Summary
How the glucocorticoid receptor (GR) activates some genes while potently repressing others remains an open question. There are three current models for suppression: trans-repression via GR ‘tethering’ to AP-1/NF-κB sites, direct GR association with inhibitory elements (nGREs), and GR recruitment of the corepressor GRIP1. To gain insights into GR suppression, we used genomic analyses and genome-wide profiling of GR, p65, and c-Jun in LPS-stimulated macrophages. We show that GR mediates both activation and repression at tethered sites, GREs, and GRIP1-bound elements, indicating that motif classification is insufficient to predict regulatory polarity of GR binding. Interestingly, sites of GR repression utilize GRIP1’s corepressor function and display reduced histone acetylation. Together, these findings suggest that while GR occupancy confers hormone responsiveness, the receptor itself may not participate in the regulatory effects. Furthermore, transcriptional outcome is not established by sequence, but is influenced by epigenetic regulators, context, and other unrecognized regulatory determinants.
Keywords: GR, NF-κB, AP-1, macrophage, inflammation, ChIP-Seq
Introduction
Inflammation is a complex immune response to injury, infection and tissue stress that is essential for survival and the maintenance of tissue homeostasis, but can greatly compromise cellular function if not controlled. Although acute inflammation is essential for wound repair and anti-microbial defense, dysregulated, chronic inflammation is now recognized to contribute to conditions that have received increasing medical and scientific attention in the past several years, such as type 2 diabetes mellitus, atherosclerosis, asthma, neurodegeneration and cancer (Medzhitov, 2010; Medzhitov and Horng, 2009).
Macrophages are crucial cellular mediators of the inflammatory response. They express pattern-recognition receptors such as Toll-like receptors that sense infectious agents and endogenous danger signals associated with tissue injury. Stimulation of TLR4 by lipopolysaccharide (LPS) results in the rapid activation of signal-dependent transcription factors, the most characterized of which are the nuclear factor-kappaB (NF-κB) and activator protein 1 (AP-1) families. These factors work in a combinatorial manner to rapidly induce the expression of numerous genes, including cytokines and chemokines, that constitute the inflammatory response, exert antimicrobial activities and mediate acquired immunity (Medzhitov and Horng, 2009; Takeda and Akira, 2007).
The glucocorticoid receptor (GR) is one of the most potent anti-inflammatory drug targets in clinical use today and one of the most powerful metabolic regulators. GR belongs to the nuclear receptor superfamily of ligand-activated transcription factors that act as important regulators of developmental, reproductive, homeostatic, metabolic and inflammatory processes (Evans, 1988; Mangelsdorf et al., 1995). Upon ligand binding, GR translocates from the cytoplasm to the nucleus where it can both positively and negatively regulate gene expression. GR has been shown to regulate transcription by binding to consensus DNA sequences known as glucocorticoid response elements (GREs), but the exact mechanisms leading to transcriptional activation versus repression are unclear (Beck et al., 2009). GR has been shown to interact with and inhibit the function of both AP-1 and NF-κB family members and thereby suppress a broad range of responses to inflammatory signaling. The general mechanism proposed for this activity is referred to as ‘trans-repression’, in which GR interferes with activation of inflammatory response genes through protein–protein interactions with co-regulatory proteins and promoter-bound transcription factors (‘tethering’), rather than direct, sequence-specific interactions with DNA. Several studies suggest that tethering of GR to AP-1 or NF-κB appears to alter the assembly of co-activator complexes that are required for gene activation, i.e. at the Il8 or Mmp13 promoters (Glass and Saijo, 2010; Luecke and Yamamoto, 2005; Ogawa et al., 2005; Rogatsky et al., 2002; Schule et al., 1990). Other potential repressive scenarios include competition for cofactors or overlapping binding sites, sequestration of transcription factors, crosstalk with other nuclear factors on cis-regulatory elements or direct binding to so-called negative GREs (nGREs), for example in the osteocalcin promoter (Beck et al., 2009; Morrison and Eisman, 1993). Though specific GRE sequences have been shown to directly affect GR conformation and function in vitro (Meijsing et al., 2009), the intrinsic features through which diverse GR responsive enhancers encode positive versus negative polarity in vivo remain unresolved.
As recent studies suggest a majority of enhancer elements are ‘promoter distal’ (Ghisletti et al., 2010; Natoli et al., 2011), we used transcriptome profiling in combination with genome-wide ChIP-Seq to explore features of negatively GR-regulated enhancers in LPS-activated macrophages. First, we find that TLR4 activation by LPS recrafts the primary ‘quiescent’ GR cistrome to a remodeled and expanded ‘inflammatory cistrome’ comprised of both GR-induced and -repressed genes. Unexpectedly, we find that a majority of both negative and positive GR enhancers are composed of canonical GREs in combination with NF-κB and AP-1 sites. Thus, despite expectations, the presence of these factors alone is not diagnostic of either a positive or negative response. Furthermore, while up to 20% of GR-dependent repression is found at nGREs and tethered sites, approximately 20% of GR-induced genes also harbor these same motifs, making cistromic motif classification insufficient to predict regulatory polarity. In exploring the epigenomic basis of these observations, we find that negative enhancers selectively utilize the corepressor function of GRIP1, antagonize IRF3 activity and display reduced levels of histone H3K9 acetylation marks. This suggests that while the presence of the GR confers hormone responsiveness, the receptor itself may not participate in its ultimate regulatory fate. Therefore, beyond motif structure, these results imply a critical role for epigenetic regulators and chromatin ‘context’ in determining the transcriptional outcome of GR controlled target genes.
Results
TLR4 signaling creates an anti-inflammatory cistrome
To gain deeper understanding of the global regulatory balance between GRE-dependent vs. -independent suppression of inflammatory signaling, we performed genome-wide mapping of GR-bound sequences in primary bone-marrow derived macrophages by ChIP-Seq. We used a dual crosslinking method to detect both tethered and directly bound chromatin, using disuccinimidyl glutarate which reacts with amino groups, and formaldehyde which crosslinks protein-DNA interactions.
