Abstract
Simian immunodeficiency virus (SIV) infection of rhesus macaques is a valuable animal model for human immunodeficiency virus (HIV)-1 vaccine development. Our laboratory recently described the immunogenicity and limited efficacy of a vif-deleted SIVmac239 proviral DNA (SIV/CMVΔvif) vaccine. The current report characterizes immunogenicity and efficacy for the SIV/CMVΔvif proviral DNA vaccine when co-inoculated with an optimized rhesus interleukin (rIL)-15 expression plasmid. Macaques co-inoculated with rIL-15 and SIV/CMVΔvif proviral plasmids showed significantly improved SIV-specific CD8 T cell immunity characterized by increased IFN-γ ELISPOT and polyfunctional CD8 T cell responses. Furthermore, these animals demonstrated a sustained suppression of plasma virus loads after multiple low dose vaginal challenges with pathogenic SIVmac251. Importantly, SIV-specific cellular responses were greater in immunized animals compared to unvaccinated controls during the initial 12 weeks after challenge. Taken together, these findings support the use of IL-15 as an adjuvant in prophylactic anti-HIV vaccine strategies.
Keywords: SIV, DNA vaccine, vif, interleukin-15
Introduction
Development of an efficacious vaccine against human immunodeficiency virus (HIV)-1 is a major priority for control of the AIDS pandemic. Simian immunodeficiency virus (SIV) and SIV/HIV (SHIV) infections of rhesus macaques have provided highly relevant animal models for HIV-1 vaccine development and valuable insights into the feasibility of developing a safe and effective vaccine against human AIDS. Due to difficulties in generating vaccines capable of eliciting high titers of broadly reactive neutralizing antibodies, efforts have focused on vaccines designed to elicit strong antigen-specific T cell responses that will significantly suppress virus loads and improve host survival. Recent accomplishments in vaccine development derived from these animal models have resulted in a focus on prime-boost strategies that incorporate priming immunization with viral antigen expression plasmids, followed by boosting with viral immunogens expressed from a viral vaccine vector (Letvin, 2006). The most extensively tested viral vectors include replicating and nonreplicating adenoviruses (Ad), with non-replicating Ad serotype 5 (Ad5) as the focus of more recent vaccine studies in both macaques and human clinical trials (Hokey and Weiner, 2006; Letvin, 2007; Robert-Guroff, 2007). However, concerns with this and other viral vectors are based on issues of pre-existing host immunity to a particular viral vector, leading to diminished and insufficient expression of the inserted immunogen and vaccine efficacy. These concerns have largely been validated with the recent failure of the Merck STEP trial whereby pre-existing immunity to the Ad vector not only down-modulated the HIV insert response, but also modulated the response to facilitate, rather than prevent infection for particular populations (Priddy et al., 2008). These results emphasize the need for continued exploration of other unique vaccine approaches that may include novel DNA vaccines and immunomodulators, or combinations thereof.
Vaccine approaches based on plasmid DNA only, or DNA plasmid priming following by a protein boost, while promising in rodents, have proven relatively inefficient in larger animal models including primates (Hokey and Weiner, 2006; Ulmer et al., 2000). However, similar vaccine approaches that include co-administration of DNA vaccines with cytokines, either as recombinant proteins or expression plasmids, have demonstrated marked improvement of vaccine-induced immune responses when tested in SIV and SHIV animal models. Cytokines that increase clonal expansion of antigen-specific T cells, including the common γ chain (γc) cytokine interleukin (IL)-15, or IL-12, have been particularly promising as adjuvants for prophylactic DNA vaccines and show some promise as potent immunomodulators when delivered in the optimal setting (Calarota and Weiner, 2004; Lori et al., 2006). IL-15 is a pleiotropic cytokine that can affect both innate and adaptive immune responses and has shown particular promise as an adjuvant for cancer vaccine design (Waldmann, 2006). IL-15 shares some properties with IL-2, including potent stimulation of proliferation of activated peripheral blood T cells, natural killer (NK) cells, and NKT cells (Becker et al., 2002; Oh et al., 2003; Surh and Sprent, 2002; Waldmann et al., 2001). IL-15 is also a potent activator of the innate immune system through mechanisms that are distinct from induction of NK cells and include promotion of dendritic cell induction and maturation. Moreover, IL-15 used either as a therapeutic or vaccine adjuvant, was shown to enhance the proliferation and longevity of memory CD8 or CD4 T cells through up-regulation of anti-apoptotic molecules (Boyer et al., 2007; Kutzler et al., 2005; Lin et al., 2005; Oh et al., 2008; Picker et al., 2006; Waldmann et al., 2001; Zhang et al., 2006). Overall, these properties of IL-15 provide strong support for further investigation of this cytokine as an adjuvant for HIV DNA vaccine approaches.
A recent report by our laboratory described the investigation of a vif-deleted SIVmac239 provirus as an attenuated proviral DNA vaccine. Inoculation of rhesus macaques with this vif-deleted provirus induced strong SIV-specific T cell proliferative responses and low antibody titers in the face of an undetectable infection (Sparger et al., 2008). Transient suppression of pathogenic SIVmac251 was observed in vaccinated animals early after vaginal challenge; however differences in virus loads between vaccinated and unvaccinated animals were not sustained. However, despite induction of strong T cell proliferative responses, SIV-specific CD8 T cell responses elicited by this DNA vaccine were either very weak or absent in vaccinated animals. Based on these observations, a second investigation was designed to test the immunogenicity and efficacy of an immunization approach that included this vif-deleted SIVmac239 provirus plasmid and an expression plasmid optimized for rhesus macaque IL-15, as a combination DNA vaccine. This report describes adjuvant effects that were conferred by IL-15 for this DNA vaccine, and that included increased frequencies of SIV-specific CD8 T cell responses and induction of antigen-specific polyfunctional CD8 T cell responses. Furthermore, animals vaccinated with proviral DNA and an IL-15 expression plasmid exhibited significantly lower virus loads compared to unvaccinated controls, during acute and chronic phases of infection after challenge with pathogenic SIVmac251.
Results
Comparison of IL-15 expression from two different rIL-15 plasmids
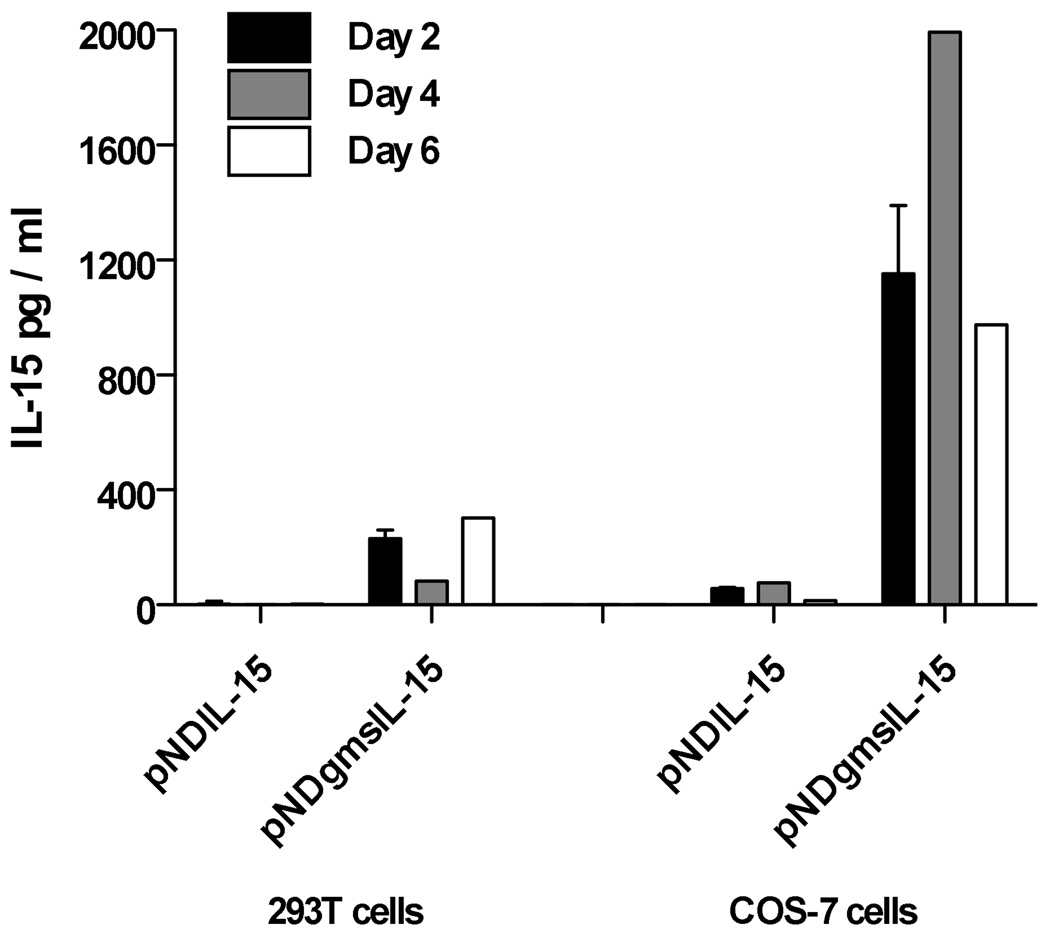
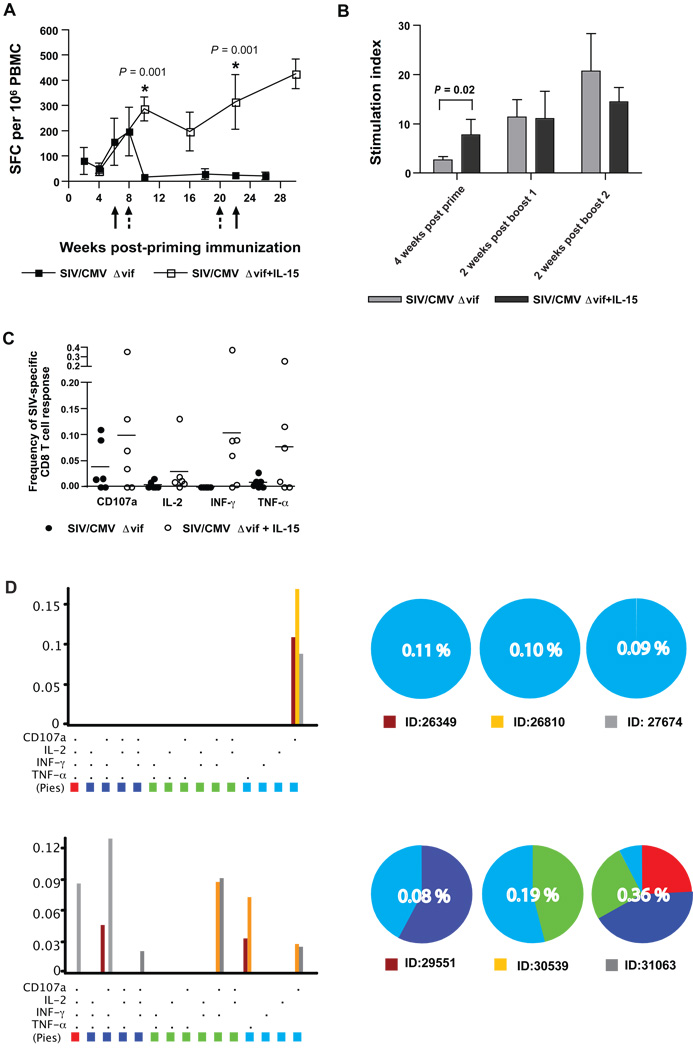
The objective of this vaccine study was to test for enhanced immunogenicity and for efficacy conferred by an IL-15 expression plasmid to a previously described vif-deleted SIVmac239 proviral DNA vaccine (SIV/CMVΔvif) (Sparger et al., 2008). Therefore, construction of a rhesus macaque IL-15 (rIL-15) expression plasmid with optimal activity was critical for accomplishing this goal. Measurements of soluble rIL-15 generated by transfection of T293 and COS-7 cells with pNDgmsRRm-IL-15 containing the Mamu IL-15 coding sequence with the granulocyte macrophage-colony stimulating factor (GM-CSF) leader revealed a 20 to 300-fold increase at every time point tested in transfected-cell culture supernatants, when compared to production after transfection with pND RRm-IL-15 encoding Mamu IL-15 coding sequence with the native IL-15 leader (Fig. 1). A recent report describing optimized rIL-15 expression plasmids, revealed a similar magnitude of increased rIL-15 expression using the same GM-CSF leader upstream of the native rIL-15 signal peptide in a different plasmid backbone, although this report revealed a slightly higher expression with use of the tissue plasminogen activator (TPA) signal peptide (Jalah et al., 2007). Furthermore, evaluation of biological activity of the released cytokine using the IL-2/15 dependent HT-2 cell line (Villinger et al., 1995) indicated that secreted cytokine was biologically active when compared to recombinant IL-15 (data not shown). These data validated use of the pNDgmsRRm-IL-15 expression plasmid for delivery of rIL-15 as a vaccine cytokine adjuvant.
