Abstract
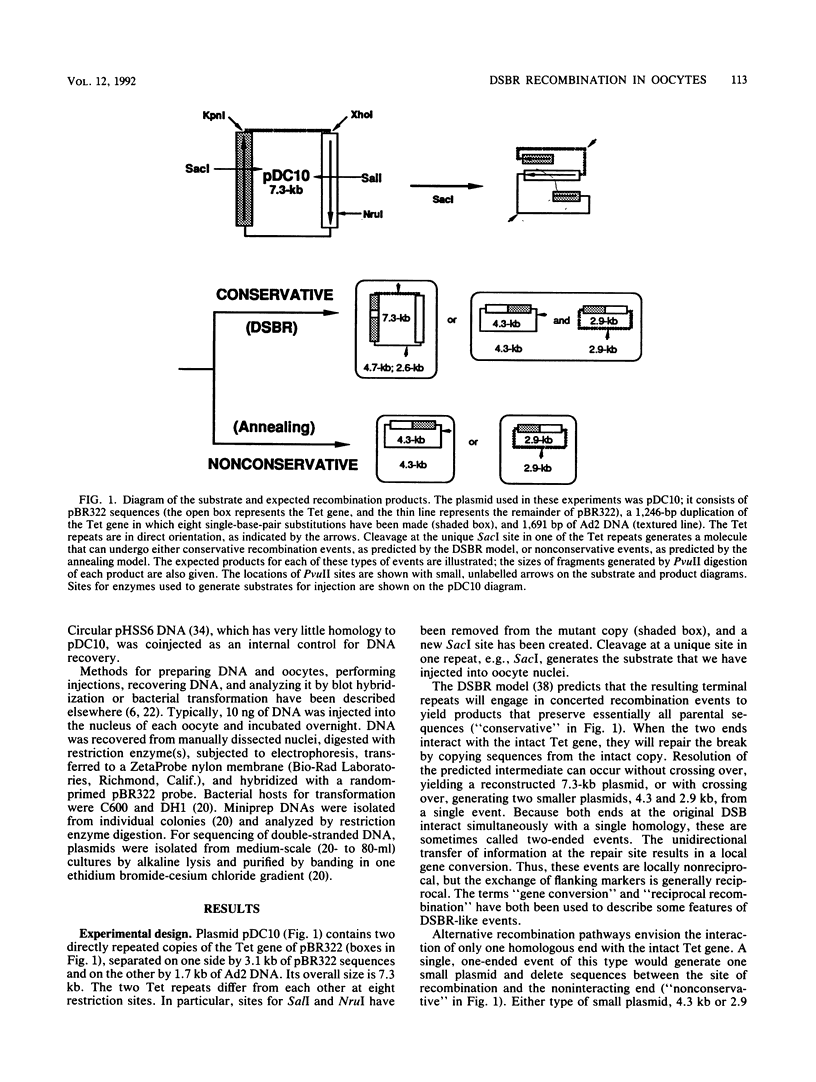
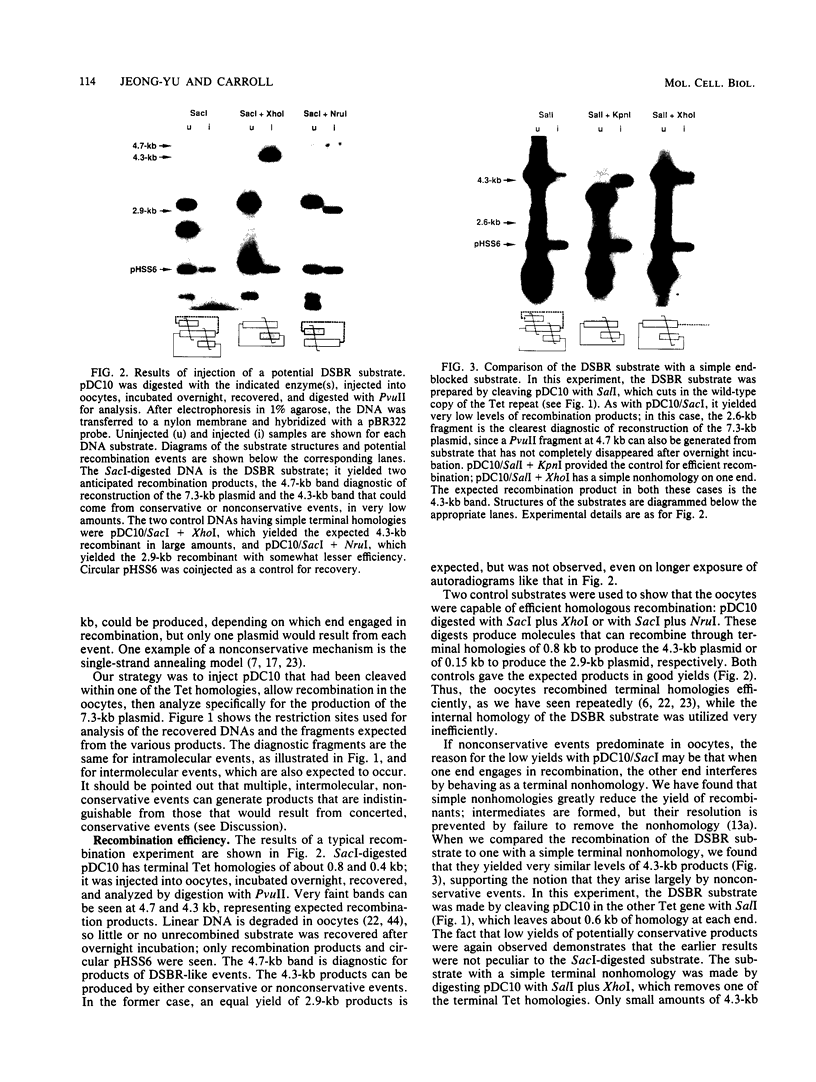
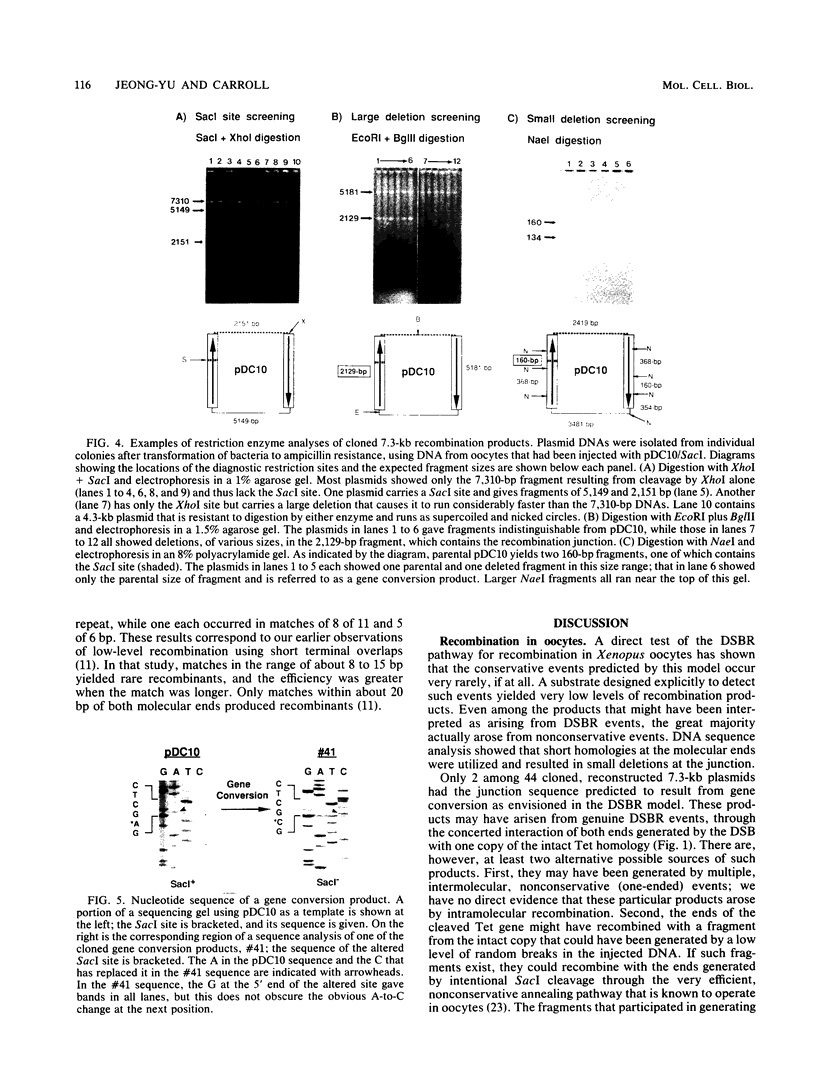
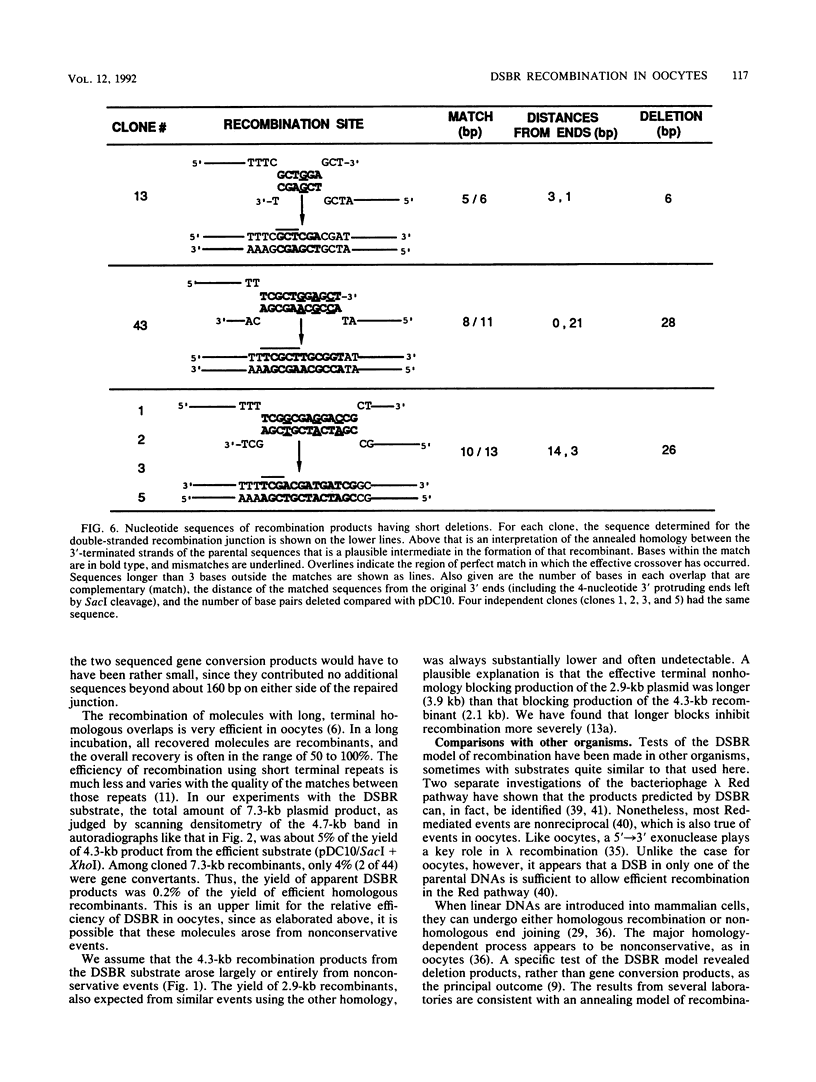
A direct test was made of predictions of the double-strand-break repair (DSBR) model of recombination in Xenopus laevis oocytes. The DNA substrate injected into oocytes had two directly repeated copies of a 1.25-kb sequence and was cleaved within one of them. Different products were expected to result from concerted, conservative events, as predicted by the DSBR model, and from nonconservative events. Only very low levels of recombination products, both conservative and nonconservative, were observed. When individual, apparent DSBR products were cloned and characterized, it emerged that the majority of them had arisen by nonconservative recombination through short, terminal homologies and not from the gene conversion events predicted for DSBR. Two cloned products among 44 tested corresponded to the predications of the DSBR model, but these could also have been generated by other processes. The most efficient recombination events in oocytes are nonconservative and are based on long, terminal homologous overlaps; when these are not available, short, imperfect overlaps support a lower level of nonconservative recombination; genuine, conservative DSBR events occur rarely, if at all.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. A., Eliason S. L. Recombination of homologous DNA fragments transfected into mammalian cells occurs predominantly by terminal pairing. Mol Cell Biol. 1986 Sep;6(9):3246–3252. doi: 10.1128/mcb.6.9.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur M., Potrykus I., Paszkowski J. Intermolecular homologous recombination in plants. Mol Cell Biol. 1990 Feb;10(2):492–500. doi: 10.1128/mcb.10.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendig M. M. Persistence and expression of histone genes injected into Xenopus eggs in early development. Nature. 1981 Jul 2;292(5818):65–67. doi: 10.1038/292065a0. [DOI] [PubMed] [Google Scholar]