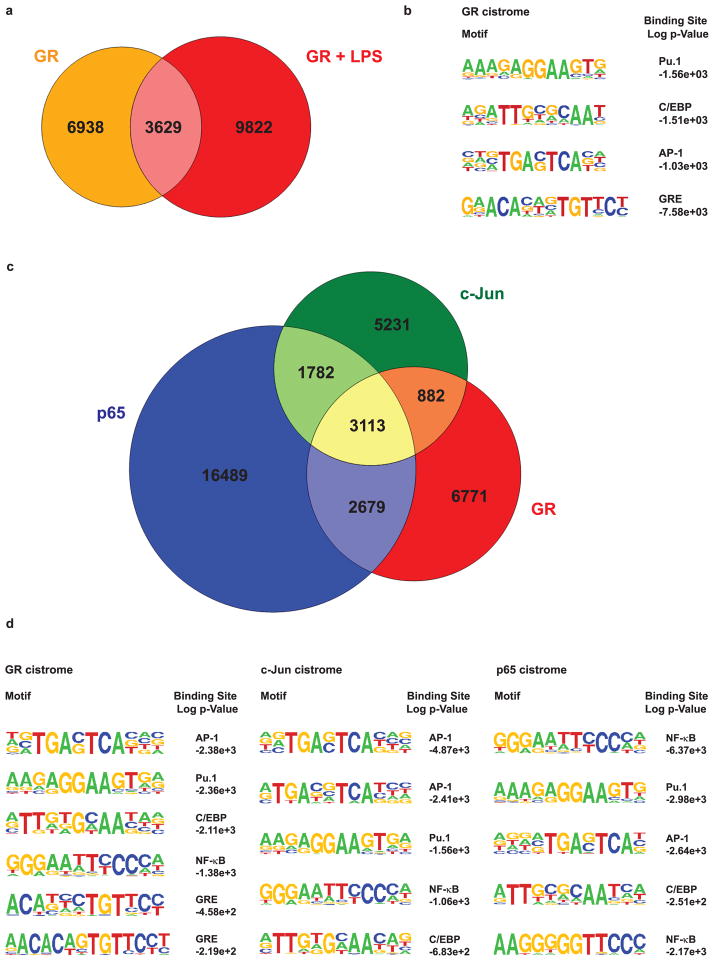
In unstimulated macrophages, we identified 10,567 genomic GR binding sites induced by Dexamethasone (Dex) treatment. Gene annotation analysis to assign ChIP-Seq peaks to the closest transcription start site (based on linear proximity) yielded a total of 5,405 GR target genes. In contrast, in LPS-stimulated macrophages treated with Dex, the GR cistrome consisted of 13,445 binding sites, corresponding to 6,406 target genes. Surprisingly, only about one-third of these sites (3,629 ChIP peaks) were bound by GR in both conditions, while the remaining two-thirds represented a de novo LPS-induced GR cistrome (Figure 1a). Accordingly, a gene ontology (GO) term analysis designating functional annotation showed significant enrichment for immune responses (i.e. cytokine production, defense response, B and T cell activation, inflammatory response) for those GR target loci bound in the presence of LPS (9,822 and 3,629 sites). In contrast, the cis-regulatory sites bound only in the absence of LPS (6,938 sites that were lost after the stimulus) correspond to genes involved in macrophage differentiation, such as regulation of MAPK, Jun Kinase and phosphatase activity, those involved in phagocytosis, proliferation and differentiation (Supplementary Table 1a), as well as ‘housekeeping’ genes.
Figure 1. GR, AP-1 and NF-κB cistromes intersect.
a) Area-proportional Venn diagram of GR cistromes in unstimulated (orange) and stimulated (red) macrophages. b) Motif analysis of the GR cistrome in macrophages treated with Dex only. c) Area-proportional Venn diagram showing the overlap between GR, c-Jun and p65 ChIP-Seq peaks in macrophages treated with LPS and Dex. Genomic regions with significant peak scores were merged and given a 100bp margin for co-occurrence. d) Globally enriched motifs within the GR, c-Jun and p65 cistromes in response to LPS. Tables show the top five to six statistically overrepresented binding sites.
As described above, current hypotheses favor GR participation in crosstalk with NF-κB and AP-1, so the LPS stimulus could conceivably lead to the signal-dependent activation of these two transcription factors which then recruit or ‘tether’ GR to their cognate binding sites. To address this issue, motif analyses were performed on the GR ChIP-sequenced DNA to globally identify statistically overrepresented transcription factor binding sites. Interestingly, in addition to GREs, the most prominent motifs in the unstimulated GR cistrome were consensus sequences for Pu.1, C/EBP and AP-1 (Figure 1b). CCAAT/enhancer-binding protein β has recently been shown to cooperate with GR during 3T3-L1 adipocyte differentiation, with a majority of GR-bound regions overlapping with the C/EBPβ adipogenic cistrome (Steger et al., 2010). In macrophages, Pu.1 and C/EBP family members act as important lineage-determining transcription or pioneer factors that shape cell type-specific responses to inflammatory stimuli (Friedman, 2007; Medzhitov and Horng, 2009).
In order to test for possible ‘tethering’ interactions, we performed ChIP-Seq experiments using antibodies against the p65 subunit of NF-κB and against c-Jun, which is part of the AP-1 complex. In LPS-stimulated Dex-treated macrophages, the c-Jun cistrome comprised 11,008 binding sites, which were annotated to the nearest 5,689 genes. We found 24,063 genomic regions that were associated with p65 binding, which potentially corresponds to 9,227 nearby genes.
Although the major part of GR’s anti-inflammatory potential has been attributed to its ability to interfere with NF-κB and AP-1 activated transcription, the extent of cistromic overlap between these signaling pathways has not yet been evaluated on a genome-wide scale. We performed bioinformatic analysis to calculate the number of co-occurring ChIP-seq peaks for all three factors. Interestingly, we identified 3,113 cis-regulatory regions that were associated with GR, p65 and c-Jun in response to LPS. An additional 2,679 sequences were bound by both GR and p65, whereas 1,782 sites showed intersecting GR and c-Jun occupancies. Our analysis found 6,771 sites which were uniquely bound by GR, along with 16,489 and 5,231 sequences that were solely bound by p65 and c-Jun, respectively (Figure 1c). Taken together, this means that 50.3% of the stimulated GR cistrome was occupied by GR alone (or in combination with unknown factors), and of the remaining fraction, 23.1%, 19.9% and 6.6% were engaged with either all three factors, GR/p65, or GR/c-Jun, respectively (Figures 1c and 3b). Accordingly, motif enrichment analysis of the LPS-reactive GR cistrome revealed AP-1, Pu.1, C/EBP, NF-κB and GRE consensus sequences. Likewise, the c-Jun cistrome displayed AP-1, Pu.1, NF-κB and C/EBP motifs as most prominent. The p65 cistrome most commonly featured NF-κB, Pu.1, AP-1 and C/EBP motifs (Figure 1d).