Figure 1. Production of rIL-15 from an optimized expression plasmid.
Expression plasmids designated pNDRRm-IL-15 and pNDgmsRRm-IL-15, were constructed as described in Materials and Methods and were distinguished by their leader sequences. Cell culture supernatants were harvested at two, four, and six days from either COS-7 or 293T cells, after transfection with a rIL-15 expression plasmid and tested for IL-15 concentrations by a commercial human IL-15 antigen-capture ELISA (Invitrogen, Biosource) as described in Materials and Methods. Error bars represent standard deviation.
Induction of SIV-specific cellular immune responses by co-immunization with SIV/CMVΔvif and pNDgmsRRm-IL-15 plasmids
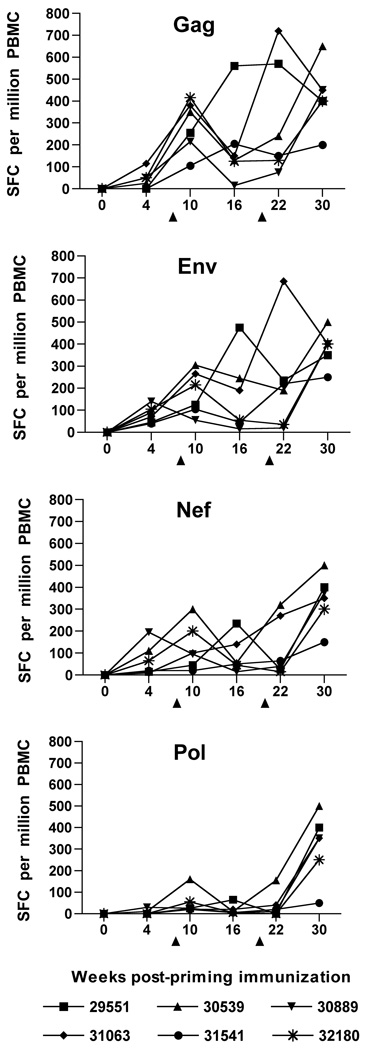
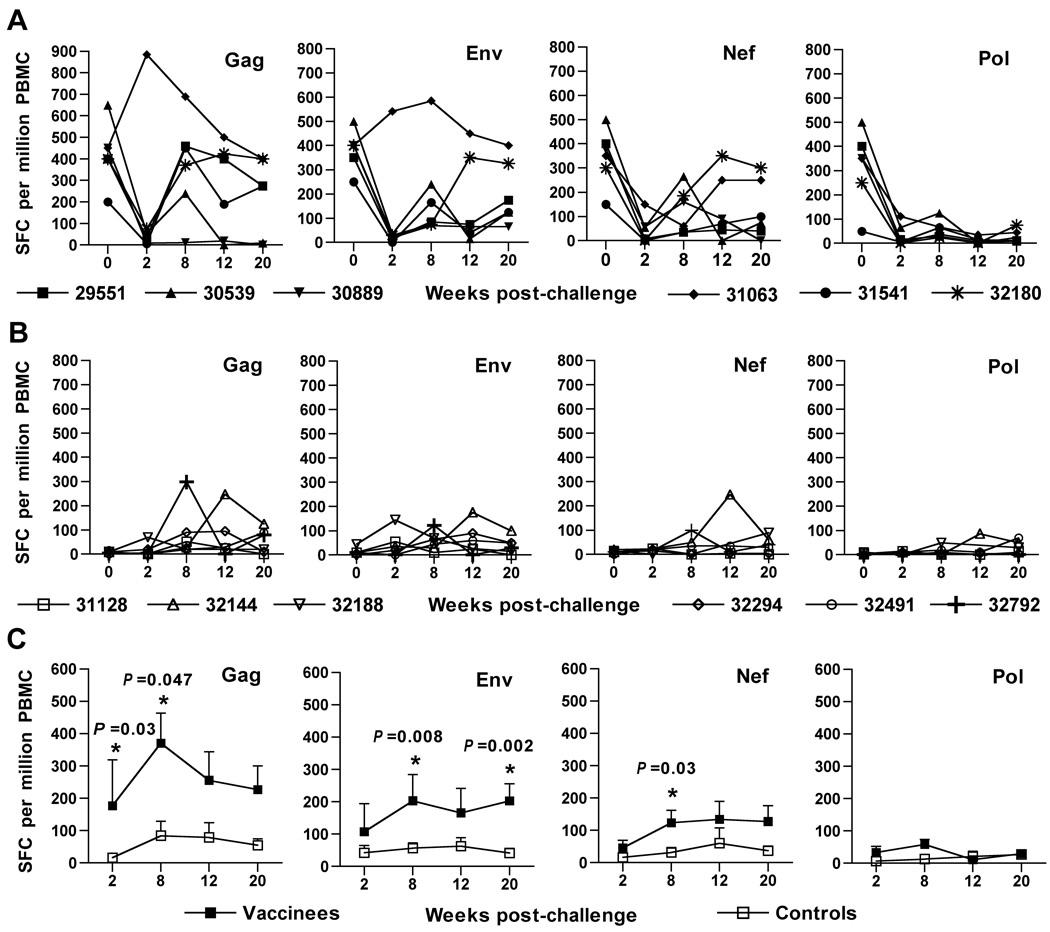
Virus-specific CD8 T cell responses have been shown to be critical for host control of plasma virus loads for both SIV and HIV-1 infections (Jin et al., 1999; Musey et al., 1997; Ogg et al., 1998; Schmitz et al., 1999; Yasutomi et al., 1993). Accordingly, a recombinant vaccinia-based interferon (IFN)-γ enzyme-linked immunospot (ELISPOT) previously shown to measure predominantly CD8 responses (Larsson et al., 1999) was utilized to examine vaccine-induced cellular responses specific to four different SIV antigens (Gag, Env, Nef, and Pol). IFN-γ ELISPOT responses were observed in PBMC from some, but not all animals, by four weeks after a priming vaccination with the SIV/CMVΔvif plus IL-15 DNA vaccine (Fig. 2). ELISPOT responses to SIV antigens were moderately improved by the first booster immunization, particularly responses to SIV Gag. IFN-γ ELISPOT responses were further augmented in a majority of the animals for all four SIV antigens, when tested at either two weeks, or ten weeks after a second booster immunization with vaccine plasmids. Vaccine-induced IFN-γ ELISPOT responses were variable over time, but were greatest for SIV Gag, Env, and Nef, with lower responses observed for SIV Pol for time points prior to challenge. Pair-wise comparisons of mean responses to either Gag, Env, and Nef with responses to Pol revealed significant differences with P values ranging from 0.002 to 0.046 for earlier time points after priming immunization. However, IFN-γ ELISPOT responses tested on the day of challenge were comparable for all four SIV antigens.
Figure 2. SIV-specific IFN-γ ELISPOT responses elicited by co-immunization with SIV/CMVΔvif and pNDgmsRRm-IL-15 plasmid DNA.
SIV-specific IFN-γ ELISPOT responses were assayed on cryopreserved PBMC harvested from animals at various time points after vaccination. Immune responses to individual SIVmac239 proteins were measured using recombinant vaccinia viruses expressing either SIV Gag, Env, Nef or Pol, with protocols described in Materials and Methods. Values less than 50 spot-forming colonies (SFC) per 106 PBMC were considered nonspecific for this assay. Arrows positioned at 8 and 20 weeks after priming immunization denoted time points of booster vaccine inoculations.
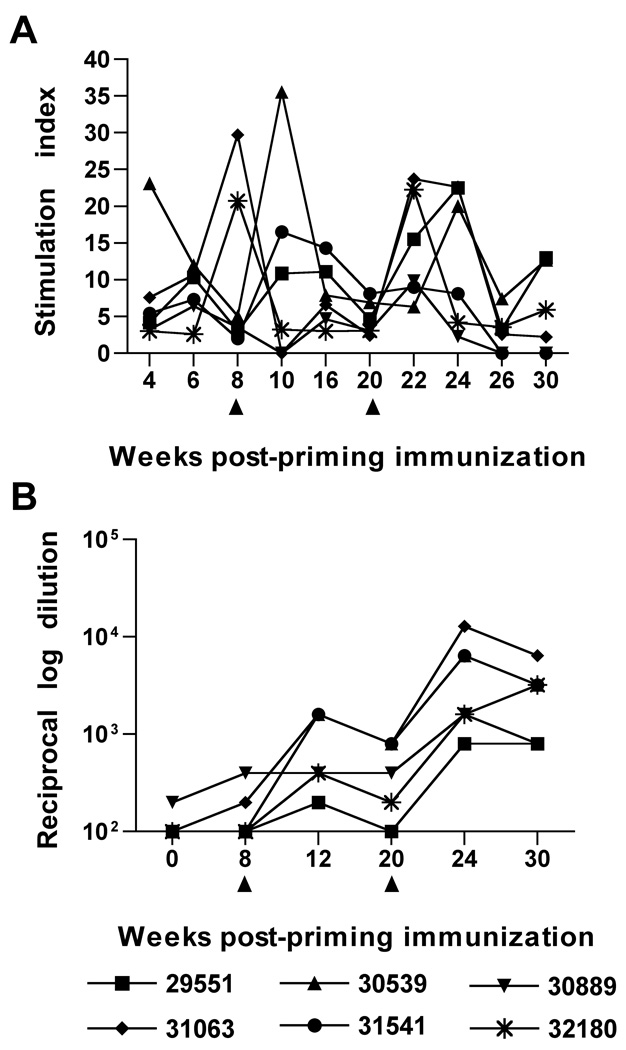
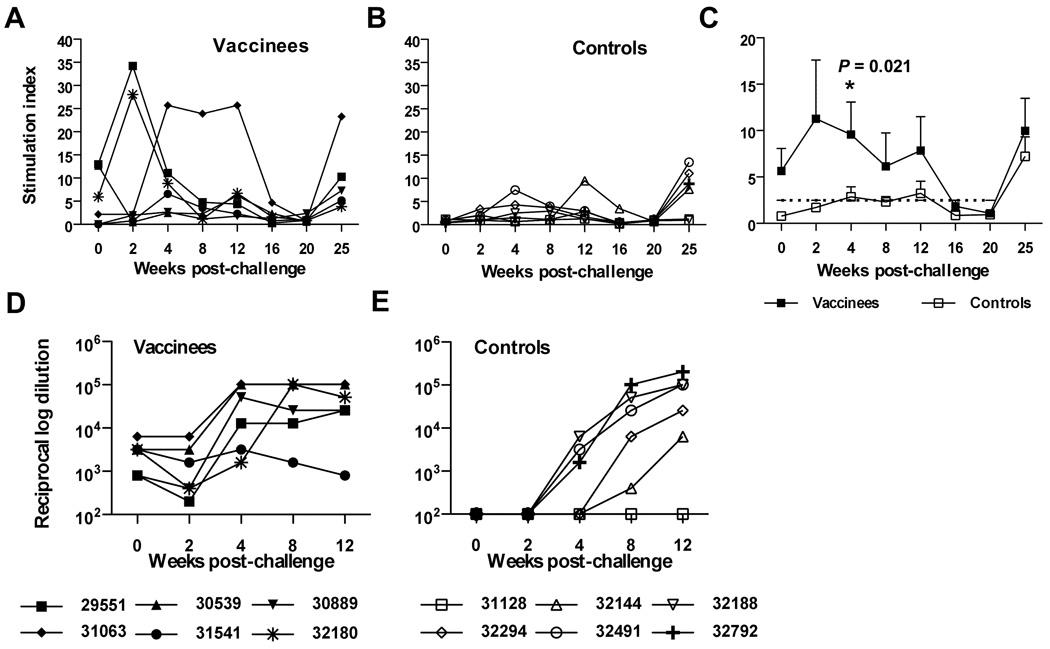
Proliferation of PBMC in response to stimulation with inactivated SIV preparations provided another measure of antiviral cellular responses that predominantly reflects CD4 activity, but may also include CD8 responses. SIV-specific proliferation responses measured in PBMC were observed for all animals at multiple time points following immunization with the SIV/CMVΔvif plus IL-15 DNA vaccine (Fig. 3A). Similar to IFN-γ ELISPOT responses, SIV-specific T cell proliferative responses were quite variable between animals and stimulation indices (SI) ranged from 2.5 to 35 over different time points. In general, proliferative responses were quite robust with SI of 15 or greater observed for all animals for at least one time point, and for at least two time points for five of six immunized macaques. Booster immunizations also resulted in enhanced antigen-specific proliferative responses for all six vaccinees. Taken together, these results revealed a strong cellular response to this proviral DNA vaccine that included a rIL-15 plasmid. Notably, inoculation of macaques with the highly attenuated vif-deleted provirus and rIL-15 plasmids did not result in detectable infection by assay of PBMC for proviral DNA by real-time PCR, similar to a previous inoculation study with SIV vif-deleted provirus alone (Sparger et al., 2008) (data not shown).
