Abstract
In many non-mammalian vertebrates, adult dentitions result from cyclical rounds of tooth regeneration wherein simple unicuspid teeth are replaced by more complex forms. Therefore and by contrast to mammalian models, the numerical majority of vertebrate teeth develop shape during the process of replacement. Here, we exploit the dental diversity of Lake Malawi cichlid fishes to ask how vertebrates generally replace their dentition and in turn how this process acts to influence resulting tooth morphologies. First, we used immunohistochemistry to chart organogenesis of continually replacing cichlid teeth and discovered an epithelial down-growth that initiates the replacement cycle via a labial proliferation bias. Next, we identified sets of co-expressed genes from common pathways active during de novo, lifelong tooth replacement and tooth morphogenesis. Of note, we found two distinct epithelial cell populations, expressing markers of dental competence and cell potency, which may be responsible for tooth regeneration. Related gene sets were simultaneously active in putative signaling centers associated with the differentiation of replacement teeth with complex shapes. Finally, we manipulated targeted pathways (BMP, FGF, Hh, Notch, Wnt/β-catenin) in vivo with small molecules and demonstrated dose-dependent effects on both tooth replacement and tooth shape. Our data suggest that the processes of tooth regeneration and tooth shape morphogenesis are integrated via a common set of molecular signals. This linkage has subsequently been lost or decoupled in mammalian dentitions where complex tooth shapes develop in first generation dentitions that lack the capacity for lifelong replacement. Our dissection of the molecular mechanics of vertebrate tooth replacement coupled to complex shape pinpoints aspects of odontogenesis that might be re-evolved in the lab to solve problems in regenerative dentistry.
Keywords: Tooth Replacement, Odontogenesis, Regeneration, Stem Cells
Introduction
Vertebrate animals differ in their capacity to renew and regenerate body parts. Various lineages have retained or evolved the ability to regenerate nervous systems (Kizil et al., 2011; Kroehne et al., 2011), limbs (Kragl et al., 2009; Nacu and Tanaka, 2011), fins (JaŸwińska et al., 2007; Singh et al., 2012) and tails (Echeverri and Tanaka, 2002; Lin and Slack, 2008), internal organs like the heart (Wang et al., 2011), as well as iterative elements like hairs, scales, taste buds and teeth (Chang et al., 2009; Harada et al., 1999; Plikus et al., 2011; Plikus et al., 2008; Wang et al., 2007). Developmental biologists are captivated by regeneration because the process may recycle well-known mechanisms of embryonic patterning and likely involves the deployment of stem cells in post-embryonic tissues. Precisely because humans (and more generally mammals) lose regenerative capacity with age, keen biomedical interest revolves around natural instances of regeneration and renewal from stem cells as exemplars for cellular reprogramming (Christen et al., 2010).
In many examples of animal regeneration, the trigger or impetus is external and unpredictable. A lizard can re-grow a tail after escaping a predator; such an interaction may be probable, but not necessary over an individual’s lifetime. By contrast, predictable programs characterize other cases of regeneration, like the shedding of hair, teeth, scales and feathers. For instance, adult cichlid fishes replace each tooth in the oral jaw approximately every 30–100 days (Tuisku and Hildebrand, 1994). When programmed regeneration is coupled with the functional requirement to maintain a particular organization of elements (feathers for flight, scales for swimming, teeth for mastication), the developmental phenomena of patterning, morphogenesis and renewal, often studied independently, must be deeply integrated across space and time. In most systems, biologists do not understand how this integration is achieved.
Vertebrate dentitions represent a seemingly apposite system in which to decipher how individual organs (teeth) develop complex shapes and inter-unit patterns, while simultaneously exhibiting programmed regeneration. Many vertebrates possess teeth in multiple rows on multiple elements of the oral jaw and pharyngeal region that are continuously replaced throughout life. In ray-finned fishes, dipnoans and urodeles, first generation teeth are small and simple unicuspids lacking blood vessels and nerves; therefore the numerical majority of vertebrate teeth develop shape and increased complexity (e.g., size, curvature, cusps) through replacement (Sire et al., 2002). Teeth likely arose in jawless vertebrates more than half a billion years ago (Fraser et al., 2009; Smith, 2003; Smith and Coates, 1998; Smith and Coates, 2000) – there is thus a long evolutionary record and broad phylogenetic distribution to bolster our understanding of how patterned dentitions are likewise regenerated.
The fact that we know very little about the coupled patterning, morphogenesis and regeneration of vertebrate dentitions can be partly explained by the peculiar biology of teeth in the mouse model. The mouse dentition is comprised of one incisor and three molars, in a single row, on each left and right quadrant of the upper and lower jaws. Incisors are separated in space from molars by a toothless diastema, and the early patterning of the incisor and molar domains is well understood (Tucker and Sharpe, 2004). Molars develop complex three-dimensional shape while incisors generally do not (Jernvall et al., 2000). Incisors exhibit self-renewal via continuous deposition of enamel on the labial surface, supported by a stem cell niche biased to the labial cervical loop (Harada et al., 1999; Wang et al., 2007); molars lack this potential. Thus, mouse molars are models of complex morphogenesis. Classic studies have demonstrated how molars develop under the influence of well-known signaling pathways (e.g., BMP, FGF, Hh, Wnt and Eda) and downstream transcription factors (i.e., Pitx, Pax, Dlx, Barx, Msx) (Åberg et al., 1997; Bei and Maas, 1998; Chen et al., 1996; Dassule et al., 2000; Jernvall et al., 1994; Peters et al., 1998; Sarkar and Sharpe, 1999; Sharpe, 1995; Thesleff and Sharpe, 1997). Recent reports point to genes that may couple these developmental pathways in the molar field (Ahn et al., 2010; Cho et al., 2011). By contrast, incisors are models of stem-based continuous growth, with context-dependent function of common pathways (BMP, FGF, Wnt) as well as novel roles for additional factors (Notch (Felszeghy et al., 2010); Follistatin (Wang et al., 2007)). Notably, neither molars nor incisors are replaced over mouse ontogeny.
Because mice do not replace their teeth, a set of new models for dental regeneration has emerged. This includes the shrew (Järvinen et al., 2008), ferret (Järvinen et al., 2009), zebrafish (Huysseune, 2006) and a cadre of reptiles (Handrigan et al., 2010). Taken together, studies suggest that tooth replacement requires (i) an epithelial connection between the functional tooth and its successor, known as successional lamina (SL), which is borne from (ii) putative dental/epithelial stem cells capable of forming a replacement tooth de novo. One limitation of these new replacement models is that the dentitions in question are relatively simple: few teeth total, often in a single row, with each tooth generally conical or spatulate in shape.
In this report, we ask how complex dentitions are shaped as they are replaced, using cichlid fishes from Lake Malawi, East Africa. The main advantage of this system is the sheer dental diversity among closely related species (Fraser et al., 2008; Streelman et al., 2003). Most cichlids endemic to Lake Malawi have evolved from a common ancestor in the last 500,000 years; their genomes are highly similar (e.g., less nucleotide diversity than observed in lab strains of zebrafish) and species share genetic polymorphism (Loh et al., 2008). Against this backdrop of genomic similarity, dental patterns and shapes vary considerably. For example, Cynotilapia afra, a rock-dwelling planktivore, possesses a small number of large, widely spaced, conical teeth in 2–3 rows while species of the algal-brushing rock-dweller genus Petrotilapia exhibit hundreds of small, tightly packed tricuspid teeth in 10–15 rows. The particular tooth pattern, that is the size and spacing of teeth as well as the extent of the tooth field in the jaw, is set with the initiation of the first generation dentition, prior to the development of tooth shapes (Fraser et al., 2008). As in other cichlids, these first generation teeth are small conical unicuspids and are not innervated (Huysseune and Sire, 1997). Complex shape and innervation are thus the phenomenological consequence of tooth replacement in cichlids, with adult shapes developing during multiple, early rounds of replacement (Streelman et al., 2003) into patterns set during initiation (Fraser et al., 2008).
We used a combination of immunohistochemistry and in situ hybridization, at multiple stages of development, to identify cell populations, putative signaling centers and molecular pathways involved in cichlid tooth replacement and morphogenesis. Armed with this information, we employed a set of small molecules to manipulate these pathways in vivo, documenting effects of treatment on both shape and replacement. The key finding from this study is that the processes of tooth morphogenesis and replacement are linked by common pathways that likely control the balance between growth, proliferation and differentiation as cusps form on tooth tips and as new dental organs initiate development from their predecessors. We suggest that this coupling of morphogenesis and lifelong regeneration is the ancestral vertebrate condition, largely lost or decoupled in the mammalian dentition. Our integrative understanding of continuous tooth replacement from nature may pinpoint features of the process to be re-evolved by bioengineers.
Materials and Methods
Fish husbandry
Species of Lake Malawi cichlids used in this analysis include: Aulonocara jacobfreibergi [AJ], Cynotilapia afra [CA], Labeotropheus fuelleborni [LF], Mchenga conophoros [MC], Metriaclima zebra [MZ], Petrotilapia chitimba [PC] Petrotilapia tridentiger [PT] and Pseudotropheus lombardoi (PL). These species were chosen to represent diversity in feeding behavior, adult tooth shape and ontogeny of tooth replacement (Table S1). Adult cichlids were maintained in re-circulating aquarium systems at 28°C (GIT). Fertilized embryos were removed from the mouths of brooding females and staged in days post-fertilization (dpf) according to a developmental series from the Nile Tilapia (Fujimura and Okada, 2007). Embryos/fry were raised to desired stages for chemical treatment or anesthetized with MS-222 for fixation in 4% paraformaldehyde followed by dehydration into MeOH.
Immunohistochemistry
Embryos were fixed in 10% NBF for 24 hours, dehydrated through ethanol, cleared with butanol and embedded in paraffin. Embryos were sectioned at 10μm and H&E stained using a Leica Autostainer XL. For proliferation assays, cichlid fry undergoing active dental replacement were incubated in 5-Bromo-2-deoxy-uridine (BrdU) for periods of 6–8 hours for nucleic acid incorporation. Fry were immediately anesthetized (MS-222), fixed, and paraffin sectioned at 10μm. We then applied the 5-Bromo-2-deoxy-uridine Labeling and Detection Kit II (Roche) according to manufacturer’s specifications (secondary antibody conjugated with AP activated NBT/BCIP, Roche). Similarly, PCNA staining was carried out on paraffin sections of wild type embryos according to manufacturer instructions (PCNA staining kit, Invitrogen), with DAB color reaction.
In situ hybridization
Digoxigenin-labeled antisense riboprobes were prepared using partial cichlid genome assemblies (Loh et al., 2008) as well as recently assembled tilapia and MZ genomes (https://www.broadinstitute.org/ftp/pub/assemblies/fish). DNA sequence diversity across the Lake Malawi assemblage is 0.28%; less than reported values for laboratory strains of zebrafish. cDNA sequences for probe design have been deposited in GenBank (accession numbers XXXX-XXXX). ISH was performed according to previously published protocols (Fraser et al., 2008; Fraser et al., 2009). Embryos were re-hydrated from MeOH and ISH was carried out in whole-mount. Digoxigenin-labeled antisense riboprobes were generated using the Riboprobe System Sp6/T7 kit (Promega). AP-conjugated anti-dig antibodies were visualized at the end of color reaction (NBT/BCIP; Roche) using light microscopy. Embryos were embedded in chick albumin cross-fixed with 2.5% gluteraldehyde and post-fixed with 4% PFA. A Leica Microsystems VT1000 vibratome was used to cut sections at 15–25μm. Histological sections were then mounted with glycerine and imaged at 10–63x using a Leica DM2500 compound microscope.
