Abstract
Controlling translation during protein synthesis is crucial for cell proliferation and differentiation. Protein translation is orchestrated by an assembly of various protein components at the ribosomal subunits. The eukaryotic translation initiation factor 4G (eIF4G) plays an important role in the formation of the translation initiation complex eIF4F consisting of eIF4G, the ATP dependent RNA helicase eIF4A and the cap binding protein eIF4E. One of the functions of eIF4G is the enhancement of the activity of eIF4A facilitated mainly through binding to the HEAT1 domain of eIF4G. In order to understand the interaction of HEAT1 with eIF4A and other components during translation initiation backbone assignment is essential. Here we report the 1H, 13C and 15N backbone assignment for the HEAT1 domain of human eIF4G isoform I (eIF4GI-HEAT1), the first of three HEAT domains of eIF4G (29 kDa) as a basis for the elucidation of its structure and interactions with its binding partners, necessary for understanding the mechanism of its biological function.
Keywords: translation initiation, human eIF4G, backbone assignment, NMR, isoform I
Biological Context
Cell proliferation and differentiation depends on the control of translation (mRNA encoded protein synthesis). More than ten eukaryotic translation initiation factors are known to be involved in the assembly of the 80S ribosome-RNA complex enabling interaction of the initiator Met-tRNA with the start codon of the mRNA.
The eukaryotic initiation factor 4G (eIF4G) plays an important role in the assembly of translation initiation factors and the small ribosomal subunit during translation initiation (Marintchev, Edmonds et al. 2009; Topisirovic and Sonenberg 2011). Together with the ATP dependent RNA helicase eIF4A and the cap-binding protein eI4E, this scaffolding protein forms the translation initiation complex eIF4F. The N-terminal domain of eIF4G contains a binding site for the poly(A) binding protein (PABP) and for eIF4E. Its C-terminal two third contain 3 HEAT repeat domains (See Fig. 1A). HEAT1 and HEAT 2 both bind to eIF4A and HEAT3 to Mnk1 (Marintchev and Wagner 2004) Additionally, HEAT1 binds to Internal Ribosomal Entry Sites (IRES) (Kolupaeva, Pestova et al. 1998). Mammals have two functional homologs of eIF4G (eIF4GI and eIF4GII) with a sequence similarity of 58.5%.
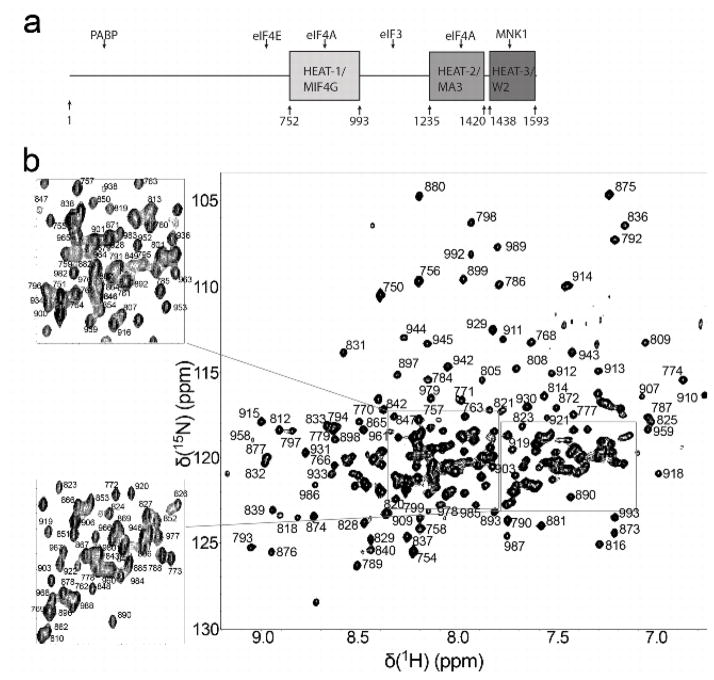
Fig. 1.
A) Domain Architecture of eIF4GI. Annotations above the domains indicate binding sites. Annotations below indicate start and end residues for the individual domains. B) 1H-15N TROSY HSQC spectrum of 2H, 15N labeled eIF4G-HEAT1 in 10 mM potassium phosphate, pH 6.5, 300 mM KCl, 2 mM DTT, 0.5 mM EDTA, 0.01% NaN3, 5% D2O, recorded on a Bruker DMX 750 MHz spectrometer at 25°C.
It has been shown that the interaction of eIF4A with the HEAT1 domain enhances the helicase catalytic activity of eIF4A by 4-fold8. Binding of HEAT2 has a modulatory role (Marintchev and Wagner 2004; Korneeva, First et al. 2005; Marintchev, Edmonds et al. 2009). The mechanism of this enhancement is not known yet. A recent crystal structure of human eIF4GII-HEAT1 domain is available (Marcotrigiano, Lomakin et al. 2001). However, it is lacking two loop regions of 9 and 30 residues, respectively (see Fig. 2). To fully characterize the interaction of the HEAT1 domain with eIF4A we performed backbone assignment of eIF4GI-HEAT1 including the important loop regions to facilitate structure-function studies of the translational initiation machinery.
Fig. 2.