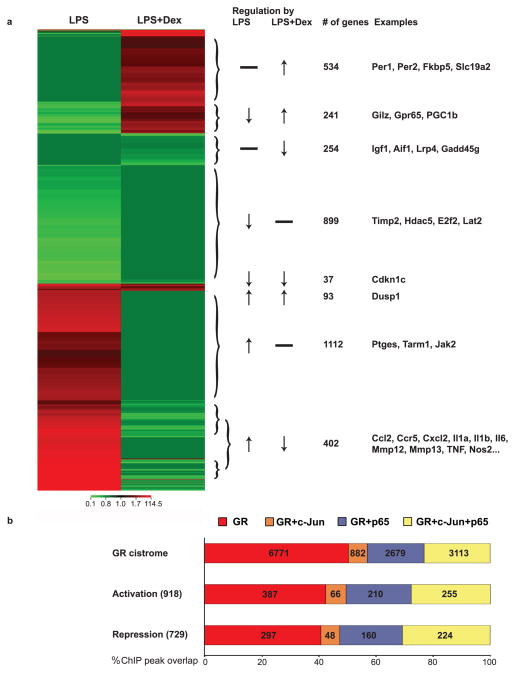
Figure 3. Correlating gene expression and recruitment in response to LPS and Dex.
a) Heat map cluster showing the numbers and overlap of microarray probe sets that were upregulated (red) or downregulated (light green) at least 1.5fold in macrophages treated with LPS and/or Dex. b) Graph showing ChIP peaks assigned to differentially expressed genes and correlating gene expression with occupancy by GR, c-Jun and p65. Percentages and numbers reflect fractions of GR ChIP peaks.
The LPS response induces convergent cistromes
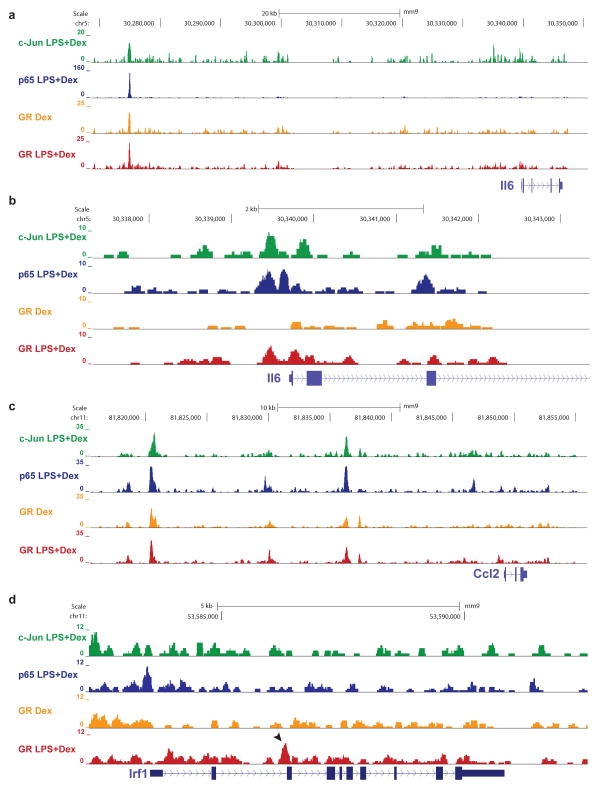
The ChIP-Seq technology per se does not allow a distinction between direct and indirect modes of DNA binding, but a comparison of the two GR cistromes in the presence and absence of the LPS stimulus and hence with and without AP-1 and NF-κB, can be used to discern between the two scenarios (Supplementary Figure 1a+b, (Barish et al., 2010)). Figure 2 shows representative examples of visualizations of these ChIP-sequencing tracks. For instance, one of the key cytokines driving the inflammatory response and a therapeutic target in chronic inflammatory conditions such as insulin resistance, inflammatory bowel disease or rheumatoid arthritis is IL6 (Naugler and Karin, 2008). Interestingly, GR binding was colocalized with p65 and c-Jun occupancy at the Il6 distant regulatory region, and it occurred in the absence of LPS (Figure 2a). Numerous early studies on GR-mediated repression of inflammatory genes have focused on proximal promoters (Luecke and Yamamoto, 2005; Schule et al., 1990). However, as shown in Figure 2b, accessibility for GR binding at the proximal Il6 promoter was dependent on LPS, when AP-1 and NF-κB are also bound, and not in cells treated with Dex alone, which could indicate a tethering situation. Nevertheless, as exemplified by Figure 2c, in many instances, GR binding at inflammatory loci was observed in both stimulated and unstimulated conditions, meaning that GR is able to find some inflammatory targets in the absence of DNA-bound NF-κB or AP-1. Moreover, as shown in Figure 2d, the LPS trigger seems to render additional sites accessible for GR binding, as depicted here for the Irf1 locus. Its intronic enhancer was engaged by GR only in response to LPS, even though it did not colocalize with c-Jun or p65. Notably, only 782 GR binding sites occurred at proximal promoters (less than 6% in stimulated cells), and about half of these appeared linked with a second distal enhancer element (data not shown), suggesting that most positive and negative GR-mediated gene regulation takes place at previously unknown long-range elements.
Figure 2. Genomic localization of GR, AP-1 and NF-κB binding in macrophages.
ChIP-sequencing tracks of regulatory regions of GR target genes in macrophages treated with LPS and/or Dex, normalized to 10 million reads. a) IL6 locus with overlapping GR, p65 and c-Jun occupancy. b) IL6 proximal promoter displaying GR binding only in the presence of c-Jun and p65. c) Ccl2 locus with overlapping GR, p65 and c-Jun binding. GR binding is LPS-independent. d) Irf1 locus with GR binding induced by LPS, but independent of p65 and c-Jun (arrow).
Cistromic intersections are linked to gene expression changes
In macrophages, a crucial point for control of inflammation by nuclear receptors and TLR ligands such as LPS is at the level of gene transcription. We therefore performed microarray expression profiling experiments on LPS- and Dex-treated macrophages to detect gene expression changes in response to the inflammatory stimulus and measure suppression or induction by GR ligand. We found 2,784 transcripts to be differentially expressed (at least 1.5 fold compared to control) upon LPS stimulation. Of these, 1,607 transcripts were induced and 1,177 were downregulated (at least 1.5 fold). Treatment with Dex resulted in 402 of the LPS-induced genes being repressed and 241 of the LPS-repressed genes being upregulated. Furthermore, GR ligand treatment resulted in suppression of an additional 254 genes and activation of 534 genes that did not show expression changes in response to LPS alone (Figure 3a). In sum, we detected changes in mRNA expression level for a total of 1,561 probes when comparing macrophages treated with LPS versus LPS plus Dex. Of these, 868 showed higher and 693 showed lower expression levels than controls (Supplementary Figure 2 and Table 2a). Assigning these genes to functional categories, we found that immune/inflammatory/defense responses were the most frequently represented GO classes (Supplementary Table 2b). For example, Dex treatment resulted in efficient repression of Ccl2, Ccl7, Ccl12, Ccr5, Cxcl2, Mmp12, Mmp13, Il1a, Il6, Il12a, Il15, and TNF mRNAs and induction of ‘classical’ GR target genes such as Per1, Gilz, Fkbp5 and Dusp1 as well as the inhibitory decoy receptor Il1r2.