Treatment with small molecules
Stock solutions were prepared for each chemical treatment experiment using Dimethyl Sulfoxide (DMSO, MP Biomedicals) or water as a solvent. Stock solutions were as follows: 5mM Cyclopamine (LC Laboratories) in DMSO, 10μm DAPT (Tocris) in DMSO, 10μm Dorsomorphin (Sigma-Alrich) in DMSO, 5mM LiCl (Alexis Biochem) in H2O, and 50μm SU5402 (see acknowledgments) in DMSO. Cichlids were raised to appropriate stages for treatment and embryos from single broods were split into small molecule and solvent control groups. All treatments were designed to evaluate perturbations to complete, fully shaped adult first-row dentitions; because species differ in the number of replacement generations (and hence time) until adult first-row tooth shape is established (Table S1), the onset of treatment varied by species accordingly (e.g., as early as 40dpf in LF). Treatment doses varied across chemicals to produce dental phenotypes without gross anodontia or fatality. All chemical and control experiments were performed in Erlenmeyer flasks at 28°C in an oscillating platform culture incubator (Barnstead Lab-Line Max 4000). After treatment, fry were washed extensively with fresh fish water and raised for 14 days prior to sacrifice, fixation, clearing and staining.
Clearing and staining
Fry previously fixed in PFA were washed with DEPC-H2O for thirty minutes. Specimens were then placed into a 1% trypsin solution for one hour. After protein digest, calcified tissues were stained using Alizarin red S solution (1g/50mL KOH). Staining averaged 30 minutes, with larger specimen requiring a longer stain time. Once the tips of the pelvic fins stained red, fry were moved to a 2% KOH solution for a period of 24 hours. Cleared and stained fishes were then graded into 100% glycerine, with thymol as a biocide.
Results
One-for-one replacement of cichlid teeth
We explored the histological events surrounding cichlid tooth replacement using both standard staining methods (i.e., hemotoxalin and eosin), as well as antibodies to proliferating cell nuclear antigen (PCNA) and incorporated bromodeoxyuridine (5-bromo-2′-deoxyuridine, BrdU). We present data for the oral dentition only, but general observations hold for oral and pharyngeal jaws, both of which house teeth in cichlids (Fraser et al., 2009). Throughout, we divide our description into three stages of replacement tooth development: (1) initiation, (2) cellular differentiation, and (3) secretion.
Cichlids exhibit intramedullary (inside the jawbone; i.e., intraosseous (Trapani, 2001)) replacement, similar to humans but distinct from other animals like reptiles (Handrigan et al., 2010), zebrafish (Danio rerio) (Huysseune, 2006) and rainbow trout (Oncorhynchus mykiss) (Fraser et al., 2006) where replacement germs develop in an extramedullary location. Cichlids replace teeth in one-for-one fashion like some other bony fishes (Bemis et al., 2005; Fraser et al., 2006; Kerr, 1960; Motta, 1984); each functional tooth serves as a placeholder, as well as a supply of epithelial cells, for subsequent replacement by a single successor tooth – a process repeated over ontogeny. This is different from the many-for-one replacement system as observed in sharks (Fraser and Smith, 2011; Smith et al., 2009), and pufferfish (Fraser et al., 2012), where many replacements form in advance of function for each tooth family.
Labial epithelial cells associated with developing first generation teeth form each successional lamina (SL, Figure 1A–C), and together with contributions from labial oral epithelium, initiate the continued supply of tooth replacements. Using BrdU and PCNA, we found that the primary stage of SL invagination is marked by high rates of proliferation (Figure 1B, C). When the lamina extends further below the existing primary tooth, proliferation continues and the lamina interacts with the receptive neural crest-derived mesenchyme to begin the process of replacement tooth organ development.
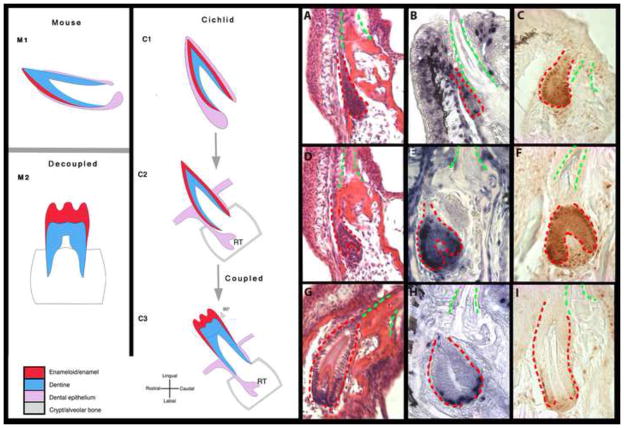
Figure 1. Dynamics of shape and (self-) renewal in mouse and cichlid dentitions.
In the mouse, the processes of self-renewal in incisors (M1) and complex shape morphogenesis in molars (M2) are decoupled in space and time. By contrast, cichlid replacement teeth form complex shapes as they regenerate (C1–C3). Cichlid tooth replacement can be broken into three stages: initiation (A–C), cellular differentiation (D–F) and secretion (G–I). We illustrate these stages by H&E histology (A, D, G); BrdU (B, E, H) and PCNA (C, F, I) immunohistochemistry. First generation teeth are outlined in green and replacement dental epithelium in red. These are paraffin sections in sagittal plane at 10μm thickness, imaged at 63x magnification. (A, D, G) - Metriaclima zebra; (B, C, E, F, H, I) - Labeotropheus fuelleborni.
As the epithelial SL interacts with the underlying mesenchyme, the bone surrounding and attached to the predecessor tooth begins to remodel and encases the newly initiated replacement organ (Figure 1D–F) in a crypt that will house it throughout maturation. We observe that the oral epithelium and SL remain connected to the developing replacement tooth by a thin epithelial stream of cells, until eruption of the replacement (Figure 1D–I). The bone forms around this ‘connector’ cell stream (the gubernacular cord (Avery and Steele, 2000)) leaving small pores called gubernacular canals, observed across vertebrates from fishes to humans (Avery and Steele, 2000; Huysseune, 2000). The SL, through the gubernacular canal, maintains a link to oral epithelia and continuity between the extramedullary epithelium and the intramedullary (crypt) mesenchyme.
The replacement tooth germ transitions to stages of cellular differentiation as the mesenchyme condenses into the dental papilla. (Figure 1D–F). The epithelium contorts into a dental bud, followed by an inward folding of the epithelium to form the cap stage tooth – the first stage of the tooth-shaping process. This epithelial folding leads to the formation of three cell layers: the inner dental epithelium (IDE), outer dental epithelium (ODE) (Fraser et al., 2008), and an intermediate layer of cells between the IDE and ODE (Figure S1), putatively analogous to the stellate reticulum of mammalian teeth (Huysseune and Thesleff, 2004; Wang et al., 2007). Epithelial and mesenchymal cells differentiate at the cap to bell transition and form enameloid-secreting ameloblasts from the IDE and dentine-secreting odontoblasts from the dental papilla. As the replacement tooth transitions from bud to cap and from cap to bell stages, we note three main sites of cell proliferation: at the tip of the developing replacement tooth, and in both cervical loops. These areas have similarly been identified in the gecko as regions of proliferation for hard tissue-secreting (enameloid and dentin) cells (Handrigan et al., 2010).
During terminal stages of cichlid replacement tooth development, the ameloblasts and odontoblasts secrete their respective hard tissue matrices. The ameloblasts continue to elongate as columnar cells and the bony crypt is remodeled to accommodate the growing tooth. As the eruption process initiates, we find that the lamina stream (gubernacular cord) that connects the oral epithelium with the successional tooth begins to break down (Figure 1G–I). BrdU and PCNA analyses during hard tissue secretion highlight a slight labial bias in proliferation at the cervical loops, presumably giving rise to additional enameloid-secreting cells on the labial surface of the tooth (Figure 1E, F, I). This asymmetry provides evidence of spatial differences in hard tissue deposition during the formation of cichlid replacement teeth.
Gene co-expression domains direct de novo tooth replacement
Cichlid one-for-one tooth replacement is initiated as an epithelial invagination, labial to the predecessor tooth (Figure 1). We sought to understand the molecular pathways that might guide this process. Because little is known about tooth replacement in vertebrates, we focused on pathways involved in the patterning and regeneration of hairs and feathers, as well as the development of mouse molars and incisors. A priori, de novo tooth replacement must combine factors providing dental competence to the epithelium and associated mesenchyme, coupled with signals of cell potency.
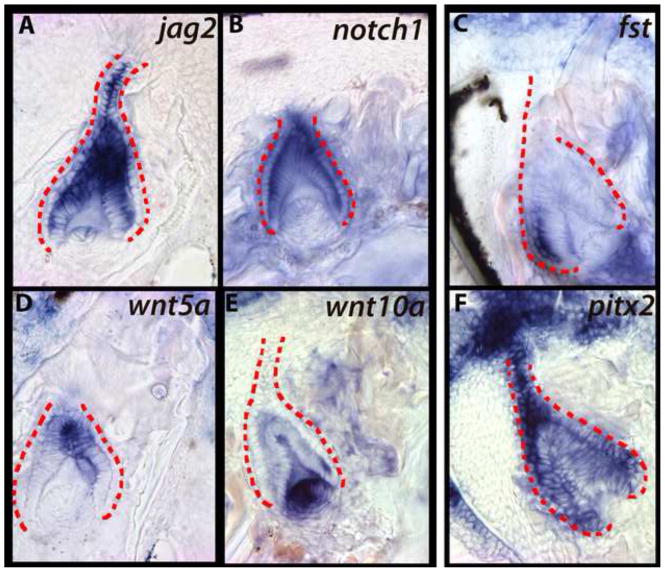
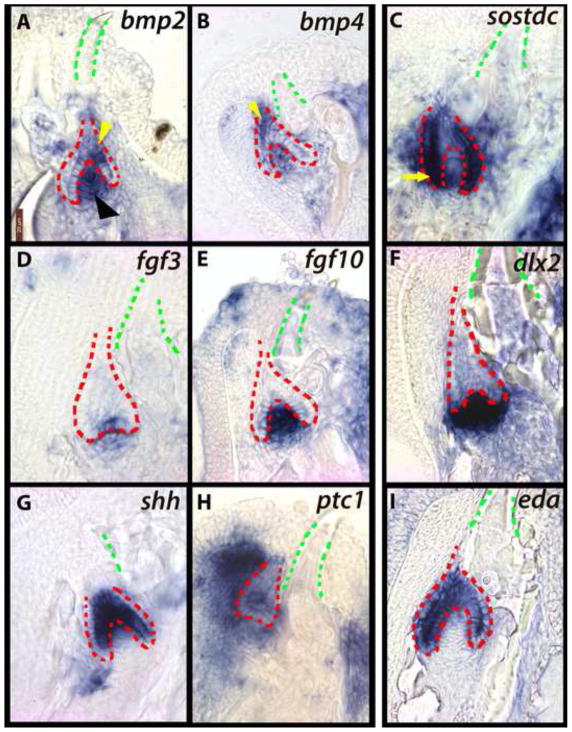
pitx2 is one of the earliest markers of dental-competent epithelium (Fraser et al., 2008). Consistent with expectation, pitx2 expression is observed within the extended SL, throughout the epithelial downgrowth (Figure 2A). The reciprocal neural crest-derived ectomesenchyme condenses and expresses a set of genes, including transcription factors such as runx2 (Figure 2B), and signaling molecules from the Wnt and FGF pathways (e.g., wnt10a, fgf10, Figure 2C, D). Genes of the BMP pathway are also recruited to the replacement tooth. The invaginating SL expresses bmp4 (Figure 2E) and bmp2 (not shown, see Figure 3A), as does the reciprocal condensing ectomesenchyme; this expression is maintained throughout the process of lamina extension and proliferation. We note a second mesenchymal domain of bmp4 expression, labial to the first generation tooth and the SL downgrowth (blue arrowhead in Figure 2E). The two BMP domains are separated in space by cells expressing osr2, a transcription factor that represses BMP expression in mouse dental mesenchyme (Zhang et al., 2009) (Figure 2F). Osr2-null mice exhibit expansion of lingual BMP expression and ultimately form lingual supernumerary teeth. It is possible that osr2 acts similarly here, to properly position the first cycle of dental replacement through restriction of odontogenic BMP.
Figure 2. Replacement teeth recruit markers of dental competence and cell potency.