Figure 3. SIV-specific T cell proliferative and antiviral antibody responses elicited by co-immunization with SIV/CMVΔvif plasmid DNA and pNDgmsRRm-IL-15 plasmid DNA.
Virus-specific T cell proliferative responses directed to purified inactivated SIV were measured in PBMC harvested from macaques at various time points after priming immunization. Values for the highest SI to either 12.5 or 125 ng of SIV p27Gag are shown (A). A SI of 2.5 or greater was considered positive. Reciprocal endpoint titers for plasma antibodies against native SIV Env were measured using a ConA ELISA with reciprocal log dilution values greater than 100 representing positive titers (B). Arrows positioned at 8 and 20 weeks after priming immunization denoted time points of booster vaccine inoculations.
SIV-specific antibody responses after immunization with SIV/CMVΔvif proviral DNA and rIL-15 plasmids
Antibody responses have proven to be very low or absent in animals inoculated with vif-deleted lentiviruses delivered either as a provirus plasmid, or as a virus preparation (Desrosiers et al., 1998; Gupta et al., 2007; Harmache et al., 1996; Lockridge et al., 2000; Sparger et al., 2008). For this reason, a sensitive concanavalin-A (Con-A) ELISA was utilized for detection of antiviral antibody induction by the SIV/CMVΔvif plus IL-15 DNA vaccine Low antibody titers to native SIV Env were detected by 8 weeks after priming immunization in two of six animals (Fig. 3B). All animals were positive for Env antibody after the first booster inoculation, although titers were low, and titers increased again after a second booster immunization with the SIV/CMVΔvif plus IL-15 DNA vaccine. SIV-specific antibody was not detected by a commercial ELISA after priming immunization (Table 1). However, antiviral antibodies were detected by this ELISA in three out of six animals after the first booster, and in five out of six after the second booster immunization. These findings of low antiviral antibody induction were consistent with previous reports describing inoculation of animals with lentiviruses encoding mutations in vif, including our previous study testing SIV/CMVΔvif as a single modality DNA vaccine (Sparger et al., 2008). These results also revealed that co-inoculation of rIL-15 expression plasmid did not result in a significant improvement of humoral responses over those induced by immunization of SIV/CMVΔvif alone (Sparger et al., 2008), at least as measured by these standard antibody assays.
TABLE 1.
Detection of SIV Antibody by ELISA
| Animal No. | Weeks after priming immunization |
|||||
|---|---|---|---|---|---|---|
| 0 | 8* | 20* | 22 | 24 | 30 | |
| 32180 | − | − | − | + | + | + |
| 31541 | − | − | + | + | + | + |
| 31063 | − | − | + | + | + | + |
| 30889 | − | − | − | + | + | − |
| 30559 | − | − | + | + | + | + |
| 29551 | − | − | − | − | − | − |
Time points for booster immunization.
Effects of vaccination with SIV/CMVΔvif proviral DNA and rIL-15 plasmids on plasma virus load after multiple low dose IVAG challenge with SIVmac251
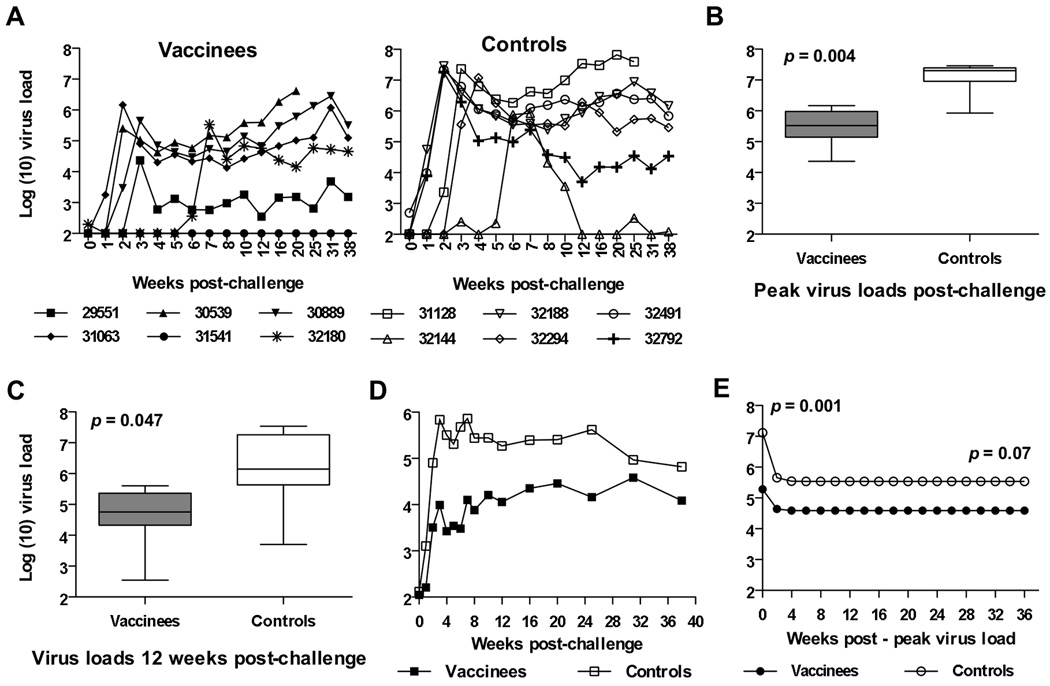
Efficacy of the SIV/CMVΔvif plus IL-15 DNA vaccine was tested by challenge with SIVmac251 by the vaginal route using a multiple low dose protocol as previously described (Ma et al., 2004). This protocol consisted of weekly intravaginal (IVAG) administrations of 103 TCID50 of SIVmac251 to each macaque until that animal became positive for SIV RNA in plasma for two consecutive weeks quantitative real-time reverse transcriptase (RT) TaqMan PCR assay. This challenge protocol was considered a more biologically relevant approach for challenge with the highly pathogenic SIVmac251. Salient findings regarding virus loads after multiple low dose IVAG challenge study included the following observations. Five of six unvaccinated control macaques were positive for plasma viral RNA between 1 to 3 weeks after initial challenge, with peak virus loads of 107 RNA copies per ml or higher (Fig. 4A). Peak virus loads and kinetics were very similar for all unvaccinated macaques with the exception of animal 32144 that required six weeks of challenge before demonstrating plasma viremia with a peak virus load of 8 × 105 RNA copies per ml. This animal subsequently controlled plasma virus load to undetectable levels in plasma by twelve weeks after the initial challenge inoculation. Importantly, plasma virus loads were maintained below 105 RNA copies per ml for four out of the six vaccinated animals, compared to two out of six unvaccinated controls up to 25 weeks after the initial challenge inoculation (Fig. 4A). A comparison of mean peak virus load for viremic vaccinees (5.03 × 105 RNA copies per ml) and unvaccinated animals (1.75 × 107 RNA copies per ml) during acute infection revealed a significant difference based on the Mann-Whitney U test (P = 0.004) (Fig. 4B). Mean virus loads for viremic vaccinated animals remained lower compared to unvaccinated controls at 12 weeks after the initial challenge time point (P = 0.047) (Fig. 4C). Furthermore, a comparison of geometric means for virus loads revealed a reduction of loads by 1 log or more for vaccinated animals compared to controls through a 25 week period after the initial challenge inoculation (Fig. 4D). In a separate analysis, a nonlinear mixed-effects model based on an exponential function was fitted to log plasma RNA values, with values recorded from first observation of peak RNA, to generate plasma virus load curves for comparison of vaccinated and unvaccinated animals. This second analysis also revealed a significantly lower mean peak plasma virus load for vaccinees (P = 0.001) during acute infection and during the early set point period of infection (up to 16 weeks after peak viremia) (P = 0.022), when compared to the virus load curve derived for unvaccinated controls (data not shown). A comparison of mean plasma virus load curves using a similar analysis that included later time points after challenge (20–36 weeks post infection), also revealed a lower mean plasma virus load for viremic vaccinees, although the difference between curves for vaccinated and control animals was not significant when later time points were included (P = 0.07) (Fig. 4E). It is important to note that these analyses included only the viremic vaccinees (5/6) and unvaccinated controls (6/6). Interestingly, one vaccinated animal (31541) remained negative for plasma virus during the entire duration of the study, with the exception of one time point (8 weeks after initial challenge) where a virus load of 60 viral RNA copies per ml was detected.
Figure 4. Plasma virus loads after multiple low dose IVAG challenge of vaccinated and unvaccinated macaques with SIVmac251.
Plasma virus loads represented as SIV RNA copies per ml of plasma, was determined after SIVmac251 challenge by a real-time RT-PCR assay for SIV gag RNA for vaccinated and unvaccinated macaques (A). Peak virus loads (B) and virus loads measured at 12 weeks after initial challenge (C) are compared between viremic vaccinated and unvaccinated macaques using the Mann-Whitney U test with the uninfected vaccinated animal excluded from the analysis. Geometric means for plasma RNA loads for all vaccinated and control macaques over time after the initial challenge inoculation are also shown (D). In a different analysis, a nonlinear mixed-effects model based on an exponential function was fitted to log RNA values, with plasma RNA concentrations recorded from the first observation of peak plasma viral RNA, to provide mean virus load curves for vaccinated and unvaccinated macaques through the duration of the study (E), also with the uninfected vaccinated animal excluded from the analysis.