Our current understanding of gene regulation is centered on a model in which transcriptional changes are the result of a combinatorial code of transcription factors bound to associated cis-regulatory regions. We therefore correlated the mRNA expression levels of differentially expressed genes with promoter/enhancer occupancy detected by ChIP-Seq. GR ChIP-seq peaks were assigned to almost half (49%) of the microarray probes that showed differential expression upon GR ligand treatment, meaning that these are most likely direct target genes. Conversely, 12.2% of GR ChIP-Seq peaks were matched to nearby up- or downregulated genes, indicating functional binding events. Altogether we identified 918 GR-bound cis-regulatory elements that were located near genes induced by Dex treatment, and 729 elements that were assigned to nearby repressed genes (Figure 3b). This affords a seemingly ideal scenario to seek differential motif assignments for Dex-induced versus -repressed genes. Unexpectedly, we found no association between ChIP peak distribution (nearby binding of all three factors / two / GR alone) and transcriptional outcome (activation vs. repression) (X-squared = 0.807, df = 6, p-value = 0.992). That is to say, whether a particular gene was up- or downregulated could not be predicted based on a combination of factors bound to cis-regulatory elements. The majority of genes encoding inflammatory cytokines, such as the IL family, CC and CXC chemokines, Nos2 and TNF, which were largely repressed, showed overlapping binding of GR, p65 and c-Jun, but not exclusively so. For instance, the Ccl12 enhancer was bound and repressed by GR alone, whereas Ccr2 appeared to be regulated by GR and p65, and Cxcl3 regulatory regions were occupied by GR together with c-Jun. GR also seemed to repress transcription of Irf5 in the absence of NF-κB or AP-1 complexes, for example. Surprisingly, the distribution of factors present as reflected by the percentage of ChIP-Seq peaks of GR together with either p65 and/or c-Jun was similar at activated/repressed loci compared to the entire cistrome (Figure 3b). For example, about 20% of the GR cistrome overlap with p65 binding, regardless of whether the associated genes were up- or downregulated. We did observe a shift of almost ten percent, however, with about 30% of cis-regulatory elements being occupied by all three factors resulting in changes in gene expression, compared to about 20% of the global cistrome. Consequently, only ~40% of activating/repressed loci are bound by GR alone, compared to 50% of total sites. It appears that cis-regulatory elements that are engaged by all three factors might be more likely to result in transcriptional changes than those enhancers that are bound only by GR.
Metabolic versus anti-inflammatory functions of GR are cell-type specific
It is believed that the anti-inflammatory actions of glucocorticoids are predominantly based on trans-repression whereas many unwanted side-effects of steroid treatment (such as hyperglycemia and obesity) are due to transcriptional activation (Beck et al., 2009; De Bosscher and Haegeman, 2009). In our study, we did not observe induction of metabolic genes such as Pck1 or G6pc in macrophages treated with Dex. While GR is widely expressed, its cistromes are likely to be cell-type specific. We therefore compared our data set to two recent reports of GR cistromes in 3T3-L1 adipocytes, 3134 mammary and AtT-20 pituitary tumor cells (John et al., 2011; Steger et al., 2010). Strikingly, pairwise cistromic comparisons reveal little overlap (<15%) between enhancers occupied by macrophage GR relative to adipocytes, pituitary and mammary cells (Supplementary Figure 5a). The four cell type comparative cistrome shows less than 1% overlap. Accordingly, there was an enrichment of Runx1 motifs in the 3134 cistrome but an enrichment of FoxA1 motifs in the AtT-20 cistrome (Supplementary Figure 5b). We performed pathway analyses to compare the four different cistromes and found that inflammatory loci are significantly enriched in macrophages but not other cell types (Supplementary Table 1b). As exemplified in Supplementary Figure 5c, GR binding at the Il1a locus only occurs in macrophages, whereas GR targets such as the circadian Per1 locus appear to be ubiquitously occupied in all cell types. In addition, the promoter of the gluconeogenic gene G6pc appears to be bound by GR only in 3134 cells, whereas no GR binding was observed at the Pck1 locus for any of the cell types examined.
Activation and repression by GR involve GREs
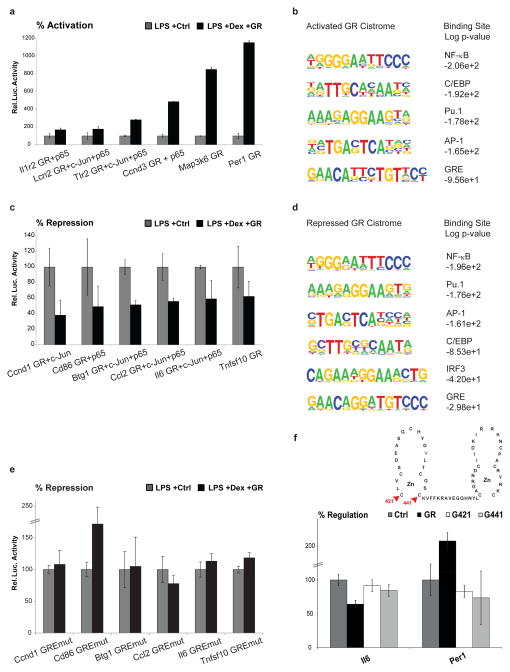
Our next goal was to identify transcriptional complexes and landmarks of GR-regulation as well as distinct motifs linked with activation versus repression. In order to test our assignment of gene expression changes to nearby ChIP peaks and to determine whether these cis-regulatory elements are functional enhancers, we cloned several of them into luciferase reporter constructs. As shown in Figure 4a, ChIP peak sequences that were discovered near upregulated genes were indeed sufficient to confer transcriptional activation in this reporter assay, performed in the RAW264.7 macrophage cell line. Again, these regulatory elements were bound by different combinations of GR/p65/c-Jun in primary macrophages. We also performed a de novo motif discovery specifically using the ChIP peak sequences that were connected to activation of gene expression (Figure 4b). Similar to the motif analysis of the global GR cistrome, these Dex-induced motifs comprised NF-κB, AP-1, Pu.1, C/EBP and GRE consensus sites. Reciprocally, luciferase reporter assays interrogating ChIP peaks detected close to downregulated genes showed that these short sequences contain all that is required for GR-mediated transcriptional repression (Figure 4c). Again, these repressive regulatory sequences were occupied by discrete combinations of the three factors in bone-marrow derived macrophages. De novo motif discovery on ChIP peak sequences categorized as repressive yielded NF-κB, AP-1, Pu.1 and C/EBP consensus sites as expected. Surprisingly, these sequences were also predicted to contain IRF3 motifs as well as classical GREs (Figure 4d). As described above, current hypotheses include a model of ‘tethered’ GRE-independent repression by protein-protein interactions (trans-repression) as a major mechanism for GR-mediated suppression of inflammation (Glass and Saijo, 2010). We therefore decided to test whether repression by GR could indeed occur via direct binding to DNA at canonical GREs by mutating the predicted GRE motifs in our luciferase reporter constructs. As shown in Figure 4e, mutation of the GRE sequence abolished the repressive action of these enhancers by GR, suggesting these GREs are crucial mediators of repression in response to glucocorticoids. In addition, cotransfecting these reporters with GR expression vectors that contain disabling point mutations of conserved cysteine residues within the zinc fingers (Hollenberg and Evans, 1988), showed that both repression and activation is dependent on DNA binding (Figure 4f). Furthermore, recombinant GR protein bound these GRE sequences in vitro in the absence of additional factors, as detected by EMSA (Supplementary Figure 4d). Finally, when performing a motif count of GREs among the different cistromic subsets, the fraction of ChIP peaks containing classical GREs always comprised about 60%, plus approximately 20% of peaks had a detectable half site, irrespective of c-Jun and p65 co-occupancy or positive versus negative regulation (Supplementary Figure 3a). If tethering was the major mechanism of gene silencing by the GR, one would expect a depletion of GRE sequences from those fractions; yet the enrichment remains about 80% across all situations, whether induced or repressed. As shown in Supplementary Figure 3, performing the same motif analyses, but restricted to ±20kb of the transcriptional start site (TSS) to be more confident about the correct assignment to up- and down-regulated genes, yielded the same results. Moreover, we found no discriminatory pattern for the genomic location (distance from the TSS), intersection with CpG islands or peak strength of positive versus negative binding sites. We further validated our motif analyses performing an independent GR ChIP-Seq replicate and taking into account a second microarray experiment performed at the same time as the ChIP studies (3 hours post stimulus), again finding Pu.1, C/EBP, AP-1, NF-κB and GRE motifs associated with both repression and activation, and IRF3 sites only with downregulation. The cistromic intersections were further confirmed by AP-1 and NF-κB ChIP-Seq replicates (Supplementary Figures 2 and 4).