The epithelial successional lamina (SL) expresses dental-commissioning pitx2 (A) in close proximity to condensing mesenchyme marked by runx2 (B), wnt10a (C), and fgf10 (D). bmp4 is active throughout epithelium and mesenchyme of the replacement tooth (E) and in a second region of labial mesenchyme (blue arrowhead) separated from the tooth germ by cells expressing the BMP inhibitor osr2 (F). shh (G) is not expressed in the invaginating SL, but is active in the oral epithelium both lingual to the replacement tooth (presumed dental lamina for lingual rows) and oral epithelium labial to the replacement germ (black arrows). ptc1 expression (H) is observed in the mesenchyme subjacent to shh-expressing oral epithelium. irx1b, a putative regulator of Hh signal, is expressed in the aboral-most region of the [Hh-negative] invagination (I). The invaginating SL contains an intermediate layer, between inner and outer dental epithelium (see Figure S1), expressing jag2 (J), notch1 (K), and sox2 (L). jag2 and sox2 are also expressed in a continuous ribbon of epithelium labial to the replacement organ (arrows). First generation teeth are outlined in green and replacement dental epithelium in red. These are vibratome sections in sagittal plane at 15μm thickness, imaged at 63x magnification. Labial is oriented to the left and oral toward the top of the page. Fishes used in this panel are ~15dpf. (A, B, D, E, F, G, H, K, J, L) - Metriaclima zebra, (C) - Labeotropheus fuelleborni, (I) - Aulonocara jacobfreibergi.
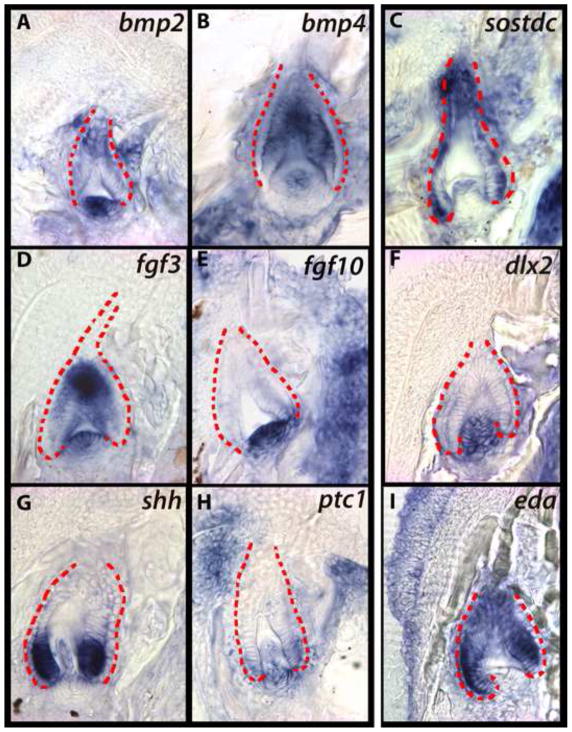
Figure 3. Expression of genes from the BMP (A–C), FGF (D–F), and Hh pathways (G–I) during the differentiation stage of cichlid replacement tooth development.
First generation teeth are outlined in green and replacement dental epithelium in red. Three expression domains are highlighted at this stage; (i) the tooth tip (A, B; yellow arrowhead), the condensing papilla (A; black arrowhead), and the cervical loops (C; yellow arrow). These are vibratome sections in sagittal plane at 15μm thickness, imaged at 63x magnification. Labial is oriented to the left and oral toward the top of the page. Fishes used in this panel are ~15–30dpf. (A, B, D, F, G, H, I) - Metriaclima zebra, (C) - Labeotropheus fuelleborni, (E) - Petrotilapia chitimba.
BMPs interact with the Hedgehog (Hh) pathway during the patterning of many organ systems, including teeth (Handrigan et al., 2010; Zhang et al., 2000); we thus examined expression of Hh ligands and receptors in the cichlid replacement program. Hedgehog is essential for tooth development in the mouse (Cobourne et al., 2004; Cobourne and Sharpe, 2004; Dassule et al., 2000), initiates the first generation dentition in cichlids (Fraser et al., 2008), but does not play a role in the initiation of the replacement dentition in the trout (Fraser et al., 2006), nor in squamates (Handrigan and Richman, 2010). Here, we did not observe activity of the extracellular ligand shh or its receptor ptc1 (Figure 2G, H) until later replacement tooth morphogenesis (Figure 3, 5). Molecules from the Iroquois homeobox family have been documented in mouse tooth development (Ferguson et al., 2001), but their roles in the process are not understood. irx1b is known to respond to Wnt signals and restrict shh in the embryonic forebrain (Scholpp et al., 2007; Scholpp et al., 2006; Sylvester et al., 2010). Here, we observe irx1b expression in the aboral-most epithelium of the extending SL (Figure 2I), in close proximity to wnt10a (Figure 2C). A putative function for irx1b in replacement tooth initiation is thus the regulation of Hh signal in the early SL downgrowth.
Figure 5. Expression of genes from the BMP (A–C), FGF (D–F), and Hh pathways (G–I) during the secretion stage of cichlid replacement tooth development.
Replacement outer dental epithelium is outlined in red. These are vibratome sections in sagittal plane at 15μm thickness, imaged at 63x magnification. Labial is oriented to the left and oral toward the top of the page. Fishes used in this panel are ~15–30dpf. (A, D, F, G, I) - Metriaclima zebra, (B, H) - Cynotilapia afra, (C) - Labeotropheus fuelleborni, (E) - Petrotilapia chitimba.
Although shh and ptc1 expression are absent from the early replacement SL, they are nonetheless active in regions relevant to tooth development (Figure 2G, H). The epithelium lingual to the outer row of erupted teeth strongly expresses shh, and as expected, the underlying mesenchyme maintains strong expression of the receptor ptc1. Cichlids continue to add posterior (lingual) rows of teeth throughout ontogeny and this lingual Hh signaling domain, in conjunction with pitx2 and BMP, may provide the potential for the initiation of first generation teeth in lingual rows (Fraser et al., 2008). Notably, there is a second region of shh-expressing epithelium and corresponding ptc1-expressing mesenchyme labial to the SL (Figure 2G, H), corresponding to the labial domain of bmp4 noted above (Figure 2E). This labial domain of BMP and Hh co-expression is maintained during subsequent stages of replacement tooth development (Figure 3). As no teeth form labial to the first generation dentition, we explored this cell population as a putative source of dental potency for replacement.
To do so, we first examined expression of Notch signaling family members. The Notch pathway is involved in the patterning of teeth (Mitsiadis et al., 2010; Mitsiadis et al., 1998; Mitsiadis et al., 2005), the stem niche of mouse incisors (Harada et al., 1999), and the general regulation of stem cells (Androutsellis-Theotokis et al., 2006). The Notch ligand jag2 is expressed in both (i) the Hh- and BMP-positive cells labial to the SL (Figure 2J, black arrow) and (ii) the intermediate cells of the SL (Figure 2J, blue arrow). These intermediate cells, between IDE and ODE, will give rise to stellate reticulum-like cells in the differentiated replacement tooth (Figure 1D–F; Figure S1). The receptor notch1 is expressed in the intermediate cells, but not labial to the SL (Figure 2K). However, the receptors notch2 and notch3 are active in both locations during subsequent developmental stages and rounds of replacement (RFB, unpublished). Next, we examined activity of the stem cell transcription factor sox2 in the replacing cichlid dentition. sox2 is expressed in both the labial domain (as well as lingual to the first tooth row) and within the intermediate cells of the SL (Figure 2L). Taken together, these patterns of gene co-expression give an mRNA signature to two distinct populations of cells (i) labial and superficial to the replacement organ and (ii) within the intermediate cells of the SL, one or both of which may enable and maintain continuous de novo dental replacement. Notably, in reptiles (Handrigan et al., 2010), stem-like cells are arranged superficially along the non-tooth forming outer layer of the dental lamina while in zebrafish (Huysseune, 2006; Huysseune and Thesleff, 2004), intermediate cells between IDE and ODE are suggested to exhibit stem-like properties.
Gene expression is evolutionarily conserved in replacement tooth differentiation
Cichlid replacement teeth undergo development within a bony crypt constantly remodeled to accommodate jaw growth and dental renewal. The replacement dental organ differentiates in a series analogous to the bud, cap and bell stages of mammalian teeth, while remaining connected to oral epithelium by a cord of cells through the gubernacular canal (Figure 1). We know very little about the molecules accompanying the morphogenesis of replacement teeth in any organism. We thus examined the expression of genes from five signaling pathways involved in the development and regeneration of vertebrate organs: BMP, FGF, Hh, Notch, and Wnt/β-catenin.
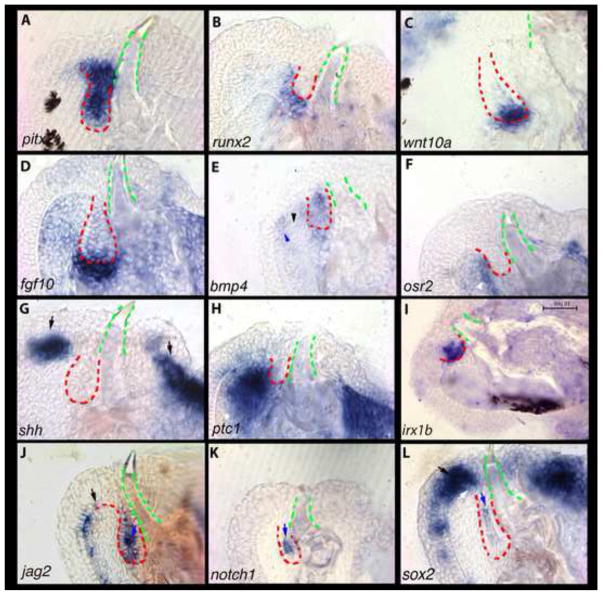
bmp2 and bmp4 are expressed in both the epithelium and mesenchyme as the developing tooth transitions from cap to bell stages (Figure 3A, B). We observe two centers of expression: one in dental mesenchyme (black arrowhead) and one at the tip of developing teeth (yellow arrowheads). While BMPs are expressed at the oral-most cap and bell stage epithelium, we note a general absence of activity in the lateral and aboral-most epithelium. Interestingly, we find the BMP antagonist, sostdc (ectodin; wise (Laurikkala et al., 2003)), expressed in the epithelium where bmp2 and bmp4 are not (Figure 3C). sostdc expression is strongest in the intermediate cells between the ODE and IDE of the cap to bell-stage replacement tooth, and less strong at the cusp tip or mesenchymal papilla (3C, yellow arrow).
sostdc influences tooth development by integrating BMP, FGF, Hh and Wnt pathways (Ahn et al., 2010; Cho et al., 2011; Kassai et al., 2005; Laurikkala et al., 2003). As cichlid replacement teeth initiate differentiation, fgf3 and fgf10 are expressed in dental mesenchyme, and fgf3 is expressed transiently in the aboral epithelium (Figure 3D, E). FGF signals induce dlx2 expression in the zebrafish pharyngeal dentition (Jackman et al., 2004; Stock et al., 2006). Similarly here, dlx2 is co-expressed with fgf10 in the papilla throughout the bud to bell stage transition (Figure 3F). shh transcripts, on the other hand, initiate expression in the invaginated SL only after the ectomesenchyme condenses, and the bud stage begins (Figure 2G; 3G). shh becomes concentrated briefly from the cap to bell stage at the tooth tip, but later is localized to the epithelium analogous to mammalian cervical loops (Figure 5G). As in the snake (Handrigan and Richman, 2010), the cichlid replacement tooth expresses the receptor ptc1 in both epithelium and mesenchyme of the differentiating tooth, implying that the mode of action of Hh signaling is both autocrine and paracrine (Figure 3H). eda, a ligand in the ectodysplasin pathway is thought to induce Hh activity in hair (Pummila et al., 2007), feathers (Houghton et al., 2005), salivary glands (Häärä et al., 2011), and teeth (Laurikkala et al., 2001). Here, we observe its expression in the epithelium of replacement teeth (Figure 3I). Expression of eda in the epithelium of cichlid replacement dental organs is notable because it is restricted to the mesenchyme during initiation of first generation teeth (Fraser et al., 2008).