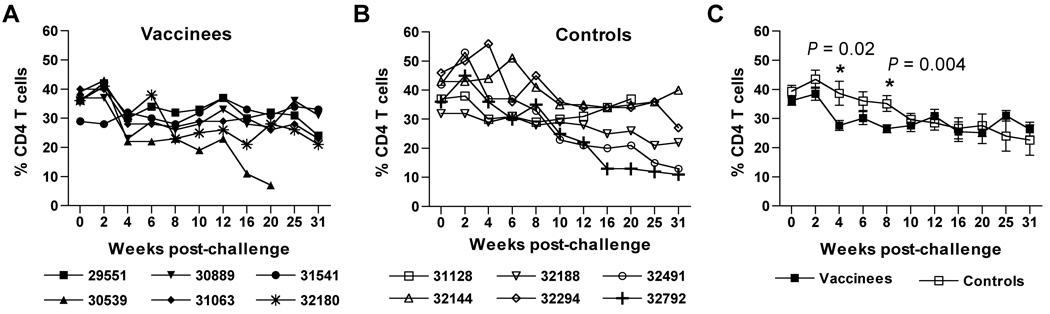
Effects of vaccination on peripheral blood CD4 T cell concentrations after challenge with SIVmac251
Although co-immunization with SIV/CMVΔvif and pNDgmsRRm-IL-15 plasmids resulted in a significant suppression of plasma virus loads after challenge, infection of vaccinated animals resulted in a general decrease of peripheral blood CD4 T cells during the acute phase of infection, even compared to infected unvaccinated controls (Fig. 5A–B). In fact, mean values for CD4 T cell percentages measured for vaccinated animals at four and eight weeks after initial challenge inoculations were moderately but significantly lower (P = 0.02 and P = 0.004, respectively) than mean values measured for unvaccinated controls (Fig. 5C). The reason for this decrease remains to be fully elucidated, but may rely on increased elimination of CCR5+ CD4 T cell targets in the periphery, or increased emigration of CD4 T cells into the tissue as a consequence of IL-15 (Picker et al., 2006). By ten weeks after initial challenge inoculation, CD4 T cell percentages for vaccinated and unvaccinated controls were comparable. Values for CD4 T cell percentages remained stable for vaccinated animals, and were comparable to those measured for unvaccinated controls for the duration of the study.
Figure 5. Quantitation of CD4 T cell percentages after challenge with SIVmac251.
Peripheral blood CD4 T cell percentages were calculated for vaccinated (A) and unvaccinated control (B) macaques after SIVmac251 challenge. Mean values with standard error for CD4 T cell percentages over time challenge are shown for both groups of animals (C). Asterisks (*) positioned over values at 4 and 8 weeks after challenge indicate a significant difference between vaccinated and SIV-infected unvaccinated controls as determined by a pair-wise comparison using a Mann-Whitney U test.
SIV-specific CD8 T cell responses after challenge
IFN-γ ELISPOT responses to all four viral antigens measured for vaccinated animals showed an immediate decline at two weeks after the initial challenge (Fig. 6A). This decline was observed for five of the six vaccinated animals, including macaque 31541 which failed to show evidence of infection. However, low to moderate responses to SIV Gag, Env, and Nef were again evident by 8 to 20 weeks after the initial challenge inoculation, for five of the six vaccinated macaques. In contrast, unvaccinated controls demonstrated very low to undetectable IFN-γ ELISPOT responses to all four SIV antigens during this same time frame (Fig. 6B). Notably, mean IFN-γ ELISPOT responses to SIV Gag, Env, and Nef for vaccinated animals were significantly greater than those measured for unvaccinated controls for multiple time points within 20 weeks after challenge (Fig. 6C). A direct correlation between IFN-γ ELISPOT responses on the day of challenge and control of virus load after challenge was not apparent for individual vaccinees. To examine an association between IFN-γ ELISPOT responses and emergence of plasma viremia after challenge, plasma virus loads were plotted against SIV Gag IFN-γ ELISPOT values over time after challenge (Supplemental Fig. 1). Interestingly, an association between virus loads and IFN-γ ELISPOT responses was suggested for five of six vaccinated macaques, including the vaccinated animal for which plasma viral RNA was detected at a very low concentration for one time point only for the duration of this vaccine study. This particular animal (31541) demonstrated IFN-γ ELISPOT responses for multiple time points after challenge despite an undetectable virus load. However, IFN-γ ELISPOT responses were not observed for another vaccinee that displayed significant suppression of plasma viral RNA concentrations (29551) over time. Higher virus loads were observed with very low or undectectable IFN-γ ELISPOT responses for four unvaccinated controls. In contrast, moderate IFN-γ ELISPOT responses were observed for one control animal (32144), which suppressed plasma virus to concentrations below the limit of detection of the real-time PCR assay. A sixth control animal (32792) that also controlled plasma virus loads to approximately 104 SIV RNA copies per ml, displayed moderate, although transient IFN-γ ELISPOT responses after challenge. Although these findings suggested a possible relationship between virus load and IFN-γ ELISPOT responses, a statistically significant relationship between IFN-γ ELISPOT responses and virus loads after challenge was not evident (R = −0.2476; P = 0.06).
Figure 6. IFN-γ ELISPOT responses in animals after challenge with SIVmac251.
Virus-specific IFN-γ ELISPOT responses were measured for vaccinated (A) and unvaccinated (B) animals over time after the initial challenge inoculation. Mean values for SFC with standard error over time after challenge are shown for both groups of animals (C). Asterisks (*) positioned over values indicate a significant difference between vaccinated and SIV-infected unvaccinated controls as determined by a pair-wise comparison using a Mann-Whitney U test.
SIV-specific T cell proliferative responses after challenge
SIV-specific proliferative responses were variable for vaccinated animals, but were observed for all vaccinees during early time points following the initial challenge inoculation with pathogenic SIVmac251 (Fig. 7A). Although the magnitude of proliferation responses declined over a 16 week time period after challenge for five out of six vaccinated animals, stimulation indices were still greater for vaccinated animals compared to unvaccinated controls (Fig. 7B), during this early time frame of infection, particularly at four weeks after the initial challenge (P = 0.02) (Fig. 7C). Positive proliferative responses during the early 12 week time period after challenge were observed for only two unvaccinated controls (Fig. 7B), which included the control (32144) most resistant to challenge and capable of suppressing plasma virus load to undetectable concentrations (Fig. 4B). Overall, these observations suggested that both proliferative responses and IFN-γ ELISPOT responses detected for vaccinated animals during early time points after challenge were vaccine-associated, rather than induced strictly from active viral infection. SIV-specific proliferation responses were detected once again for vaccinated and unvaccinated animals by 25 weeks after the initial challenge, with the exception of one vaccinee (30359) that did not survive beyond 20 weeks after challenge, and two unvaccinated macaques (31128, 32188), one of which did not survive beyond 25 weeks after challenge. Of note, proliferative responses detected on the day of challenge did not correlate with control of virus load for individual vaccinees (Fig. 4A). Also of interest was the overall transient loss of proliferative responses in vaccinated animals at 16 to 20 weeks after challenge; a time period for which significant differences in set point viral loads were also diminished between vaccinees and controls.
Figure 7. SIV-specific T cell proliferative responses and SIV Env antibody responses in animals after challenge with SIVmac251.
Virus-specific T cell proliferative responses are shown for vaccinated (A) and unvaccinated control (B) animals over time after the initial challenge inoculation. Mean values for SI with standard error over time after challenge are shown for both groups of animals (C). A SI of 2.5 or greater was considered positive (see dashed cut-line). Asterisks (*) positioned over values indicate a significant difference between vaccinated and SIV-infected unvaccinated controls as determined by a pair-wise comparison using a Mann-Whitney U test. Reciprocal endpoint titers for plasma antibodies against native SIV Env are described for vaccinated (D) and unvaccinated control (E) animals over consecutive time points after challenge. Reciprocal endpoint titers > 100 were considered positive.
Antiviral antibody responses after challenge
Anamnestic antibody responses to SIVenv were noted by four weeks after challenge for all vaccinated animals with the exception of vaccinee 31541 that remained negative for plasma viral RNA after multiple challenges (Fig. 7D), confirming the virus load data. Positive SIV Env antibody titers were found for three out of six unvaccinated control animals by week 4 after initial challenge and for five controls by eight weeks after challenge (Fig. 7E). One unvaccinated control animal (31128) failed to develop detectable antiviral plasma antibodies when assayed by EIA (data not shown) or ConA ELISA (Fig. 7E), as well as detectable SIV-specific IFN-γ ELISPOT or proliferative responses, and did not survive beyond 25 weeks after challenge. By 12 weeks after infection Env antibody titers were comparable in controls and vaccines, with the exception of uninfected vaccinee 31541 and the rapid progressor control 31128.
Quality of the cellular immune responses conferred by a rIL-15 expression plasmid to a SIV/CMVΔvif proviral DNA vaccine
To further examine changes in immunogenicity associated with co-immunization of SIV/CMVΔvif DNA vaccine with an rIL-15 expression plasmid, cellular immune responses induced by immunization with SIV/CMVΔvif DNA alone, as previously described (Sparger et al., 2008) were compared to those induced by co-immunization with SIV/CMVΔvif and pNDgmsRRm-IL-15 plasmids. The SIV-specific IFN-γ ELISPOT and proliferation assays used for both vaccine studies were identical and conducted by the same laboratories. Although IFN-γ ELISPOT responses to SIV Gag measured at early time points after priming immunization were comparable for macaques vaccinated with SIV/CMVΔvif DNA alone or with pNDgmsRRm-IL-15 plasmid, responses after the first and second booster immunizations were significantly greater for those animals vaccinated with a SIV/CMVΔvif DNA vaccine that included the rIL-15 expression plasmid (Fig. 8A). In contrast, SIV-specific T cell proliferative responses induced by the priming immunization were significantly improved by addition of the rIL-15 expression plasmid, although improvement was not observed for subsequent time points after booster immunizations (Fig. 8B). These observations suggested that incorporation of rIL-15 into this DNA vaccine regimen affected both CD4 and CD8 T cell responses induced by a SIV/CMVΔvif DNA vaccine, although the effects on CD8 responses were more long-lasting and striking. Alternatively, effects on SIV-specific CD4 T cell responses may have appeared short-lived due to IL-15-induced preferential emigration of antigen-specific effector memory T cells from the periphery to tissues (Picker et al., 2006).
Figure 8. Adjuvant effects on SIV-specific cellular immune responses conferred by IL-15.
Mean values for SIV-specific IFN-γ ELISPOT (SFC) to SIV Gag with standard error over time after priming immunization are shown for animals vaccinated with either SIV/CMVΔvif DNA alone, or with both SIV/CMVΔvif and pNDgmsRRm-IL-15 plasmid DNA (A). Arrows with a solid line denote booster immunization time points for animals vaccinated with SIV/CMVΔvif DNA alone. Arrows with a dashed line denote booster immunization time points for animals vaccinated with SIV/CMVΔvif DNA and pNDgmsRRm-IL-15 plasmids. Asterisks (*) positioned over values indicate a significant difference between values measured for the two different vaccine groups, as determined by a pair-wise comparison using a Mann-Whitney U test. Similarly, mean values for peripheral blood SIV-specific T cell proliferation responses (SI) over time after priming and booster immunizations are shown for animals vaccinated with SIV/CMVΔvif DNA alone or also with pNDgmsRRm-IL-15 plasmid (B). Frequencies of peripheral blood CD8 T cells expressing cytokines and a degranulation marker CD107a after exposure to a SIV capsid peptide pool were measured by CFC at 2 weeks after a second DNA vaccine booster immunization and compared between the two vaccine groups (C). Significant differences between frequencies for each immune response marker for the two vaccine groups were not observed using a pair-wise comparison using a Mann-Whitney U test. Frequencies of peripheral blood CD8 T cells expressing multiple immune response markers, also at 2 weeks after a second DNA vaccine booster immunization, were compared between the two vaccine groups (D). Bar graphs on the left show the percentages of SIV Gag-specific CD8 T cells in each of 15 categories representing all possible combinations of the cytokines and the degranulation marker analyzed. Cells positive for all four markers (i.e., IL-2, IFN-γ, TNF-α and CD107) are indicated in red. Cells positive for three markers are shown in dark blue; those positive for two functions in green; and those positive for a single function in light blue. Pie charts convert the total capsid-specific CD8 T-cell response in each macaque to 100%, with 4-, 3-, 2-, and 1-function cells shown as individual slices of the pie. Pie slices are color-coded to match the bar graphs. Numbers in the center of each pie indicate the total capsid-specific CD8 T-cell response, with all functional categories combined. ID denotes animal number and bar color for that particular animal represented on the graph.
