristofaro M, Manganelli F, Di Salle F, Tedeschi G. Familial aggregation of white matter lesions in myotonic dystrophy type 1. Neuromuscul Disord. 2008;18:299–305. doi: 10.1016/j.nmd.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 143.Leroy O, Wang J, Maurage CA, Parent M, Cooper T, Buee L, Sergeant N, Andreadis A, Caillet-Boudin ML. Brain-specific change in alternative splicing of Tau exon 6 in myotonic dystrophy type 1. Biochim Biophys Acta. 2006;1762:460–467. doi: 10.1016/j.bbadis.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 144.Lee G, Leugers CJ. Tau and tauopathies. Prog Mol Biol Transl Sci. 2012;107:263–293. doi: 10.1016/B978-0-12-385883-2.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Marteyn A, Maury Y, Gauthier MM, Lecuyer C, Vernet R, Denis JA, Pietu G, Peschanski M, Martinat C. Mutant human embryonic stem cells reveal neurite and synapse formation defects in type 1 myotonic dystrophy. Cell Stem Cell. 2011;8:434–444. doi: 10.1016/j.stem.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 146.Anderson KN, Baban D, Oliver PL, Potter A, Davies KE. Expression profiling in spinal muscular atrophy reveals an RNA binding protein deficit. Neuromuscul Disord. 2004;14:711–722. doi: 10.1016/j.nmd.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 147.Halgren C, Bache I, Bak M, Myatt MW, Anderson CM, Brondum-Nielsen K, Tommerup N. Haploinsufficiency of CELF4 at 18q12.2 is associated with developmental and behavioral disorders, seizures, eye manifestations, and obesity. Eur J Hum Genet. 2012 doi: 10.1038/ejhg.2012.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yang Y, Mahaffey CL, Berube N, Maddatu TP, Cox GA, Frankel WN. Complex seizure disorder caused by Brunol4 deficiency in mice. PLoS Genet. 2007;3:e124. doi: 10.1371/journal.pgen.0030124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Colby DW, Prusiner SB. Prions. Cold Spring Harb Perspect Biol. 2011;3:a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lee EB, Lee VM, Trojanowski JQ. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat Rev Neurosci. 2012;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Udan M, Baloh RH. Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion. 2011;5:1–5. doi: 10.4161/pri.5.1.14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chang ET, Donahue JM, Xiao L, Cui Y, Rao JN, Turner DJ, Twaddell WS, Wang JY, Battafarano RJ. The RNA-binding protein CUG-BP1 increases survivin expression in oesophageal cancer cells through enhanced mRNA stability. The Biochemical journal. 2012;446:113–123. doi: 10.1042/BJ20120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Rodriguez FJ, Giannini C, Asmann YW, Sharma MK, Perry A, Tibbetts KM, Jenkins RB, Scheithauer BW, Anant S, Jenkins S, Eberhart CG, Sarkaria JN, Gutmann DH. Gene expression profiling of NF-1-associated and sporadic pilocytic astrocytoma identifies aldehyde dehydrogenase 1 family member L1 (ALDH1L1) as an underexpressed candidate biomarker in aggressive subtypes. J Neuropathol Exp Neurol. 2008;67:1194–1204. doi: 10.1097/NEN.0b013e31818fbe1e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Geh JL, Moss AL. Multiple pilomatrixomata and myotonic dystrophy: a familial association. Br J Plast Surg. 1999;52:143–145. doi: 10.1054/bjps.1998.3036. [DOI] [PubMed] [Google Scholar]
- 156.Gadalla SM, Lund M, Pfeiffer RM, Gortz S, Mueller CM, Moxley RT, 3rd, Kristinsson SY, Bjorkholm M, Shebl FM, Hilbert JE, Landgren O, Wohlfahrt J, Melbye M, Greene MH. Cancer risk among patients with myotonic muscular dystrophy. JAMA. 2011;306:2480–2486. doi: 10.1001/jama.2011.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Win AK, Perattur PG, Pulido JS, Pulido CM, Lindor NM. Increased cancer risks in myotonic dystrophy. Mayo Clin Proc. 2012;87:130–135. doi: 10.1016/j.mayocp.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mueller CM, Hilbert JE, Martens W, Thornton CA, Moxley RT, 3rd, Greene MH. Hypothesis: neoplasms in myotonic dystrophy. Cancer Causes Control. 2009;20:2009–2020. doi: 10.1007/s10552-009-9395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang GL, Salisbury E, Shi X, Timchenko L, Medrano EE, Timchenko NA. HDAC1 promotes liver proliferation in young mice via interactions with C/EBPbeta. J Biol Chem. 2008;283:26179–26187. doi: 10.1074/jbc.M803545200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Arnal-Estape A, Tarragona M, Morales M, Guiu M, Nadal C, Massague J, Gomis RR. HER2 silences tumor suppression in breast cancer cells by switching expression of C/EBPss isoforms. Cancer Res. 2010;70:9927–9936. doi: 10.1158/0008-5472.CAN-10-0869. [DOI] [PubMed] [Google Scholar]