- Brenner D. A., Smigocki A. C., Camerini-Otero R. D. Effect of insertions, deletions, and double-strand breaks on homologous recombination in mouse L cells. Mol Cell Biol. 1985 Apr;5(4):684–691. doi: 10.1128/mcb.5.4.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990 Jun 15;61(6):1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Carroll D., Wright S. H., Wolff R. K., Grzesiuk E., Maryon E. B. Efficient homologous recombination of linear DNA substrates after injection into Xenopus laevis oocytes. Mol Cell Biol. 1986 Jun;6(6):2053–2061. doi: 10.1128/mcb.6.6.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto E., Radding C. M. Mechanism for the action of lambda exonuclease in genetic recombination. Nat New Biol. 1971 Jan 6;229(1):13–16. doi: 10.1038/newbio229013a0. [DOI] [PubMed] [Google Scholar]
- Certa U., Bannwarth W., Stüber D., Gentz R., Lanzer M., Le Grice S., Guillot F., Wendler I., Hunsmann G., Bujard H. Subregions of a conserved part of the HIV gp41 transmembrane protein are differentially recognized by antibodies of infected individuals. EMBO J. 1986 Nov;5(11):3051–3056. doi: 10.1002/j.1460-2075.1986.tb04605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Seidman M. M. Intramolecular recombination between transfected repeated sequences in mammalian cells is nonconservative. Mol Cell Biol. 1986 Jul;6(7):2520–2526. doi: 10.1128/mcb.6.7.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folger K. R., Thomas K., Capecchi M. R. Nonreciprocal exchanges of information between DNA duplexes coinjected into mammalian cell nuclei. Mol Cell Biol. 1985 Jan;5(1):59–69. doi: 10.1128/mcb.5.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiuk E., Carroll D. Recombination of DNAs in Xenopus oocytes based on short homologous overlaps. Nucleic Acids Res. 1987 Feb 11;15(3):971–985. doi: 10.1093/nar/15.3.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M., de Villiers J., Weber F., Schaffner W. High frequency of homologous recombination in mammalian cells between endogenous and introduced SV40 genomes. Cell. 1985 Dec;43(3 Pt 2):695–703. doi: 10.1016/0092-8674(85)90242-9. [DOI] [PubMed] [Google Scholar]