Enhancement of SIV-specific CD8 T cell responses was also substantiated by a subsequent study testing cryopreserved PBMC samples harvested from animals vaccinated with SIV/CMVΔvif DNA alone or with SIV/CMVΔvif and rIL-15 vaccine plasmids, by a cytokine flow cyotometry (CFC) assay testing for antigen-specific expression of cytokines IL-2, TNF-α, IFN-γ, and the degranulation marker CD107a. Based on findings for IFN-γ ELISPOT responses for these two vaccine groups, frequencies of SIV capsid-specific T cells expressing either single or multiple immune response markers at two weeks after a second booster immunization were compared between the groups. Frequencies of SIV-specific CD8 T cells expressing either the degranulation marker CD107a, or TNF-α, and IFN-γ were appreciably higher at this time point for macaques co-immunized with SIV/CMVΔvif DNA plus rIL-15 compared to animals immunized with SIV/CMVΔvif DNA alone (Fig. 8C), although the differences between mean frequencies were not statistically significant. More importantly, evidence of polyfunctional CD8 T cells was shown for three out of the six macaques co-immunized with SIV/CMVΔvif DNA plus rIL-15, whereas polyfunctional responses were not observed for any animals vaccinated with SIV/CMVΔvif DNA alone at this same time point after booster immunizations (Fig. 8D). In contrast, notable differences in SIV-specific CD4 T cell responses between the two vaccine groups at this same time point were not observed by this assay (data not shown). These findings generated from the multi-parameter CFC assay provided further evidence of an enhancement of vaccine-induced CD8 T cell responses by the inclusion of rIL-15 as an immunomodulator for this DNA vaccine approach.
Discussion
Our previous study testing immunogenicity and efficacy of the SIV/CMVΔvif proviral DNA vaccine, revealed that immunization with this vif-deleted SIV provirus resulted in robust SIV-specific T cell proliferative responses, but very weak antigen-specific CD8 T cell responses (Sparger et al., 2008). Furthermore, suppression of virus after vaginal challenge with SIVmac251 in vaccinated animals was transient and apparent only for the first eight weeks after challenge. Long-term immunological protection is determined by both the quantity and quality of memory T cells produced following initial antigen exposure (Sarkar et al., 2008). As the number of memory T cells generated is largely dependent on the magnitude of the primary immune response, T cell-based vaccine strategies have focused on expanding antigen-specific CD8 T cell populations. Earlier reports suggested promise for the γc chain cytokine IL-15 as a vaccine adjuvant capable of promoting amplification and survival of antigen-specific CD8 T cells (Becker et al., 2002; Oh et al., 2003; Surh and Sprent, 2002; Waldmann et al., 2001). Accordingly, we hypothesized that co-immunization of the SIV/CMVΔvif DNA vaccine with an optimized rIL-15 expression plasmid would result in SIV-specific CD8 T cell responses augmented for both frequency and polyfunctionality. We also hypothesized that rIL-15-amplified antigen-specific CD8 T cell responses would result in sustained suppression of virus loads after vaginal challenge with pathogenic SIVmac251. Notably, findings from this current study confirmed that co-administration of a rIL-15 expression vector with the SIV/CMVΔvif DNA vaccine was capable of substantial adjuvant activity for the SIV/CMVΔvif DNA vaccine, based on results from both immunogenicity and efficacy studies.
One objective of this vaccine study was to examine SIV-specific immune responses elicited by the SIV/CMVΔvif proviral DNA vaccine regimen that incorporated a rIL-15 expression plasmid, and to compare these responses to those elicited by immunization with SIV/CMVΔvif DNA alone as reported previously (Sparger et al., 2008). Given that IL-15 would be expected to primarily impact T cell responses, similarities in antibody responses that were observed for the two different vaccine protocols were not unexpected (data not shown). Interestingly, robust vaccine-induced SIV-specific peripheral T cell proliferative responses were comparable for SIV/CMVΔvif DNA-vaccinated animals with, or without rIL-15 expression plasmid co-inoculation. Similarly, assessment of SIV Gag-specific CD4 responses tested at a single time point, two weeks after a second booster immunization, by the multi-parameter CFC assay also revealed comparable activity for animals immunized with SIV/CMVΔvif DNA alone or with rIL-15 plasmid (data not shown).
In contrast to findings for antigen-specific T cell proliferative and CD4 responses, observations from this vaccine study suggested that the principal IL-15 adjuvant effect was to increase function and frequency of peripheral SIV-specific CD8 T cell responses. Although IL-15 was shown by other investigators to enhance both CD4 and CD8 antigen-specific responses (Boyer et al., 2007; Oh et al., 2008; Picker et al., 2006; Villinger et al., 2004), previous reports focused on IL-15-mediated effects on antigen-specific CD8 T cell populations, demonstrated increased antigen-specific CD8 T cell frequency, survival, and independence from the requirement for CD4 T cell help (Boyer et al., 2007; Halwani et al., 2008; Melchionda et al., 2005; Mueller et al., 2005; Oh et al., 2008; Picker et al., 2006; Surh et al., 2006; Villinger et al., 2004; Waldmann et al., 2001; Wallace et al., 2006; Zhang et al., 2006). A comparison of SIV-specific IFN-γ ELISPOT after immunization with either SIV/CMVΔvif DNA alone, or with SIV/CMVΔvif DNA and rIL-15, revealed significant increases in ELISPOT responses to SIV Gag for animals immunized with vaccine including rIL-15 expression plasmid, particularly for time-points after the second booster immunization. A trend towards increased frequencies of SIV Gag-specific CD8 T cells capable of expressing either IFN-γ, TNF-α, or CD107a, was also observed at this same time point. Moreover, animals immunized with an SIV/CMVΔvif DNA vaccine that included rIL-15, were more likely to develop polyfunctional CD8 T cell responses, as shown by data from the CFC assay. These data revealed that at least half of the animals co-immunized with SIV/CMVΔvif and rIL-15 DNA, demonstrated polyfunctional SIV Gag-specific CD8 T cells capable of expressing two to three cytokines including IFN-γ, TNF-α, and IL-2, as well as a degranulation marker CD107a. Conversely, none of the animals vaccinated with SIV/CMVΔvif alone exhibited SIV Gag-specific polyfunctional CD8 T cell responses. Recent reports have shown that polyfunctional antigen-specific CD8 and CD4 T cell responses may be important correlates for control of HIV infection and for vaccine efficacy (Almeida et al., 2007; Duvall et al., 2008; Genescà et al., 2007; Harari et al., 2007; Makedonas and Betts, 2006; Precopio et al., 2007). Frequencies of SIV Gag-specific CD8 T cells measured by these CFC assays and observed for animals immunized with SIV/CMVΔvif and rIL-15 DNA were modest compared to values observed for SIV or HIV-infection or combination immunization approaches that including both DNA and viral vector modalities. However, these responses were very significant compared to those generated after immunization with HIV-1 and SIV DNA vaccines alone (Catanzaro et al., 2007; Kwissa et al., 2007; Tavel et al., 2007). Accordingly, these findings from the CFC studies provide strong evidence of IL-15’s capacity to improve both the quality and quantity of antigen-specific CD8 T cell responses induced by a SIV/CMVΔvif DNA vaccine.
The possibility that IL-15 may have increased viral gene expression or virus replication from the provirus vaccine plasmid via activation of the transduced target cells cannot be ruled out and warrants consideration. Reports describing investigation of IL-15 as a therapeutic in SIV-infected animals have generated conflicting observations regarding the benefits of such treatment with this cytokine. Two reports described beneficial effects for IL-15 on either CD4 or CD8 T cell populations, without adverse effects on virus load (Mueller et al., 2005; Picker et al., 2006). Other reports described adverse effects on disease and increased virus loads when IL-15 was tested as a therapeutic agent during acute SIV infection (Mueller et al., 2008), or was used as an adjuvant for a therapeutic vaccine for SIV infection (Hryniewicz et al., 2007). Both reports described significant proliferation or activation of the memory effector CD4 T cell subset associated with IL-15 treatment that possibly caused, or at least contributed to the enhanced virus loads observed with use of this cytokine in a therapeutic regimen. However, testing of IL-15 as a therapeutic during infection as described by these reports, is quite different from use of an IL-15 expression plasmid as an adjuvant for a prophylactic DNA vaccine. The CMV/SIVΔvif DNA vaccine is not an absolutely defective provirus; replication of virus generated from this provirus is possible only in cells that do not express critical APOBEC3 proteins including APOBEC3F and APOBEC3G (Chiu and Greene, 2008). These cellular restriction factors eliminate most, if not all of the viable target cell populations capable of propagating this viral mutant in vivo. Therefore, the nature of the pre-integration infectivity defect conferred by a vif deletion generates an essentially single cycle virus, and rules out the likelihood of actual increased virus replication or virion infectivity induced by IL-15, regardless of CD4 T cell or immune activation events. Importantly, real-time PCR assays for peripheral blood virus load, including plasma viral RNA and PBMC viral DNA, did not detect viral nucleic acid after multiple proviral DNA inoculations with IL-15 for the current study (data not shown), similar to previous findings with multiple inoculations of proviral DNA without IL-15 (Sparger et al., 2008). Furthermore, evidence of increased virus expression was not supported by increased antiviral antibody titers after immunization when compared to vaccination with CMV/SIVΔvif alone. Thus, augmented CD8 T cell response most likely resulted from immunomodulatory properties associated with the rIL-15, rather than increased proviral DNA vaccine expression.
Another critical objective for this vaccine study was to test the efficacy of a SIV/CMVΔvif proviral DNA vaccine approach that incorporated rIL-15 plasmid as a cytokine adjuvant. Vaccinated and unvaccinated control macaques were challenged with highly pathogenic SIVmac251 by the IVAG route using a previously described multiple low dose protocol (Ma et al., 2004). Although virus load kinetics were comparable for vaccinated and unvaccinated controls, peak plasma virus loads and more importantly viral load set points, were significantly lower for vaccinated animals compared to unvaccinated controls for an extended period of time, a result that has been challenging for many vaccine studies using SIVma251 as the challenge isolate (Feinberg and Moore, 2002; Watkins et al., 2008). Moreover, this control of viremia in vaccinated animals correlated with moderate SIV IFN-γ ELISPOT and T cell proliferative responses that were absent in unvaccinated controls during the first twelve weeks after the initial challenge inoculation. These observations suggest that vaccine-induced cellular immune responses were preserved during the acute phase of infection in the face of peak virus replication and may have contributed to suppression of virus loads in vaccinated animals. Plotting of SIV Gag IFN-γ ELISPOT responses against virus loads over the first 20 weeks after challenge suggested an association between T cell responses and host control of virus replication although a significant correlation was not detected. These data therefore lend rationale for the inclusion of a T cell-targeted immunomodulator such as IL-15 for HIV-1 prophylactic immunization strategies.