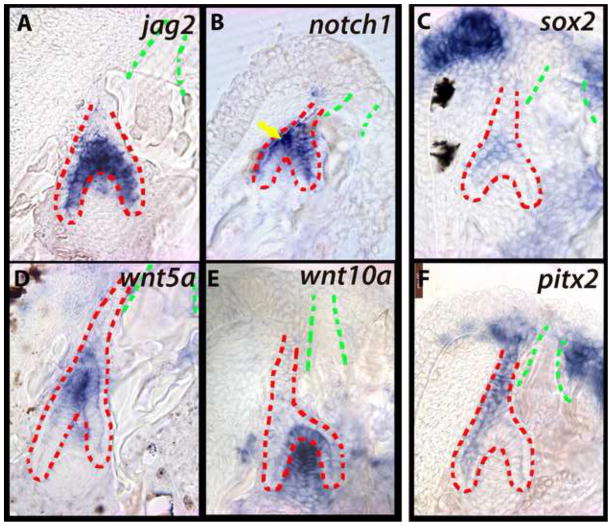
In mouse incisors, FGF signaling from the mesenchyme maintains Notch activity in cervical loop epithelium (Harada et al., 1999) and presumptive stem cells in the stellate reticulum (Harada et al., 2002). Consistent with our observations from the initiation of dental replacement, jag2 and notch1 are expressed in localized cells of the epithelium and at the tooth tip (Figure 4A, B, yellow arrow in B). Because we observed FGF signal in dental mesenchyme and Notch activity in the epithelium, we evaluated whether the stem cell marker sox2 was expressed at this stage. sox2 expression is maintained in the epithelium labial to the replacement tooth organ and is also observed in discrete epithelial cells favoring the labial side of the tooth (Figure 4C).
Figure 4. Expression of genes from the Notch (A–C) and Wnt pathways (D–F) during the differentiation stage of cichlid replacement tooth development.
First generation teeth are outlined in green and replacement dental epithelium in red. Genes expressed in the intermediate cell layer include notch1 (B; yellow arrow) and sox2 (C) at differentiation stage. These are vibratome sections in sagittal plane at 15μm thickness, imaged at 63x magnification. Labial is oriented to the left and oral toward the top of the page. Fishes used in this panel are ~15–30dpf. (A, E, F) - Metriaclima zebra, (B) - Mchenga conophoros, (C) - Petrotilapia chitimba, (D) - Labeotropheus fuelleborni.
The Wnt/β-catenin pathway is similarly active in differentiating replacement teeth. We observe lef1 and β-catenin expression throughout the dental epithelium during the bud to cap transition (Figure S2A, B). wnt5a (Figure 4D) exhibits a local focus of expression at presumptive tooth tips; in the mouse, this gene is active in dental epithelium (including enamel knots) as well as in mesenchyme (Cai et al., 2011). wnt10a continues to be expressed in dental mesenchyme (Figure 4E). In the mouse dentition, Pitx2 and β-catenin directly interact to regulate Lef1 (Vadlamudi et al., 2005). Here, pitx2 marks a distinct set of labial epithelial cells that connect the oral epithelium to the replacement tooth (Figure 4F). Overall, the molecular events that choreograph the progression from dental bud to bell stage tooth development are highly conserved between single generation mouse teeth and the continuously replacing cichlid dentition. Conservation of the genetic toolkit for individual tooth differentiation is particularly notable in this context of continuous one-for-one dental replacement.
Gene expression domains sharpen during secretion stage
By secretion stage, the replacement organ has begun to deposit hard tissues and nears eruption (Figure 1G–I). We observe the same set of pathways active in replacement teeth at this stage, although expression domains for certain molecules have shifted with tooth maturation. bmp4 and bmp2 are expressed in the replacement organ at the tooth tip and the dental papilla (Figure 5A, B) but are nearly absent from the putative cervical loops; sostdc is expressed in these cervical loops as well as a region far oral to the bmp4-positive tooth tip (Figure 5C). This complimentary pattern of expression between signal and antagonist is also observed for members of the FGF pathway. fgf3 is expressed in the epithelium, including the tooth tip, and the dental papilla (Figure 5D), but is absent from the aboral-most cervical loops; fgf10 is strongly expressed in the papilla (Figure 5E); the receptor fgfr2, which transduces FGF signal in teeth (Parsa et al., 2010), is active throughout (Figure S3A). The FGF inhibitor spry4 (Boran et al., 2009; Charles et al., 2011) is concentrated along the cervical loop epithelium, also expressed in the dental papilla (Figure S3B).
Ligands and receptors of the Hh pathway are dynamically expressed through the sequence of replacement tooth development. shh is initially absent from the invaginating SL (Figure 2G), then is active throughout the bud to bell stage epithelium (Figure 3). At mature stages of replacement tooth morphogenesis, shh is strongly localized to the basal cervical loops (Figure 5G). The receptors ptc1 (epithelium and mesenchyme, Figure 2H, 3H) and ptc2 (mesenchyme only), as well as the Hh activator eda, are likewise confined to cervical loop epithelium and contact mesenchyme (Figure 5H, I; Figure S3C). These aboral domains of activity for the Hh pathway are similar to observations from mouse incisors, where Hh is required for the differentiation of ameloblasts from dental stem cells within the stellate reticulum (Parsa et al., 2010). In contrast to the changing patterns of activity exhibited by Hh, molecules in the Notch pathway are consistently expressed across the stages of replacement tooth development, localized to the intermediate stellate reticulum-like cells between IDE and ODE, as well as at the tooth tips (Figure 6A, B).
Figure 6. Expression of genes from the Notch (A–C) and Wnt pathways (D–F) during the secretion stage of cichlid replacement tooth development.
Replacement outer dental epithelium is outlined in red. These are vibratome sections in sagittal plane at 15μm thickness, imaged at 63x magnification. Labial is oriented to the left and oral toward the top of the page. Fishes used in this panel are ~15–30dpf. (A, F) - Metriaclima zebra, (B) - Cynotilapia afra, (C) - Petrotilapia chitimba, (D, E) - Labeotropheus fuelleborni.
We note an intriguing and strong bias in the expression of follistatin (fst) during the final stage of replacement tooth development. Fst is expressed with a lingual bias in mouse incisors where, by antagonizing BMP activity, it contributes to the reduction in enamel-secreting ameloblasts on the lingual surface (Wang et al., 2004). Cichlids, however, express fst with a labial bias overlapping Notch-expressing intermediate cells (Figure 6C). Such biases might contribute to asymmetries in cell proliferation and differentiation to give cichlid teeth their characteristic slight lingual curvature (Figure 1), but the molecular mechanism would then be distinct from that explaining the same curve of mouse incisors.
wnt5a and wnt10a continue to be expressed at the tooth tip (Figure 6D) and the dental papilla (Figure 6E), as in earlier stages and pitx2 maintains expression linking the dental epithelium of the replacement organ to the oral epithelium (Figure 6F). lef1 expression spans the epithelium and mesenchyme of the replacement tooth unit (Figure S3D). Notably, axin2, an effector of Wnt/β-catenin signaling, is active in dental mesenchyme, as well as dental epithelium including the cervical loops and the intermediate cells between IDE and ODE (Figure S3E). This is in contrast to mouse incisors, where Axin2 is only weakly expressed in cervical loop epithelium and is absent from the stellate reticulum (Suomalainen and Thesleff, 2010). Irx family members irx1b (not shown) and irx2 also localize to the cervical loop domains during late replacement tooth morphogenesis (Figure S3F).
The data presented here in conjunction with previous reports of cichlid tooth initiation (Fraser et al., 2008; Fraser et al., 2009) demonstrate that five signaling pathways (BMP, FGF, Hh, Notch, Wnt/β-catenin) are sequentially active in specific cell populations (the SL as an extension of oral epithelium, the tooth tip, the dental papilla, and intermediate cells of the cervical loops) during the process of replacement tooth development. It is likely that the signaling pathways we highlight (i) set the precise dental pattern (the size and spacing of teeth, (Fraser et al., 2008), (ii) requisition one-for-one replacement, which maintains that pattern, and (iii) build the tooth organ for every new generation. The spatial domains of pathway gene co- and complementary expression are thus presumed to confine odontogenesis to precise locations and to accurately reset the process of tooth development for regeneration.
Treatment with small molecules affects replacement and shape
Cichlids replace (shaped) teeth in one-for-one fashion, using a set of common signaling pathways throughout (Figures 2–6). The spatial proximity of patterned tooth families and the temporal continuity of morphogenesis plus regeneration suggest to us that these processes are deeply integrated. One prediction, then, is that manipulation of key pathways should result in altered replacement and shape phenotypes. To test this prediction, we exposed cichlid individuals to temporally precise, non-lethal doses of small molecules known to agonize or antagonize the BMP, FGF, Hh, Notch and Wnt/β-catenin pathways.
A typical treatment experiment involved (i) culture of replicate cichlid juveniles at the appropriate stage (e.g., 40–100dpf) in fish water with the small molecule or delivery control (1% DMSO) for 24 hours, (ii) a 14-day recovery period under standard conditions and (iii) finally, sacrifice for phenotypic analysis (see Materials and Methods). There are two major advantages of this approach in the cichlid system. First, treatment over such a brief temporal window will affect only those teeth at sensitive stages of development; there are thus ‘control’ individuals that did not receive small molecule baths, as well as ‘control’ teeth in the jaws of experimental animals. Second, non-lethal in vivo treatment, subsequent growth in fish water and post-sacrifice clearing and staining with Alizarin red allows us to examine the presence or absence of replacement teeth in the jaw’s bony crypt, as well as additional effects of treatment (i.e., jaw hypertrophy) that might influence tooth development. A general caveat holds for all of these experiments; effects differ depending upon the duration and concentration of the dose applied.
Manipulation of two of the five signaling pathways did not affect cichlid tooth replacement. Treatment with the Hh antagonist cyclopamine strongly disrupts the patterning of the cichlid dentition when administered during the initiation of first generation teeth (Fraser et al., 2008) but does not interfere with dental replacement in snakes and lizards (Handrigan and Richman, 2010). Our results are similar here upon treatment at tooth replacement stages; cyclopamine (25μm) affects the shaping and morphogenesis of teeth (below), but does not abrogate the replacement process (Table 1). This is consistent with our gene expression data (Figures 2, 3, 5) wherein Hh molecules are not active in the replacement tooth until differentiation begins. Treatment with the FGF antagonist SU5402 does not interfere with the production of replacement teeth per se (we observe replacement tooth development deep to the predecessor), but seems to interfere in some animals with the process of functional tooth shedding, thus an indirect effect on replacement (Table 1). In some tooth positions in treated animals, we noted that functional teeth were elongated and that bony crypt morphology was disrupted, perhaps the result of jaw hypertrophy (data not shown).
Table 1.