- Kitamura Y., Yoshikura H., Kobayashi I. Homologous recombination in a mammalian plasmid. Mol Gen Genet. 1990 Jul;222(2-3):185–191. doi: 10.1007/BF00633816. [DOI] [PubMed] [Google Scholar]
- Kucherlapati R. S., Eves E. M., Song K. Y., Morse B. S., Smithies O. Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3153–3157. doi: 10.1073/pnas.81.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Goyon C., Schultes N. P., Treco D., Szostak J. W., Haber J. E., Nicolas A. Detection of heteroduplex DNA molecules among the products of Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7653–7657. doi: 10.1073/pnas.87.19.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol Cell Biol. 1990 Jan;10(1):103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Repair of double-stranded DNA breaks by homologous DNA fragments during transfer of DNA into mouse L cells. Mol Cell Biol. 1990 Jan;10(1):113–119. doi: 10.1128/mcb.10.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Characterization of recombination intermediates from DNA injected into Xenopus laevis oocytes: evidence for a nonconservative mechanism of homologous recombination. Mol Cell Biol. 1991 Jun;11(6):3278–3287. doi: 10.1128/mcb.11.6.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryon E., Carroll D. Degradation of linear DNA by a strand-specific exonuclease activity in Xenopus laevis oocytes. Mol Cell Biol. 1989 Nov;9(11):4862–4871. doi: 10.1128/mcb.9.11.4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Fungal recombination. Microbiol Rev. 1985 Mar;49(1):33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenberger B. A., Roeder G. S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991 Mar;11(3):1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer P., Vielmetter W. Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Res. 1988 Feb 11;16(3):907–924. doi: 10.1093/nar/16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976 Jun;59(1):97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Rudin N., Haber J. E. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1988 Sep;8(9):3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin N., Sugarman E., Haber J. E. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989 Jul;122(3):519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusconi S., Schaffner W. Transformation of frog embryos with a rabbit beta-globin gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):5051–5055. doi: 10.1073/pnas.78.8.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman M. M. Intermolecular homologous recombination between transfected sequences in mammalian cells is primarily nonconservative. Mol Cell Biol. 1987 Oct;7(10):3561–3565. doi: 10.1128/mcb.7.10.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert H. S., Chen E. Y., So M., Heffron F. Shuttle mutagenesis: a method of transposon mutagenesis for Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Feb;83(3):735–739. doi: 10.1073/pnas.83.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. Homologous recombination in procaryotes. Microbiol Rev. 1988 Mar;52(1):1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Treco D., Szostak J. W. Extensive 3'-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991 Mar 22;64(6):1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Takahashi N., Kobayashi I. Evidence for the double-strand break repair model of bacteriophage lambda recombination. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2790–2794. doi: 10.1073/pnas.87.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler D. S., Stahl M. M., Stahl F. W. Double-chain-cut sites are recombination hotspots in the Red pathway of phage lambda. J Mol Biol. 1987 May 5;195(1):75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- Thaler D. S., Stahl M. M., Stahl F. W. Tests of the double-strand-break repair model for red-mediated recombination of phage lambda and plasmid lambda dv. Genetics. 1987 Aug;116(4):501–511. doi: 10.1093/genetics/116.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. I., Haber J. E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990 Mar;9(3):663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie A. H., Laskey R. A., Finch J., Gurdon J. B. Selective DNA conservation and chromatin assembly after injection of SV40 DNA into Xenopus oocytes. Dev Biol. 1978 May;64(1):178–188. doi: 10.1016/0012-1606(78)90069-6. [DOI] [PubMed] [Google Scholar]