A comparison of vaccine efficacy generated by co-immunization with CMV/SIVΔvif DNA and rIL-15 as an adjuvant with efficacy afforded by immunization with CMV/SIVΔvif proviral DNA alone (Sparger et al., 2008), was somewhat complicated by the use of two different SIVmac251 IVAG challenge protocols for each vaccine study. Interestingly, virus load kinetics for the unvaccinated controls challenged with multiple low doses in the current study were comparable to those for unvaccinated controls receiving a single day high dose vaginal challenge in our previous CMV/SIVΔvif DNA vaccine study (data not shown). However, a decreased median survival for unvaccinated controls was associated with the high dose IVAG challenge used in the previous study; this difference in responses to challenge by the unvaccinated controls precluded a direct comparison of outcomes for animals vaccinated with either CMV/SIVΔvif DNA alone or with rIL-15. Thus, while we are not proposing a direct comparison of challenge outcomes between both studies, our current data suggest improvement in vaccine-induced cellular immune responses, conservation of these responses after infection, and antiviral control upon addition of IL-15 to the CMV/SIVΔvif DNA vaccine. These findings support other recent reports showing improved efficacy elicited by addition of a rIL-15 expression plasmid to a SHIV DNA vaccine (Boyer et al., 2007; Yin et al., 2008), but conflict with other recent studies showing no efficacy for different SIV DNA vaccine approaches incorporating a rIL-15 expression plasmid when tested against either SHIV89.6P or SIVmac251 challenges (Chong et al., 2007; Demberg et al., 2008). However, differences in multiple variables between these studies and our current study including the specific SIV DNA vaccine tested, IL-15 plasmid dose, DNA delivery schedule, sites for IL-15 plasmid delivery, and challenge routes, may be responsible for the dissimilarity in challenge outcomes. Clearly, future studies with larger numbers of animals and immune response assays that address antigen-specific responses within different T cell memory populations as well as expression of additional T cell functions, will be needed to more clearly define immune correlates for this vaccine approach based on the combination of vif-deleted proviral DNA and IL-15 expression plasmid. Regardless, our current findings illustrate the potential for this cytokine to modulate immunogenicity and efficacy of a prophylactic anti-SIV DNA vaccine. Overall, these findings reveal important implications for the design of future HIV-1 vaccines and support further exploration of IL-15 as an immunomodulator for a prophylactic HIV-1 vaccine.
Materials and methods
Vaccine plasmids
The SIV/CMVΔvif vaccine plasmid containing a vif-deleted SIVmac239 proviral DNA genome and a hybrid SIVmac239 LTR encoding the enhancer region of the human cytomegalovirus (CMV) immediate early (IE) gene promoter, was previously described (Sparger et al., 2008; Zou and Luciw, 1996). All SIV/CMVΔvif plasmid preparations were tested for stability by proviral plasmid transfection of 293 T cells followed by assay of transfected-cell culture supernatants for virus particles and for infectivity for MM221 cells, a rhesus T cell line (provided by Ronald Desrosiers, New England Primate Research Center) as previously described (Sparger et al., 2008).
Two approaches were taken to generate a rIL-15 expression plasmid for testing as a cytokine adjuvant for the SIV/CMVΔvif proviral DNA vaccine. One plasmid designated pNDRRm-IL-15, was constructed by insertion of Mamu (rhesus macaque) IL-15 sequence including the long native 48 amino acid leader sequence (Villinger et al., 1995), into expression vector pND (kindly provided by Dr. G Rhodes, University of California Davis) (Lena et al., 2002). The insert of a second rhesus rIL-15 pND plasmid (pNDgmsRRm-IL-15) contained the 17 amino acid Mamu GM-CSF leader sequence fused to the mature Mamu IL-15 coding sequence to replace the native IL-15 leader sequence. Both rIL-15 expression plasmids were confirmed by nucleotide sequencing and assayed for activity as described below. All vaccine plasmid DNA was prepared by standard alkaline lysis and centrifugation to equilibrium twice in cesium chloride-ethidium bromide gradients.
In vitro analysis of the rhIL-15 expression plasmid
Plasmids pNDRRm-IL-15 and pNDgmsRRm-IL-15 were tested for production of cell-free IL-15 by transfection of COS-7 and 293T cells using DOTAP (Roche) according to the manufacturer’s instructions. Transfected-cell culture supernatants were harvested at two, four, and six days after transfection, passed through a 0.22 µ filter and tested by a commercial human IL-15 antigen-capture ELISA (Invitrogen, Biosource) that is cross-reactive with Mamu IL-15. Supernatants from cells transfected with IL-15 expression plasmids were also tested for IL-15 biological activity using the IL-2/15-dependent HT-2 cell line as previously described (Villinger et al., 1995).
Vaccination and Challenge
Adult colony-bred, multiparous female rhesus macaques (Macaca mulatta) were housed at the California National Primate Research Center (CNPRC) in accordance with the regulations of the American Association for Accreditation of Laboratory Animal Care standards. All animals were negative for antibodies to HIV-2, SIV, type D retrovirus, and simian T-cell lymphotropic virus type 1 prior to the start of the study. Tissue typing revealed that all animals were negative for the major histocompatibility complex (MHC) class I allele Mamu-A*01, whereas one animal (30889) within the vaccinated group was positive for MHC class I allele Mamu-B*17. Animals were examined at regular time intervals for body weight, lymphadenopathy, splenomegaly, and opportunistic infections. Criteria for euthanasia included three or more of the following: a greater than 10% loss of body weight, persistent infections unresponsive to therapy, chronic diarrhea unresponsive to therapy, persistent and marked hematologic abnormalities, and persistent dehydration without fluid supplementation.
Six female macaques were immunized by co-inoculation of 1 mg SIV/CMVΔvif and 1 mg pNDgmsRRm-IL-15 distributed between intramuscular (IM) and intradermal (ID) routes of delivery. Specifically, each animal received 600 µg of SIV/CMVΔvif plasmid DNA and 600 µg of pNDgmsRRm-IL-15 plasmid as two separate IM injections. Each animal also received 400 µg of SIV/CMVΔvif plasmid DNA and 400 µg of pNDgmsRRm-IL-15 plasmid via separate ID inoculations. Animals received booster inoculations of vaccine plasmids in identical concentrations and by the same routes at 8 and 20 weeks after the priming immunization. At 30 weeks after priming immunization, a multiple low dose SIVmac251 challenge by the vaginal route was performed weekly as previously described (Ma et al., 2004). Briefly, each individual animal received two, 1 ml inoculations of 103 50% tissue culture infectious doses 50 (TCID50) of SIVmac251 intravaginally within a four hour time period over one day, at weekly intervals until that animal demonstrated a plasma viremia for two consecutive weeks, or until all control animals tested positive for plasma SIV RNA. Peripheral blood was collected at two to four week intervals after immunization and challenge, for assay of virus-specific immune responses and hematologic alterations by complete blood cell count and CD4/CD8 subset analysis.
Virus load determination
A previously described quantitative real-time RT TaqMan PCR assay (Applied Biosystems, Foster City, CA) (Leutenegger et al., 2001) with a detection limit of 100 RNA copies/ml, was used for measurement of plasma SIV RNA concentrations. The presence of PBMC-associated SIV proviral DNA was also tested using previously described protocols for DNA extraction (Sparger et al., 2008) and a real-time TaqMan PCR assay that included assay for rhesus IL-2 as a house-keeping gene for all reactions (Leutenegger et al., 2001). All PCR reactions were performed in an ABI PRISM Sequence Detector (Applied Biosystems).
Antiviral antibody detection
Plasma SIV-specific antibody was assayed by a commercial HIV-1/HIV-2 ELISA (Genetic Systems Corporation, Redmond, WA) following the manufacturer’s instructions. Plasma antibody to native SIV envelope proteins was assayed by a highly sensitive Con-A ELISA as previously described (Cole et al., 1997) with the following modification. ELISA plates were coated with a four-fold increase in concentration of SIVmac239 virus to enhance the sensitivity of this ELISA.
IFN-γ ELISPOT assay
SIV-specific IFN-γ-producing PBMC were quantitated from cryopreserved PBMC by a previously described recombinant vaccinia virus-based ELISPOT assay after immunization and virus challenge (Larsson et al., 1999; Shacklett et al., 2002; Sparger et al., 2008). Antigens used for this IFN-γ ELISPOT included recombinant vaccinia viruses expressing SIVmac251 Gag (vAbT252), Env (vAbT2531), Pol (vAbt258), and Nef, or control r-VV (Therion Biological, Cambridge, MA). Spot-forming cells (SFC) detected on a commercial 96-well ELISPOT plate (U-Cytech, Utrecht, The Netherlands) coated with monoclonal antibody specific for macaque IFN-γ were counted using an automated ELISPOT reader (AID, Automimmun Diagnostika, Cell Technology Inc., Columbia MD). Results were presented as spot-forming cells (SFC) per 106 cells after subtraction of background SFC measured for control vaccinia virus and values greater than 50 SFC per 106 cells, were considered positive.
SIV-specific T cell proliferation assay
SIV-specific T cell proliferative responses were measured using a previously described traditional 3H-labeled thymidine incorporation assay (Abel et al., 2003; McChesney et al., 1998). SIV inactivated by treatment with aldrithiol-2 (AT-2) (kindly provided by Jeff Lifson, Laboratory of Retroviral Pathogenesis, SAIC Frederick, Bethesda, MD) at concentrations of 12.5 and 125 ng of SIV p27Gag/well, provided the viral antigen used for this T cell proliferation assay. The stimulation index (SI) was calculated as the ratio between counts per minute (CPM) with antigen and CPM in the absence of antigen and a SI of 2.5 or greater was considered positive. All proliferation assays were performed by the CNPRC Immunology Core.
Cytokine flow cytometry
SIV-specific T cell responses after vaccination were examined for phenotype and multiple functions by a multiparameter cytokine flow cytometry assay. All antibodies were purchased from BD Biosciences and titrated prior to use. The LIVE/DEAD Aqua Fixable Cell Stain Kit was purchased from Invitrogen/Molecular Probes. Cryopreserved rhesus macaque PBMC prepared from vaccinated animals from the current study and a previous SIV/CMVΔvif DNA vaccine study described below (Sparger et al., 2008), were thawed and rested overnight in RPMI 1640 containing 20% FBS at 37C with 5% CO2. PBMC were tested for SIV Gag responses using a pool of SIV capsid 15-mer peptides overlapping by 11 amino acids (NIH AIDS Research and Reference Reagent Program). Stimulation with mitogen SEB (100 ug/ml) and DMSO (1.5%) were used as positive and negative controls, respectively. During the 6-hour stimulation in R15 (15% FBS) media, CD49d (clone L25) and CD28 (clone L293) antibodies (1 ug/ml), Brefeldin A (5 ug/ul, Sigma), GolgiStop (1 ul, BD Biosciences) and CD107a PE-Cy7 (clone H4A3) were included. Cells were harvested, washed, and further processed using Cytofix/Cytoperm Kit (BD Biosciences) according to manufacturers recommendations with the following modifications. Cells were re-suspended in 0.5mM EDTA and stained for cell surface markers with CD4-FITC (clone L200), CD8-APC (clone SK1), CD3-PacBlue (clone SP34-2), and for viability with Aqua dye. Cells were next washed with PBS, and fixed and permeabilized using cytofix/cytoperm reagent from the commercial kit. Cells were then stained for intracellular cytokine expression with monoclonal antibodies reactive to TNF-α-Alexa700 (clone MAb11), INF-γ-PE-Cy5 (clone B27), and IL-2-PE (clone MQ1-17H12). Data from 100,000 lymphocyte-gated events were acquired using a LSRII flow cytometer (BD Biosciences) and analyzed using FlowJo software (Treestar, Inc). Data was reported after subtraction of values for DMSO treatments alone. Responses with frequencies equal to or greater than 0.02% were considered positive for single and multiple markers. Software programs SPICE v4.15 and PESTLE v1.5.2 (provided by Mario Roederer, Vaccine Research Center, NIAID, NIH) were used to characterize polyfunctional T cell responses and to generate pie charts representative of individual animal’s CD8 T cell responses.