Effective non-lethal doses of small molecules are arranged in rows. 1A: Numbers of affected and treated cichlid broods and species used; MZ, Metriaclima zebra; LF, Labeotropheus fuelleborni; PL, Pseudotropheus lombardoi; PT, Petrotilapia tridentiger. 1B: Phenotypes are recorded with respect to numbers of teeth; mean and standard deviations (SD) of both affected and total teeth are presented. Phenotypes are reported as having an effect on shape (S) and/or replacement (R). Higher concentrations elicited multiple phenotypes in a dose dependent manner. NB, unaffected individuals developed complete dentitions with no patterning defects despite being exposed to the same chemicals at the same time as their affected siblings. A single animal may have more than one phenotype.
| Chemical | Concentration | Affect/Treat – Brood | Affect/Treat - Individual | Species | Mean tooth No. Affected/individual | Mean Tooth No. Total/individual | %Affected Phenotype |
|---|---|---|---|---|---|---|---|
| DAPT | 100um | 5/6 | 6/11 | LF; MZ; PT | 2.5 (SD 1.77) | 10.0 (SD 2.80) | R/S – 35/65% |
| DAPT | 75um | 5/5 | 9/10 | LF; MZ; PT | 4.75 (SD 2.42) | 9.0 (SD 2.49) | R/S – 22/78% |
| DAPT | 50um | 3/3 | 9/9 | LF; MZ | 6.25 (SD 2.05) | 9.67 (SD 1.30) | S – 100% |
| DAPT | 40um | 2/2 | 3/3 | MZ | 4.5 (SD 1.38) | 9.5 (SD 0.84) | S – 100% |
| Dorsomorphin | 1.0mM | 1/1 | 3/3 | PL | 4.0 (SD 2.94) | 10.5 (SD 4.04) | R/S – 21/79% |
| Dorsomorphin | 0.5mM | 6/6 | 12/14 | PL | 4.07 (SD 1.62) | 10.07 (SD 2.12) | R/S – 3/97% |
| Cyclopamine | 25um | 4/5 | 7/9 | LF; MZ; PT | 4.44 (SD 2.04) | 9.28 (SD 2.19) | S – 100% |
| LiCl | 250um | 5/5 | 10/10 | LF; MZ | 3.15 (SD 1.07) | 8.69 (SD 1.97) | R/S – 10/90% |
| LiCl | 500um | 2/2 | 4/4 | LF | 5.0 (SD 0.89) | 8.33 (SD 0.52) | R/S – 37/63% |
| SU5402 | 50um | 2/2 | 5/5 | MZ | 3.5 (SD 1.51) | 8.57 (SD 2.82) | R/S–14/86% |
Small molecules targeting any of three pathways, BMP, Notch and Wnt/β-catenin, affected the process of cichlid tooth replacement. Treatment with the BMP antagonist dorsomorphin (BML-275), at 1mM concentration, results in tooth positions that do not undergo natural replacement on both upper and lower jaws (Figure 7A–C; Table 1). Furthermore, there is no evidence of replacement teeth (at any stage of development) in the underlying bony crypt of affected positions. Dorsomorphin exposure thus has a major effect on the replacement dentition and uniquely (among our treatments) perturbs adjacent tooth positions. We also observed a replacement phenotype after DAPT exposure (100μM), which inhibits the Notch signaling pathway. Treatment with DAPT produces a number of tooth positions that lack replacements (at any developmental stage) across multiple tooth rows (Figure 7D–F). Notably, this manipulation differs from treatment of the BMP pathway in that the disrupted tooth families tend not to be nearest neighbors, and are mirrored across the jaw symphysis. Lastly, treatment with LiCl, an agonist of Wnt/β-catenin signaling, has only modest effects on tooth replacement at low concentration (0.25mM; Table 1), but results in cusp and replacement phenotypes at higher (0.5mM) concentration (Figure 7G–I and below). This regeneration phenotype is intriguing because it is not a complete knockout of the replacement tooth unit. Rather, treatment appears to affect the rate and/or timing of replacement cycles, such that the phasing of tooth replacement in even vs. odd positions and across the symphysis is disrupted, compared to control. Together, these experiments demonstrate that the BMP, Notch and Wnt/β-catenin pathways are necessary for the proper initiation, rate and/or timing of continuous tooth replacement cycles in cichlid fishes. Given the expression of molecules from these pathways at early stages (Figure 2–4), our treatments have likely affected the invagination or potency of the epithelial SL and/or the responsive mesenchyme that facilitates dental replacement. We have yet to conduct molecular analysis of treated morphants; therefore effects from individual small molecules might be the indirect result of interactions between BMP, Notch and Wnt/β-catenin pathways, well known from other systems (Mitsiadis et al., 2010; Mustonen et al., 2002; Plikus et al., 2008).
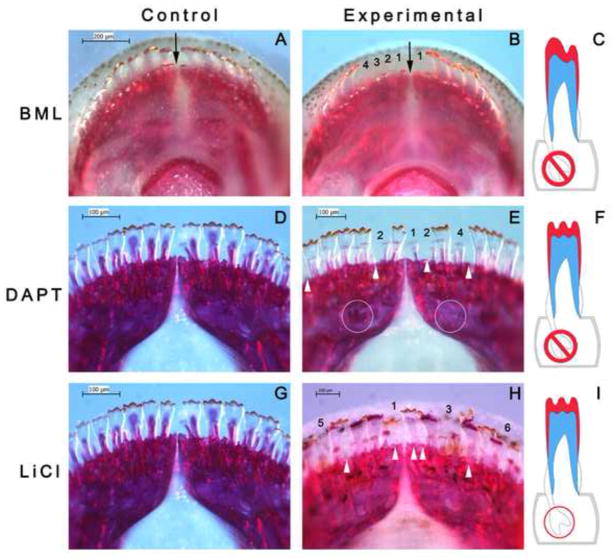
Figure 7. Small molecules targeting the BMP, Notch and Wnt/β-catenin pathways modulate tooth regeneration.
We present dorsal views of Alizarin red stained upper (A–B) and lower (D–E, G–H) oral jaws from a variety of Malawi cichlid species. All individuals received small molecule treatments or vehicle controls for 24 hours, followed by 14 days of recovery in fish water prior to sacrifice and analysis. A–B, Pseudotropheus lombardoi, 100dpf at the start of treatment. D–E and G–H, Labeotropheus fuelleborni, 40dpf at the start of treatment. A, D and G are vehicle controls and show the normal tooth formula. In B, after treatment with the BMP pathway inhibitor BML275 (dorsomorphin, 1mM), teeth from positions 1–4 of the first row, right quadrant and position 1, left quadrant, are not replaced; black arrow indicates the symphysis of the upper jaw. E, after treatment with the Notch pathway inhibitor DAPT (100μM), teeth from positions 1, 2, and 4 of the first row (right quadrant) and tooth positions 2 and 7 (left quadrant) are not replaced. White circles show bony crypt space deep to functional teeth, with a replacement tooth present in the left circle and absent at right. H, after treatment with the Wnt/β-catenin pathway agonist, LiCl (0.5mM), teeth in multiple positions are delayed in eruption (red circle in I) and/or are out of phase in the replacement cycle (white arrowheads). Red circle in I refers to the white arrowhead positions (in H) showing functional positions without teeth; however tooth replacements are present in the crypts – hence a delay to the replacement process rather than a loss of tooth positions. Colors in schematics (C, F, and I): red=enameloid; blue=dentine; grey=bony crypt.
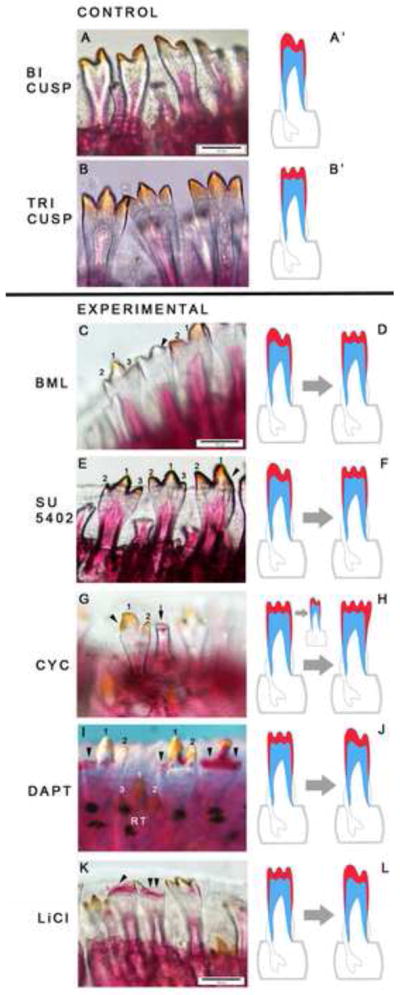
Notably, manipulation of all five signaling pathways for brief durations, and typically lower concentrations, produced tooth shape phenotypes (Figure 8; Table 1). For instance, treatment with 0.5mM dorsomorphin (BMP antagonist) results in transformation of outer row bicuspid to tricuspid teeth in Pseudotropheus lombardoi (Figure 8C–D). Similarly, treatment with 50μM SU5402 (antagonist of FGF signaling) results in triscuspid teeth in bicuspid first-row positions of Metriaclima zebra (Figure 8E–F). Thus, inhibition of both BMP and FGF pathways affects bicuspid teeth in the same way – through the addition of a medial cusp. Treatment with cyclopamine (Hh antagonist) for only 6 hours has a dramatic effect on tooth morphogenesis in Petrotilapia tridentiger (Figure 8G–H), a species with an exclusively tricuspid dentition. Inhibition of Hh signaling interferes with natural cusp formation, resulting in teeth with no cusps, unevenly patterned enameloid, and even an unusual four-cusp phenotype (not shown). The variation in shape phenotypes after cyclopamine treatment matches the dynamic patterns of Hh gene expression during replacement tooth development (Figures 2, 3, 5). Treatment with DAPT, a Notch antagonist, impacts cusp development in Labeotropheus fuelleborni, a species with tricuspid teeth (Figure 8I–J). We observe mineralization defects in both the dentine and enameloid of lateral and central tooth cusps, implying that Notch signaling is essential for correct hard-tissue biogenesis and cusp formation. Similar mineralization defects are observed in fishes treated with the Wnt/β-catenin agonist LiCl (Figure 8K–L). Small-molecule treatment effects on replacement as well as shape (i) are reproducible in replicate individuals and experiments across multiple broods of different species (Table 1) and (ii) are often matched in upper and lower jaws and on each side of the symphysis. Such replacement phenotypes and shape transformations have not been observed in healthy fishes from natural populations (Streelman et al., 2007). Our in vivo manipulations demonstrate an essential role for these five signaling pathways in the proper morphogenesis and shaping of cichlid teeth. Particularly exciting are treatments of the BMP and FGF pathways that transform tooth type from bicuspid to tricuspid, mimicking ecologically relevant differences among closely related species (Fraser et al., 2008; Streelman and Albertson, 2006; Streelman et al., 2003). Taken together, these experiments (Figures 7–8) provide evidence for a model linking tooth morphogenesis to tooth replacement through the function of key signaling pathways.
Figure 8. Small molecules targeting five signaling pathways modulate tooth shape.
Control bicuspid (A-A′; Metriaclima zebra) and tricuspid (B-B′; Labeotropheus fuelleborni) dentitions are shown above the solid black line and small molecule treated dentitions are indicated below. All individuals received small molecule treatments or vehicle controls for 24 hours, followed by 14 days of recovery in fish water prior to sacrifice and analysis. C, BML275 treatment, (dorsomorphin, BMP inhibitor, 0.5mM) results in transformation of bicuspid to tricuspid teeth (Metriaclima zebra). E, similarly, treatment with the FGF inhibitor SU5402 (50μM) transforms teeth from bicuspid to tricuspid (Metriaclima zebra). In each of these cases, ‘control’ bicuspid teeth are present next to those sensitive to the temporal window of small molecule application. G, treatment with cyclopamine (Hh antagonist, 25μM) elicits numerous effects on shape, primarily through variation in enameloid patterning (arrows and arrowheads; Petrotilapia tridentiger). I, inhibition of the Notch pathway with DAPT (25μM) affects cusp development and mineralization (arrowheads; Labeotropheus fuelleborni). K, treatment with the Wnt/β-catenin agonist LiCl (0.25mM and 0.5mM) results in mineralization defects with increasing dose (Labeotropheus fuelleborni). Colors in schematics (D, F, H, J, L): red=enameloid; blue=dentine; grey=bony crypt. Mineralization defects are inferred in treated individuals when the dentine (alizarin red stained) and/or the enameloid (yellow-orange color from Fe deposition) are abnormal, compared to controls.
Discussion
The cichlid dentition integrates tooth replacement and shape
The ‘homeobox code’ for the mammalian dentition posits that tooth shape is the product of linear position along the jaw margin (Sharpe, 1995; Tucker and Sharpe, 2004). Mouse teeth represent the extreme condition of this general model, wherein only incisors and molars develop under distinct gradients of BMP-Msx and FGF-Barx1, respectively. In the mouse (and more generally the mammalian) dentition, molars undergo complex morphogenesis and develop cusps, which are absent and perhaps suppressed (Ohazama et al., 2010) from incisors. Mouse incisors, by contrast, exhibit enamel renewal via a labial stem cell niche. Thus for the mouse model, the phenomena of (i) complex cusp development and (ii) stem-based (self-)renewal are decoupled in space and in time (Figure 1). Many other vertebrate dentitions do not follow such binary rules. For instance, cichlid teeth are shaped through rounds of replacement (all first generation teeth are conical) such that tricuspid teeth may replace unicuspid teeth in the same jaw position (Streelman et al., 2003). Once adult tooth shapes are present, tooth replacement continues through ontogeny, maintaining shape and pattern fidelity. Tooth shape therefore is not correlated with position within a row and teeth with complex shapes undergo regeneration. The key finding from this study is that common signaling pathways are active and essential during the coupled phenomena of replacement and morphogenesis of cichlid dentitions, forcing us to think differently about the integration of these processes in odontogenesis (Jernvall and Thesleff, 2012). Our data inspire a model implicating specific signaling pathways (BMP, FGF, Hh, Notch, Wnt/β-catenin) in the process of cichlid tooth regeneration, explicitly coupled to tooth morphogenesis and shape. Genes from these families are sequentially co- and complimentarily expressed in spatial domains throughout tooth development and replacement. Our model posits a mechanistic connection between replacement and shape, as these signaling pathways likely mediate proliferation and differentiation at both the tooth tips and in presumed stem cell populations for renewal (Figure 9).