- 161.Guerzoni C, Bardini M, Mariani SA, Ferrari-Amorotti G, Neviani P, Panno ML, Zhang Y, Martinez R, Perrotti D, Calabretta B. Inducible activation of CEBPB, a gene negatively regulated by BCR/ABL, inhibits proliferation and promotes differentiation of BCR/ABL-expressing cells. Blood. 2006;107:4080–4089. doi: 10.1182/blood-2005-08-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vlasova-St Louis I, Bohjanen PR. Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1. Curr Opin Genet Dev. 2011;21:444–451. doi: 10.1016/j.gde.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Nguyen-Chi M, Morello D. RNA-binding proteins, RNA granules, and gametes: is unity strength? Reproduction. 2011;142:803–817. doi: 10.1530/REP-11-0257. [DOI] [PubMed] [Google Scholar]
- 165.Dev A, Nayernia K, Meins M, Adham I, Lacone F, Engel W. Mice deficient for RNA-binding protein brunol1 show reduction of spermatogenesis but are fertile. Mol Reprod Dev. 2007;74:1456–1464. doi: 10.1002/mrd.20742. [DOI] [PubMed] [Google Scholar]
- 166.Gautier-Courteille C, Le Clainche C, Barreau C, Audic Y, Graindorge A, Maniey D, Osborne HB, Paillard L. EDEN-BP-dependent post-transcriptional regulation of gene expression in Xenopus somitic segmentation. Development. 2004;131:6107–6117. doi: 10.1242/dev.01528. [DOI] [PubMed] [Google Scholar]
- 167.Cibois M, Gautier-Courteille C, Vallee A, Paillard L. A strategy to analyze the phenotypic consequences of inhibiting the association of an RNA-binding protein with a specific RNA. RNA. 2010;16:10–15. doi: 10.1261/rna.1742610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Matsui T, Sasaki A, Akazawa N, Otani H, Bessho Y. Celf1 regulation of dmrt2a is required for somite symmetry and left-right patterning during zebrafish development. Development. 2012;139:3553–3560. doi: 10.1242/dev.077263. [DOI] [PubMed] [Google Scholar]
- 169.Milne CA, Hodgkin J. ETR-1, a homologue of a protein linked to myotonic dystrophy, is essential for muscle development in Caenorhabditis elegans. Curr Biol. 1999;9:1243–1246. doi: 10.1016/s0960-9822(99)80504-1. [DOI] [PubMed] [Google Scholar]
- 170.Loria PM, Duke A, Rand JB, Hobert O. Two neuronal, nuclear-localized RNA binding proteins involved in synaptic transmission. Curr Biol. 2003;13:1317–1323. doi: 10.1016/s0960-9822(03)00532-3. [DOI] [PubMed] [Google Scholar]
- 171.Yan N, Macdonald PM. Genetic interactions of Drosophila melanogaster arrest reveal roles for translational repressor Bruno in accumulation of Gurken and activity of Delta. Genetics. 2004;168:1433–1442. doi: 10.1534/genetics.104.033985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tani H, Akimitsu N. Genome-wide technology for determining RNA stability in mammalian cells: Historical perspective and recent advantages based on modified nucleotide labeling. RNA Biol. 2012;9 doi: 10.4161/rna.22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Adeli K. Translational control mechanisms in metabolic regulation: critical role of RNA binding proteins, microRNAs, and cytoplasmic RNA granules. Am J Physiol Endocrinol Metab. 2011;301:E1051–1064. doi: 10.1152/ajpendo.00399.2011. [DOI] [PubMed] [Google Scholar]
- 175.Scheu S, Stetson DB, Reinhardt RL, Leber JH, Mohrs M, Locksley RM. Activation of the integrated stress response during T helper cell differentiation. Nat Immunol. 2006;7:644–651. doi: 10.1038/ni1338. [DOI] [PubMed] [Google Scholar]
- 176.Lin JC, Tarn WY. RNA-binding motif protein 4 translocates to cytoplasmic granules and suppresses translation via argonaute2 during muscle cell differentiation. J Biol Chem. 2009;284:34658–34665. doi: 10.1074/jbc.M109.032946. [DOI] [PMC free article] [PubMed] [Google Scholar]