Comparison of immunogenicity afforded by an IL-15 expression plasmid to a vif-deleted SIVmac239 proviral DNA vaccine
Cellular immune responses measured by SIV-specific IFN-γ ELISPOT, proliferation, and CFC assays were compared for vaccine groups immunized with SIV/CMVΔvif proviral DNA alone as previously described (Sparger et al., 2008), or co-immunized with SIV/CMVΔvif and pNDRRm-IL-15 plasmids as described for the current study. For the previously reported SIV/CMVΔvif DNA vaccine study, a group of six female rhesus macaques was immunized with SIV/CMVΔvif DNA plasmid at similar concentrations (1 mg) distributed between ID and IM routes as described for the current study. The boosting schedule used for this previous study was slightly different with a first booster SIV/CMVΔvif DNA inoculation administered at 6, instead of 8, weeks after priming immunization, and a second booster SIV/CMVΔvif DNA at 22, instead of 20, weeks after priming immunization. SIV/CMVΔvif DNA plasmid concentrations and routes of delivery for booster immunizations were identical to those described above for SIV/CMVΔvif DNA co-immunized with pNDgmsRRm-IL-15 plasmid.
Statistics
A nonlinear mixed-effects model based on an exponential function was fitted to log RNA values using software SAS version 9, with RNA recorded from the first observation of peak plasma viral RNA, to compare virus loads between vaccinated and unvaccinated animals after challenge. Virus loads at specific time points were also compared using the Mann-Whitney U test (GraphPad Prism). Other statistical analysis comparing values for SIV-specific IFN-γ ELISPOT, T cell proliferation responses, and CD8 T cell frequencies for single immune response markers at specific time points, was also based on the Mann-Whitney U test. Correlation between IFN-γ ELISPOT responses and virus loads after challenge was assayed by the Spearman rank-order correlation test (GraphPad Prism). P values < 0.05 were considered significant.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge the expert technical assistance of Hung Kieu, Lou Adamson, Joanne Higgins, and Ding Lu (CNPRC). We would also like to thank Jonathon Steckbeck (University of Pittsburgh). The authors acknowledge helpful discussions and consultations with Drs. Kristina Abel, Meritxell Genesca, and Christopher Miller. We thank Dr. Mario Roedoerer (NIH, VRC) for providing SPICE and PESTLE software. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: Human rIL-2 from Dr. Maurice Gately, Hoffmann-La Roche Inc. This research was supported by NIH/NIAID grants R01AI49703 (E. E. Sparger), P30 AI49366 (R. Pollard), R0152058 (K. S. Cole), T32 AI060555 (J. Solnick), and in part by the Base Grant (RR00169) to the California National Primate Research Center (P. A. Luciw) and the Resource for Nonhuman Primate Immune Reagents (F. Villinger, NIH R24-RR016988).
References
- Abel K, Compton L, Rourke T, Montefiori D, Lu D, Rothaeusler K, Fritts L, Bost K, Miller CJ. Simian-human immunodeficiency virus SHIV89.6-induced protection against intravaginal challenge with pathogenic SIVmac239 is independent of the route of immunization and is associated with a combination of cytotoxic T-lymphocyte and alpha interferon responses. J. Virol. 2003;77:3099–3118. doi: 10.1128/JVI.77.5.3099-3118.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida JR, Price DA, Papagno L, Arkoub ZA, Sauce D, Bornstein E, Asher TE, Samri A, Schnuriger A, Theodorou I, Costagliola D, Rouzioux C, Agut H, Marcelin AG, Douek D, Autran B, Appay V. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 2007;204:2473–2485. doi: 10.1084/jem.20070784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker TC, Wherry EJ, Boone D, Murali-Krishna K, Antia R, Ma A, Ahmed R. Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. J. Exp. Med. 2002;195:1541–1542. doi: 10.1084/jem.20020369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer JD, Robinson TM, Kutzler MA, Vansant G, Hokey DA, Kumar S, Parkinson R, Wu L, Sidhu MK, Pavlakis GN, Felber BK, Brown C, Silvera P, Lewis MG, Monforte J, Waldmann TA, Eldridge J, Weiner DB. Protection against simian/human immunodeficiency virus (SHIV) 89.6P in macaques after coimmunization with SHIV antigen and IL-15 plasmid. Proc. Natl. Acad. Sci. USA. 2007;104:18648–18653. doi: 10.1073/pnas.0709198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calarota SA, Weiner DB. Approaches for the design and evaluation of HIV-1 DNA vaccines. Expert Rev. Vaccines. 2004;3:S135–S149. doi: 10.1586/14760584.3.4.s135. [DOI] [PubMed] [Google Scholar]
- Catanzaro AT, Roederer M, Koup RA, Bailer RT, Enama ME, Nason MC, Martin JE, Rucker S, Andrews CA, Gomez PL, Mascola JR, J NG, Graham BS, Team VS. Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine. 2007;25:4085–4092. doi: 10.1016/j.vaccine.2007.02.050. [DOI] [PubMed] [Google Scholar]
- Chiu YL, Greene WC. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. [DOI] [PubMed] [Google Scholar]
- Chong SY, Egan MA, Kutzler MA, Megati S, Masood A, Roopchard V, Garcia-Hand D, Montefiori DC, Quiroz J, Rosati M, Schadeck EB, Boyer JD, Pavlakis GN, Weiner DB, Sidhu MK, Eldridge JH, Israel ZR. Comparative ability of plasmid IL-12 and IL-15 to enhance cellular and humoral immune responses elicited by a SIVgag plasmid DNA vaccine and alter disease progression following SHIV(89.6P) challenge in rhesus macaques. Vaccine. 2007;25:4967–4982. doi: 10.1016/j.vaccine.2006.11.070. [DOI] [PubMed] [Google Scholar]
- Cole KS, Rowles JL, Jagerski BA, Murphey-Corb M, Unangst T, Clements JE, Robinson J, Wyand MS, Desrosiers RC, Montelaro RC. Evolution of envelope-specific antibody responses in monkeys experimentally infected or immunized with simian immunodeficiency virus and its association with the development of protective immunity. J. Virol. 1997;71:5069–5079. doi: 10.1128/jvi.71.7.5069-5079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demberg T, Boyer JD, Malkevich N, Patterson LJ, Venzon D, Summers EL, Kalisz I, Kalyanaraman VS, Lee EM, Weiner DB, Robert-Guroff M. Sequential priming with SIV DNA vaccines, with or without encoded cytokines, and a replicating Ad-SIV recombinant followed by protein boosting does not control a pathogenic SIVmac251 mucosal challenge. J. Virol. 2008 doi: 10.1128/JVI.01129-08. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers RC, Lifson JD, Gibbs JS, Czajak SC, Howe AY, Arthur LO, Johnson RP. Identification of highly attenuated mutants of simian immunodeficiency virus. J. Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall MG, Precopio ML, Ambrozak DA, Jaye A, McMichael AJ, Whittle HC, Roederer M, Rowland-Jones SL, Koup RA. Polyfunctional T cell responses are a hallmark of HIV-2 infection. Eur. J. Immunol. 2008;38:350–363. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg MB, Moore JP. AIDS vaccine models: challenging challenge viruses. Nat. Med. 2002;8:207–210. doi: 10.1038/nm0302-207. [DOI] [PubMed] [Google Scholar]
- Genescà M, Rourke T, Li J, Bost K, Chohan B, McChesney MB, Miller CJ. Live attenuated lentivirus infection elicits polyfunctional simian immunodeficiency virus Gag-specific CD8+ T cells with reduced apoptotic susceptibility in rhesus macaques that control virus replication after challenge with pathogenic SIVmac239. J. Immunol. 2007;179:4732–4740. doi: 10.4049/jimmunol.179.7.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Leutenegger CM, Dean GA, Steckbeck JD, Cole KS, Sparger EE. Vaccination of cats with attenuated feline immunodeficiency virus proviral DNA vaccine expressing gamma interferon. J. Virol. 2007;81:465–473. doi: 10.1128/JVI.00815-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halwani R, Boyer JD, Yassine-Diab B, Haddad EK, Robinson TM, Kumar S, Parkinson R, Wu L, Sidhu MK, Phillipson-Weiner R, Pavlakis GN, Felber BK, Lewis MG, Shen A, Siliciano RF, Weiner DB, Sekaly RP. Therapeutic Vaccination with Simian Immunodeficiency Virus (SIV)-DNA+IL-12 or IL-15 Induces Distinct CD8 Memory Subsets in SIV-Infected Macaques. J. Immunol. 2008;180:7969–7979. doi: 10.4049/jimmunol.180.12.7969. [DOI] [PubMed] [Google Scholar]
- Harari A, Cellerai C, Enders FB, Köstler J, Codarri L, Tapia G, Boyman O, Castro E, Gaudieri S, James I, John M, Wagner R, Mallal S, Pantaleo G. Skewed association of polyfunctional antigen-specific CD8 T cell populations with HLA-B genotype. Proc. Natl. Acad. Sci. USA. 2007;104:16233–16238. doi: 10.1073/pnas.0707570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmache A, Russo P, Guiguen F, Vitu C, Vignoni M, Bouyac M, Hieblot C, Pepin M, Vigne R, Suzan M. Requirement of caprine arthritis encephalitis virus vif gene for in vivo replication. Virology. 1996;224:246–255. doi: 10.1006/viro.1996.0526. [DOI] [PubMed] [Google Scholar]
- Hokey DA, Weiner DB. DNA vaccines for HIV: challenges and opportunities. Springer Semin. Immunopathol. 2006;28:267–279. doi: 10.1007/s00281-006-0046-z. [DOI] [PubMed] [Google Scholar]
- Hryniewicz A, Price DA, Moniuszko M, Boasso A, Edghill-Spano Y, West SM, Venzon D, Vaccari M, Tsai WP, Tryniszewska E, Nacsa J, Villinger F, Ansari AA, Trindade CJ, Morre M, Brooks D, Arlen P, Brown HJ, Kitchen CM, Zack JA, Douek DC, Shearer GM, Lewis MG, Koup RA, Franchini G. Interleukin-15 but not interleukin-7 abrogates vaccine-induced decrease in virus level in simian immunodeficiency virus mac251-infected macaques. J. Immunol. 2007;178:3492–3504. doi: 10.4049/jimmunol.178.6.3492. [DOI] [PubMed] [Google Scholar]
- Jalah R, Rosati M, Kulkarni V, Patel V, Bergamaschi C, Valentin A, Zhang GM, Sidhu MK, Eldridge JH, Weiner DB, Pavlakis GN, Felber BK. Efficient systemic expression of bioactive IL-15 in mice upon delivery of optimized DNA expression plasmids. DNA Cell Biol. 2007;26:827–840. doi: 10.1089/dna.2007.0645. [DOI] [PubMed] [Google Scholar]
- Jin X, Bauer DE, Tuttleton SE, Lewin S, Gettie A, Blanchard J, Irwin CE, Safrit JT, Mittler J, Weinberger L, Kostrikis LG, Zhang L, Perelson AS, Ho DD. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 1999;189:991–998. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzler MA, Robinson TM, Chattergoon MA, Choo DK, Choo AY, Choe PY, Ramanathan MP, Parkinson R, Kudchodkar S, Tamura Y, Sidhu M, Roopchand V, Kim JJ, Pavlakis GN, Felber BK, Waldmann TA, Boyer JD, Weiner DB. Coimmunization with an optimized IL-15 plasmid results in enhanced function and longevity of CD8 T cells that are partially independent of CD4 T cell help. J. Immunol. 2005;175:112–123. doi: 10.4049/jimmunol.175.1.112. [DOI] [PubMed] [Google Scholar]
- Kwissa M, Amara RR, Robinson HL, Moss B, Alkan S, Jabbar A, Villinger F, Pulendran B. Adjuvanting a DNA vaccine with a TLR9 ligand plus Flt3 ligand results in enhanced cellular immunity against the simian immunodeficiency virus. J. Exp. Med. 2007;204:2733–2746. doi: 10.1084/jem.20071211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson M, Jin X, Ramratnam B, Ogg GS, Engelmayer J, Demoitie MA, McMichael AJ, Cox WI, Steinman RM, Nixon D, Bhardwaj N. A recombinant vaccinia virus based ELISPOT assay detects high frequencies of Pol-specific CD8 T cells in HIV-1-positive individuals. AIDS. 1999;13:767–777. doi: 10.1097/00002030-199905070-00005. [DOI] [PubMed] [Google Scholar]