Figure 4. Activation versus repression by GR.
a) Luciferase assays using reporters containing ChIP peak sequences that were linked to activated genes. Black bars represent percent induction in response to Dex. Bottom legend lists occupancy of GR/c-Jun/p65 as determined by ChIP-Seq in primary macrophages. b) De novo motif discovery on ChIP peak sequences that were assigned to genes activated by Dex. c) Luciferase assays using reporters containing ChIP peak sequences that were assigned to repressed genes. Black bars represent percent repression in response to Dex. Bottom legend lists occupancy of GR/c-Jun/p65 as determined by ChIP-Seq in primary macrophages. d) De novo motif discovery on ChIP peak sequences that were assigned to genes repressed by Dex. e) Luciferase assays using reporters that were repressed by GR in c) and that now contain a point mutation in the GRE motif. f) Luciferase assays performed in CV-1 cells (devoid of endogenous GR) using GR point mutants that abolish DNA binding (C421G and C441G). Error bars represent S.E.M.
The coregulator GRIP1 is recruited by GR
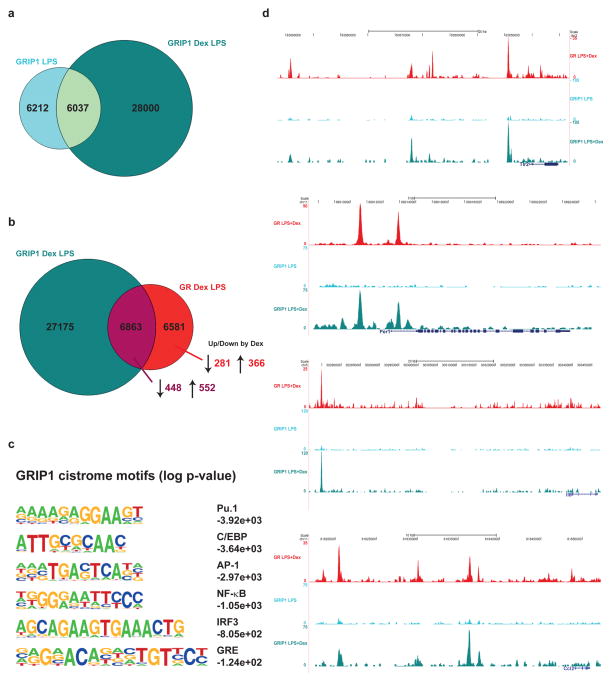
As shown in Figure 4d, we observed IRF3 motifs specifically enriched at enhancers that are bound and repressed by GR. Interference with IRF3 function has previously been implicated in GR-controlled transcriptional repression, and several mechanisms have been proposed. One scenario entails disruption of a p65/IRF3 complex that is required for TLR4-dependent transcriptional activation of inflammatory genes (Ogawa et al., 2005), whereas another proposes competition for the cofactor GRIP1 by GR and IRF3 downstream of TLR3 signaling. GRIP1 is a member of the NCoA/SRC/p160 family of coregulators that has been shown to interact with IRF3 and act as a corepressor for GR. (Reily et al., 2006; Rogatsky et al., 2002). We therefore performed ChIP-Seq experiments using an antibody against GRIP1. As depicted in Figure 5a, treatment with Dex in comparison to LPS alone induced a large number of de novo GRIP1 binding sites. This ligand-dependent GRIP-1 cistrome showed overlapping binding events with the GR-occupied genomic landscape (Figure 5b). Unpredictably, the percentages of GR ChIP peaks colocalizing with GRIP1 and associated with transcriptional activation (66%) or with repression (63%) are strikingly similar. As expected from a GR-associated factor, bioinformatic motif analysis of the GRIP1 cistrome revealed enrichment for Pu.1, C/EBP, AP-1, NF-κB, IRF3 and GRE sites (Figure 5c). As depicted in Figure 5d, GRIP1 binding to GR target genes was induced by ligand treatment, but nearly undetectable in macrophages treated with LPS alone. Interestingly, GRIP1 was equally recruited to activated (Tlr2, Per1) and to repressed (Il6, Ccl2) genes. Notably, we did not observe reduced IRF3 or p65/c-Jun enhancer binding in the presence of GR ligand (Supplementary Figures 1c and 6). In fact, large parts of the c-Jun/p65-occupied landscapes were unaffected by Dex, indicating that GR acts by blocking transcriptional activation rather than cistromic inhibition of DNA binding or chromatin remodeling.
Figure 5. GR recruits GRIP1 to activated and repressed genes.
a) Area-proportional Venn diagram of GRIP1 cistromes in LPS-stimulated (light blue) and Dex-treated (cyan) macrophages. b) Area-proportional Venn diagram showing the overlap between GRIP1 and GR ChIP-Seq peaks in macrophages treated with LPS and Dex. Arrows and numbers on bottom right represent ChIP peaks associated with GR-repressed versus –activated genes. c) Globally enriched motifs found in the GRIP1 cistrome in response to LPS+Dex. d) ChIP-sequencing tracks of regulatory regions of GR target genes in macrophages treated with LPS plus or minus Dex, normalized to 10 million reads. Ligand-dependent GRIP1 recruitment can be observed at activated (Tlr2, Per1) and repressed (IL6, Ccl2) enhancers.