Figure 9. Cichlid teeth integrate tooth shape and lifelong regeneration, a linkage lost in mammals.
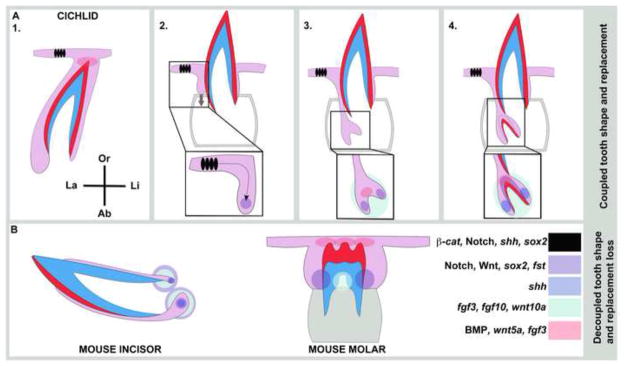
Cichlid dental organs simultaneously coordinate shape morphogenesis and regeneration developmental programs, in the same tooth position (also Figure 1 schematic). This is achieved via tight control of gene co-expression in zones of differentiation and zones of renewal. We identify cell populations and putative signaling centers that may regulate cichlid tooth shape and regeneration (A1–4), including (i) a population of epithelial cells (black) labial to the predecessor tooth superficial to the sucessional lamina of the replacement organ (grey arrow), (ii) cells of the intermediate layer between IDE and ODE (purple), (iii) cervical loop regions (blue), (iv) dental papilla (green) and (v) putative enameloid knots at the tips of teeth (pink). These cellular domains and putative signaling centers are color-coded based on empirical measure of gene co-expression (Figures 2–7). In B, we show comparable gene activity in mouse incisors (Felszeghy et al., 2010; Harada et al., 1999; Harada et al., 2002; Jernvall and Thesleff, 2012; Wang et al., 2007; Wang et al., 2004) capable of self-renewal and mouse molars (Jernvall et al., 2000; Jernvall et al., 1994; Kettunen et al., 2000; Zhang et al., 2012) that develop complex 3D shapes. Throughout, tooth tissue colors are as in Figure 1.
Cichlid teeth retain the capacity for lifelong de novo renewal. Our data suggest that at least two cellular domains may be important for tooth replacement. The first is similar in location to that housing putative stem-like cells in the zebrafish (Handrigan et al., 2010; Huysseune, 2006; Huysseune and Thesleff, 2004). These cells are found in an intermediate layer between IDE and ODE that may be analogous to the stellate reticulum of mouse incisor cervical loops. These intermediate cells in cichlids co-express a common set of markers throughout replacement tooth development: sostdc, an inhibitor of BMP expression; Notch pathway ligands and receptors; β-catenin, and the stem transcription factor sox2. Molecules active at discrete, later stages of tooth replacement in the intermediate cells include spry4, an inhibitor of FGF signaling, shh, fst and axin2. A second population of cells that may contribute stem potential to tooth replacement in cichlids is the labial oral epithelium, superficial to each invaginating SL. This epithelium co-expresses shh, Notch pathway ligands and receptors, β-catenin and sox2 throughout the stages of replacement tooth development; as noted, this labial epithelial domain is matched with mesenchymal expression of ptc1 and bmp4. In reptiles, stem-like cells are located superficially, along the the non-tooth forming regions of the dental lamina (Handrigan et al., 2010). It is notable that the pathways (BMP, Notch and Wnt/β-catenin) active throughout cichlid tooth development in both of these domains are those where small molecule manipulation elicit the strongest tooth replacement phenotypes (Figure 7, Table 1). Our immunohistochemical, gene expression and small molecule experiments do not definitively prove that either of these cell populations contains dental stem cells. Yet, based on gene co-expression signatures and the anatomical position of these cellular domains with respect to regenerating dental organs in other species, it is tempting to speculate that each of these populations contributes to and/or regulates the stem niche for cichlid tooth replacement (Figure 9).
Cichlid teeth are shaped as they are replaced. First generation teeth are conical; generally, the first shaped replacement teeth have sharper cusps and more rapid replacement cycles than those that follow (Streelman et al., 2003). Once adult tooth shape is reached, teeth continue to be replaced with shape fidelity, roughly every 30–100 days. This means that the molecular signals that determine tooth shape do so later in the life of an individual cichlid than say, in the life of an individual mouse, whose first and only set of molars are shaped during embryogenesis. Two aspects of tooth shape are relevant given the diversity among cichlid species and the data we report here. The first is the degree of lingual curvature of the tooth, taken to the extreme in some algal brushing species that exhibit a near 90° angle between the long and flexible tooth ‘stalk’ and the multicuspid ‘brush’ at the tip (Fryer and Illes, 1972). We note from our histological data that cichlid replacement teeth begin as a downward extension of the SL on the labial side of the functional tooth, and that a labial bias in cell proliferation persists into hard tissue secreting stages (Figure 1). Both the labial and lingual surfaces of cichlid teeth are covered with enameloid, but a slight bias in the production and proliferation of ameloblasts on the labial side might be facilitated by slight differences in molecular signaling in the labial vs. lingual cervical loops. BMPs and FGFs seem to be largely absent from both cervical loop locations while the Hh, Notch and Wnt/β-catenin pathways are active. We observe a striking labial bias in the expression of fst that might contribute to different rates of ameloblast production and/or proliferation on the labial surface; this is a prime focus of future research because the bias is opposite that observed in mouse incisors (Wang et al., 2007).
The second relevant aspect of tooth shape is the number of cusps on each tooth, which in cichlids ranges from one to three with dramatic variation in the relative size and pattering of individual cusps (Fryer and Illes, 1972; Streelman et al., 2003). Our data from ISH and small molecule treatments illustrate that (i) genes from all five pathways studied are active in putative signaling centers associated with cusp morphogenesis and (ii) manipulation of these pathways, individually, is sufficient to modulate shape. Strikingly, we observe a suite of molecules (bmp2/4, fgf3, shh, Notch ligands and receptors, wnt5a) active at the tooth tip in expression foci with similarity to mammal enamel knots (Jernvall et al., 1994). We suggest then that fishes (and perhaps all vertebrates with complex cusp shapes) possess primitive enamel knot-like signaling centers that function to control cusp number, sharpness and size.
One-for-one replacement of a complex dentition requires simultaneous activation of molecular programs for morphogenesis and regeneration within the same tooth. Our analysis has focused on specific cell populations and putative signaling centers that co- and sequentially express stem and dental markers because it is likely that the spatio-temporal complementarity of gene activity is what allows a dental organ to tune proliferation, growth and differentiation all at once. In this sense, the coordination of these processes may depend as much on excluding molecular signals from a specific domain at a specific time as it does on the integration of signaling. Our data may be particularly useful in understanding how this segregation of gene expression is regulated in space and time. For instance, shh expression is absent from the initial downgrowth of the SL; this observation is consistent with data from other bony fishes and reptiles (Fraser et al., 2006; Handrigan et al., 2010). We observe the expression of irx1b, a known mediator of Wnt signaling, antagonist of shh in the embryonic forebrain (Houweling et al., 2001; Scholpp et al., 2007; Scholpp et al., 2006; Sylvester et al., 2010), and antagonist of Bmp4 at gastrulation (Gomez-Skarmeta et al., 2001), in the aboral-most epithelium of the SL invagination. The irx1/2 genes are later active in the cervical loop regions, this time co-expressed with Hh molecules and complementarily expressed with bmp2/4. Irx molecules have been noted in mouse teeth, but function is unknown (Ferguson et al., 2001). Our data suggest that these transcription factors may couple signals from the BMP, Hh and Wnt pathways and may be important negative regulators of Hh in the early SL. Similarly, osr2 is expressed with a complementary pattern to bmp4 at the initiation of primary replacement and may facilitate delineation of odontogenic cell populations across the jaw. At later stages of replacement tooth development, genes from the BMP and FGF pathway are invariably confined to activity in the dental papilla and the putative enameloid-knot signaling centers at the tooth tip, and in particular are largely absent from the cervical loops and intermediate cells between IDE and ODE. We observe antagonists in each of these pathways (sostdc, spry4) expressed precisely in those cells where BMPs and FGFs are absent. Taken together, our data suggest that cichlid dental organs integrate shape and replacement by sometimes combining and other times segregating differentiation signals (BMPs, FGFs, Hh) from renewal and regeneration signals (Notch, Wnt, sox2), with temporal and spatial precision. It is likely that these interactions, necessary to pattern regenerating dentitions with complex shapes, prefigure the molecular programs found within multicuspid molars and self-renewing incisors (Figure 9) (Jernvall and Thesleff, 2012). We speculate that the difference between organisms with lifelong regeneration of complex dentitions (i.e., cichlids, reptiles) and those without (e.g., mammals) lies in the continued maintenance and repeated activation of stem-like cells in positions superficial to successional lamina (Handrigan et al., 2010).
Stem cells and programmed evolvability of patterned elements
The majority of patterned dentitions in the long evolutionary history of vertebrates have been capable of continuous replacement (Huysseune and Thesleff, 2004), and yet we do not understand for any dentition how the processes of patterning, morphogenesis and regeneration are integrated in space and time. From first principles, we see that the replacement of dentitions de novo in a one-for-one fashion, while maintaining shape fidelity of individual units and inter-unit pattern, requires (i) signals of dental competence to specify tooth vs. non-tooth, (ii) a morphogenesis program that can be recruited again and again within a tooth family, (iii) signals of renewal that can provide cell potency, and (iv) a clock mechanism to coordinate timing. Our study addresses the first three of these a priori requirements. Cichlid teeth carry out largely conserved morphogenesis programs coupled to regeneration via the co- and complementary expression of key signaling molecules (Figure 9). Notable among these signals of cell potency is the transcription factor sox2 that, among other functions, acts to maintain the undifferentiated stem state in embryonic and adult stem cells (Avilion et al., 2003) and specifically marks stem cells of the mouse incisor (Juuri et al., 2012). We observe sox2 expression in two cellular domains that may mark the location of stem-like for cichlid tooth replacement. One of these domains shares anatomical features with dental stellate reticulum-like cells in other vertebrates. The second domain, labial to tooth rows, is particularly interesting because it may shuttle cells to the developing tooth unit (Figure 9). A recent report of SOX2 anophthalmia syndrome in humans documented multiple dental phenotypes including supernumerary impacted teeth and the persistence of deciduous teeth (Numakura et al., 2010). It is likely then that sox2/Sox2/SOX2 plays (and has played) a central role in tooth replacement across vertebrates.
It is not clear what factors contributed to evolutionary modifications of the dentition in mammals, including the reduction in tooth number, tooth rows and lifelong replacement cycles. What is clear, however, is that this latter contingency has constrained the plasticity of mammalian teeth over an individual’s ontogeny (particularly so for molars) and has limited the developmental window available for evolutionary tinkering over a lineage’s phylogeny. The recently noted ‘difficulty of increasing dental complexity’ in mammals (Harjunmaa et al., 2012) may be a direct result of this constraint. By contrast, we suggest that the phenotypic plasticity and dramatic shape diversity observed in cichlid teeth is facilitated by the potential for evolvability afforded by lifelong replacement. It is particularly this feature of cichlid teeth, the simultaneous integration of morphogenesis and regeneration programs, which might galvanize bio-inspired advancement in the field of regenerative dentistry.