- Lena P, Villinger F, Giavedoni L, Miller CJ, Rhodes G, Luciw P. Co-immunization of rhesus macaques with plasmid vectors expressing IFN-gamma, GM-CSF, and SIV antigens enhances anti-viral humoral immunity but does not affect viremia after challenge with highly pathogenic virus. Vaccine. 2002;20(Suppl 4):A69–A79. doi: 10.1016/s0264-410x(02)00391-2. [DOI] [PubMed] [Google Scholar]
- Letvin NL. Progress and obstacles in the development of an AIDS vaccine. Nat. Rev. Immunol. 2007;6:930–939. doi: 10.1038/nri1959. [DOI] [PubMed] [Google Scholar]
- Letvin NL. Correlates of immune protection and the development of a human immunodeficiency virus vaccine. Immunity. 2007;27:366–369. doi: 10.1016/j.immuni.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Leutenegger CM, Higgins J, Matthews T, Tarantal AF, Luciw PA, Pedersen NC, North TW. Real-time TaqMan® PCR as a specific and more sensitive alternative to the branched-chain DNA assay for quantitation of simian immunodeficiency virus RNA. AIDS Res. Hum. Retroviruses. 2001;17:243–251. doi: 10.1089/088922201750063160. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Cheng PJ, Hsiao SS, Lin HH, Hung PF, Kuo ML. Differential effect of IL-15 and IL-2 on survival of phytohemagglutinin-activated umbilical cord blood T cells. Am. J. Hematol. 2005;80:106–112. doi: 10.1002/ajh.20431. [DOI] [PubMed] [Google Scholar]
- Lockridge K, Chien M, Luciw PA, Sparger EE. Protective immunity against feline immunodeficiency virus inducedby inoculation with vif-deleted proviral DNA. Virology. 2000;273:67–79. doi: 10.1006/viro.2000.0395. [DOI] [PubMed] [Google Scholar]
- Lori F, Weiner DB, Calarota SA, Kelly LM, Lisziewicz J. Cytokine-adjuvanted HIV-DNA vaccination strategies. Springer Semin. Immunopathol. 2006;28:231–238. doi: 10.1007/s00281-006-0047-y. [DOI] [PubMed] [Google Scholar]
- Ma ZM, Abel K, Rourke T, Wang Y, Miller CJ. A period of transient viremia and occult infection precedes persistent viremia and antiviral immune responses during multiple low-dose intravaginal simian immunodeficiency virus inoculations. J. Virol. 2004;78:14048–14052. doi: 10.1128/JVI.78.24.14048-14052.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makedonas G, Betts MR. Polyfunctional analysis of human t cell responses: importance in vaccine immunogenicity and natural infection. Springer Semin. Immunopathol. 2006;28:209–219. doi: 10.1007/s00281-006-0025-4. [DOI] [PubMed] [Google Scholar]
- McChesney MB, Collins JR, Lu D, Lü X, Torten J, Ashley RL, Cloyd MW, Miller CJ. Occult systemic infection and persistent SIV-specific CD4+ T cell proliferative responses in rhesus macaques that were transiently viremic after intravaginal inoculation of SIV. J. Virol. 1998;72:10029–10035. doi: 10.1128/jvi.72.12.10029-10035.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchionda F, Fry TJ, Milliron MJ, McKirdy MA, Tagaya Y, Mackall CL. Adjuvant IL-7 or IL-15 overcomes immunodominance and improves survival of the CD8+ memory cell pool. J. Clin. Invest. 2005;115:1177–1187. doi: 10.1172/JCI23134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller YM, Do DH, Altork SR, Artlett CM, Gracely EJ, Katsetos CD, Legido A, Villinger F, Altman JD, Brown CR, Lewis MG, Katsikis PD. IL-15 treatment during acute simian immunodeficiency virus (SIV) infection increases viral set point and accelerates disease progression despite the induction of stronger SIV-specific CD8+ T cell responses. J. Immunol. 2008;180:350–360. doi: 10.4049/jimmunol.180.1.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller YM, Petrovas C, Bojczuk PM, Dimitriou ID, Beer B, Silvera P, Villinger F, Cairns JS, Gracely EJ, Lewis MG, Katsikis PD. Interleukin-15 increases effector memory CD8+ t cells and NK Cells in simian immunodeficiency virus-infected macaques. J. Virol. 2005;79:4877–4885. doi: 10.1128/JVI.79.8.4877-4885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musey L, Hughes J, Schacker T, Shea T, Corey L, McElrath MJ. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N. Engl. J. Med. 1997;337:1267–1274. doi: 10.1056/NEJM199710303371803. [DOI] [PubMed] [Google Scholar]
- Ogg GS, Jin X, Bonhoeffer S, Dunbar PR, Nowak MA, Monard S, Segal JP, Cao Y, Rowland-Jones SL, Cerundolo V, Hurley A, Markowitz M, Ho DD, Nixon DF, McMichael AJ. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- Oh S, Berzofsky JA, Burke DS, Waldmann TA, Perera LP. Coadministration of HIV vaccine vectors with vaccinia viruses expressing IL-15 but not IL-2 induces long-lasting cellular immunity. Proc. Natl. Acad. Sci. USA. 2003;100:3392–3397. doi: 10.1073/pnas.0630592100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Perera LP, Terabe M, Ni L, Waldmann TA, Berzofsky JA. IL-15 as a mediator of CD4+ help for CD8+ T cell longevity and avoidance of TRAIL-mediated apoptosis. Proc. Natl. Acad. Sci. USA. 2008;105:5201–5206. doi: 10.1073/pnas.0801003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picker LJ, Reed-Inderbitzin EF, Hagen SI, Edgar JB, Hansen SG, Legasse A, Planer S, Piatak MJ, D LJ, Maino VC, Axthelm MK, Villinger F. IL-15 induces CD4 effector memory T cell production and tissue emigration in nonhuman primates. J. Clin. Invest. 2006;116:1514–1524. doi: 10.1172/JCI27564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Precopio ML, Betts MR, Parrino J, Price DA, Gostick E, Ambrozak DR, Asher TE, Douek DC, Harari A, Pantaleo G, Bailer R, Graham BS, Roederer M, Koup RA. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J. Exp. Med. 2007;204:1405–1416. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priddy FH, Brown D, Kublin J, Monahan K, Wright DP, Lalezari J, Santiago S, Marmor M, Lally M, Novak RM, Brown SJ, Kulkarni P, Dubey SA, Kierstead LS, Casimiro DR, Mogg R, DiNubile MJ, Shiver JW, Leavitt RY, Robertson MN, Mehrotra DV, Quirk E. Safety and immunogenicity of a replication-incompetent adenovirus type 5 HIV-1 clade B gag/pol/nef vaccine in healthy adults. Clin. Infect. Dis. 2008;46:1769–1781. doi: 10.1086/587993. [DOI] [PubMed] [Google Scholar]
- Robert-Guroff M. Replicating and non-replicating viral vectors for vaccine development. Curr. Opin. Biotechnol. 2007;374:322–337. doi: 10.1016/j.copbio.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz JE, Kuroda MJ, Santra S, Sasseville VG, Simon MA, Lifton MA, Racz P, Tenner-Racz K, Dalesandro M, Scallon BJ, Ghrayeb J, Forman MA, Montefiori DC, Rieber EP, Letvin NL, Reimann KA. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- Shacklett BL, Shaw KE, Adamson LA, Wilkens DT, Cox CA, Montefiori DC, Gardner MB, Sonigo P, Luciw PA. Live, attenuated simian immunodeficiency virus SIVmac-M4, with point mutations in the Env transmembrane protein intracytoplasmic domain, provides partial protection from mucosal challenge with pathogenic SIVmac251. J. Virol. 2002;76:11365–11378. doi: 10.1128/JVI.76.22.11365-11378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparger EE, Dubie RA, Shacklett BL, Cole KS, Chang WL, Luciw PA. Vaccination of rhesus macaques with a vif-deleted simian immunodeficiency virus proviral DNA vaccine. Virology. 2008;374:261–272. doi: 10.1016/j.virol.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surh CD, Boyman O, Purton JF, Sprent J. Homeostasis of memory T cells. Immunol. Rev. 2006;211:154–163. doi: 10.1111/j.0105-2896.2006.00401.x. [DOI] [PubMed] [Google Scholar]
- Surh CD, Sprent J. Regulation of naïve and memory T-cell homeostasis. Microbes Infect. 2002;4:51–56. doi: 10.1016/s1286-4579(01)01509-x. [DOI] [PubMed] [Google Scholar]
- Tavel JA, Martin JE, Kelly GG, Enama ME, Shen JM, Gomez PL, Andrews CA, Koup RA, Bailer RT, Stein JA, Roederer M, Nabel GJ, Graham BS. Safety and immunogenicity of a Gag-Pol candidate HIV-1 DNA vaccine administered by a needle-free device in HIV-1-seronegative subjects. J. Acquir. Immune Defic. Syndr. 2007;44:601–605. doi: 10.1097/QAI.0b013e3180417cb6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer JB, Wahren B, Liu MA. Gene-based vaccines: recent technical and clinical advances. Trends Mol. Med. 2000;12:216–222. doi: 10.1016/j.molmed.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Villinger F, Brar SS, Mayne A, Chikkala N, Ansari AA. Comparative sequence analysis of cytokine genes from human and nonhuman primates. J. Immunol. 1995;155:3946–3954. [PubMed] [Google Scholar]
- Villinger F, Miller R, Mori K, Mayne AE, Bostik P, Sundstrom JB, Sugimoto C, Ansari AA. IL-15 is superior to IL-2 in the generation of long-lived antigen specific memory CD4 and CD8 T cells in rhesus macaques. Vaccine. 2004;22:3510–3521. doi: 10.1016/j.vaccine.2003.07.022. [DOI] [PubMed] [Google Scholar]
- Waldmann TA. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat. Rev. Immunol. 2006;6:595–601. doi: 10.1038/nri1901. [DOI] [PubMed] [Google Scholar]
- Waldmann TA, Dubois S, Tagaya Y. Contrasting roles of IL-2 and IL-15 in the life and death of lymphocytes:implications for immunotherapy. Immunity. 2001;14:105–110. [PubMed] [Google Scholar]
- Wallace DL, Bérard M, Soares MV, Oldham J, Cook JE, Akbar AN, Tough DF, Beverley PC. Prolonged exposure of naïve CD8+ T cells to interleukin-7 or interleukin-15 stimulates proliferation without differentiation or loss of telomere length. Immunology. 2006;119:243–253. doi: 10.1111/j.1365-2567.2006.02429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins DI, Burton DR, Kallas EG, Moore JP, Koff WC. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat. Med. 2008;14:617–621. doi: 10.1038/nm.f.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasutomi Y, Reimann KA, Lord CI, Miller MD, Letvin NL. Simian immunodeficiency virus-specific CD8+ lymphocyte response in acutely infected rhesus monkeys. J. Virol. 1993;67:1707–1711. doi: 10.1128/jvi.67.3.1707-1711.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Dai A, Kutzler MA, Shen A, Lecureux J, Lewis MG, Waldmann T, Weiner DB, Boyer JD. Sustained suppression of SHIV89.6P replication in macaques by vaccine-induced CD8+ memory T cells. AIDS. 2008;22:1739–1748. doi: 10.1097/QAD.0b013e32830efdae. [DOI] [PubMed] [Google Scholar]
- Zhang W, Dong SF, Sun SH, Wang Y, Li GD, Qu D. Coimmunization with IL-15 plasmid enhances the longevity of CD8 T cells induced by DNA encoding hepatitis B virus core antigen. World J. Gastroentol. 2006;12:4727–4735. doi: 10.3748/wjg.v12.i29.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JX, Luciw PA. The requirement for Vif of SIVmac is cell-type dependent. J. Gen. Virol. 1996;77:427–434. doi: 10.1099/0022-1317-77-3-427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.