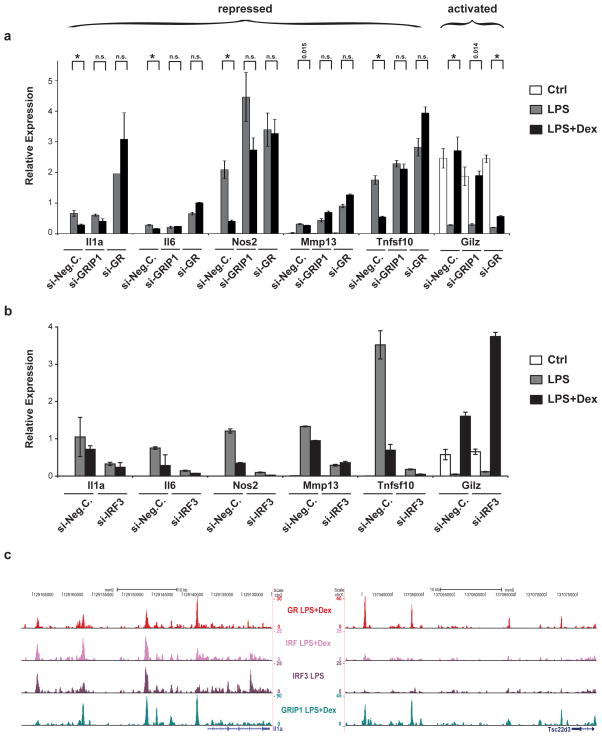
Repression by GR may serve to antagonize IRF3 activity
As described above, GRIP1 recruitment has been linked to disturbance of IRF3 function, and indeed, knockdown of GRIP1 expression affects GR-mediated repression of LPS-induced target genes such as Il1a, Il6, Nos2, Mmp13 and Tnfsf10, but not positive transcripts such as Gilz (Figure 6a), despite the observed equal recruitment to both groups (Figure 5b,d). The most likely explanation for this phenomenon is compensation of coactivator function by other redundant SRC family members, whereas the corepressor property appears to be unique to GRIP1. These repressive loci may act by disruption of IRF3 activity, as shown in Figure 6b. In RAW264.7 macrophage cells, knockdown of IRF3 expression affects expression of these LPS-induced GR target genes, but not of other genes such as Gilz. We therefore performed ChIP-sequencing with an α-IRF3 antibody in LPS-treated primary macrophages to identify these potentially common target genes. Indeed, IRF3 colocalizes with GR and GRIP1 at negative enhancers, for example at Il1a and Ccl2 loci, but not at positive loci such as Gilz and Per1 (Figure 6c and Supplementary Figure 6). Accordingly, motif analysis of the IRF3 ChIP-sequences identifies AP-1, NF-κB and GRE motifs among others. Indeed, hypergeometric testing (p=5.87e-06) confirmed a statistically significant enrichment of IRF3-bound sequences overlapping with negative GR enhancers. (We also detected some overlap with positive sites, but not to a significant extent.) As presented in Supplementary Figure 6, GR ligand treatment in the presence of LPS changes the IRF3-bound cistrome, thereby increasing the number of bound sites colocalizing with GRIP1, but it does not appear to result in competition for binding of cis-regulatory elements.
Figure 6. GR repression includes GRIP-1 corepressor function and opposition of IRF3 activity.
a) siRNA knockdown of GR and GRIP-1 in RAW264.7 macrophages treated with LPS and/or Dex. Reduction of both GR and GRIP-1 expression levels impairs repression of LPS-induced genes Il1a, Il6, Nos2, Mmp13 and Tnfsf10 by Dex, but only GR knockdown affects the positively regulated gene Gilz. Brackets above bars show p-values for differential expression in LPS vs. LPS+Dex treated cells, * p <0.01, n.s. not significant. b) siRNA knockdown experiments in RAW264.7 cells. Knockdown of IRF3 affects expression of LPS-induced and Dex-repressed GR target genes, but not Dex-activated genes such as Gilz. Error bars represent S.T.D. c) Representative examples of IRF3 ChIP-Seq tracks. IRF3 binding co-occurs with the presence of GR and GRIP1 at repressed genes such as Il1a, but not at activated ones like Gilz.
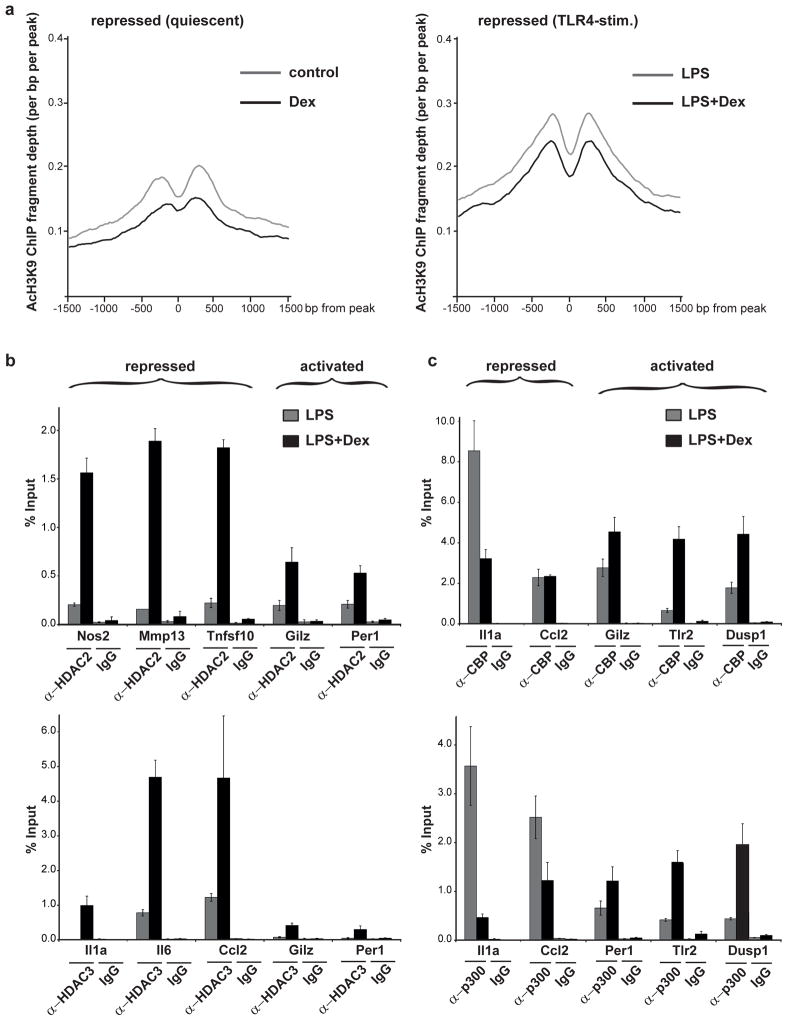
GR-mediated suppression of inflammation involves histone deacetylation
Nuclear receptor coactivators such as the SRC family members stimulate transcriptional initiation by recruitment of chromatin-remodeling and -modifying complexes that mark active chromatin and attract other proteins to the complex, as exemplified by the engagement of CBP/p300 histone acetyltransferases (Bulynko and O’Malley, 2010). Corepressors, on the other hand, are believed to oppose the actions of coactivators, for example via recruitment of histone deacetylases, and mediate the assembly of transcriptional repressor complexes (Perissi et al., 2010; Rosenfeld et al., 2006). In order to detect potentially discriminating features between activated and repressed loci, we performed ChIP experiments using antibodies against acetylated histone H3K9 as a readout for actively transcribed chromatin. As depicted in Figure 7a, GR ligand treatment reduces acetyl-H3K9 patterns at repressed enhancers in both quiescent and in LPS-stimulated primary macrophages, effectively unmarking active chromatin. Conversely, activated enhancers display increased H3K9 acetylation profiles in response to Dex (Supplementary Figure 7a). These differential acetylation profiles correlate with increased recruitment of HDACs at repressed, but not at activated loci (Figure 7b) in response to Dex. Concomitantly, hormone treatment results in p300/CBP HAT recruitment to positive, but not to negative enhancers (Figure 7c).