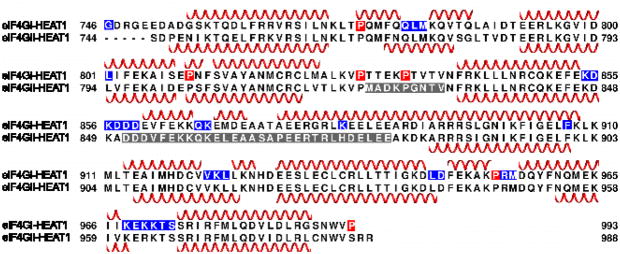
Sequence alignment and secondary structure elements of eIF4G-HEAT1 of isoform I (top) and isoform II (bottom). The secondary structure of isoform II was obtained from the crystal structure, PDB code 1HU3 (Marcotrigiano, Lomakin et al. 2001). Residues highlighted in gray were missing in the crystal structure. The secondary structure of isoform I was predicted based on the chemical shifts using TALOS+ (Shen, Delaglio et al. 2009). The secondary structure of residues in gray could not be predicted. Residues marked in blue could not be assigned. Proline residue are marked in red.
Methods and Experiments
Sample preparation
Human eIF4GI (747–994) was expressed in E. coli Bl21(DE3) with N-terminal protein G domain (Gb1) and His-tags. 15N, 2D labeled eIF4GI-HEAT1 was expressed in M9 media dissolved in D2O with 15NH4Cl as the sole nitrogen source and 13C-glucose as a carbon source. Purification was performed using metal immobilized affinity chromatography using TALON resin (Clontech, USA) followed by overnight Tobacco etch virus (TEV) protease cleavage at 4°C. The cleaved product was further purified by size exclusion chromatography. The purified protein was concentrated to 0.7 mM and exchanged into buffer containing 10 mM phosphate buffer pH 6.5, 300 mM KCl, 2 mM DTT, 0.5 mM EDTA, 0.01% NaN3, 5% D2O at pH 6.5 for NMR measurements.
NMR experiments
All NMR measurements were performed at 25°C on Bruker DMX 750 MHz and Bruker DRX 600 MHz spectrometers equipped with TXI cryoprobes with Z gradient. Data were processed and analyzed using NMRPipe (Delaglio, Grzesiek et al. 1995) and NMRView (Johnson and Blevins 1994). 2D [1H,15N] TROSY HSQC experiments were acquired at 750 MHz using 256 t1 increments with a sweep width of 2659 Hz and 2048 t2 points with a sweep width of 9014 Hz. The TROSY versions of the following triple resonance experiments were utilized for sequential backbone assignment of eIF4G-HEAT1 (numbers in parentheses indicate the number of real points and sweep width in Hertz for each dimension): HNCO (H: 1024/10504; C: 50/2262; N: 64/3419), HNCA (H: 1024/13515; C: 50/1976; N: 64/6031), HNCACB (H: 1024/10504; C: 64/12077; N: 64/3418), HNCOCA (H: 1024/10504; C: 64/6031; N: 48/2203). A 3D 15N TROSY NOESY was measured with the following parameters H: 1024/9615; N: 50/2190; Hind: 128/9603 and a mixing time of 90 ms.
Assignment and Data Deposition
The [1H,15N]-HSQC spectrum of 15N,D-eIF4GI-HEAT1 is show in Fig. 1. Due to the helical character of the construct, there is significant overlap on the center region of the spectrum. Assignments were obtained for 85% of the backbone nuclei (15N, 1HN, 13Cα) and 13Cβ. Out of 242 expected backbone amide N-H pairs (5 prolines), 207 were assigned. Several intense cross peaks could not be assigned either due to lacking cross peaks in the triple resonance experiments or due to extensive overlap of either the cross peak in question or potential neighboring cross peaks. Two main regions that lack assignment are residues 854–859 (sequence KDKDDD) and residues 968–973 (sequence KEKKTS). Due to the high repetition rate of amino acids no unambiguous assignment was possible.
The secondary structure elements of eIF4GI-HEAT1 were predicted on the basis of the chemical shifts using the program Talos+ (Shen, Delaglio et al. 2009). Structural elements are in agreement with the published crystal structure of isoform 2, (PDB code 1HU3, Fig. 2) (Marcotrigiano, Lomakin et al. 2001). The crystal structure does not show structural propensity for two loop regions of eIF4GII-HEAT1 (residues 802–910 and 851–880). These loops show significant structural content in eIF4GI-HEAT1. All chemical shifts were deposited in the BioMagResBank (http://www.bmrb.wisc.edu) under accession number 18738.
References
- Delaglio F, Grzesiek S, et al. NMRPIPE - A MULTIDIMENSIONAL SPECTRAL PROCESSING SYSTEM BASED ON UNIX PIPES. Journal of Biomolecular Nmr. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMR VIEW - A COMPUTER-PROGRAM FOR THE VISUALIZATION AND ANALYSIS OF NMR DATA. Journal of Biomolecular Nmr. 1994;4(5):603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, et al. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J Biol Chem. 1998;273(29):18599–18604. doi: 10.1074/jbc.273.29.18599. [DOI] [PubMed] [Google Scholar]
- Korneeva NL, First EA, et al. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. Journal of Biological Chemistry. 2005;280(3):1872–1881. doi: 10.1074/jbc.M406168200. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano J, I, Lomakin B, et al. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell. 2001;7(1):193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Edmonds KA, et al. Topology and Regulation of the Human eIF4A/4G/4H Helicase Complex in Translation Initiation. Cell. 2009;136(3):447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Quarterly Reviews of Biophysics. 2004;37(3–4):197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, et al. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topisirovic I, Sonenberg N. Translational control by the eukaryotic ribosome. Cell. 2011;145(3):333–334. doi: 10.1016/j.cell.2011.04.006. [DOI] [PubMed] [Google Scholar]