Supplementary Material
Cichlid fishes replace teeth in one for one fashion throughout life.
Replacement teeth recruit BMP, FGF, Hh, Notch and Wnt pathways to putative signaling centers.
Manipulation of signaling pathways affects both tooth shape and tooth renewal.
We present a model of integrated renewal and morphogenesis, via common pathways.
This linkage has been lost or decoupled in mammalian dentitions.
Acknowledgments
We thank members of the Streelman lab and two anonymous reviewers for critical comments on previous versions of this manuscript; Anthony Graham for the gift of SU5402; and the US National Institutes of Health (R01DE019637) for funding.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Åberg T, Wozney J, Thesleff I. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev Dyn. 1997;210:383–396. doi: 10.1002/(SICI)1097-0177(199712)210:4<383::AID-AJA3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Ahn Y, Sanderson BW, Klein OD, Krumlauf R. Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development. 2010;137:3221–3231. doi: 10.1242/dev.054668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae SK, Kittappa R, McKay RD. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. doi: 10.1038/nature04940. [DOI] [PubMed] [Google Scholar]
- Avery JK, Steele PF. Essentials of oral histology and embryology: a clinical approach. C. V. Mosby; St. Louis, MO: 2000. [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bei M, Maas R. FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth development. Development. 1998;125:4325–4333. doi: 10.1242/dev.125.21.4325. [DOI] [PubMed] [Google Scholar]
- Bemis WE, Giuliano A, McGuire B. Structure, attachment, replacement and growth of teeth in bluefish, Pomatomus saltatrix (Linnaeus, 1776), a teleost with deeply socketed teeth. Zoology (Jena) 2005;108:317–327. doi: 10.1016/j.zool.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Boran T, Peterkova R, Lesot H, Lyons DB, Peterka M, Klein OD. Temporal analysis of ectopic enamel production in incisors from sprouty mutant mice. J Exp Zool B Mol Dev Evol. 2009;312B:473–485. doi: 10.1002/jez.b.21254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Mutoh N, Shin JO, Tani-Ishii N, Ohshima H, Cho SW, Jung HS. Wnt5a plays a crucial role in determining tooth size during murine tooth development. Cell and tissue research. 2011;345:367–377. doi: 10.1007/s00441-011-1224-4. [DOI] [PubMed] [Google Scholar]
- Chang C, Wu P, Baker RE, Maini PK, Alibardi L, Chuong CM. Reptile scale paradigm: Evo-Devo, pattern formation and regeneration. Int J Dev Biol. 2009;53:813–826. doi: 10.1387/ijdb.072556cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles C, Hovorakova M, Ahn Y, Lyons DB, Marangoni P, Churava S, Biehs B, Jheon A, Lesot H, Balooch G, Krumlauf R, Viriot L, Peterkova R, Klein OD. Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase signaling. Development. 2011;138:4063–4073. doi: 10.1242/dev.069195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Bei M, Woo I, Satokata I, Maas R. Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development. 1996;122:3035–3044. doi: 10.1242/dev.122.10.3035. [DOI] [PubMed] [Google Scholar]
- Cho SW, Kwak S, Woolley TE, Lee MJ, Kim EJ, Baker RE, Kim HJ, Shin JS, Tickle C, Maini PK, Jung HS. Interactions between Shh, Sostdc1 and Wnt signaling and a new feedback loop for spatial patterning of the teeth. Development. 2011;138:1807–1816. doi: 10.1242/dev.056051. [DOI] [PubMed] [Google Scholar]
- Christen B, Robles V, Raya M, Paramonov I, Izpisua Belmonte JC. Regeneration and reprogramming compared. BMC biology. 2010;8:5. doi: 10.1186/1741-7007-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobourne MT, Miletich I, Sharpe PT. Restriction of sonic hedgehog signalling during early tooth development. Development. 2004;131:2875–2885. doi: 10.1242/dev.01163. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Sharpe PT. Sonic hedgehog signaling and the developing tooth. Curr Top Dev Biol. 2004;65:255–287. doi: 10.1016/S0070-2153(04)65010-1. [DOI] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- Echeverri K, Tanaka EM. Ectoderm to mesoderm lineage switching during axolotl tail regeneration. Science. 2002;298:1993–1996. doi: 10.1126/science.1077804. [DOI] [PubMed] [Google Scholar]
- Felszeghy S, Suomalainen M, Thesleff I. Notch signalling is required for the survival of epithelial stem cells in the continuously growing mouse incisor. Differentiation. 2010;80:241–248. doi: 10.1016/j.diff.2010.06.004. [DOI] [PubMed] [Google Scholar]
- Ferguson CA, Tucker AS, Heikinheimo K, Nomura M, Oh P, Li E, Sharpe PT. The role of effectors of the activin signalling pathway, activin receptors IIA and IIB, and Smad2, in patterning of tooth development. Development. 2001;128:4605–4613. doi: 10.1242/dev.128.22.4605. [DOI] [PubMed] [Google Scholar]
- Fraser GJ, Berkovitz BK, Graham A, Smith MM. Gene deployment for tooth replacement in the rainbow trout (Oncorhynchus mykiss): a developmental model for evolution of the osteichthyan dentition. Evol Dev. 2006;8:446–457. doi: 10.1111/j.1525-142X.2006.00118.x. [DOI] [PubMed] [Google Scholar]
- Fraser GJ, Bloomquist RF, Streelman JT. A periodic pattern generator for dental diversity. BMC biology. 2008;6:32. doi: 10.1186/1741-7007-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GJ, Britz R, Hall A, Johanson Z, Smith MM. Replacing the first-generation dentition in pufferfish with a unique beak. Proc Natl Acad Sci U S A. 2012;109:8179–8184. doi: 10.1073/pnas.1119635109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GJ, Hulsey CD, Bloomquist RF, Uyesugi K, Manley NR, Streelman JT. An ancient gene network is co-opted for teeth on old and new jaws. PLoS biology. 2009;7:e31. doi: 10.1371/journal.pbio.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GJ, Smith MM. Evolution of developmental pattern for vertebrate dentitions: an oro-pharyngeal specific mechanism. J Exp Zool B: MDE. 2011;316B:99–112. doi: 10.1002/jez.b.21387. [DOI] [PubMed] [Google Scholar]
- Fryer G, Illes TD. The cichlid fishes of the Great Lakes of Africa: Their biology and evolution. Oliver and Boyd; Edinburgh: 1972. [Google Scholar]
- Fujimura K, Okada N. Development of the embryo, larva and early juvenile of Nile tilapia Oreochromis niloticus (Pisces: Cichlidae). Developmental staging system. Development, growth & differentiation. 2007;49:301–324. doi: 10.1111/j.1440-169X.2007.00926.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta J, de La Calle-Mustienes E, Modolell J. The Wnt-activated Xiro1 gene encodes a repressor that is essential for neural development and downregulates Bmp4. Development. 2001;128:551–560. doi: 10.1242/dev.128.4.551. [DOI] [PubMed] [Google Scholar]
- Häärä O, Fujimori S, Schmidt-Ullrich R, Hartmann C, Thesleff I, Mikkola ML. Ectodysplasin and Wnt pathways are required for salivary gland branching morphogenesis. Development. 2011;138:2681–2691. doi: 10.1242/dev.057711. [DOI] [PubMed] [Google Scholar]
- Handrigan GR, Leung KJ, Richman JM. Identification of putative dental epithelial stem cells in a lizard with life-long tooth replacement. Development. 2010;137:3545–3549. doi: 10.1242/dev.052415. [DOI] [PubMed] [Google Scholar]
- Handrigan GR, Richman JM. Autocrine and paracrine Shh signaling are necessary for tooth morphogenesis, but not tooth replacement in snakes and lizards (Squamata) Dev Biol. 2010;337:171–186. doi: 10.1016/j.ydbio.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Mitsuyasu T, Toyono T, Toyoshima K. Epithelial stem cells in teeth. Odontology/the Society of the Nippon Dental University. 2002;90:1–6. doi: 10.1007/s102660200000. [DOI] [PubMed] [Google Scholar]
- Harjunmaa E, Kallonen A, Voutilainen M, Hämäläinen K, Mikkola ML, Jernvall J. On the difficulty of increasing dental complexity. Nature. 2012;483:324–327. doi: 10.1038/nature10876. [DOI] [PubMed] [Google Scholar]
- Houghton L, Lindon C, Morgan BA. The ectodysplasin pathway in feather tract development. Development. 2005;132:863–872. doi: 10.1242/dev.01651. [DOI] [PubMed] [Google Scholar]
- Houweling AC, Dildrop R, Peters T, Mummenhoff J, Moorman AF, Rüther U, Christoffels VM. Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech Dev. 2001;107:169–174. doi: 10.1016/s0925-4773(01)00451-8. [DOI] [PubMed] [Google Scholar]
- Huysseune A. Developmental plasticity in the dentition of a heterodont polyphyodont fish species. In: Teaford MF, Smith MM, Ferguson MWJ, editors. Development, Function and evolution of teeth. Cambridge University Press; Cambridge: 2000. pp. 231–241. [Google Scholar]
- Huysseune A. Formation of a successional dental lamina in the zebrafish (Danio rerio): support for a local control of replacement tooth initiation. Int J Dev Biol. 2006;50:637–643. doi: 10.1387/ijdb.052098ah. [DOI] [PubMed] [Google Scholar]
- Huysseune A, Sire JY. Structure and development of first-generation teeth in the cichlid Hemichromis bimaculatus (Teleostei, Cichlidae) Tissue & cell. 1997;29:679–697. doi: 10.1016/s0040-8166(97)80044-4. [DOI] [PubMed] [Google Scholar]
- Huysseune A, Thesleff I. Continuous tooth replacement: the possible involvement of epithelial stem cells. Bioessays. 2004;26:665–671. doi: 10.1002/bies.20039. [DOI] [PubMed] [Google Scholar]
- Jackman WR, Draper BW, Stock DW. Fgf signaling is required for zebrafish tooth development. Dev Biol. 2004;274:139–157. doi: 10.1016/j.ydbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Järvinen E, Tummers M, Thesleff I. The role of the dental lamina in mammalian tooth replacement. J Exp Zoolog B Mol Dev Evol. 2009;312B:281–291. doi: 10.1002/jez.b.21275. [DOI] [PubMed] [Google Scholar]
- Järvinen E, Välimäki K, Pummila M, Thesleff I, Jernvall J. The taming of the shrew milk teeth. Evol Dev. 2008;10:477–486. doi: 10.1111/j.1525-142X.2008.00258.x. [DOI] [PubMed] [Google Scholar]
- JaŸwińska A, Badakov R, Keating MT. Activin-betaA signaling is required for zebrafish fin regeneration. Curr Biol. 2007;17:1390–1395. doi: 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Keranen SV, Thesleff I. Evolutionary modification of development in mammalian teeth: quantifying gene expression patterns and topography. Proc Natl Acad Sci U S A. 2000;97:14444–14448. doi: 10.1073/pnas.97.26.14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int J Dev Biol. 1994;38:463–469. [PubMed] [Google Scholar]
- Jernvall J, Thesleff I. Tooth shape formation and tooth renewal: evolving with the same signals. Development. 2012;139:3487–3497. doi: 10.1242/dev.085084. [DOI] [PubMed] [Google Scholar]
- Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, Klein OD, Thesleff I, Michon F. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev Cell. 2012;23:317–328. doi: 10.1016/j.devcel.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassai Y, Munne P, Hotta Y, Penttilä E, Kavanagh K, Ohbayashi N, Takada S, Thesleff I, Jernvall J, Itoh N. Regulation of mammalian tooth cusp patterning by ectodin. Science. 2005;309:2067–2070. doi: 10.1126/science.1116848. [DOI] [PubMed] [Google Scholar]
- Kerr T. Development and structure of some actinopterygian and urodele teeth. Proc Zool Soc London. 1960;133:401–423. [Google Scholar]