Figure 7. Repression by GR involves selective HDAC recruitment and histone deacetylation.
a) ChIP-Seq experiments in primary macrophages show a reduction in acetylated histone H3K9 marks at sites repressed in response to Dex, in both unstimulated and in TLR4-activated conditions. b) At repressed enhancers such as Nos2, Il6 etc., Dex treatment leads to increased recruitment of HDAC2 and/or HDAC3, as determined by ChIP experiments. c) Conversely, HAT activity is increased in response to Dex at activated enhancers (i.e. Dusp1), as determined by ChIP experiments using α-CBP and α-p300 antibodies, but not at repressed loci. Error bars represent S.T.D.
As a result, negative response elements appear to be characterized by the corepressor as opposed to the coactivator function of GRIP1, which leads to chromatin deactivation. The epigenetic specification of GR polarity at these repressive enhancers may be predetermined prior to hormone action by context-specific factors (such as IRF3) or might depend on other unknown regulators.
Discussion
How the glucocorticoid receptor, other nuclear receptors or transcription factors in general can activate certain genes while at the same time repressing others remains an open question. A general presumption for the GR in the macrophage is that negative regulation is associated with ‘trans-repression’ such that GR can oppose the positive action of AP-1 or NF-κB via tethering. Unexpectedly, using genome-wide profiling in LPS-activated macrophages, we find that the positive and negative GR cistromes are predominantly composed of classical GREs in close proximity to NF-κB and AP-1 binding sites. While up to 20% of GR-dependent repression is found at nGREs and tethered sites, the identical motifs seem to direct positive regulation at about the same frequency. Accordingly, cistromic motif classification is insufficient to predict regulatory fate. We show that GR-dependent repression of inflammatory genes involves selective utilization of the nuclear receptor corepressor function of GRIP1, which may in part serve to antagonize IRF3 activity. In exploring the epigenomic basis of these observations, we find that these negative enhancers are characterized by HDAC recruitment and a depletion of histone acetylation marks. Accordingly, although recruitment of GR to chromatin confers hormone responsiveness, the receptor itself many not decide its regulatory fate. These results imply a critical role for epigenetic regulators and chromatin ‘context’ in predetermining the transcriptional polarity of GR controlled target genes.
In regards to the GR cistrome, we find that TLR4-activation by LPS recrafts ‘primary chromatin’ to create an inflammatory cistrome that comprises both GR-induced and repressed target genes. Previous studies have shown that TLR4 activation results in chromatin reorganization (Smale, 2011), a process, based on this work, that presumably creates new domains of GR accessibility. In addition, LPS has been shown to increase GR expression levels (Barish et al., 2005), which could enhance the anti-inflammatory response and potentially revert chromatin to its pre-activated state. TLR4 signaling thus not only triggers the transition from quiescent to activated chromatin landscape but simultaneously primes or prepares it for the subsequent termination of the inflammatory response.
The enrichment of Pu.1 and C/EBP binding sites throughout the entire GR cistrome suggests these pioneer factors provide points of chromatin access for the recruitment of collaborating transcription factors, but do not directly establish whether a particular enhancer will be induced or repressed by glucocorticoids (Ghisletti et al., 2010; Heinz et al., 2010). Furthermore, the frequent proximity of GR binding sites with AP-1, NF-κB and/or GRIP1 binding at both induced and repressed inflammatory enhancers, emphasizes that the transcriptional outcome cannot be predicted based on the simple combination of these factors.
In this study we identified 918 GR-bound cis-regulatory elements that were linked to positive gene expression, and 729 elements that served a negative function. Comparison of motif signatures within these regulatory sites reveals remarkably similar features at both induced and repressed cis elements. These findings reverse several established models and reveal the lack of evidence for the dominance of prevailing concepts for GR-mediated repression. Tethering of GR to NF-κB/AP-1 (whether positive or negative) cannot explain our observed lack of a difference in motif content between induced or repressed genes. Thus, GR does not always seem to ‘naturally antagonize’ AP-1/NF-κB, but rather appears to frequently cooperate to facilitate hormone induction. Similarly, GR recruits GRIP1 in a comparable fashion to both up- and downregulated genes, where it may act as a coactivator or corepressor, depending on the context. Based on prevalence of motifs, we show that both activation and repression by GR entail classical GREs, and that ‘cis-repression’ by GR occurs most commonly via direct DNA binding. Hormone-induced repression at enhancers that contain no discernible GRE motif, i.e. the IL12a locus, suggest that GR is tethered at up to one-fifth of negatively regulated sites. However, an equal portion of tethered sites can be detected at activated target genes. Again this illustrates that motif categorization alone does not express a regulatory mechanism and that positive or negative polarity likely requires additional, unrecognized, regulatory components. The above argument also extends to the recently described negative response elements (called IR nGREs), which in the context of specific genes has been shown to result in the formation of repressive complexes at loci distinct from those shared with p65 or c-Jun (Surjit et al., 2011). However, we were not able to detect any enrichment of IR nGRE motifs in our data set (Supplementary Figure 3a), again confirming that repression of inflammatory AP-1 and NF-κB signaling occurs through a mechanism that is not intrinsic to the motif itself.