- Kettunen P, Laurikkala J, Itäranta P, Vainio S, Itoh N, Thesleff I. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev Dyn. 2000;219:322–332. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1062>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Kizil C, Kaslin J, Kroehne V, Brand M. Adult neurogenesis and brain regeneration in zebrafish. Dev Neurobiol. 2011;72:429–461. doi: 10.1002/dneu.20918. [DOI] [PubMed] [Google Scholar]
- Kragl M, Knapp D, Nacu E, Khattak S, Maden M, Epperlein HH, Tanaka EM. Cells keep a memory of their tissue origin during axolotl limb regeneration. Nature. 2009;460:60–65. doi: 10.1038/nature08152. [DOI] [PubMed] [Google Scholar]
- Kroehne V, Freudenreich D, Hans S, Kaslin J, Brand M. Regeneration of the adult zebrafish brain from neurogenic radial glia-type progenitors. Development. 2011;138:4831–4841. doi: 10.1242/dev.072587. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Kassai Y, Pakkasjärvi L, Thesleff I, Itoh N. Identification of a secreted BMP antagonist, ectodin, integrating BMP, FGF, and SHH signals from the tooth enamel knot. Dev Biol. 2003;264:91–105. doi: 10.1016/j.ydbio.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Laurikkala J, Mikkola M, Mustonen T, Åberg T, Koppinen P, Pispa J, Nieminen P, Galceran J, Grosschedl R, Thesleff I. TNF signaling via the ligand-receptor pair ectodysplasin and edar controls the function of epithelial signaling centers and is regulated by Wnt and activin during tooth organogenesis. Dev Biol. 2001;229:443–455. doi: 10.1006/dbio.2000.9955. [DOI] [PubMed] [Google Scholar]
- Lin G, Slack JM. Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol. 2008;316:323–335. doi: 10.1016/j.ydbio.2008.01.032. [DOI] [PubMed] [Google Scholar]
- Loh YHE, Katz LS, Mims MC, Kocher TD, Yi S, Streelman JT. Comparative analysis reveals signatures of differentiation amid genomic polymorphism in Lake Malawi cichlids. Genome Biology. 2008;9 (7):R113. doi: 10.1186/gb-2008-9-7-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiadis TA, Graf D, Luder H, Gridley T, Bluteau G. BMPs and FGFs target Notch signalling via jagged 2 to regulate tooth morphogenesis and cytodifferentiation. Development. 2010;137:3025–3035. doi: 10.1242/dev.049528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiadis TA, Hirsinger E, Lendahl U, Goridis C. Delta-notch signaling in odontogenesis: correlation with cytodifferentiation and evidence for feedback regulation. Dev Biol. 1998;204:420–431. doi: 10.1006/dbio.1998.9092. [DOI] [PubMed] [Google Scholar]
- Mitsiadis TA, Regaudiat L, Gridley T. Role of the Notch signalling pathway in tooth morphogenesis. Arch Oral Biol. 2005;50:137–140. doi: 10.1016/j.archoralbio.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Motta PJ. Tooth attachment, replacement, and growth in the butterflyfish, (Chaetodon miliaris) Chaetodontidae, Perciformes. Can J Zool. 1984;62:183–189. [Google Scholar]
- Mustonen T, Tümmers M, Mikami T, Itoh N, Zhang N, Gridley T, Thesleff I. Lunatic fringe, FGF, and BMP regulate the Notch pathway during epithelial morphogenesis of teeth. Dev Biol. 2002;248:281–293. doi: 10.1006/dbio.2002.0734. [DOI] [PubMed] [Google Scholar]
- Nacu E, Tanaka EM. Limb regeneration: a new development? Annu Rev Cell Dev Biol. 2011;27:409–440. doi: 10.1146/annurev-cellbio-092910-154115. [DOI] [PubMed] [Google Scholar]
- Numakura C, Kitanaka S, Kato M, Ishikawa S, Hamamoto Y, Katsushima Y, Kimura T, Hayasaka K. Supernumerary impacted teeth in a patient with SOX2 anophthalmia syndrome. Am J Med Genet A. 2010;152A:2355–2359. doi: 10.1002/ajmg.a.33556. [DOI] [PubMed] [Google Scholar]
- Ohazama A, Blackburn J, Porntaveetus T, Ota MS, Choi HY, Johnson EB, Myers P, Oommen S, Eto K, Kessler JA, Kondo T, Fraser GJ, Streelman JT, Pardinas UF, Tucker AS, Ortiz PE, Charles C, Viriot L, Herz J, Sharpe PT. A role for suppressed incisor cuspal morphogenesis in the evolution of mammalian heterodont dentition. Proc Natl Acad Sci U S A. 2010;107:92–97. doi: 10.1073/pnas.0907236107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsa S, Kuremoto K, Seidel K, Tabatabai R, Mackenzie B, Yamaza T, Akiyama K, Branch J, Koh CJ, Al Alam D, Klein OD, Bellusci S. Signaling by FGFR2b controls the regenerative capacity of adult mouse incisors. Development. 2010;137:3743–3752. doi: 10.1242/dev.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Baker RE, Chen CC, Fare C, de la Cruz D, Andl T, Maini PK, Millar SE, Widelitz R, Chuong CM. Self-organizing and stochastic behaviors during the regeneration of hair stem cells. Science. 2011;332:586–589. doi: 10.1126/science.1201647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, de la Cruz D, Baker RE, Maini PK, Maxson R, Chuong CM. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature. 2008;451:340–344. doi: 10.1038/nature06457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pummila M, Fliniaux I, Jaatinen R, James MJ, Laurikkala J, Schneider P, Thesleff I, Mikkola ML. Ectodysplasin has a dual role in ectodermal organogenesis: inhibition of Bmp activity and induction of Shh expression. Development. 2007;134:117–125. doi: 10.1242/dev.02708. [DOI] [PubMed] [Google Scholar]
- Sarkar L, Sharpe PT. Expression of Wnt signalling pathway genes during tooth development. Mech Dev. 1999;85:197–200. doi: 10.1016/s0925-4773(99)00095-7. [DOI] [PubMed] [Google Scholar]
- Scholpp S, Foucher I, Staudt N, Peukert D, Lumsden A, Houart C. Otx1l, Otx2 and Irx1b establish and position the ZLI in the diencephalon. Development. 2007;134:3167–3176. doi: 10.1242/dev.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholpp S, Wolf O, Brand M, Lumsden A. Hedgehog signalling from the zona limitans intrathalamica orchestrates patterning of the zebrafish diencephalon. Development. 2006;133:855–864. doi: 10.1242/dev.02248. [DOI] [PubMed] [Google Scholar]
- Sharpe PT. Homeobox genes and orofacial development. Connect Tissue Res. 1995;32:17–25. doi: 10.3109/03008209509013701. [DOI] [PubMed] [Google Scholar]
- Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev Cell. 2012;22:879–886. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sire JY, Davit-Beal T, Delgado S, Van Der Heyden C, Huysseune A. First-generation teeth in nonmammalian lineages: evidence for a conserved ancestral character? Microscopy research and technique. 2002;59:408–434. doi: 10.1002/jemt.10220. [DOI] [PubMed] [Google Scholar]
- Smith MM. Vertebrate dentitions at the origin of jaws: when and how pattern evolved. Evol Dev. 2003;5:394–413. doi: 10.1046/j.1525-142x.2003.03047.x. [DOI] [PubMed] [Google Scholar]
- Smith MM, Coates MI. Evolutionary origins of the vertebrate dentition: phylogenetic patterns and developmental evolution. Eur J Oral Sci. 1998;106(Suppl 1):482–500. doi: 10.1111/j.1600-0722.1998.tb02212.x. [DOI] [PubMed] [Google Scholar]
- Smith MM, Coates MI. Evolutionary origins of teeth and jaws: developmental models and phylogenetic patterns. In: Teaford MF, Smith MM, Ferguson MWJ, editors. Development, Function and Evolution of Teeth. Cambridge University Press; Cambridge: 2000. pp. 133–151. [Google Scholar]
- Smith MM, Fraser GJ, Chaplin N, Hobbs C, Graham A. Reiterative pattern of sonic hedgehog expression in the catshark dentition reveals a phylogenetic template for jawed vertebrates. Proc Biol Sci. 2009;276:1225–1233. doi: 10.1098/rspb.2008.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock DW, Jackman WR, Trapani J. Developmental genetic mechanisms of evolutionary tooth loss in cypriniform fishes. Development. 2006;133:3127–3137. doi: 10.1242/dev.02459. [DOI] [PubMed] [Google Scholar]
- Streelman JT, Albertson R, Kocher T. Variation in body size and trophic morphology within and among genetically differentiated populations of the cichlid fish, Metriaclima zebra, from Lake Malawi. Freshwater Biology. 2007;52:525–538. [Google Scholar]
- Streelman JT, Albertson RC. Evolution of novelty in the cichlid dentition. J Exp Zool B: Mol Dev Evol. 2006;306:216–226. doi: 10.1002/jez.b.21101. [DOI] [PubMed] [Google Scholar]
- Streelman JT, Webb JF, Albertson RC, Kocher TD. The cusp of evolution and development: a model of cichlid tooth shape diversity. Evol Dev. 2003;5:600–608. doi: 10.1046/j.1525-142x.2003.03065.x. [DOI] [PubMed] [Google Scholar]
- Suomalainen M, Thesleff I. Patterns of Wnt pathway activity in the mouse incisor indicate absence of Wnt/beta-catenin signaling in the epithelial stem cells. Dev Dyn. 2010;239:364–372. doi: 10.1002/dvdy.22106. [DOI] [PubMed] [Google Scholar]
- Sylvester JB, Rich CA, Loh YH, van Staaden MJ, Fraser GJ, Streelman JT. Brain diversity evolves via differences in patterning. Proc Natl Acad Sci U S A. 2010;107:9718–9723. doi: 10.1073/pnas.1000395107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I, Sharpe P. Signalling networks regulating dental development. Mech Dev. 1997;67:111–123. doi: 10.1016/s0925-4773(97)00115-9. [DOI] [PubMed] [Google Scholar]
- Trapani J. Position of developing replacement teeth in teleosts. Copeia. 2001;2001:35–51. [Google Scholar]
- Tucker A, Sharpe P. The cutting-edge of mammalian development; how the embryo makes teeth. Nat Rev Genet. 2004;5:499–508. doi: 10.1038/nrg1380. [DOI] [PubMed] [Google Scholar]
- Tuisku F, Hildebrand C. Evidence for a neural influence on tooth germ generation in a polyphyodont species. Dev Biol. 1994;165:1–9. doi: 10.1006/dbio.1994.1228. [DOI] [PubMed] [Google Scholar]
- Vadlamudi U, Espinoza HM, Ganga M, Martin DM, Liu X, Engelhardt JF, Amendt BA. PITX2, beta-catenin and LEF-1 interact to synergistically regulate the LEF-1 promoter. J Cell Sci. 2005;118:1129–1137. doi: 10.1242/jcs.01706. [DOI] [PubMed] [Google Scholar]
- Wang J, Panáková D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, Werdich AA, Yelon D, Macrae CA, Poss KD. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, Maas RL, Chuong CM, Schimmang T, Thesleff I. An integrated gene regulatory network controls stem cell proliferation in teeth. PLoS biology. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XP, Suomalainen M, Jorgez CJ, Matzuk MM, Werner S, Thesleff I. Follistatin Regulates Enamel Patterning in Mouse Incisors by Asymmetrically Inhibiting BMP Signaling and Ameloblast Differentiation. Dev Cell. 2004;7:719–730. doi: 10.1016/j.devcel.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Zhang L, Yuan G, Liu H, Lin H, Wan C, Chen Z. Expression pattern of Sox2 during mouse tooth development. Gene expression patterns : GEP. 2012;12:273–281. doi: 10.1016/j.gep.2012.07.001. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang Z, Zhao X, Yu X, Hu Y, Geronimo B, Fromm SH, Chen YP. A new function of BMP4: dual role for BMP4 in regulation of Sonic hedgehog expression in the mouse tooth germ. Development. 2000;127:1431–1443. doi: 10.1242/dev.127.7.1431. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lan Y, Chai Y, Jiang R. Antagonistic actions of Msx1 and Osr2 pattern mammalian teeth into a single row. Science. 2009;323:1232–1234. doi: 10.1126/science.1167418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.