In conclusion, our objective genome-wide analysis reveals that prevailing models (including tethering, nGREs or GRIP1 recruitment) are insufficient to explain the anti-inflammatory actions of GR. In contrast to previous predictions, the lack of a discernible difference in motif content between activation and repression indicates a mechanism that is context-dependent. This context may be established by the existence of predetermined inflammatory sites in the genome, for example those that are marked by IRF3 occupancy, or by other unknown factors that might not bind directly to DNA. Thus, while the presence of GR confers hormone responsiveness, the mechanisms switching GRIP1 from coactivator to corepressor, ultimately leading to histone acetylation versus deacetylation, remain to be elucidated. Interestingly, in support of the observed deacetylation mechanism, we have preliminary evidence showing the nuclear receptor corepressors NCoR and SMRT present at repressive sites in the genome, independently of and before GR ligand treatment. The epigenetic specification of GR polarity by SMRT, NCoR, IRF3 or other unknown factors may predetermine these loci as repressive enhancers prior to hormone action, implying the existence of a yet unrecognized regulatory determinant (Supplementary Figure 7b).
Experimental Procedures
Primary bone-marrow derived macrophages from male C57BL/6 mice were isolated and differentiated in culture as previously described (Barish et al., 2005). Cells were treated overnight with 1μM Dex (Sigma) or ethanol and/or LPS (100ng/ml, Sigma) for 3 hours. RNA isolation and qPCR were performed as previously described (Barish et al., 2005). Microarrays were performed with Illumina Mouse Ref-8v2.0 Expression BeadChips after overnight incubation with Dex and 6 hours LPS treatment.
ChIP assays were performed as described elsewhere (Barish et al., 2010). Antibodies used for ChIP were anti- GR (Santa Cruz), p65 (Abcam), c-Jun (Santa Cruz), GRIP1 (Abcam), P-IRF3 (Cell Signaling) and IgG (Santa Cruz). ChIP-Seq libraries were made per manufacturer’s instructions and sequenced using an Illumina Genome Analyzer II. Data analysis was performed using HOMER (Heinz et al., 2010).
Luciferase assays were carried out as previously described (Wan et al., 2007). RAW264.7 or CV-1 cells were treated overnight with 1μM Dex or ethanol and/or LPS. Cis-regulatory elements were cloned into pGL4.23 (Promega) and point mutations were inserted using the Quick Change II Site Directed Mutagenesis kit (Agilent). siRNA knockdowns were achieved using Dharmacon SMARTpool mouse siRNAs (Thermo Scientific) according to standard protocols. The genomic data have been deposited in NCBI’s GEO and are accessible through accession numbers GSE31793 (microarray), GSE31796 (ChIP-Seq), GSM419051 (ChIP input) and GSM611117 (ChIP p65 +LPS). See Supplemental Methods and Table 3 for details.
Supplementary Material
Highlights.
TLR4 activation dramatically remodels the GR cistrome in macrophages
Both negative and positive GR enhancers include GREs, NF-κB & AP-1 sites
Classical models fail to predict positive or negative polarity of GR regulation
Hormone treatment marks negative enhancers by histone deacetylation
Acknowledgments
We thank E. Ong, S. Ganley and C. Brondos for administrative assistance, J. Nery for DNA sequencing, C. Benner for assistance with HOMER software, and H. Juguilon, M. Heinig and A. Bauerfeind for technical assistance. N.H.U. was supported by an EMBO fellowship, EMBO ALTF 686-2010, G.D.B. by K08HL092298. This work was funded by grants from the National Institutes of Health (HL105278, HL088093, DK057978, DK090962, CA014195 and ES010337), the Helmsley Charitable Trust, and the Samuel Waxman Cancer Research Foundation. R.M.E. is an investigator of the Howard Hughes Medical Institute and March of Dimes Chair in Molecular and Developmental Biology at the Salk Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, Mangelsdorf DJ, Evans RM. A Nuclear Receptor Atlas: macrophage activation. Mol Endocrinol. 2005;19:2466–2477. doi: 10.1210/me.2004-0529. [DOI] [PubMed] [Google Scholar]
- Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-kappaB cistromes mediate opposing regulation of the innate immune response. Genes Dev. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck IM, Vanden Berghe W, Vermeulen L, Yamamoto KR, Haegeman G, De Bosscher K. Crosstalk in inflammation: the interplay of glucocorticoid receptor-based mechanisms and kinases and phosphatases. Endocr Rev. 2009;30:830–882. doi: 10.1210/er.2009-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulynko YA, O’Malley BW. Nuclear Receptor Coactivators: Structural and Functional Biochemistry. Biochemistry. 2010 doi: 10.1021/bi101762x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bosscher K, Haegeman G. Minireview: latest perspectives on antiinflammatory actions of glucocorticoids. Mol Endocrinol. 2009;23:281–291. doi: 10.1210/me.2008-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, Evans RM. Multiple and cooperative trans-activation domains of the human glucocorticoid receptor. Cell. 1988;55:899–906. doi: 10.1016/0092-8674(88)90145-6. [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev. 2005;19:1116–1127. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell. 2010;140:771–776. doi: 10.1016/j.cell.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison N, Eisman J. Role of the negative glucocorticoid regulatory element in glucocorticoid repression of the human osteocalcin promoter. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 1993;8:969–975. doi: 10.1002/jbmr.5650080810. [DOI] [PubMed] [Google Scholar]
- Natoli G, Ghisletti S, Barozzi I. The genomic landscapes of inflammation. Genes Dev. 2011;25:101–106. doi: 10.1101/gad.2018811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naugler WE, Karin M. The wolf in sheep’s clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends Mol Med (England) 2008:109–119. doi: 10.1016/j.molmed.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nature reviews Genetics. 2010;11:109–123. doi: 10.1038/nrg2736. [DOI] [PubMed] [Google Scholar]
- Reily MM, Pantoja C, Hu X, Chinenov Y, Rogatsky I. The GRIP1:IRF3 interaction as a target for glucocorticoid receptor-mediated immunosuppression. Embo J. 2006;25:108–117. doi: 10.1038/sj.emboj.7600919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci U S A. 2002;99:16701–16706. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Schule R, Rangarajan P, Kliewer S, Ransone LJ, Bolado J, Yang N, Verma IM, Evans RM. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell (United States) 1990:1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- Smale ST. Hierarchies of NF-kappaB target-gene regulation. Nature immunology. 2011;12:689–694. doi: 10.1038/ni.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Jr, Lazar MA. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24:1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- Takeda K, Akira S. Toll-like receptors. Curr Protoc Immunol. 2007;Chapter 14(Unit 14):12. doi: 10.1002/0471142735.im1412s77. [DOI] [PubMed] [Google Scholar]
- Wan Y, Chong LW, Evans RM. PPAR-gamma regulates osteoclastogenesis in mice. Nat Med. 2007;13:1496–1503. doi: 10.1038/nm1672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.